Abstract
Colorectal cancer (CRC) is one of the most common cancers in the developed countries, and nearly 70% of patients with CRC develop colorectal liver metastases (CRLMs). During the last decades, several scores have been proposed to predict recurrence after CRLM resection. However, these risk scoring systems do not accurately reflect the prognosis of these patients. Therefore, this investigation was designed to identify a proteomic profile in human hepatic tumor samples to classify patients with CRLM as “mild” or “severe” based on the 5-year survival. The study was performed on 85 CRLM tumor samples. Firstly, to evaluate any distinct tumor proteomic signatures between mild and severe CRLM patients, a training group of 57 CRLM tumor samples was characterized by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry, and a classification and regression tree (CART) analysis was subsequently performed. Finally, 28 CRLM tumor samples were used to confirm and validate the results obtained. Based on all the protein peaks detected in the training group, the CART analysis was generated, and four peaks were considered to be the most relevant to construct a diagnostic algorithm. Indeed, the multivariate model yielded a sensitivity of 85.7% and a specificity of 86.1%, respectively. In addition, the receiver operating characteristic (ROC) curve showed an excellent diagnostic accuracy to discriminate mild from severe CRLM patients (area under the ROC: 0.903). Finally, the validation process yielded a sensitivity and specificity of 68.8% and 83.3%, respectively. We identified a proteomic profile potentially useful to determine the prognosis of CRLM patients based on the 5-year survival.
Introduction
Colorectal cancer (CRC) is the third most common cancer in the United States in terms of incidence and mortality [1], and similar results are also observed in other developed countries [2]. Nearly 70% of patients with CRC develop colorectal liver metastases (CRLMs), and 10% to 25% are diagnosed at the time of resection of the primary tumor [3], [4]. At present, surgical resection still remains the most effective procedure to lengthen patient survival and is the only curative treatment [5], [6], with a 5-year survival rate of CRLM after hepatic resection ranging from 30% to 60% [7]. Although some patients are not surgical candidates, recent advances in chemotherapy, radiofrequency ablation, and thermoablative methods have increased the number of patients eligible for surgical resection [8]. Several scores to predict recurrence after CRLM resection have been proposed to improve the diagnosis and prognosis of these patients [9], [10], [11], [12], with the Fong score being one of the most known tools used for this purpose. However, current risk scoring systems do not accurately reflect the prognosis, and patient survival can be very similar even with very different score values [13]. Thus, identification of a more accurate system to predict patient survival is still of major relevance. In this regard, the use of biomarkers that could, independently or associated with classical prognostic indices, predict the neoplastic evolution of the patient after surgical CRLM resection is a well-defined strategy to enhance the reliability of predicting the prognosis of CRLM [14], [15]. Furthermore, biomarkers could improve the selection of the optimal treatment for each patient to enhance their prognosis.
In the current investigation, high-throughput proteomic techniques were used to identify a potential protein panel in human hepatic tumor samples to detect CRLM patients with a poor prognosis. Ultimately, we aimed to validate whether the protein panel profile identified could be useful as a prognostic tool in CRLM patients.
Patients and Methods
Patients and Routine Laboratory Tests
Patients admitted to the Liver Transplantation Unit to undergo CRLM resection from April 2005 to March 2012 were prospectively considered for this study and were followed according to institutional guidelines [16]. We obtained all clinical and pathological data from the Liver Surgery Unit prospective database and clinical charts and selected only the patients who could be included in a specific prognostic group according to survival time. Thus, a total of 85 CRLMs were studied. Exclusion criteria were previous CRLM resection or refusal to participate in the study. Chemotherapy regimens used before hepatic resections were based on 5-fluorouracil alone or associated with irinotecan or oxaliplatin according to local protocols. Patients were classified based on their long-term outcome. Thus, patients were considered as “mild” if their survival time was longer than 5 years or “severe” if it was lower. Venous blood samples from all CRLM patients were obtained after fasting. Extracted serum samples were kept at −80°C, and several serum parameters, including alanine transaminase, aspartate aminotransferase, albumin, bilirubin, or creatinine, among others, were measured with the ADVIA 2400 Instrument (Siemens Healthcare Diagnostics, Tarrytown, NY). CEA serum levels were determined using the ADVIA Centaur XP Immunoassay System (Siemens Healthcare Diagnostics).
The design of the study was two-fold. Firstly, in a training set of 57 CRLM patients, we assessed whether the tissue proteomic profile of the mild patients differed from that of severe patients. Routine liver and renal function tests were also analyzed in these patients. Thereafter, liver tissue samples from 28 CRLM patients were also collected and included in the blinded validation group to confirm the results obtained.
Liver Resection of Metastatic Colorectal Cancer and Specimen Collection
All metastatic tissue samples from patients undergoing curative resection of CRLM were collected and immediately cryopreserved in liquid nitrogen and kept until further analysis.
