Abstract
The phage T4Dam and EcoDam DNA-[adenine-N6] methyltransferases (MTases) methylate GATC palindromic sequences, while the BamHI DNA-[cytosine-N4] MTase methylates the GGATCC palindrome (which contains GATC) at the internal cytosine residue. We compared the ability of these enzymes to interact productively with defective duplexes in which individual elements were deleted on one chain. A sharp decrease in kcat was observed for all three enzymes if a particular element of structural symmetry was disrupted. For the BamHI MTase, integrity of the ATCC was critical, while an intact GAT sequence was necessary for the activity of T4Dam, and an intact GA was necessary for EcoDam. Theoretical alignment of the region of best contacts between the protein and DNA showed that in the case of a palindromic interaction site, a zone covering the 5′-symmetric residues is located in the major groove versus a zone of contact covering the 3′-symmetric residues in the minor groove. Our data fit a simple rule of thumb that the most important contacts are aligned around the methylation target base: if the target base is in the 5′ half of the palindrome, the interaction between the enzyme and the DNA occurs mainly in the major groove; if it is in the 3′ half, the interaction occurs mainly in the minor groove.
INTRODUCTION
Post-replicative DNA methylation has been found in members of virtually every major biological group, from viruses through mammals. This reaction is carried out by DNA methyltransferases (MTases), which catalyze methyl group transfer from donor S-adenosyl-l-methionine (AdoMet), producing S-adenosyl-l-homocysteine (AdoHcy) and methylated cytosine or adenine bases within specific nucleotide sequences. The most common role of MTases in prokaryotes and their phages is to protect DNA from the cell's restriction enzymes. However, some also act as regulators of gene expression and affect other critical functions (1). In this connection, of great interest is the elucidation of the mechanism of action of these enzymes.
Three types of prokaryotic MTases are known: [C5-cytosine], [N4-cytosine], and [N6-adenine] MTases (2). Generally, these enzymes recognize short palindromic DNA sequences. The catalytic mechanism of methyl group transfer from the donor AdoMet in the case of C5-MTases includes a covalent binding of the enzyme to the target cytidine residue (3). In contrast, [N6-adenine] and [N4-cytosine] MTases transfer the methyl group onto the exocyclic NH2-group without the formation of a transient covalent bond (2). Despite a significant variety of recognition sites, all classes show structural similarity in the catalytic domains comprising the overall fold and topology, the AdoMet binding site and in the location of the catalytic center (4). In addition, flipping of the target residue out of the DNA double helix appears to be a common property of the reaction catalyzed by all DNA MTases (5).
One of the unresolved questions on the mechanism of the interaction between MTases and their DNA target sites concerns the nature of contacts between amino acid residues and chemical groups that are included in the recognition site. While X-ray crystallographic analysis of protein–DNA complexes gives the most direct answer, such data for DNA (amino)-MTases are very limited. The structures of specific MTase–DNA complexes are known at the present time only for the [cytosine-C5] HhaI (6) and HaeIII (7) MTases, and for [adenine-N6] TaqI (8) and T4Dam MTases [(9); J. R. Horton, S. Hattman and X. Cheng, manuscript in preparation]. Other informative approaches, such as the different variants of footprinting (10), base substitution (11) and ‘dissected duplex’ analysis (12) have been used for a limited number of DNA MTases.
Earlier we showed that the phage T4 DNA MTase (T4Dam), which catalyzes methylation of the GATC palindromic sequence, has the ability to interact productively with the defective sites provided that the GAT portion is intact in both DNA strands (13). In contrast, the bacterial EcoDam MTase, which also methylates GATC, is less stringent because it requires only the GA to be intact in both strands (14). Both T4Dam and EcoDam are functional monomers at low catalytic concentrations (15,16) and they do not have symmetric structural domains (17). Hence, the nature of the requirement for intactness of symmetric elements in both DNA strands reflects the region of best contacts between each enzyme and its target. Given the structural similarity of [adenine-N6]and [cytosine-N4] MTases as well as the similarity of the active-site sequence motifs of both enzyme families, the question arises as to what the nucleotide residues are that form the region of best contacts between a [cytosine-N4] MTase and DNA. To investigate this problem, we used the BamHI DNA MTase, which recognizes GGATCC (containing an internal GATC) and modifies the internal cytosine residue in this sequence (18).
