Abstract
ZAM is an long terminal repeat (LTR) retrotransposon from Drosophila melanogaster that bears striking resemblance to the vertebrate retroviruses, in their structure and replication cycle. This element transposes via an RNA intermediate and its reverse transcription, and ultimately inserts copies within the germ line. In this paper, we show that intercellular communication established between the germ line cells and the somatic follicle cells is used to initiate the replication cycle of ZAM. ZAM has been shown to be transcribed in the follicle cells located at the posterior pole of the oocyte. Here, we determine the cis-regulatory elements necessary for its somatic expression, and show that they respond to the EGF-receptor signaling pathway and its activation by the ligand Gurken emitted by the germ line. We further show that the ETS-transcription factor Pointed2 acting downstream of this pathway acts as a trans-regulatory factor and targets a specific cis-regulatory binding site located within the ZAM LTR. Our data give an insight into the molecular mechanism for how intercellular communications between germ cells and somatic cells may be used by endogenous retroviruses to control their replication, and thereby specify their intrinsic and highly restricted expression in the reproductive apparatus.
INTRODUCTION
LTR retrotransposons share structural and functional homologies with the proviral form of vertebrate retroviruses and form a large and widespread family of mobile genetic elements present in all eukaryotic genomes studied so far. When active, these elements propagate by reverse transcription of RNA intermediates. The synthesized cDNA copies insert themselves within their host genome, which results in the inactivation or activation of genes located in the vicinity. These transpositions can then be a major source of mutations and as such contribute to the induction and/or enhancement of pathological processes. Nevertheless, retrotransposons are ancient constituents of eukaryotic genomes, indicating that their transposition is tightly controlled so as not to be too deleterious, and that they have now reached a status and a life cycle compatible with their own host's life cycle. In that respect, they constitute an adequate system to study interactions that should exist between these potentially mutagenic, parasite-like elements and their host.
Although their control at different steps of their replication cycle can limit transposition, transcriptional initiation is the first rate-limiting step in this process. Thereby, LTR retrotransposons display a strict pattern of expression, confining their transcription to some very specific tissues (1–5). Such specificities involve interactions between host-encoded, trans-regulatory proteins and complementary cis-regulatory protein binding sites located within the retrotransposon (6).
For the past few years, our laboratory has been engaged in studying the control of an LTR retrotransposon, ZAM from Drosophila melanogaster, that is similar in structure and replication cycle to mammalian retroviruses (1,7). This element is generally present with a very low copy number (not exceeding two copies) in the lines studied. However, one line called Rev exists in which this control has been disturbed, resulting in a high expression of ZAM and an increased mobilization rate. In the process of analyzing this control, we have found that ZAM expression is subject to two types of controls: one restricts its expression to specific somatic cells in the ovaries; the other silences this expression in most of the lines tested but not in ‘Rev’. We previously reported that this latter control, which depends on the genetic background of the line, is controlled by an heterochromatic locus located in 20A2-3 on the X chromosome (8). However, we had not investigated the molecular basis of its control that restricts expression to a specific subset of somatic cells located at the posterior pole of the oocyte and that was only observed in the Rev line when silencing is abolished. In this study, we carried out an in-depth analysis of the intrinsic regulatory properties of ZAM and the host-encoded factor that defines this strict tissue specificity.
The expression pattern of LTR retrotransposons is partly controlled by sequences located in cis, on the retrotransposon proper and generally on the LTR. The LTRs are composed of a U3 region, a central R and a U5 region. The transcription initiation site defines the boundary between U3 and R regions. The polyadenylation site corresponds to the boundary between R and U5 regions. We have found that the first 190 bp of the U3 region of ZAM can drive expression of reporter genes in a pattern that reproduces the ovary expression pattern of endogenous ZAM elements. This establishes this 190 bp as a short and defined enhancer. We used this accessible system to identify factors involved in the transcriptional specificity. Since ZAM is transcribed in the follicle cells located at the posterior pole of the oocyte, we focused our attention on Drosophila-encoded factors known to be necessary for the differentiation of these cells.
During Drosophila oogenesis, cell–cell signaling occurs between the germ-line-derived oocyte and the somatically-derived follicle cells that surround it. The follicle cell layer becomes polarized along the anterior–posterior axis when the oocyte induces the follicle cells at one end of the egg chamber to adopt a posterior rather than an anterior fate (9). This induction requires Gurken in the germ line and the Drosophila homologue of the epidermal growth factor receptor, EGFR, in the soma (10,11). Gurken is thought to bind directly to the EGFR to activate a typical receptor tyrosine kinase signal transduction cascade that specifies posterior fate (12,13). Some of the effectors and target genes of EGF receptor pathway have been identified (14–16). One of them is an ETS transcription factor encoded by the pointed gene that is expressed in response to activation of the EGF receptor pathway, and brings about its negative regulation (17).
The present study has revealed a connection between EGF receptor signaling and regulation of ZAM expression. Indeed, we have shown that the EGF receptor pathway is necessary for the expression of ZAM in the posterior follicle cells. The transcriptional factor involved has been identified as the Pointed2 protein, an ETS-binding protein that is able to bind an ETS-binding site located at position 79 on the ZAM sequence.