Hepatic Protein Extraction and Protein Fractionation from Liver Homogenates
Approximately 50 mg of tumor hepatic tissue was ground to fine powder in dry ice and solubilized by pestle homogenization in 500 μl of urea buffer (9.5 M urea, 2% CHAPS, 1% DTT, 50 mM Tris-HCl, pH). Thereafter, tissue homogenates were incubated in a rotating mixer for 1 hour at 4°C, and insoluble material was removed by centrifugation (16,000 ×g, 4°C for 15 minutes). Afterwards, samples were aliquoted and kept at −80°C until protein fractionation was performed. In short, samples were fractionated by pH using a 96-well filtration plate (Pall Corp., Port Washington, NY) with 200 μl of a macro-prep high Q anion exchange support (Bio-Rad Laboratories, Hercules, CA) in each well. Flow-through was discarded by vacuum filtration. Subsequently, each well was washed twice with 200 μl of distilled water and equilibrated with 200 μl of rehydration buffer (50 mM Tris-HCl, pH). Wells were incubated for 1 hour at room temperature. Thereafter, each well was equilibrated three times with 200 μl of an equilibration buffer [1 M urea, 0.2% (w/v) CHAPS, 50 mM Tris-HCl, pH]. Prior to fractionation, 20 μl of tissue homogenates was mixed with 30 μl of urea buffer [9 M urea, 2% (w/v) CHAPS, 50 mM Tris-HCl, pH] in a 96-well V-bottom plate for 20 minutes at 4°C in a horizontal orbital microplate shaker. Samples were then diluted with 50 μl of equilibration buffer, transferred to each well, and incubated for 30 minutes at 4°C with shaking. All samples were eluted in a stepwise manner by altering the pH of the wash buffer. Six different fractions were obtained. Flow-through was collected by vacuum filtration into V-bottom microplates, and all were stored at −80°C until proteomic analysis.
Proteomic Processing of Hepatic Tissue Samples
Protein profiling was analyzed by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) using the eight-spot format ProteinChip array (Bio-Rad). In a preliminary study to optimize the experimental conditions, two pooled samples from mild CRLM patients and severe CRLM patients were fractionated, and the six fractions obtained were loaded onto three different types of ProteinChip arrays that had different protein binding affinity; weak cation exchange arrays (CM10), immobilized metal affinity chromatography arrays (IMAC30), and hydrophobic/reverse-phase arrays (H50). The resulting protein profile from each pool was compared, and the fraction number 3 using the CM10 ProteinChip array showed the highest number of peaks detected, the highest total signal intensity, and the major differences between groups when compared with all other fractions and ProteinChip arrays. Thus, fraction number 3 and CM10 ProteinChip array were selected for the subsequent studies. Prior to sample loading, spots were equilibrated twice with 200 μl of a sodium acetate buffer (0.1 M sodium acetate, pH). Meanwhile, 40 μl of the fractionated sample was mixed with 60 μl of the sodium acetate buffer, and this mixture (100 μl) was subsequently loaded randomly and incubated for 1 hour on a shaker at room temperature. Afterward, CM10 ProteinChip arrays were washed three times with 200 μl of the sodium acetate buffer for 5 minutes on a shaker at room temperature and twice with deionized water to remove unbound proteins. Thereafter, arrays were air-dried, and 1 μl of energy-absorbing matrix (saturated sinapinic acid in an aqueous solution containing 50% acetonitrile and 0.5% trifluoroacetic acid) was added twice to each spot. The surface was allowed to air dry between each application. Finally, ProteinChips were read in the ProteinChip PBS II reader (Bio-Rad).
Data Acquisition and Analysis
All data were processed as described elsewhere [17]. Briefly, each spot was analyzed at three different energy laser intensities: 2500 nJ, 3000 nJ, and 3500 nJ, and the mass-to-charge ratio (m/z) was set from m/z 1000 to 25,000 for the low-energy laser intensity, between m/z 2500 and 200,000 for the medium-energy laser intensity, and from m/z 5000 to 200,000 for the high-energy laser intensity. In addition, peak resolution was focused at m/z 5000, 12,000, or 19,000 according to low-, medium-, or high-energy laser intensity, respectively.
Protein spectra were calibrated using two different external calibration standards (all-in-one peptide standard and all-in-one protein standard, Bio-Rad Laboratories), and they were subsequently normalized by the average total ion current across the group. To minimize outliers, all spectra differing by at least twice the standard deviation from the mean were deleted. Baseline adjustments, peak selection parameters such as the minimal signal-to-noise ratio, valley depth, and m/z error were also defined as described previously [17]. Afterward, all protein peak clusters automatically identified were manually verified. Relabeling, removal, or addition of peaks was performed when necessary.
Statistical Analysis
Statistical analyses were performed by the nonparametric Mann-Whitney U test or the Fisher test when appropriate. Results are expressed as mean ± SE, and P values lower than .05 were considered significant. All statistical clinical analyses were performed with the PASW Statistical Package Version 18.0 for Windows (SPSS, Chicago, IL). Quantitative proteomic data were analyzed using GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA). Finally, a classification and regression tree (CART) analysis was performed using the Biomarker Patterns (Bio-Rad) to detect the protein peaks with the greatest contribution to discriminate both groups.