MATERIALS AND METHODS
Enzymes and chemicals
[3H-CH3]-AdoMet (15 Ci/mmol; 1 mCi/ml) was purchased from Amersham. Unlabeled AdoMet (Sigma) was purified further by chromatography on a C18-reversed-phase column as described previously (19). Oligodeoxynucleotides (ODNs; Table 1) were synthesized using ‘Applied Biosystem 380A/380B’ and their concentrations were determined spectrophotometrically. The duplexes were obtained by heating and annealing complementary ODN chains from 90 to 20°C over 7–12 h. Homogeneous BamHI MTase was kindly provided by W. Lindstrom and N. Reich (University of California, Santa Barbara, CA).
Table 1. Synthetic oligodeoxynucleotides.
| Oligonucleotides | 5′-sequence-3′ | Length |
|---|---|---|
| n1 | CAGTTTAGGATCCATTTCAC | 20 |
| n1m | CAGTTTAGGMTCCATTTCAC | 20 |
| n2 | GTGAAATGGATCCTAAACTG | 20 |
| n3 | CTAAACTG | 8 |
| n4 | GTGAAATGGATC | 12 |
| n5 | CCTAAACTG | 9 |
| n6 | GTGAAATGGAT | 11 |
| n7 | TCCTAAACTG | 10 |
| n8 | GTGAAATGGA | 10 |
| n9 | ATCCTAAACTG | 11 |
| n10 | GTGAAATGG | 9 |
| n11 | GATCCTAAACTG | 12 |
| n12 | GTGAAATG | 8 |
| n13 | GTGAAATGGTACCTAAACTG | 20 |
| n14 | CGCGGATCCGCG | 12 |
Underlined bases denote residues that constitute all or part of the GGATCC hexamer.
DNA MTase assay
DNA MTase assay conditions were similar to those previously reported for steady-state reactions (13). BamHI MTase reactions were carried out at 25°C in buffer containing 100 mM Tris–HCl, pH 8.0, 1 mM EDTA, 1 mM DTT, 2% glycerol and 0.2 mg/ml BSA. Concentrations of BamHI MTase and [3H]-AdoMet were 5 nM and 10 μM, respectively. The concentrations of substrate DNAs were varied according to their reaction capabilities. Reaction times were selected to ensure a substrate conversion of not >15% for the measurement of initial reaction rates. Different substrate variants of ODN duplexes were obtained by the combination of 20mer ODN n1 (upper strand in all duplexes) with different ODNs (Table 1). The substrates and the corresponding structures of the combined sites are presented in Table 2. Icosanucleotides n1 and n2 are complementary and form a stable duplex d1, which contains the specific GATC palindrome in the center of GGATCC.
Table 2. Steady-state kinetic parameters of BamHI MTase methylation of variant duplexes containing a canonical or defective recognition site.