MATERIALS AND METHODS
Fly stocks
The fly strains used were w1118 an S-line in which ZAM is silenced, and Rev, a U-line in which ZAM expression is active (both from the collection of INSERM U384); [hs-FLP 12, FRT 42D topCO, FRT 42D hs-myc] (18); pntΔ88 (19); UASpntP1, UASpntP2 (20); GAL 4 line 55B (15); [w1118 70FLP; cu kar2 Sb/TM6, Ubx eS] (kindly provided by Kent Golic); [w1118; P{w + mC = UAS-GFP.nls}14], [w1118; P{ry + t7.2 = neoFRT}82B P{w + t* ry + t* = white-un1}90E], [w*; P{ry + t7.2 = neoFRT}82B P{w + mC = Ubi-GFP}83] (Bloomington Stock Center). To obtain the pntΔ88 allele together with an FRT site on the third chromosome, meiotic recombination assays were generated in a set of crosses involving two fly stocks: the pntΔ88 stock from the name of its mutated allele and [w*; P{ry + t7.2 = neoFRT}82B P{w + t* ry + t* = white-un1}90E]. The pZ499, pZ310 and pZ190 transgenic lines were obtained by injection of pZ499, pZ310 and pZ190 transformation vectors into w1118 flies. These transgenic lines were established with the X chromosome of Rev unstable line and autosomes of w1118 line. Flies used for analysis of expression were raised and kept at 25°C.
Generation of clones
We used FRT/FLP techniques to generate clones of follicle cells homozygous for a null mutation in the EGFR gene, topCO (18,21,22), or a null allele of pointed, pntΔ88. Females with genotypes [hs-FLP 12; FRT 42D topCO/FRT 42D hs-myc; ZAM-LTR/+] and [hs-FLP 70; ZAM-LTR/+; FRT 82B pntΔ88/FRT 82B Ubi-GFP], respectively, were fed yeast for 24 h, heat shocked for 1 h in a 37°C water bath and placed back in yeasted vials at 25°C for 60 h. Females [hs-FLP 12; FRT 42D topCO/FRT 42D hs-myc; ZAM-LTR/+] were then heat shocked again for 1 h at 37°C to induce expression of the myc tag. Females were dissected and fixed 1.5–2 h later.
DNA constructs
The pZ499 also called ZAM-LacZ has been described by Desset et al. (8). Sub-fragments of ZAM-LTR were amplified by PCR using primers Cliqt35 (5′-AGTTACCGACCCATCG-3′) and ZAM190CiBam (5′-CGGATCCGTATGCGTTGTTCTGTCTGAG-3′) for pZ190 construct, and Cliqt35 and ZAM 8271Ci (5′-CGAATTCGTCTTAAATGGGCTAACAG-3′) for pZ310 construct. The ZAM-LTR fragments extending, respectively, from nucleotides 1 to 190 and 1 to 310 were inserted into the EcoRI site of the pLacZ vector carrying the Escherichia coli LacZ gene and the minimum Hsp70 promotor. These constructs called pZ190 and pZ310 were injected into w1118 flies, and a minimum of two independent transgenics lines were established. Then, by crosses with the Rev line, these transgenes were put in a genetic background allowing ZAM expression.
Histochemical staining for β-galactosidase
Ovaries were dissected in 1× phosphate-buffered saline (PBS), fixed in 0.5% glutaraldehyde in PBS for 4 min at room temperature and rinsed twice in 1× PBS and in Fe/NaP buffer (0.003 M Na2HPO4, 0.072 M NaH2NPO4, 0.003 M K3Fe(CN)6, K4Fe(CN)6, 0.15 M NaCl, 0.001 M MgCl2). Staining was performed in Fe/NaP buffer with X-Gal (0.2 mg/ml) final concentration at 37°C. All samples were stained simultaneously and for the same length of time (2 h). Stained tissues were washed 4 times in 1× PBS and examined under an Axiophot microscope (Zeiss) using Nomarski optics.
Immunohistochemical procedures and microscopy
Ovaries were dissected in PBS and fixed in 4% formaldehyde for 15 min. After washing in PBT, tissues were permeabilized and blocked for 4 h in PBT at room temperature. The anti-LacZ primary polyclonal antibody (Sigma) was diluted to 1/2000. The Cy3 conjugated anti-rabbit secondary antibody (Molecular Probes) was diluted to 1/300. The anti α-tubulin (Sigma) was diluted to 1/2000. The Cy5 conjugated anti-mouse secondary antibody (Molecular Probes) was diluted to 1/300. Light and fluorescence microscopy were performed on Axiophot (Zeiss) and confocal microscopy was performed on an Olympus confocal microscope.
Preparation of Pointed fusion protein
The GST-PntC construct was kindly provided by R. Carthew (23). The GST–PntC fusion protein contains the DNA binding domain of Pointed fused to GST. Expression of the GST–PntC fusion protein was induced in the E.coli BL21 by addition of 0.1 mM IPTG during the log-phase bacterial culture for 4 h at 30°C. Bacteria were collected by centrifugation and the pellet was resuspended in a lysis buffer containing an anti-proteinase cocktail. Cells were broken by brief sonication and cleared from insoluble material by centrifugation. Gag fusion protein was collected from the supernatant with 500 μl of glutathione agarose (GSH) beads (Sigma) for 1 h at room temperature. Fusion protein was eluted from the beads by gentle shaking in 50 mM Tris–HCl, pH 9, 5 mM reduced glutathione for 10 min at 4°C. Fusion protein yield was estimated by Bradford spectrophotometric analysis and the quality of each fraction was tested by SDS-PAGE.
