Abstract
Werner syndrome (WS) is a genetic premature aging disorder in which patients appear much older than their chronological age. The gene mutated in WS encodes a nuclear protein (WRN) which possesses 3′–5′ exonuclease and ATPase-dependent 3′–5′ helicase activities. The genomic instability associated with WS cells and the biochemical characteristics of WRN suggest that WRN plays a role in DNA metabolic pathways such as transcription, replication, recombination and repair. Recently we have identified poly(ADP-ribose) polymerase-1 (PARP-1) as a new WRN interacting protein. In this paper, we further mapped the interacting domains. We found that PARP-1 bound to the N-terminus of WRN and to the C-terminus containing the RecQ-conserved (RQC) domain. WRN bound to the N-terminus of PARP-1 containing DNA binding and BRCA1 C-terminal (BRCT) domains. We show that unmodified PARP-1 inhibited both WRN exonuclease and helicase activities, and to our knowledge is the only known WRN protein partner that inactivates both of the WRN's catalytic activities suggesting a biologically significant regulation. Moreover, this dual inhibition seems to be specific for PARP-1, as PARP-2 did not affect WRN helicase activity and only slightly inhibited WRN exonuclease activity. The differential effect of PARP-1 and PARP-2 on WRN catalytic activity was not due to differences in affinity for WRN or the DNA substrate. Finally, we demonstrate that the inhibition of WRN by PARP-1 was influenced by the poly(ADP-ribosyl)ation state of PARP-1. The biological relevance of the specific modulation of WRN catalytic activities by PARP-1 are discussed in the context of pathways in which these proteins may function together, namely in the repair of DNA strand breaks.
INTRODUCTION
Werner syndrome (WS) is a premature aging disease caused by mutations in the Werner gene (WRN). WRN belongs to the human RecQ family of DNA helicases, which also includes RecQ1, Bloom (BLM), Rothmund–Thompson (RTS) and RecQ5 (1). Mutations in BLM and RTS are also associated with heritable human diseases (2). Cells from WS patients display defects in replication, altered telomere dynamics and genome instability (3), suggesting an important role for WRN in DNA metabolic pathways. In addition, many WRN-interacting proteins participate in transcription, recombination, replication, repair and/or telomere maintenance (3).
WRN possesses ATPase-dependent helicase activity and is the only member of the human RecQ family to possess exonuclease activity (4). WRN's catalytic activities are structure specific and are active on DNA substrates resembling replication forks (long forked substrates), recombination intermediates (Holliday junctions), DNA repair intermediates (recessed and nicked substrates) and telomere structures (D-loops) (3), further supporting a role for WRN in these processes. Interestingly, both the exonuclease and the helicase activities of WRN are modulated by several interacting proteins and post-translational modifications (3).
The RecQ-conserved (RQC) domain of WRN plays a central role in WRN's functions. It contains a nucleolar targeting sequence (5), possesses DNA binding properties (6), stimulates flap endonuclease-1 (FEN-1) cleavage (7) and mediates binding to many of WRN's protein partners (7–11). Recently we have identified poly(ADP-ribose) polymerase-1 (PARP-1) as the most prominent WRN RQC binding protein (11).
Following DNA damage, PARP-1 catalyzes the sequential transfer of ADP-ribose monomers onto protein acceptors, including itself, using NAD+ as a substrate (12,13). The poly(ADP-ribosyl)ation state of PARP-1 differentially affects its binding to other proteins, such as X-ray repair cross-complementing 1 (XRCC1) (14–16), DNA ligase III (lig III) (17), and WRN (11). This poly(ADP-ribosyl)ation reaction is one of the earliest responses to DNA damage in the cell, and is mainly catalyzed by PARP-1 (12). We have recently observed that the WRN/PARP-1 interaction is important for the poly(ADP-ribosyl)ation pathway following oxidative stress-induced DNA damage (11).
Mice deficient in PARP-1 displayed a residual poly(ADP-ribosyl)ation activity following DNA damage, and this observation led to the discovery of PARP-2 (18,19). PARP-1 and PARP-2 are involved in different aspects of genomic stability, such as modulation of chromatin structure, replication, transcription and single-strand break/base excision repair (SSBR/BER) (14,20,21). Thus far, and supporting a role in DNA metabolic pathways, PARP-1 and PARP-2 are the sole PARP family members whose catalytic activities are stimulated in vitro and in vivo by DNA strand breaks (19,22,23). Interestingly, the effect on DNA strand break repair is as dramatic in the absence of PARP-2 as it is in the absence of PARP-1 (23). This was surprising due to the fact that the absence of PARP-2 had only a small effect on the total cellular PARP activity stimulated by DNA breaks (23), and may be explained by the fact that PARP-1 and PARP-2 form both homo- and heterodimers (23). If PARP-1 and PARP-2 act as a heterodimer in SSBR/BER, then the absence of either would have similar consequences on repair efficiency (23).