| Substrate | ODN combination | Structure of recognition site | Km (nM) | kcat (× 10−3 s−1) | kcat/Km(M−1 s−1) | Relative kcat/Kma |
|---|---|---|---|---|---|---|
| d1 | n1 + n2 | –G–G–A–T–C–C– | 38.8 ± 4.2 | 17.7 ± 0.3 | 460 | (1.00) |
| –C–C–T–A–G–G– | ||||||
| d2 | n2 + n1m | –G–G–A–T–C–C– | 38.0 ± 4.0 | 16.0 ± 0.3 | 410 | 0.90 |
| –C–C–T–M–G–G– | ||||||
| d3 | n14b | –G–G–A–T–C–C– | 304 ± 53 | 9.2 ± 0.5 | 30 | 0.07 |
| –C–C–T–A–G–G– | ||||||
| d4 | n1 | –G–G–A–T–C–C– | 310 ± 43 | 5.8 ± 0.3 | 19 | 0.04 |
| d5 | n1 + n13 | –G–G–A–T–C–C– | 1730 ± 390 | 0.58 ± 0.07 | 0.34 | 0.0007 |
| –C–C–A–T–G–G– | ||||||
| d6 | n1 + n3 + n4 | –G–G–A–T–C–C– | 95.6 ± 11.8 | 3.0 ± 0.1 | 31 | 0.07 |
| –C∧C–T–A–G–G– | ||||||
| d7 | n1 + n5 + n6 | –G–G–A–T–C–C– | 198 ± 46 | 0.14 ± 0.01 | 0.69 | 0.002 |
| –C–C∧T–A–G–G– | ||||||
| d8 | n1 + n7 + n8 | –G–G–A–T–C–C– | 274 ± 31 | 5.8 ± 0.2 | 21 | 0.05 |
| –C–C–T∧A–G–G– | ||||||
| d9 | n1 + n9 + n10 | –G–G–A–T–C–C– | 35.0 ± 3.7 | 14.0 ± 0.3 | 400 | 0.88 |
| –C–C–T–A∧G–G– | ||||||
| d10 | n1 + n11 + n12 | –G–G–A–T–C–C– | 18.2 ± 5.1 | 17.8 ± 0.8 | 980 | 2.2 |
| –C–C–T–A–G∧G– | ||||||
| d11 | n1 + n4 | –G–G–A–T–C–C– | 68.0 ± 11.0 | 2.3 ± 0.1 | 33 | 0.07 |
| … C–T–A–G–G– | ||||||
| d12 | n1 + n3 + n6 | –G–G–A–T–C–C– | — | 0 | — | — |
| –C … T–A–G–G– | ||||||
| d13 | n1 + n5 + n8 | –G–G–A–T–C–C– | — | 0 | — | — |
| –C–C … A–G–G– | ||||||
| d14 | n1 + n7 + n10 | –G–G–A–T–C–C– | 386 ± 100 | 0.43 ± 0.02 | 1.1 | 0.002 |
| –C–C–T … G–G– | ||||||
| d15 | n1 + n9 + n12 | –G–G–A–T–C–C– | 16.7 ± 2.9 | 12.5 ± 0.3 | 750 | 1.6 |
| –C–C–T–A … G– | ||||||
| d16 | n1 + n11 | –G–G–A–T–C–C– | 16.4 ± 4.4 | 22.0 ± 0.8 | 1300 | 3.0 |
| –C–C–T–A–G … | ||||||
| d17 | n1 + n9 | –G–G–A–T–C–C– | 11.8 ± 4.1 | 15.0 ± 1.3 | 1200 | 2.8 |
| –C–C–T–A …… |
‘∧’Signifies the absence of an internucleotide phosphate; ‘…’ signifies absence of a nucleotide residue.
M = N6-methyladenine.
akcat/Km values are relative to that for canonical duplex 1.
b12mer self-complementary oligonucleotide.
The ODNs n4, n6, n10 and n12 are complementary to the 3′ end, while the oligonucleotides n3, n5, n7, n9 and n11 are complementary to the 5′ end of upper strand. Using equimolar mixtures of oligonucleotides n3 + n12 in different combinations with upper strand n1, duplexes were obtained lacking some internucleotide phosphate(s) or nucleotide(s) at different positions of the recognition site. Combination of the ODNs n1 and n13 formed duplex d5, which contains two adjacent mismatches, A:A and T:T. The ODN n1m contains an N6-methyladenine (M) in place of adenine in the sequence GGATCC. The 12mer ODN n14 is self-complementary. Standard kinetic analyses (13) were carried out to determine the kinetic parameters, kcat and Km, which are presented in Table 2.