Mobility shift assays
The 190 bp fragment of ZAM was excised from plasmid pZ190 by an EcoRI digestion, and purified. Oligonucleotides used are as follows: ZAMets 5′-CGAACCGGGAAGCTT-3′ (forward) and 5′-AAGCTTCCCGGTTCG-3′ (reverse). The sequences of the Mut oligonucleotides were 5′-CGAACCGCCTTGCTT-3′ (forward) and 5′-AAGCAAGGCGGTTCG-3′ (reverse) (underlined nucleotides are mutated). Double-stranded fragment probes were prepared with the Invitrogen T4 polynucleotide kinase Kit and [γ-32P]dATP at 3000 Ci/mmol. The DNA binding reaction (20 μl) is carried out by incubating 44, 88 and 264 ng purified GST fusion protein or GST alone with 20 000 c.p.m. of labelled dsDNA fragment probe in 10 mM HEPES (pH 7.9), 1 mM DTT, 100 mM KCl, 10% (v/v) glycerol, 1 μg poly(dI–dC), 10 μg BSA for 20 min at 25°C. DNA–protein complexes are resolved on 4% non-denaturing polyacrylamide gels (19:1) containing 0.5× TBE. In competition experiments, a 25- or 100-fold excess of unlabeled oligonucleotides was added in binding reactions.
RESULTS
Reporter constructs containing the ZAM U3 region reproduce the follicle cell-specific expression of ZAM
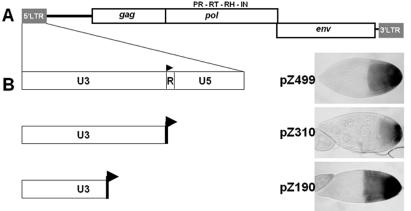
The expression pattern of LTR retrotransposons is generally governed by sequences contained within the retrotransposon itself. We previously reported that indeed, the tissue specificity and a strain control are determined by the ZAM LTR (8). In order to focus on the tissue specificity of its transcription and better localize sequences involved, we constructed plasmids containing ZAM sequences and a LacZ reporter gene which could mimic ZAM expression in vivo. To this end, a PCR-amplified ZAM fragments containing the U3 region (1–310) was fused to the E.coli LacZ gene in a P-element transformation vector. Since the 310 bp do not contain the ZAM promoter (7), they have been cloned in a transgenic vector upstream of a minimum hsp70 promoter fused to LacZ. This construct has been called pZ310. It has been injected into w1118 flies, and five independent transgenic lines were established. Using a histochemical assay for β-galactosidase activity, we have studied the expression of the LacZ reporter gene controlled by ZAM U3 sequences. In two independent transgenic lines, β-galactosidase activity is detected in the somatic follicle cells located at the posterior pole of follicles all along the ovarioles (Figure 1). This transcription of LacZ exactly reproduces the expression pattern of endogenous ZAM elements (1) and of the full length LTR of ZAM when fused to LacZ [(Figure 1A and (8)]. To define the sequence involved more precisely, we then tested the first 190 bp from the N-terminal part of U3 (1–190) fused to LacZ (pZ190). The established transgenic line obtained with such a transgene expressed the LacZ reporter in the follicle cells located at the posterior pole of the oocyte. This indicated that the enhancer specific to this patch of cells is contained within the 190 bp of U3. The transgenic approaches used in this experiment demonstrate that the tissue-specific expression of ZAM is only dependent on sequences present in the U3 region of ZAM LTR and more precisely in its first 190 bp.
Figure 1.
ZAM expression in the ovaries of adult females. (A) Molecular structure of ZAM. (B) On the left, ZAM fragments placed upstream of LacZ. Transgenes contain the full length 499 bp LTR (pZ499), the U3 region of 310 bp (pZ310), or the 5′ part of U3 up to nucleotide 190 (pZ190). For each construct, an example of histochemical detection of β-galactosidase activity in the follicle cells located at the posterior pole of stages 9 and 10 follicles is presented on the right.
The EGF receptor signaling pathway controls the tissue-specific expression of ZAM
The identity of the terminal follicle cells where ZAM expression is detected, is determined by an oocyte signal transmitted to the follicle cells by the EGF receptor localized at the surface of the follicle cells (18). We therefore investigated whether the EGF receptor pathway is also responsible for the tissue specificity of ZAM. For this purpose, we tested the responsiveness of the U3 region through the pZ310 transgenes to signaling molecules of the EGF pathway, and searched whether mutations affecting this signaling also affect the expression of the transgenes.
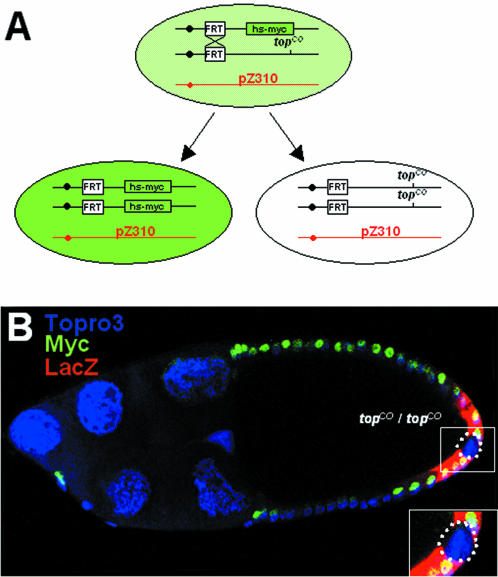
In the first set of experiments, we generated small marked mosaic clones homozygous for topCO [a null allele of the EGF receptor (Egfr) gene, also called torpedo in D.melanogaster] in the posterior follicle cells during oogenesis, and followed their ZAM control by staining for the β-galactosidase activity of the pZ310 transgene. The generation of Egfr−/− follicle cell clones were induced through FLP mitotic recombination in females trans-heterozygous for hs-myc and topCO. One daughter of a recombinant follicle cell carries two copies of myc, whereas the other lacks myc and is homozygous for topCO (Figure 2A).