Ethical Approval
We obtained written informed consent from all patients included in the study when they were proposed for surgery, and the investigation was approved by the Investigation and Ethics Committee of the Hospital Clinic of Barcelona following the ethical guidelines of the 1975 Declaration of Helsinki.
Results
The main demographic variables of the patients included in the training set are summarized in Table 1. Of note is that nearly 40% of the CRLM patients included in the training group showed discrepancies between their theoretical risk factor, obtained by Fong's prognostic scoring system, and their actual survival.
Table 1.
Patient Characteristics and Differences between Training and Validation Groups
| Overall Group (n = 85) | Training Group (n = 57) | Validation Group (n = 28) | P Value | |
|---|---|---|---|---|
| Gender (M/F) | 65/20 | 43/14 | 22/6 | NS |
| Age (years) | 65.0 ± 0.9 | 65.2 ± 1.1 | 64.6 ± 1.8 | NS |
| Node positive in primary tumor [n (%)] | 58 (68.2%) | 41 (71.9%) | 17 (60.7%) | NS |
| Disease-free interval <12 months [n (%)] | 58 (68.2%) | 43 (75.4%) | 15 (53.6%) | NS |
| CEA (ng/ml) | 34.2 ± 8.3 | 43.4 ± 12.2 | 15.8 ± 3.8 | <.05 |
| CEA <200 ng/ml | 84 (94.1%) | 52 (91.2%) | 28 (100%) | NS |
| Number of tumors | 2.7 ± 0.3 | 2.5 ± 0.2 | 3.0 ± 0.6 | NS |
| Multinodularity [n (%)] | 56 (65.9%) | 38 (66.7%) | 18 (64.3%) | NS |
| Tumor size (cm) | 3.0 ± 0.2 | 2.6 ± 0.2 | 3.9 ± 0.6 | <.05 |
| Largest tumor >5 cm [n (%)] | 10 (11.8%) | 5 (8.8%) | 5 (17.9%) | NS |
| Fong score | 2.2 ± 0.9 | 2.3 ± 0.1 | 1.9 ± 0.2 | NS |
| Fong score ≤2 (low risk) | 51 | 32 | 19 | – |
| Fong score >3 (high risk) | 34 | 25 | 9 | – |
NS, not significant.
Reproducibility of SELDI-TOF-MS Protein Profiling in Hepatic Tumor Samples
Experiments combining hepatic protein extraction and protein fractionation were performed to evaluate intraexperiment variation. A pooled hepatic tumor sample from the 27 CRLM patients included in the training group was obtained and treated as the other samples. Basically, it was processed independently 15 times and subsequently measured in a CM ProteinChip array (Bio-Rad). Finally, the SELDI-TOF-MS spectra were accurately analyzed. Based on the protein peaks detected in the m/z 1000 to 25,000 range, the intraassay (spot to spot) maximum coefficient of variation (CV) was 44.70% for peak intensity and 0.04% for mass accuracy. In addition, the day-to-day variation was also evaluated. The pooled hepatic tumor sample was processed and analyzed independently in three different days, and the maximum CV for peak intensity and mass accuracy was 39.56% and 0.12%, respectively.
Differential Proteomic Profiles Between Mild CRLM Patients and Severe CRLM Patients
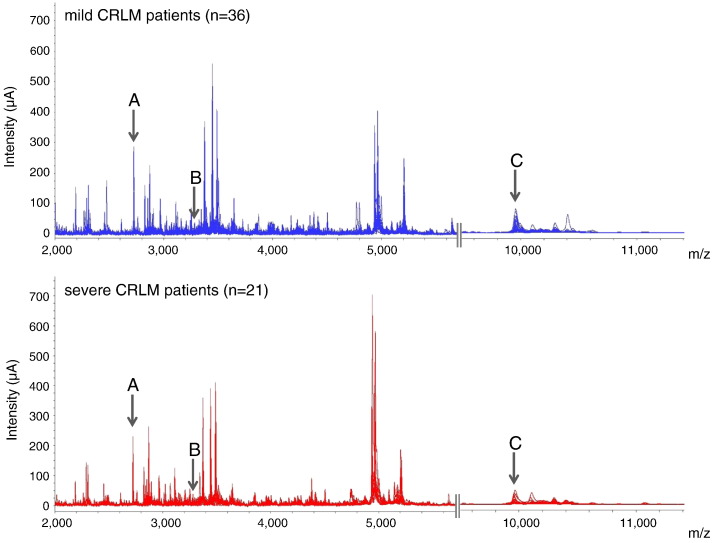
Twenty-one severe CRLM patients and 36 mild CRLM patients constituted the training set (Table 1). Two technical replicates were analyzed for each sample. We detected 118 protein peaks over a range of 1 to 25 kDa. Among these, only proteins showing a peak intensity of at least 2 μA and a m/z standard deviation below 11 were considered for further analysis. Moreover, and based on the results obtained in the pooled hepatic tumor sample, proteins with a CV greater than 30% for peak intensity and 0.02% for mass accuracy were also excluded. According to this protein peak exclusion strategy, 39 protein peaks were selected (see supplementary data 1). Figure 1 shows the protein spectra obtained by SELDI-TOF-MS which ranged between m/z 2000 and 11,000 in these subjects. Several protein differences were detected in the hepatic tumor proteomic profile between the two groups. In particular, three protein peaks with m/z values of 2726, 3251, and 9950 and shown in the Figure 1 as A, B, and C, respectively, were the most statistically significant. The intensity of the m/z 2726 and m/z 3251 protein peaks was downregulated in mild compared with severe CRLM patients. On the other hand, the m/z 9950 protein peak was upregulated in the mild group (Table 2). However, no single protein peak was able to clearly discriminate between mild and severe CRLM patients.