RESULTS AND DISCUSSION
Determination of BamHI MTase kinetic constants with various ODN duplexes
Steady-state studies of BamHI MTase methylation of the canonical 20mer duplex d1 yielded values of Km = 38.8 nM and kcat = 0.018 s−1. These values are practically identical for the 20mer duplex d2, which contains a methylated adenine residue (M) in the lower strand (GGATCC/GGMTCC; Table 2). Shortening of the substrate DNA length from a 20mer to a 12mer (d3) resulted in a 7.5-fold increase in Km and a 2-fold decrease in kcat. Interestingly, the single-stranded ODN n1 (listed as d4 in Table 2) exhibited approximately the same kinetic constants as the 12mer duplex d3. Earlier it was shown that the EcoDam MTase is capable of methylating ODN n1, which forms a short double-stranded palindromic structure GGATCC/GGATCC, flanked by single-stranded regions (14). It is likely that BamHI reacts with ODN n1 in a similar fashion. The sharpest decline in kinetic constants was seen with 20mer duplex d5, in which there is a repositioning of the central bases A and T in the bottom strand resulting in double mismatches.
For defective duplexes d6, d7 and d8, the absence of an internucleotide phosphate between bottom strand residues C and C, T and C, or A and T resulted in an increase in Km with a concomitant decrease in kcat. Consequently, the specificity coefficient, kcat/Km, for the interaction of these duplexes with BamHI was reduced 14- to 670-fold relative to native duplex d1 (Table 2). In contrast, BamHI catalytic activity was not appreciably reduced if an internucleotide phosphate was absent between the bottom strand residues A and G (duplex d9), while the absence of an internucleotide phosphate between the two bottom strand G residues (duplex d10) resulted in a 2-fold decrease in Km. Surprisingly, deletion of the internal G residue (duplex d15) resulted in a 60% increase in the kcat/Km value. Moreover, 3-fold increases in the specificity coefficient were registered for the duplexes where the external G (duplex d16) or both the G residues (duplex 17) were deleted; in these instances, the increased specificity coefficients were mainly due to a lowering in Km. In contrast, BamHI MTase activity was lost completely when either the T or the internal C residue was deleted (duplexes d12 and d13, respectively), even though the upper strand presented an intact target sequence.
Comparison of the BamHI, T4Dam and EcoDam MTases
As the same approach of dissected DNA substrates was used earlier for the investigation of EcoDam and T4Dam (13,14), it is possible to compare the influence of different defects in the DNA substrate structure on the activities of all the three MTases. In all variant defective duplexes, one chain (upper) was intact, while individual elements were deleted in the other (bottom) strand. Figure 1 shows the influence on the kcat of each deletion in the specific target GATC (for T4Dam and EcoDam) and GGATCC (for BamHI). It is evident that a sharp decrease in activity of all three MTases occurred if a particular element of structural symmetry was disrupted. For BamHI, integrity of the ATCC (including internucleotide phosphates) is critical, while an intact GAT sequence is necessary for the activity of T4Dam, and an intact GA is necessary for EcoDam. These sequences partially overlap and are centered about the principal element of recognition site modification (adenine in the case of the Dam MTases and the internal cytosine residue in the case of BamHI).
Figure 1.
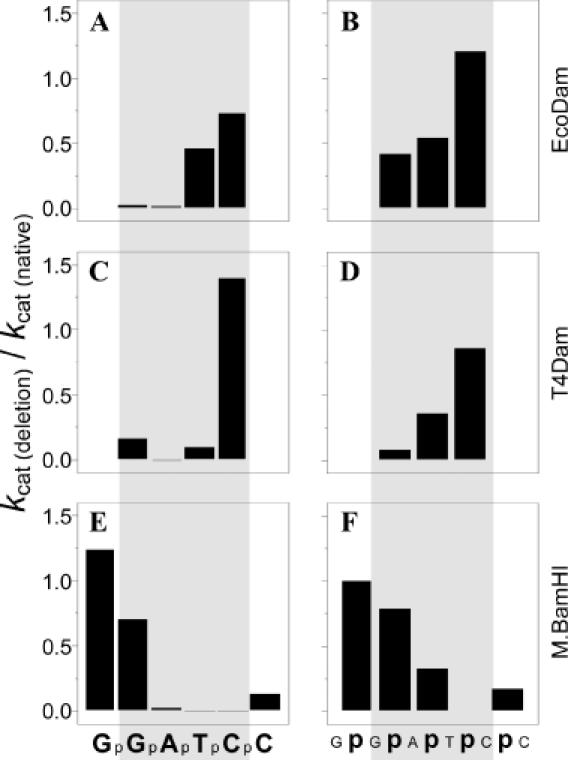
Effect of deleting an individual nucleotide (A, C and E) or phosphate (B, D and F) in the (bottom strand) recognition site on the kinetics of methyl transfer by the BamHI (A and B) EcoDam (C and D) and T4Dam (E and F) MTases. The data for BamHI were taken from Table 2; the data for EcoDam and T4Dam were taken from previous studies (13,14) in our laboratory.