Figure 2.
The tissue-specific expression of ZAM depends on the EGF receptor pathway. (A) Scheme for the generation of Egfr−/− follicle cell clones. Flp-mediated recombination was induced in females trans-heterozygous for hs-myc and topCO, a null allele of the EGF receptor of Drosophila, torpedo. One daughter of a recombinant follicle cell carries two copies of myc, whereas the other lacks myc and is homozygous for topCO. (B) A stage 10 egg chamber carrying mutant topCO follicle cell clones at the posterior pole of the oocyte. In these mutant clones highlighted by broken lines, no LacZ expression (red) is detected. The wild-type clones express the myc marker (green) and LacZ at the posterior pole. In the whole structure, nuclei are in blue due to the ToPro3 marker.
In homozygous topCO clones at the posterior follicle cells of stage 9 to 10 of oogenesis, we found that ZAM expression is abolished (Figure 2B). The LacZ staining is specifically absent in the mutant topCO cells whereas it is detected in the surrounding cells expressing myc. This pattern of expression has been found in all the four clones generated in this experiment. Since removal of the Egfr causes an absence of ZAM expression, we conclude that ZAM expression is dependent on the EGF signaling pathway.
The Pointed transcription factor activates ZAM expression
In order to identify the transcription factor involved in ZAM expression, we next investigated whether a transcription factor acting downstream of the EGF receptor signaling pathway could be involved in transducing the EGF-type signal to the ZAM promoter (17,18). Candidate factors potentially involved are encoded by the pointed gene that is transcribed as ZAM in the follicle cells at the posterior pole of the oocyte. Pointed (pnt) encodes two transcription factors, PntP1 and PntP2, containing the same ETS domain that mediates sequence-specific DNA binding (24,25).
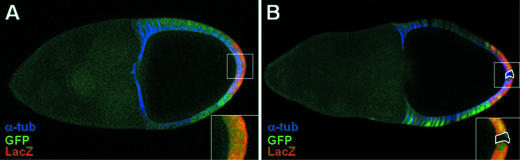
In flies bearing the pZ190 transgene, we generated marked mosaic clones, homozygous for pntΔ88, a null allele for both PntP1 and PntP2 (19), in the posterior follicle cells. A wild-type chromosome was marked with the GFP gene controlled by a ubiquitin promotor which directs expression in all the follicle cells. The chromosome bearing the pntΔ88 null allele does not carry the GFP marker. Thus pntΔ88 homozygous mutant cells can be easily detected due to a lack of GFP expression. On another hand, ZAM expression is followed by staining for the β-galactosidase activity of the pZ190 transgene. Eleven mosaic clones were generated in posterior follicle cells. LacZ staining was never detected in these 11 clones, indicating that pZ190 is only expressed in cells in which Pointed is expressed (Figure 3). Thus, we conclude that ZAM expression depends on the Pointed transcription factors.
Figure 3.
A loss of Pointed expression results in a loss of ZAM expression in the posterior follicle cells. (A) Wild-type follicle cells at the posterior pole of the oocyte. The wild-type cells express the GFP marker (green) and LacZ at the posterior pole. (B) In a pntΔ88 mutant clone (highlighted by a broken line) at the posterior pole of the oocyte, no LacZ expression (red) is detected. The overall structure of the ovariole is labelled for α-tubulin (blue).
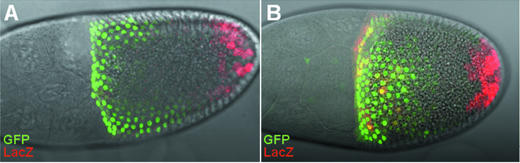
To further investigate the implication of Pointed in the restricted expression of ZAM in follicle cells with a posterior fate, we ectopically expressed pointed in another cell lineage owing to the GAL4/UAS system (26) and examined the pZ190 expression. The GAL4 driver present in line 55B was used to induce expression of UAS-target genes in the anterior follicle cells at the ventral and dorsal sides of oocytes in stages 8 to 10, which is illustrated by staining for GFP expressed from an UAS-GFP target transgene [Figure 4 and (15)]. The Gal4 line 55B carrying the UAS-GFP transgene as a marker, was then crossed to lines bearing Gal4-responsive pntP1 or pntP2 genes: UAS-pntP1 or UAS-pntP2. This cross led to two independent lines with the following genotypes: [UAS-GFP/CyO; UAS-pntP1/55-Gal4] and [UAS-GFP/CyO; UAS-pntP2/55-Gal4] in which GFP and PntP1 or PntP2 are co-expressed in the anterior follicle cells. Submitted to the Gal4 driver, expression of these transgenes is thus driven in the same cells as the UAS-GFP transgene. Their expression is then indirectly followed through staining for GFP (Figure 4). In a final set of crosses, pZ190 was introduced within the genome leading to flies with the following genotypes: [UAS-GFP/pZ190; UAS-pntP1/55-Gal4] and [UAS-GFP/pZ190; UAS-pntP2/55-Gal4]. This allowed testing of the effects of ectopic expression of PntP1 and PntP2, respectively, on ZAM transcription by staining for β-galactosidase activity.
Figure 4.