Figure 1.
Differential proteomic signature of mild and severe CRLM patients. Two segments of the SELDI-TOF-MS spectra ranging from m/z 2000 to m/z 5500 and from m/z 9500 to m/z 11,000 of all the tumor tissue samples from CRLM patients comprising the training group. The upper figure shows all the overlapped spectra of mild CRLM patients (n = 36). The bottom figure shows the spectra of severe CRLM patients (n = 21). Arrows show the three protein peaks with statistical significance between groups. Upper letters correspond to the peptide identification peak noted in Table 2.
Table 2.
Main Characteristics of the Protein Peaks Displaying Statistically Different Intensities on Comparing Mild and Severe CRLM Patients
| Intensity (μA) |
||||
|---|---|---|---|---|
| Peptide | m/z (Da) | Mild | Severe | P Value |
| A | 2726 | 59.0 ± 8.8 | 87.6 ± 12.6 | .04 |
| B | 3251 | 26.2 ± 2.9 | 34.3 ± 3.3 | .03 |
| C | 9950 | 22.7 ± 3.0 | 13.8 ± 2.2 | .04 |
Statistical analysis was performed by Mann-Whitney U test. Results are given as mean ± SE.
Classification of the Severity of CRLM Patients by CART Analysis
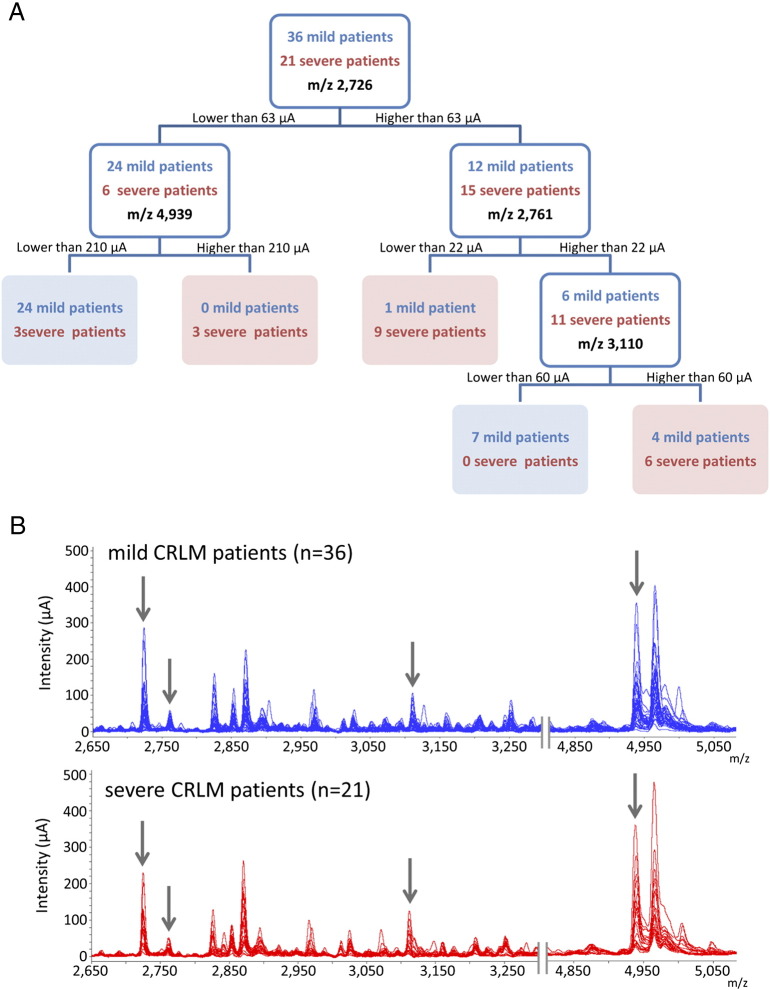
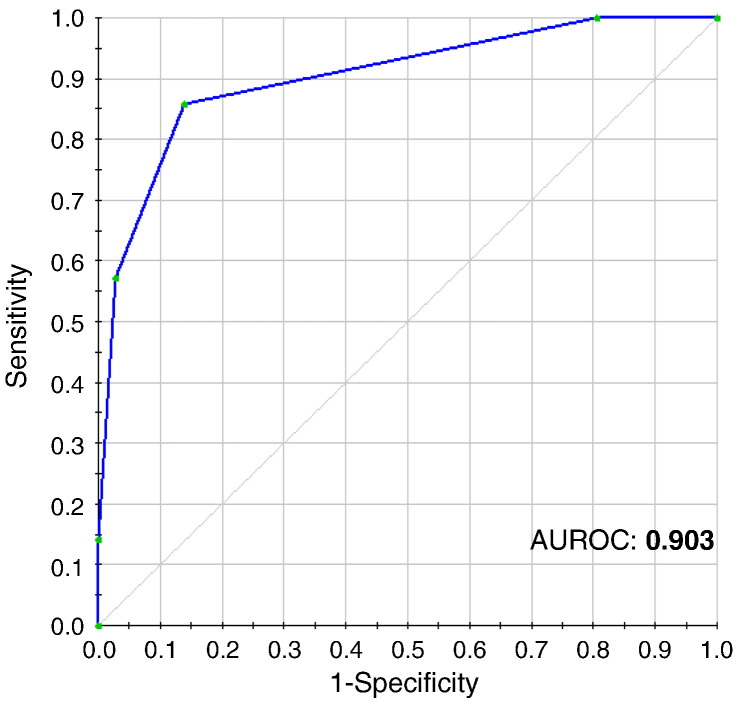
To measure the impact of all the protein peaks detected and identify those with the greatest discriminatory power, a nonparametric procedure was used based on the normalized data obtained by SELDI-TOF-MS. A CART with five terminal nodes was generated, and the combination of four different protein peaks was considered as the most relevant to construct the diagnostic algorithm (Figure 2A). The protein peaks which best differentiated mild from severe CRLM patients were m/z 2726, m/z 2761, m/z 3110, and m/z 4939. Figure 2B shows the proteomic spectra of these protein peaks. The classification tree yielded a sensitivity of 85.7%, a specificity of 86.1%, and a negative predictive value (NPV) and a positive predictive value (PPV) of 91.2% and 78.3%, respectively. To further test the diagnostic accuracy of the classification tree in CRLM patients, a receiver operating characteristic (ROC) curve was constructed. Figure 3 shows the excellent diagnostic accuracy of the classification tree to differentiate mild from severe CRLM patients (area under the ROC: 0.903).