Why are symmetrical elements in the specific palindromic site necessary for the productive interaction of a DNA substrate with DNA MTases that are functional monomers? It becomes clearer if one represents the DNA unrolled in a ladder format (20) (Figure 2). For a protein lying in the major groove, which runs from top left to bottom right (Figure 2, Dam), the rectangle shows the theoretical left-to-right alignment for the region of best contacts. Formally, the major groove has a zone of contact of up to 3 bp and gives a left-to-right alignment that is offset by four ribose phosphates. It can be seen that in the case of a palindromic recognition site, a zone of contact covers the symmetric residues, although a protein may curve somewhat to expand or narrow the region of contact. The rectangular area of best contacts in the major groove corresponds fairly well to the actual regions of best contacts between the Dam MTases and recognition site 5′-GATC/5′-GATC (compare Figures 1 and 2, Dam). In contrast, the regions of best contacts between the BamHI MTase and the palindromic site 5′-GGATCC/5′-GGATCC form a rectangle having an opposite direction relative to the major groove (compare Figures 1 and 2, M.BamHI). This direction suggests an arrangement of BamHI MTase in the DNA minor groove. However, this would be less favorable for direct readout recognition compared with the major groove, where direct access to the four combinations C–G, G–C, A–T and T–A is unobstructed in B-DNA (20).
Figure 2.
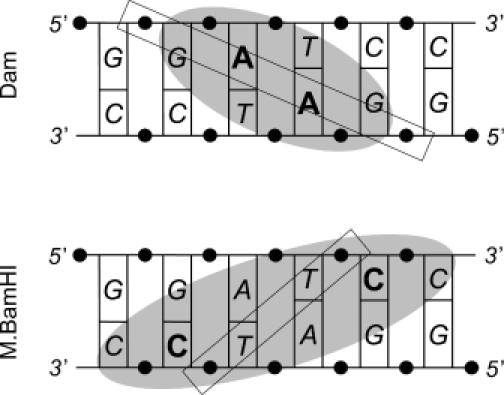
Theoretical (rectangles) and experimental (ovals) left-to-right alignment of the region of best contacts between the enzyme and the substrate DNA shown in ladder format.