Ectopic expression of Pointed P2 results in ectopic expression of ZAM. (A) and (B) display X-Gal staining (red) resulting from the expression of the pZ190 transgene when Pointed P1 and Pointed P2, respectively, are expressed in the anterior follicle cells using the 55B-Gal4 driver. The ectopic expression of UAS-Pointed P1 and UAS-Pointed P2 is followed due to an UAS-GFP transgene used as a marker. PntP1 and pZ190 are never co-expressed in the anterior follicle cells, while a clear expression of pZ190 is detected in numerous cells of this lineage when PntP2 is expressed.
When ovaries dissected from females [UAS-GFP/pZ190; UAS-pntP1/55B-Gal4] were examined no expression of pZ190 over 10 independent clones examined could ever be detected in the anterior follicle cells where PntP1 is ectopically expressed (Figure 4A). In contrast, in ovaries dissected from females with the [UAS-GFP/pZ190; UAS-pntP2/55B-Gal4] genotype, LacZ staining due to an ectopic expression of pZ190 generated was clearly detected in numerous cells located at the anterior pole of the oocyte, and expressing PntP2 (Figure 4B). As illustrated in Figure 4B, LacZ staining due to the expression of pZ190 is not observed in all the cells expressing GFP. Several reasons can be considered. First, the PntP2 transgene may not be expressed in all the cells in which the UAS-GFP transgene is present. Second, the incomplete rescuing of LacZ expression could be due to a lower level of PntP2 expression compared to its endogenous expression within the posterior follicle cells. Third, PntP2 is active as a phosphorylated protein (27). When ectopically expressed, PntP2 may be less phosphorylated and therefore less efficient. Fourth, the transcriptional activation of ZAM by PntP2 may require a specific co-factor expressed at a limited rate in these anterior cells. Nevertheless, these experiments clearly show that activation of ZAM transcription may be partially recovered, in follicle cells ectopically expressing PntP2. These data indicate a functional requirement of PntP2 for proper patterning of ZAM expression in follicle cells with a posterior fate.
Pnt interacts with an ETS binding site located within the first 190 bp of ZAM
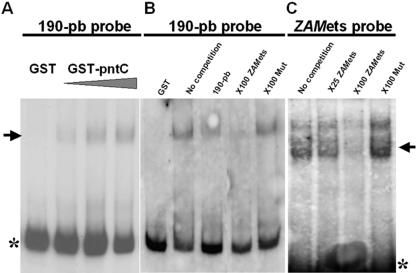
As Pointed is a transcription factor, there remains the question of whether it binds to the 190 bp of ZAM directly or indirectly. We expressed the DNA binding domain of Pointed tagged with GST in bacteria, and performed gel-shift mobility assays using the 190 bp fragment as a probe (see Materials and Methods). We have found that the GST–PntC fusion protein does indeed bind the 190 bp long probe of ZAM (Figure 5A) while a control lysate programmed with a GST peptide alone, with no Pointed sequence, is not. Additionally, the ability of an unlabeled 190 bp fragment to outcompete the DNA binding complex confirmed this result (Figure 5B).
Figure 5.
The ETS-transcription factor, Pointed, binds to an ETS-binding site located 79 bp within ZAM LTR. The DNA binding domain of Pointed fused to GST (GST–PntC) was used in these assays. (A) The GST–PntC protein specifically binds the first 190 bp of ZAM. Increasing amounts of GST-PntC used in this gel-shift assay are 44, 88 and 264 ng. In comparison, no shift is observed with 250 ng of GST protein alone. (B) Competition experiments: binding is tested with a 264 ng of unlabeled GST–PntC, 100-fold excess of ZAMets and 100-fold excess of Mut an oligonucleotide mutated for the EBS (see Materials and Methods). (C) Competition experiments: the GST–PntC specifically binds the ZAMets oligonucleotide (from nucleotides 72 to 86 on ZAM sequence) which contains the presumptive EBS. A 100-fold excess of unlabelled ZAMets competes with the labelled oligonucleotide for the binding. A 100-fold excess of unlabelled oligonucleotide Mut, mutated for the EBS, is unable to compete. The asterisk and the arrow indicate the free probe and the gift, respectively.
We then proceeded to determine which site within the 190 bp is responsible for this binding, and scanned the ZAM promoter region in silico for putative ETS binding sites (EBS). One site potentially recognized by a Drosophila ETS transcription factor (5′-GGAA-3′) was found at position 79 according to ZAM sequence (28). If the ETS transcription factor is Pointed, then it should associate directly with this ETS-binding site, and one would expect to see competition when an excess of an oligonucleotide encompassing the domain is used. This is indeed what has been obtained. As shown in Figure 5B, the ZAMets oligonucleotide (from nucleotides 72 to 86 on ZAM sequence, see Material and Methods) efficiently competed with the complex. Additionally, an oligonucleotide such as Mut identical to ZAMets but with a mutated ETS binding site (see Materials and methods) is then unable to compete with the complex (Figure 5B). These results indicate that the transcription factor Pointed directly binds an ETS-binding site present at position 79 within the ZAM sequence.
If Pointed associates with the ETS-binding site of ZAM, it should bind the ZAMets oligonucleotide in gel-shift assays. We performed a gel-shift assay using the ZAMets oligonucleotide as a probe. We have found that the GST–PntC fusion protein does indeed bind ZAMets (Figure 5C), while a control lysate with a GST peptide alone does not. Additionally, an unlabeled ZAMets primer added as a 25- or 100-fold excess is able to outcompete the DNA binding complex while Mut, the mutated primer for the EBS is not (Figure 5C). Overall, these results indicate that Pointed associates with an ETS binding site located at positions 79 to 82 within the ZAM LTR.