Figure 2.
CART analysis to classify and differentiate mild from severe CRLM patients based on the proteomic profile obtained from the hepatic tumor samples. (A) Decision tree model for the classification of liver tumor samples from mild and severe CRLM patients. Node 0 (the upper white square of the decision tree) is known as the root node, which indicates the starting point of the decision tree construction and included all the patients. This node is divided based on the intensity value of the m/z 2726. If the intensity is lower than 63 μA, then these patients are put in a primary node (left white square) and are subsequently split based on the intensity value of the m/z 4939. However, if the intensity is higher than this cutoff (63 μA), patients are placed in the right primary node (white square) and further split based on the m/z 2761 protein peak. This procedure is subsequently repeated until a terminal node is reached (blue and red squares). As observed, the majority of mild CRLM patients are clustered in the green terminal nodes, whereas severe CRLM patients are grouped in the red terminal node. Only three severe CRLM patients and five mild CRLM patients were misclassified.
(B) Two sections of the SELDI-TOF-MS spectra. The first ranges from m/z 2650 to 3250, and the second is comprised between m/z 4850 and 5050. The upper figure shows all the overlapped spectra of the mild CRLM patients (n = 36). The bottom figure shows the spectra of the severe CRLM patients (n = 21). Arrows indicate the four protein peaks used to construct the diagnostic algorithm based on the CART analysis.
Figure 3.
Diagnostic accuracy of the CART analysis. Diagnostic accuracy of the classification tree to differentiate mild and severe CRLM patients. The area under the ROC obtained from the CART analysis was 0.903.