Targets of modification for DNA MTases of type alpha and gamma are located in the 5′ half and 3′ half of palindromes, respectively, while targets of type beta enzymes show no preference (4). The DNA MTases that modify adenine or cytosine residues in the 5′ half of the sequence must contact the target base in the major groove and have contact with the symmetrical bases in the 5′ half of the sequence. The HhaI MTase, which recognizes the 5′-GCGC/5′-GCGC sequence and modifies the first C residue, contacts the DNA in the major groove (6). Integrity of 5′-GC sequences in both strands is critical for catalytic activity of HhaI (21), while single-base substitutions in other positions of this palindrome were not significant. In fact, certain modifications in the 3′ half of palindrome 5′-GCGC/5′-GCGC (e.g. 5′-GCGC/5′-GMAG and 5′ GCGC/5′-GMTG) actually increased methylation (22). Analysis of the protein–DNA interface of the EcoRV MTase bound to its recognition sequence, 5′-GATATC (the first Ade residue is the methylation target), showed that the enzyme makes most of its contacts in the major groove (23). Contacts are made to the four phosphates NpNpNpGpA and the three bases GAT (the 5′ half of the GATATC site) on both strands. The phosphates and bases in the 3′ ATC half are much less important. In contrast, the crystal structure of another [adenine-N6] MTase, M.TaqI (recognizes palindrome 5′-TCGA) in complex with a specific DNA and a non-reactive cofactor analog revealed that direct specific contacts between amino acid residues of the catalytic domain and the bases of the recognition sequence were formed within the widened minor groove (8), although specific contacts involving amino acid residues of the smaller domain were made via the major groove. Interaction of the [adenine-N6] KpnI MTase (which recognizes the palindrome 5′-GGTACC) with DNA was studied by in vivo footprinting with 1,10-phenantroline-copper complex, (OP)2Cu (24). A 28 bp region was protected encompassing the KpnI recognition sequence. Since (OP)2Cu binds to the minor groove [reviewed in (25)], it suggests that KpnI MTase also forms specific contacts between amino acid residues and bases within the minor groove.
The data on the Dam and BamHI MTases discussed above fit the simple rule of thumb that the most important contacts to the symmetric fragments of the palindrome are aligned around the methylation target base. If the target base is in 5′ half of the palindrome, the interaction between the enzyme and the DNA occurs mainly in the major groove; if it is in the 3′ half of the palindrome, the interaction occurs mainly in the minor groove. In this regard, recent studies with T4Dam confirm that specific protein–DNA base interactions occur in the major groove (J. R. Horton, S. Hattman and X. Cheng, manuscript in preparation). However, considering the flexibility of proteins, this principle is not a strict rule, but it affords a useful indication of the probable contacts between a DNA MTase and its target site groove. The region of best contacts can be extended if the protein is flexible enough to curve along the line of the groove. On the other hand, the bound DNA shows extensive distortions compared to B-form DNA. The most prominent examples of distortions are the complete rotation of the target base out of the DNA helix (6–8) and, in some cases, significant bending of the bound DNA [reviewed in (26)]. However, our results indicate that the Dam and BamHI MTases obey this simple rule.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a US Public Health Service grant from the Fogarty International Center (No. TW05755), a grant from the Russian Foundation for Fundamental Researches (No. 99-04-49868) and a US Public Health Service grant GM29227 from the National Institutes of Health (to S.H.).
REFERENCES
- 1.Barras F. and Marinus,M.G. (1989) The great GATC: DNA methylation in E. coli. Trends Genet., 5, 139–143. [DOI] [PubMed] [Google Scholar]
- 2.Cheng X. (1995) Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct., 24, 293–318. [DOI] [PubMed] [Google Scholar]
- 3.Wu J.C. and Santi,D.V. (1987) Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem., 262, 4778–4786. [PubMed] [Google Scholar]
- 4.Malone Th., Blumenthal,R.M. and Cheng,X. (1995) Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- 5.Jeltsch A. (2002) Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem, 3, 274–293. [DOI] [PubMed] [Google Scholar]