DISCUSSION
Oogenesis involves direct interaction between germline cells and somatic follicle cells for the successful development of fertile eggs (29). We have found that such intercellular communications from germ line to somatic line in the first instance, and from somatic line to germ line in the second also play crucial roles in the replication cycle of an LTR retrotransposon ZAM from D.melanogaster. We previously demonstrated the necessity of a somatic cell to germ cell communication for the transport of ZAM particles. Indeed, ZAM particles formed within the follicle cells take benefit of the vittelogenin traffic to sort out of the follicle cells and enter the closely apposed oocyte (1). Here, we show that in the first steps of its replication cycle, ZAM requires another signal transmitted by the germ line and received by the somatic line.
The somatic transcription of ZAM depends on the Pointed factor acting downstream of the EGF receptor pathway
We previously reported that ZAM transcripts are detected in a group of somatically derived cells, which are the follicle cells surrounding the posterior part of the oocyte (1). This transcription of ZAM implies that a tissue-specific promoter controls its expression during oogenesis. By analyzing the long terminal repeat and specifially introducing the U3 region (5′ part of the LTR) upstream of the reporter gene LacZ into flies (leading to the so-called pZ310 transgenic flies), we demonstrate that this region contains the tissue-specific enhancer of ZAM. Furthermore, a similarly restricted pattern of expression was found in another series of transgenic flies called pZ190 that we have generated using 190 bp of the 5′ end of U3. LacZ expression was found to be similar to that observed when the full-length LTR or the U3 region was fused to LacZ. These transgenic lines indicate that the essential cis-regulatory elements for the transcriptional regulation of the ZAM promoter are included within its first 190 bp.
It has been well described that the EGF receptor pathway is necessary for the differentiation of these very specific follicle cells where ZAM is expressed. Through a genetic approach, we further demonstrated that the first 190 bp of ZAM are also the direct target of the EGF receptor pathway. Mosaic clones of cells homozygous for a null allele of the torpedo gene, the Drosophila EGF receptor, were generated within the posterior follicle cells. LacZ staining due to the pZ310 (U3–LacZ) or pZ190 (190 bp–LacZ) transgene expression was undetected in the clones, whereas it was present within surrounding cells with a wild-type genotype.
Various factors have been shown to be involved in mediating a transcriptional response upon the reception of signals from the EGF receptor pathway. Two of them are the isoforms encoded by the pointed genes: Pointed1 and Pointed2. Ectopic expression of Pointed2 in a subset of follicle cells with an anterior fate of differentiation (instead of a posterior fate as necessary for ZAM expression) has shown that the presence of Pointed2 causes an ectopic expression of the pZ190 transgene in these anterior follicle cells. This result contrasts with what is observed in a wild-type background or with ectopic expression of Pointed1 in the anterior follicle cells, which does not allow ZAM expression within those follicle cells with an anterior fate. Our data therefore indicate that Pointed2 is a necessary positive effector in ZAM expression. Pnt2 is required for proper cell determination during various stages of Drosophila development: in the eyes or during the development of renal tubules (30–32). Nevertheless, expression of endogenous ZAM elements has never been detected in these tissues. Similarly, when we ectopically expressed Pointed2 in the eyes under control of a Glass-Gal4 driver, no LacZ staining resulting from the ectopic expression of pZ190 could be detected (data not shown). These results indicate that the expression of PntP2 is necessary but not sufficient to activate ZAM. Additional co-factors are almost certainly required for a refined regulation of its transcription in the specific tissue, i.e. the ovaries. Such co-factors have not been identified yet, however, we have to stress the fact that, at this time of our study, Pointed1 cannot be definitely discarded as a potential candidate. Indeed, if PntP2 acts in parallel or downstream of PntP1, our experiments would not have allowed us to detect its involvement in ZAM transcription.
Whatever the multiple factors involved, we have provided further evidence that Pointed, and its ETS-binding site common to PntP1 and PntP2, is able to activate ZAM transcription via its direct binding to cis-regulatory elements present in the responsive target of 190 bp. In silico we have found that an ETS binding site (5′-GGAA-3′), located within the 190 bp at position 79 according to ZAM sequence, is indeed the precise target DNA sequence recognized by Pointed. Overall, these data indicate that Pointed binds ZAM LTR and is a key factor in determining the strict expression of ZAM in the follicle cell layer in response to a signal emitted by the oocyte.
An outcome of the present investigation is that genes that are regulated by the Pointed factor and participate in differentiation of the posterior lineage of follicle cells could be identified by using the characterized target DNA sequence of ZAM as an easily detectable marker. Thus it provides an entry point to monitor the EGF receptor pathway and study mechanisms of posterior follicle cell induction and patterning in Drosophila at the transcriptional level. Additionally, our study provides an experimental tool, the ZAM promoter, which can be specifically used to direct expression of any transgene to the posterior pole of the oocyte.
Biological significance of a retroviral cycle starting within a somatic tissue, whereas its ultimate insertion occurs within the germ line
At the biological level, an intriguing question yet to be answered is why ZAM expression takes place in a somatic tissue in response to a signal (Gurken) transmitted by the oocyte while novel insertions are precisely known to occur in the germ line (7,33). We can hypothesize that it is advantageous for the retrotransposon to produce its transcripts in somatic cells to circumvent transcriptional arrest of the oocyte during meiosis. Alternatively, a high expression of ZAM occurring directly within the germ line might generate too many proviral copies of ZAM and be deleterious for the next generation. In the course of evolution, an adaptive strategy to limit the extent of ZAM insertion within the germ line could have led the element to be expressed outside of the germ line. Expression within a somatic lineage and its subsequent transfer to the germ line could decrease the efficiency of its insertion and confer a selective advantage. Alternatively, some defence mechanism may specifically protect the germ line from the presence of any retroviral/retrotransposon RNA. This constraint may have led ZAM to adopt a novel strategy to invade the germ line. It starts its replication cycle and assembles viral-like particles (VLP) in a somatic tissue of the reproductive apparatus. While encapsidated in particles, the RNA genome is converted into a cDNA. When VLPs enter the closely apposed germ line by virtue of retroviral infection, DNA copies of the retroelement are no more recognized by the host defence selectively addressed against RNA invasions. The retroviral element can then complete its replication cycle leading to insertion within the host germ-line.