CART Analysis Assessment in the Validation Group
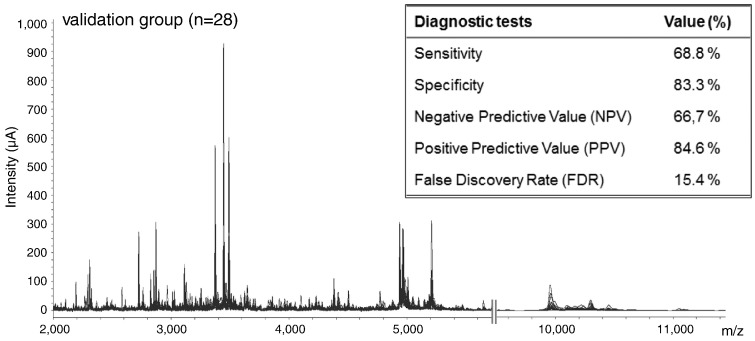
To confirm that the classification tree could be used as a prognostic tool, an independent group of 28 CRLM patients was tested as the blinded validation set. All hepatic tumor samples were collected and processed as described in the training group. As noticed in the MS spectra, very similar patterns were observed in the validation group when compared with the training set (Figure 4). In particular, the four protein peaks used to construct the classification tree showed an extremely high mass accuracy. The maximum m/z error was detected in the 3.1-kDa protein peak, despite the mass accuracy being below 505 ppm. After performing the CART analysis, 15 CRLM patients were clustered as “mild,” whereas 13 were considered “severe.” Subsequently, the proteomic data were compared with the clinical outcome of each subject. The principal demographic characteristics of the patients included in the validation set are shown in Table 1. Only two mild patients were misclassified, whereas this figure was five in the group of severe patients. These results yielded a sensitivity and specificity of 68.8% and 83.3%, respectively. Finally, the NPV as well as the PPV and the false discovery rate (1 − PPV) were also determined (Figure 4). The figures were 66.7%, 84.6%, and 15.4%, respectively.
Figure 4.
SELDI-TOF-MS proteomic profile and diagnostic tests from the validation group. Two portions of the SELDI-TOF-MS spectra from the 28 CRLM patients that comprised the blinded validation group. The first segment ranges from m/z 2000 to 5500, and the second is comprised between m/z 9500 and 11,000. As observed, the proteomic signature was very similar to that of the training group. The insert shows the diagnostic tests (sensitivity, specificity, NPV, and PPV) performed to further strengthen the clinical relevance of the diagnostic algorithm.
Discussion
Early detection of CRLM patients with poor prognosis is fundamental to improve therapeutic interventions and thus increase their life expectancy. At present, the neoplastic prognosis after CRLM resection is mainly based on several clinical indicators [9], [10], with tumor-free interval being one of the most determinant. Indeed, it is known that the development of neoplastic recurrence within the first year after liver resection is associated with a worse outcome [10]. However, mortality and the potential degree of severity based on all these different risk scoring systems are not always closely related [13], as other variables may influence the prognosis of these patients. In this study, we observed that there was not a clear correlation between the theoretical risk factor obtained from Fong's score system and the survival time. Particularly, 22 of the 57 CRLM patients included in the training group showed discrepancies. Therefore, the search for new tools capable of predicting survival still remains an open challenge.
In the present study, we used a high-throughput proteomic technique to unveil a differential proteomic signature among CRLM patients based on their survival time. SELDI-TOF-MS, which is based on matrix-assisted laser desorption/ionization TOF-MS, has been used as a proteomic strategy in several studies to detect and identify potential biomarkers related to many different diseases including colorectal cancer [17], [18], [19], [20], [21]. It combines a solid-phase chromatographic surface with TOF-MS that enables the analysis of crude samples such as tissue lysate, saliva, serum, urine, etc. Sample fractionation is performed previously to reduce sample complexity, thus enabling the detection of low-abundance proteins. In the current study, tumor tissue samples were fractionated by anion exchange chromatography, and the third eluate was analyzed by MS. The solid-phase chromatographic surface was selected based on the highest number of protein peaks detected and the highest total signal intensity analyzed in a preliminary study. Using this procedure, we detected 118 protein peaks over a range of 1 to 25 kDa. Three protein peaks with m/z values of 2726, 3251, and 9950 were statistically significant. However, none of these peaks clearly differentiated mild from severe CRLM patients. Therefore, we performed a CART analysis which showed that the expression of four different protein peaks was the most relevant protein combination to classify these patients according to survival. Among these peaks, only the one located at m/z 2726 was statistically significant. The other three protein peaks selected in the CART analysis did not coincide with any of those that were statistically significant. This feature is not surprising because the protein peaks which best separate the different smaller subsets obtained during the CART analysis are not necessarily the same as those that are globally statistically significant between both groups.
There are few studies focused on the proteomic profiles of CRLM patients [22], [23]. These investigations are mainly related to predicting neoplastic recurrence after CRLM resection and metachronous liver metastasis from colorectal cancer in serum samples. However, none of these investigations has explored the tumor proteomic signature to predict survival. Therefore, we first performed a proteomic analysis in 57 liver tumor samples from CRLM patients to assess whether there is a differential proteomic profile between patients with a greater or less than 5-year survival. As mentioned above, CART analysis revealed four protein peaks enabling the classification of CRLM patients and thus their outcome. These results were further validated in a new group of CRLM patients possessing similar clinical characteristics to those observed in the training group. Of note was that the specificity and the PPV values obtained in the validation set (83.3% and 84.6%, respectively) were very similar to those previously determined in the training group. However, those corresponding to sensitivity and NPV were lower (68.8% and 66.7%, respectively).