- 6.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 7.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 8.Goedecke K., Pignot,M., Goody,R.S., Scheidig,A.J. and Weinhold,E. (2001) Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nature Struct. Biol., 8, 121–125. [DOI] [PubMed] [Google Scholar]
- 9.Yang Z., Horton,J.R., Zhou,L., Zhang,X.J., Dong,A., Zhang,X., Schlagman,S.L., Kossykh,V., Hattman,S. and Cheng,X. (2003) Structure of the bacteriophage T4 DNA adenine methyltransferase. Nature Struct. Biol., 10, 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dryden D.T. (1999) Bacterial DNA methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-adenosylmethionine-dependent Methyltransferases: Structures and Functions. World Scientific, Singapore, pp. 283–340. [Google Scholar]
- 11.Thielking V., Dubois,S., Eritja,R. and Guschlbauer,W. (1997) Dam methyltransferase from Escherichia coli: kinetic studies using modified DNA oligomers: nonmethylated substrates. Biol. Chem., 378, 407–415. [DOI] [PubMed] [Google Scholar]
- 12.Buryanov Ya.I., Zinoviev,V.V., Gorbunov,Yu.A., Tuzikov,F.V., Rechkunova,N.I., Malygin,E.G. and Bayev,A.A. (1988) Interaction of the EcoDam methyltransferase with synthetic oligodeoxyribonucleotides. Gene, 74, 67–69. [DOI] [PubMed] [Google Scholar]
- 13.Zinoviev V.V., Evdokimov,A.A., Gorbunov,Yu.A., Malygin,E.G., Kossykh,V.G. and Hattman,S. (1998) Phage T4 DNA [N6-adenine] methyltransferase: kinetic studies using oligonucleotides containing native or modified recognition sites. Biol. Chem., 379, 481–488. [DOI] [PubMed] [Google Scholar]
- 14.Malygin E.G. and Zinoviev,V.V. (1989) Studies on the role of symmetry in the specific recognition of natural and synthetic DNA by type II restriction and modification enzymes. Sov. Sci. Rev. D. Physiochem. Biol., 9, 87–142. [Google Scholar]
- 15.Malygin E.G., Evdokimov,A.A., Zinoviev,V.V., Ovechkina,L.G., Lindstrom,W.M., Reich,N.O., Schlagman,S.L. and Hattman,S.M. (2001) A dual role for substrate S-adenosyl-l-methionine in the methylation reaction with bacteriophage T4 Dam DNA-[N6-adenine]-methyltransferase. Nucleic Acids Res., 29, 2361–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modrich P. (1982) Studies on sequence recognition by type II restriction and modification enzymes. CRC Crit. Rev. Biochem., 13, 287–323. [DOI] [PubMed] [Google Scholar]
- 17.Kossykh V.G., Schlagman,S.L. and Hattman,S.M. (1993) Conserved sequence motif DPPY in region IV of the phage T4 Dam DNA-[N6-adenine]-methyltransferase is important for S-adenosyl-l-methionine binding. Nucleic Acids Res., 21, 4659–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hattman S., Keister,T. and Gottehrer,A. (1978) Sequence specificity of DNA methylases from Bacillus amyloliquefaciens and Bacillus brevis. J. Mol. Biol., 124, 701–711. [DOI] [PubMed] [Google Scholar]
- 19.Kossykh V.G., Schlagman,S.L. and Hattman,S.M. (1995) Phage T4 DNA [N6-adenine]-methyltransferase: overexpression, purification and characterization. J. Biol. Chem., 270, 14389–14393. [DOI] [PubMed] [Google Scholar]
- 20.Seeman N.C., Rosenberg,J.M. and Rich,A. (1976) Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl Acad. Sci. USA, 73, 804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renbaum P. and Razin,A. (1995) Interaction of M.SssI and M.HhaI with single-base mismatched oligodeoxynucleotide duplexes. Gene, 157, 177–179. [DOI] [PubMed] [Google Scholar]
- 22.Klimasauskas S. and Roberts,R.J. (1995) M.HhaI binds tightly to substrates containing mismatches at the target base. Nucleic Acids Res., 23, 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szczelkun M.D., Jones,H. and Connolly,B.A. (1995) Probing the protein–DNA interface of the EcoRV modification methyltransferase bound to its recognition sequence, GATATC. Biochemistry, 34, 10734–10743. [DOI] [PubMed] [Google Scholar]
- 24.Basak S. and Nagaraja,V. (2001) A versatile in vivo footprinting technique using 1,10-phenanthroline-copper complex to study important cellular processes. Nucleic Acids Res., 29, e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papavassiliou A.G. (1995) Chemical nucleases as probes for studying DNA–protein interactions. Biochem. J., 305 (Pt 2), 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gromova E.S. and Khoroshaev,A.V. (2003) Prokaryotic DNA methyltransferases: the structure and the mechanism of interaction with DNA. Mol. Biol., 37, 300–314. [PubMed] [Google Scholar]