Recent studies implicate the biological role of dsRNA-mediated silencing as a transposon repression and antiviral mechanism (34–37). This host surveillance system is known to be active in the germ line (38,39) and could certainly account for such a protection against LTR retrotransposons. However, numerous questions remain to be addressed to demonstrate whether this mechanism could have acted as an evolutionary constraint, resulting in ZAM, and potentially some related elements being transcribed in the somatic lineage.
This expression of ZAM within a somatic tissue apposed to the germ line is reminiscent of what is observed for some mouse or chicken viruses (40,41), or for endogenous retroviral particles which are massively expressed in the human placenta (42,43). A question raised by our data is: could some HERVs be expressed in placental tissues because of a signal emitted by the fetus? The fetal tissue would then be responsible for the acquisition of specific trophoblastic functions that have been attributed potentially to HERVs, such as protection against retroviral infection, protection of the fetus against maternal immune system, or placenta morphogenesis through fusogenic effects (44,45). Although the question is open, it would be worth studying signals coming from the fetus, that could ultimately be responsible for endogenous retroviruses activation in the placenta.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Muriel Grammont, François Mallet, Ruth Bancewicz and Sharron Vass for helpful comments and critical review of the manuscript. This work was supported by grants from INSERM (U384) and CNRS (GDR2157), and by a project grant from Association pour la Recherche contre le Cancer (ARC 3441) to C.V. C.M. received a grant from the Ministère de l'Enseignement Supérieur et de la Recherche (MESR) and the Fondation pour la Recherche Médicale (FRM).
REFERENCES
- 1.Leblanc P., Desset,S., Giorgi,F., Taddei,A.R., Fausto,A.M., Mazzini,M., Dastugue,B. and Vaury,C. (2000) Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J. Virol., 74, 10658–10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelisson A., Song,S.U., Prud'homme,N., Smith,P.A., Bucheton,A. and Corces,V.G. (1994) Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J., 13, 4401–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matyunina L.V., Jordan,I.K. and McDonald,J.F. (1996) Naturally occurring variation in copia expression is due to both element (cis) and host (trans) regulatory variation. Proc. Natl Acad. Sci. USA, 93, 7097–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lammel U. and Klambt,C. (2001) Specific expression of the Drosophila midline-jumper retro-transposon in embryonic CNS midline cells. Mech. Dev., 100, 339–342. [DOI] [PubMed] [Google Scholar]
- 5.Vogel A.M. and Gerster,T. (1999) Promoter activity of the zebrafish bhikhari retroelement requires an intact activin signaling pathway. Mech. Dev., 85, 133–146. [DOI] [PubMed] [Google Scholar]
- 6.Cavarec L., Jensen,S., Casella,J.F., Cristescu,S.A. and Heidmann,T. (1997) Molecular cloning and characterization of a transcription factor for the copia retrotransposon with homology to the BTB-containing lola neurogenic factor. Mol. Cell. Biol., 17, 482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leblanc P., Desset,S., Dastugue,B. and Vaury,C. (1997) Invertebrate retroviruses: ZAM a new candidate in D.melanogaster. EMBO J., 16, 7521–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desset S., Meignin,C., Dastugue,B. and Vaury,C. (2003) COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics, 164, 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Reyes A. and St Johnston,D. (1994) Role of oocyte position in establishment of anterior–posterior polarity in Drosophila. Science, 266, 639–642. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Reyes A., Elliott,H. and St Johnston,D. (1995) Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature, 375, 654–658. [DOI] [PubMed] [Google Scholar]
- 11.Roth S., Neuman-Silberberg,F.S., Barcelo,G. and Schüpbach,T. (1995) Cornichon and the EGF receptor signalling process are necessary for both anterior–posterior and dorsal–ventral pattern formation in Drosophila. Cell, 81, 967–978. [DOI] [PubMed] [Google Scholar]
- 12.Neuman-Silberberg F.S. and Schupbach,T. (1993) The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell, 75, 165–174. [PubMed] [Google Scholar]
- 13.Neuman-Silberberg F.S. and Schupbach,T. (1996) The Drosophila TGF-alpha-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech. Dev., 59, 105–113. [DOI] [PubMed] [Google Scholar]
- 14.Lu X., Melnick,M.B., Hsu,J.C. and Perrimon,N. (1994) Genetic and molecular analyses of mutations involved in Drosophila raf signal transduction. EMBO J., 13, 2592–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brand A.H. and Perrimon,N. (1994) Raf acts downstream of the EGF receptor to determine dorsoventral polarity during Drosophila oogenesis. Genes Dev., 8, 629–639. [DOI] [PubMed] [Google Scholar]