CART analysis has proven to be a relevant strategy to predict the response of cancer patients to treatment [24] and the prognosis of disorders other than colorectal cancer, including chronic myelomonocytic leukemia or pulmonary complications after lung resection [25], [26]. One of the major advantages of CART analysis is the simple and intuitive nature of this algorithm. In comparison to traditional regression methods, in which the prognostic score is obtained as a weighted average of several biochemical and clinical factors, CART classifies patients into groups based on simple combinations of the patients' characteristics [27]. In the current investigation, CART analysis resulted in the identification of 4 protein peaks able to discriminate CRLM patients with a survival time of longer than 5 years from those with shorter survival periods. Therefore, by using the algorithm obtained, it is possible to predict the survival time of these subjects at the time of tumor resection with a remarkable sensitivity and specificity.
A major issue emerging from these results is related to the identification of the different peaks included in the CART analysis. However, although this information could be very relevant to better understand the mechanisms involved in colorectal carcinoma metastases, the identification of these peaks would not result in an improvement in the performance of the algorithm as a prognostic indicator of survival.
In conclusion, the results presented in the current investigation indicate that proteomic analysis of liver tumor samples from CRLM patients may be useful to determine their 5-year survival. The use of a classification tree allowed the identification of four protein peaks with clear clinical relevance. This finding was validated in another set of CRLM patients, further strengthening its diagnostic value. These results, therefore, set the rationale to perform future studies focused on the identification of these four protein peaks. In addition, they could provide valuable insight into the altered molecular signaling pathways that are involved in the survival of patients with CRLM.
Ethical Conduct of Research
The authors state that they have obtained written informed consent from all patients included in the study when they were proposed for surgery, and the investigation was approved by the Investigation and Ethics Committee of the Hospital Clinic of Barcelona following the ethical guidelines of the 1975 Declaration of Helsinki.
Competing Interests
The authors do not have any more disclosures to report.
Footnotes
Funding: This work has been supported by grants from the Ministerio de Economía y Competitividad (SAF 2012-35979 and SAF 2015-64126-R to W. Jiménez and BES-2010-035452 to S. Marfà) and from the Agència de Gestió d'Ajuts Universitaris i de Recerca (SGR 2009/1496 and SGR 2014/219). This work has also been cofinanced by the European Union through the European Regional Development Fund, “A way of making Europe.” CIBEREHD is funded by the Instituto de Salud Carlos III.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2016.08.002.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Siegel R., Desantis C., Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Sheth K.R., Clary B.M. Management of hepatic metastases from colorectal cancer. Clin Colon Rectal Surg. 2005;18:215–223. doi: 10.1055/s-2005-916282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruers T., Bleichrodt R.P. Treatment of liver metastases, an update on the possibilities and results. Eur J Cancer. 2002;38:1023–1033. doi: 10.1016/s0959-8049(02)00059-x. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham D., Atkin W., Lenz H.J., Lynch H.T., Minsky B., Nordlinger B., Starling N. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 6.Khatri V.P., Petreli N.J., Belghiti J. Extending the frontiers of surgical therapy for hepatic colorectal metastases. Is there a limit? J Clin Onc. 2005;23:8490–8499. doi: 10.1200/JCO.2004.00.6155. [DOI] [PubMed] [Google Scholar]
- 7.Mentha G., Terraz S., Andres A., Toso C., Rubbia-Brandt L., Majno P. Operative management of colorectal liver metastases. Semin Liver Dis. 2013;33:262–272. doi: 10.1055/s-0033-1351785. [DOI] [PubMed] [Google Scholar]
- 8.Vibert E., Canedo L., Adam R. Strategies to treat primary unresectable colorectal liver metastases. Semin Oncol. 2005;32:33–39. doi: 10.1053/j.seminoncol.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Nordlinger B., Guiguet M., Vaillant J.C., Balladur P., Boudjema K., Bachellier P., Jaeck D. Surgical resection of colorectal carcinoma metastases to the liver: a prognostic scoring system to improve case selection, based on 1568 patients. Association Francaise de Chirurgie. Cancer. 1996;77:1254–1262. [PubMed] [Google Scholar]