Based on our findings that WRN interacts with PARP-1 and that the WRN/PARP-1 interaction is important for the poly(ADP-ribosyl)ation pathway following oxidative DNA damage (11), we examined the effects of PARP-1 on WRN catalytic activities. We demonstrate here that unmodified PARP-1 inhibits both WRN exonuclease and helicase activities. This inhibition was specific for PARP-1, because PARP-2 did not affect WRN helicase activity and only slightly inhibited WRN exonuclease activity. However, the inhibition of WRN catalytic activities was relieved in the presence of auto-poly(ADP-ribosyl)ated PARP-1. We discuss the importance of these results in the context of DNA metabolic pathways in which these proteins participate.
MATERIALS AND METHODS
Recombinant proteins
His-tagged WRN (24), his-tagged N-WRN (amino acids 1–368) (25), human PARP-1 (26) and mouse PARP-2 (19) were purified as described previously. Glutathione S transferase (GST)–WRN fragments were expressed and purified as described previously (8). The heterotrimer of human replication protein A (RPA 70, RPA 32 and RPA 14) was a generous gift from Mark Kenny (Albert Einstein Cancer Center, Bronx, NY). The recombinant human Ku 70/80 heterodimer was kindly provided by Dale Ramsden (University of North Carolina, Chapel Hill, NC).
In vitro GST pull-down assay
The pull-down assay with various GST–WRN fragments and purified recombinant PARP-1 was performed basically as described previously (8). Recombinant PARP-1 (5 μg per sample) was incubated with GST–WRN fragments bound to glutathione sepharose beads (40 μl per sample) for 2 h at 4°C. Bound PARP-1 was detected using an anti-PARP-1 monoclonal antibody (1:2000, Sigma, clone C-2-10).
In vivo GST pull-down assay
HeLa S3 cells (2 × 106 cells) were transfected by calcium-phosphate co-precipitation with 10 μg of recombinant DNA (14,23). Cells were lysed 48 h later in 50 mM Tris–HCl, pH 8, 150 mM NaCl, 0.1% NP-40, 0.5 mM PMSF and protease inhibitors (Complete Mini, Roche, Mannheim, Germany). Lysates were cleared by centrifugation and incubated for 3 h with glutathione sepharose beads (Pharmacia, Uppsala, Sweden). Beads were washed 3 times with 50 mM Tris–HCl, pH 8, 250 mM NaCl, 0.25% NP-40 and 0.5 mM PMSF. The samples were resuspended in Laemmli buffer, boiled for 4 min and analyzed by 10% SDS–PAGE and immunoblotting. Blots were subsequently incubated with rabbit polyclonal anti-WRN antibodies (1:1000, Abcam, Cambridge, UK) and mouse monoclonal anti-GST antibodies (1:10 000, IGBMC, Illkirch, France). Blots were then probed with horseradish peroxidase-coupled secondary antibodies (goat anti-rabbit, 1:20 000 or sheep anti-mouse, 1:20 000, Sigma, St Louis, MO), and immunoreactivity was detected by enhanced chemiluminescence (NEN, Boston, MA) according to the manufacturer.
Enzyme-linked immunosorbent assay
The enzyme-linked immunosorbent assay (ELISA) was performed as described previously (11). Briefly, equal molar concentrations (80 nM per well) of PARP-1 and PARP-2 were coated onto the wells for 16 h at 4°C. Serial dilutions of recombinant WRN (ranging between 0.15 and 40 nM per well) were then added to the corresponding wells for 90 min at 37°C. The binding reaction was performed in 1× phosphate-buffered saline (PBS), 2% BSA, 0.1% Tween-20 and 20 μg/ml of ethidium bromide. Bound WRN was detected with anti-WRN antibodies (1:1000, Novus Biologicals).
Helicase assay
Helicase reactions were performed essentially as described previously (10). Briefly, oligonucleotide 22Fork3 (Table 1) was 5′ end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs) and annealed to its unlabeled complementary oligonucleotide 22Fork4. Reactions (20 μl) were performed in helicase reaction buffer (40 mM Tris–HCl, pH 8.0, 4 mM MgCl2, 5 mM DTT, 2 mM ATP and 0.1 mg/ml BSA) and contained DNA substrate (0.5 nM) and WRN, PARP-1 or PARP-2 as indicated. Samples were incubated at 37°C for 15 min and terminated by the addition of stop dye (50 mM EDTA, 40% glycerol, 0.9% SDS, 0.05% bromophenol blue and 0.05% xylene cyanol). Products were run on a 12% native polyacrylamide gel and visualized using a PhosphorImager (Molecular Dynamics).