- 16.Schnorr J.D. and Berg,C.A. (1996) Differential activity of Ras1 during patterning of the Drosophila dorsoventral axis. Genetics, 144, 1545–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morimoto A.M., Jordan,K.C., Tietze,K., Britton,J.S., O'Neill,E.M. and Ruohola-Baker,H. (1996) Pointed, an ETS domain transcription factor, negatively regulates the EGF receptor pathway in Drosophila oogenesis. Development, 122, 3745–3754. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Reyes A. and St Johnston,D. (1998) The Drosophila AP axis is polarised by the cadherin-mediated positioning of the oocyte. Development, 125, 3635–3644. [DOI] [PubMed] [Google Scholar]
- 19.Scholz H., Deatrick,J., Klaes,A. and Klambt,C. (1993) Genetic dissection of pointed, a Drosophila gene encoding two ETS-related proteins. Genetics, 135, 455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klaes A., Menne,T., Stollewerk,A., Scholz,H. and Klambt,C. (1994) The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell, 78, 149–160. [DOI] [PubMed] [Google Scholar]
- 21.Clifford R.J. and Schupbach,T. (1989) Coordinately and differentially mutable activities of torpedo, the Drosophila melanogaster homolog of the vertebrate EGF receptor gene. Genetics, 123, 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu T. and Rubin,G.M. (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development, 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
- 23.Kauffmann R.C., Li,S., Gallagher,P.A., Zhang,J. and Carthew,R.W. (1996) Ras1 signaling and transcriptional competence in the R7 cell of Drosophila. Genes Dev., 10, 2167–2178. [DOI] [PubMed] [Google Scholar]
- 24.Klambt C. (1993) The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development, 117, 163–176. [DOI] [PubMed] [Google Scholar]
- 25.Kodandapani R., Pio,F., Ni,C.Z., Piccialli,G., Klemsz,M., McKercher,S., Maki,R.A. and Ely,K.R. (1996) A new pattern for helix–turn–helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature, 380, 456–460. [DOI] [PubMed] [Google Scholar]
- 26.Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 27.Baonza A., Murawsky,C.M., Travers,A.A. and Freeman,M. (2002) Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat. Cell. Biol., 4, 976–980. [DOI] [PubMed] [Google Scholar]
- 28.Macleod K., Leprince,D. and Stehelin,D. (1992) The ets gene family. Trends Biochem. Sci., 17, 251–256. [DOI] [PubMed] [Google Scholar]
- 29.Goode S., Wright,D. and Mahowald,A.P. (1992) The neurogenic locus brainiac cooperates with the Drosophila EGF receptor to establish the ovarian follicle and to determine its dorsal–ventral polarity. Development, 116, 177–192. [DOI] [PubMed] [Google Scholar]
- 30.Brunner D., Ducker,K., Oellers,N., Hafen,E., Scholz,H. and Klambt,C. (1994) The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature, 370, 386–389. [DOI] [PubMed] [Google Scholar]
- 31.O'Neill E.M., Rebay,I., Tjian,R. and Rubin,G.M. (1994) The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell, 78, 137–147. [DOI] [PubMed] [Google Scholar]
- 32.Sudarsan V., Pasalodos-Sanchez,S., Wan,S., Gampel,A. and Skaer,H. (2002) A genetic hierarchy establishes mitogenic signalling and mitotic competence in the renal tubules of Drosophila. Development, 129, 935–944. [DOI] [PubMed] [Google Scholar]
- 33.Desset S., Conte,C., Dimitri,P., Calco,V., Dastugue,B. and Vaury,C. (1999) Mobilization of two retroelements, ZAM and Idefix, in a novel unstable line of Drosophila melanogaster. Mol. Biol. Evol., 16, 54–66. [DOI] [PubMed] [Google Scholar]
- 34.Aravin A.A., Naumova,N.M., Tulin,A.V., Vagin,V.V., Rozovsky,Y.M. and Gvozdev,V.A. (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol., 11, 1017–1027. [DOI] [PubMed] [Google Scholar]
- 35.Mourrain P., Beclin,C., Elmayan,T., Feuerbach,F., Godon,C., Morel,J.B., Jouette,D., Lacombe,A.M., Nikic,S., Picault,N. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- 36.Waterhouse P.M., Wang,M.B. and Lough,T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- 37.Plasterk R.H., Izsvak,Z. and Ivics,Z. (1999) Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet., 15, 326–332. [DOI] [PubMed] [Google Scholar]
- 38.Kennerdell J.R., Yamaguchi,S. and Carthew,R.W. (2002) RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev., 16, 1884–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sijen T. and Plasterk,R.H. (2003) Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature, 426, 310–314. [DOI] [PubMed] [Google Scholar]
- 40.Lock L.F., Keshet,E., Gilbert,D.J., Jenkins,N.A. and Copeland,N.G. (1988) Studies of the mechanism of spontaneous germline ecotropic provirus acquisition in mice. EMBO J., 7, 4169–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Stefano H.S. and Dougherty,R.M. (1966) Mechanisms for congenital transmission of avian leukosis virus. J. Natl Cancer Inst., 37, 869–883. [PubMed] [Google Scholar]
- 42.Villarreal L.P. and Villareal,L.P. (1997) On viruses, sex, and motherhood. J. Virol., 71, 859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris J.R. (1998) Placental endogenous retrovirus (ERV): structural, functional, and evolutionary significance. Bioessays, 20, 307–316. [DOI] [PubMed] [Google Scholar]
- 44.Blaise S., de Parseval,N., Benit,L. and Heidmann,T. (2003) Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl Acad. Sci. USA, 100, 13013–13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallet F., Bouton,O., Prudhomme,S., Cheynet,V., Oriol,G., Bonnaud,B., Lucotte,G., Duret,L. and Mandrand,B. (2004) The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl Acad. Sci. USA, 101, 1731–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]