- 10.Fong Y., Fortner J., Sun R.L., Brennan M.F., Blumgart L.H. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–321. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwatsuki S., Dvorchik I., Madariaga J.R., Marsh J.W., Dodson F., Bonham A.C., Geller D.A., Gayowski T.J., Fung J.J., Starzl T.E. Hepatic resection for metastatic colorectal adenocarcinoma: a proposal of a prognostic scoring system. J Am Coll Surg. 1999;189:291–299. doi: 10.1016/s1072-7515(99)00089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schindl M., Wigmore S.J., Currie E.J., Laengle F., Garden O.J. Prognostic scoring in colorectal cancer liver metastases: development and validation. Arch Surg. 2005;140:183–189. doi: 10.1001/archsurg.140.2.183. [DOI] [PubMed] [Google Scholar]
- 13.Zakaria S., Donohue J.H., Que F.G., Farnell M.B., Schleck C.D., Ilstrup D.M., Nagorney D.M. Hepatic resection for colorectal metastases: value for risk scoring systems? Ann Surg. 2007;246:183–191. doi: 10.1097/SLA.0b013e3180603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martí J., Fuster J., Solà A.M., Hotter G., Molina R., Pelegrina A., Ferrer J., Deulofeu R., Fondevila C., García-Valdecasas J.C. Prognostic value of serum neutrophil gelatinase-associated lipocalin in metastatic and nonmetastatic colorectal cancer. World J Surg. 2013;37:1103–1109. doi: 10.1007/s00268-013-1930-z. [DOI] [PubMed] [Google Scholar]
- 15.Tejpar S. The multidisciplinary management of gastrointestinal cancer. The use of molecular markers in the diagnosis and treatment of colorectal cancer. Best Pract Res Clin Gastroenterol. 2007;21:1071–1087. doi: 10.1016/j.bpg.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Martí J., Modolo M.M., Fuster J., Comas J., Cosa R., Ferrer J., Molina V., Romero J., Fondevila C., Charco R. Prognostic factors and time-related changes influence results of colorectal liver metastases surgical treatment: a single-center analysis. World J Gastroenterol. 2009;15:2587–2594. doi: 10.3748/wjg.15.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marfà S., Crespo G., Reichenbach V., Forns X., Casals G., Morales-Ruiz M., Navasa M., Jiménez W. Lack of a 5.9 kDa peptide C-terminal fragment of fibrinogen α chain precedes fibrosis progression in patients with liver disease. PLoS One. 2014;9:e109254. doi: 10.1371/journal.pone.0109254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Wu J., Zhang W., Zhang N., Guo H. Identification of serum CCL15 in hepatocellular carcinoma. Br J Cancer. 2013;108:99–106. doi: 10.1038/bjc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung L., Moore K., Phillips L., Boyle F.M., Marsh D.J., Baxter R.C. Novel serum protein biomarker panel revealed by mass spectrometry and its prognostic value in breast cancer. Breast Cancer Res. 2014;16:R63. doi: 10.1186/bcr3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asano T., Koizumi S., Takagi A., Hatori T., Kuwabara K., Fujino O., Fukunaga Y. Identification of a novel biomarker candidate, a 4.8-kDa peptide fragment from a neurosecretory protein VGF precursor, by proteomic analysis of cerebrospinal fluid from children with acute encephalopathy using SELDI-TOF-MS. BMC Neurol. 2011;11:101. doi: 10.1186/1471-2377-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Y., Tan C.W., Shen H., JK Y., Fang X.F., Jiang W.Z., Zheng S. Identification of the biomarkers for the prediction of efficacy in first-line chemotherapy of metastatic colorectal cancer patients using SELDI-TOF-MS and artificial neural networks. Hepatogastroenterology. 2012;59:2461–2465. doi: 10.5754/hge12127. [DOI] [PubMed] [Google Scholar]
- 22.Martí J., Fuster J., Estanyol J.M., Fernández F., Deulofeu R., Ferrer J., Pelegrina A., Reyes A., Fondevila C., García-Valdecasas J.C. Clinical and prognostic usefulness of serum proteomic profile in hepatic colorectal metastases: a pilot prospective study. Clin Transl Oncol. 2013;15:691–697. doi: 10.1007/s12094-012-0990-0. [DOI] [PubMed] [Google Scholar]
- 23.Zhu D., Zhong Y., Wu H., Ye L., Wang J., Li Y., Wei Y., Ren L., Xu B., Xu J. Predicting metachronous liver metastasis from colorectal cancer using serum proteomic fingerprinting. J Surg Res. 2013;184:861–866. doi: 10.1016/j.jss.2013.04.065. [DOI] [PubMed] [Google Scholar]
- 24.Cecchin E., Agostini M., Pucciarelli S., De Paoli A., Canzonieri V., Sigon R., De Mattia E., Friso M.L., Biason P., Visentin M. Tumor response is predicted by patient genetic profile in rectal cancer patients treated with neo-adjuvant chemo-radiotherapy. Pharmacogenomics J. 2011;11:214–226. doi: 10.1038/tpj.2010.25. [DOI] [PubMed] [Google Scholar]
- 25.Onida F., Kantarjian H.M., Smith T.L., Ball G., Keating M.J., Estey E.H., Glassman A.B., Albitar M., Kwari M.I., Beran M. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99:840–849. doi: 10.1182/blood.v99.3.840. [DOI] [PubMed] [Google Scholar]
- 26.Kim J.Y., Hildebrandt M.A., Pu X., Ye Y., Correa A.M., Vaporciyan A.A., Wu X., Roth J.A. Variations in the vascular endothelial growth factor pathway predict pulmonary complications. Ann Thorac Surg. 2012;94:1079–1084. doi: 10.1016/j.athoracsur.2012.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savas S., Liu G., Xu W. Special considerations in prognostic research in cancer involving genetic polymorphisms. BMC Med. 2013;11:149. doi: 10.1186/1741-7015-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.