Table 1. Oligonucleotides used in this study.
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| 22Fork3 | TTTTTTTTTTTTTTTGAGTGTGGTGTACATGCACTAC |
| 22Fork4 | GTAGTGCATGTACACCACACTCTTTTTTTTTTTTTTT |
| 34ForkA | TTTTTTTTTTTTTTTTTAGGGTTAGGGTTAGGGTTAGGGCATGCACTAC |
| 34ForkB | GTAGTGCATGCCCTAACCCTAACCCTAACCCTAATTTTTTTTTTTTTTT |
| 15P | CTGCAGCTGATGCGC |
| 34G | GTACCCGGGGATCCGTACGGCGCATCAGCTGCAG |
| 19D | CGTACGGATCCCCGGGTAC |
| 17D | TACGGATCCCCGGGTAC |
Exonuclease assay
Exonuclease activity was detected as described previously (10). Briefly, oligonucleotide 34ForkA was 5′ end-labeled and annealed to unlabeled 34ForkB. Reactions (10 μl) were performed in helicase reaction buffer (see above) and contained DNA substrate (0.5 nM) and WRN, N-WRN, PARP-1, and PARP-2 as indicated. Samples were incubated for 15 min at 37°C and terminated by the addition of an equal volume of formamide stop dye (80% formamide, 0.5× TBE buffer, 0.1% bromophenol blue and 0.1% xylene cyanol). Products were heat-denatured at 90°C for 5 min and run on a 14% denaturing polyacrylamide gel. Radioactive products were visualized using a PhosphorImager.
Electrophoretic mobility shift assay
The electrophoretic mobility shift assays (EMSAs) were performed basically as previously described (6). Oligonucleotide 34ForkA was 5′ end-labeled and annealed to unlabeled complementary oligonucleotide 34ForkB. Reactions (10 μl) were performed in binding buffer (40 mM Tris, pH 7.0, 1 mM EDTA, 20 mM NaCl, 20 μg/ml BSA and 8% glycerol) and contained DNA substrate (3 nM) and Ku 70/80, RPA, PARP-1 and PARP-2 as indicated. Samples were incubated at 4°C for 30 min. Loading buffer (40% glycerol and 0.25% bromophenol blue) was added only to the no-enzyme control sample, and all samples were loaded onto a 5% polyacrylamide (37:1) gel and electrophoresed at 200 V at 4°C in 1× TAE (40 mM Tris–acetate, pH 8.3, and 1 mM EDTA). Products were visualized using a PhosphorImager.
In vitro auto-poly(ADP-ribosyl)ation of PARP-1
Reactions (50 μl) were performed in helicase buffer (see above) and contained 500 ng of PARP-1, 100 μM NAD+ and 250 fmol of unlabeled DNA substrate (34ForkA/34ForkB). Samples were incubated at room temperature for 10 min. For western blotting, 11 μl was analyzed by SDS–PAGE and probed with mouse anti-PARP-1 (1:1000, Santa Cruz) or mouse anti-PAR (1:1000, Alexis, clone 10H) antibodies. For exonuclease assays, 6 μl from the ADP-ribosylation reaction (corresponding to 60 ng PARP-1 and 30 fmol unlabeled substrate) were added to helicase buffer (see above), 10 fmol radiolabeled DNA substrate (34ForkA/34ForkB) and WRN (7.5 nM) in a total volume of 10 μl. Reactions were analyzed as described above (exonuclease assay). For helicase assays, 2 μl from the ADP-ribosylation reaction (corresponding to 20 ng PARP-1 and 10 fmol unlabeled substrate) were added to helicase buffer (see above), 20 fmol radiolabeled DNA substrate (22Fork3/22Fork4) and WRN (1 nM) in a total volume of 20 μl. Reactions were analyzed as described above (helicase assay).
RESULTS
Identification of WRN domains involved in the interaction with PARP-1
We have recently demonstrated that PARP-1 is a prominent binder to GST–WRN RQC (amino acids 949–1092) (11). Thus far, most of the WRN-interacting proteins bind to more than one WRN region (8,9,27,28). To more precisely map the PARP-1 binding domain(s) of WRN, we used various GST–WRN fragments and recombinant PARP-1 for in vitro pull-down assays. As shown in Figure 1A, recombinant PARP-1 bound to three WRN regions: the N-terminus (amino acids 1–120), the helicase domain (amino acids 500–946) and the C-terminus (amino acids 949–1432). Neither GST alone nor the WRN acidic region (amino acids 239–499) demonstrated any significant binding activity (Figure 1A). As we have shown previously that GST–WRN949–1092 also pulls down PARP-1 (11), PARP-1 binding to the C-terminal domain of WRN is likely mediated through the RQC domain.
Figure 1.
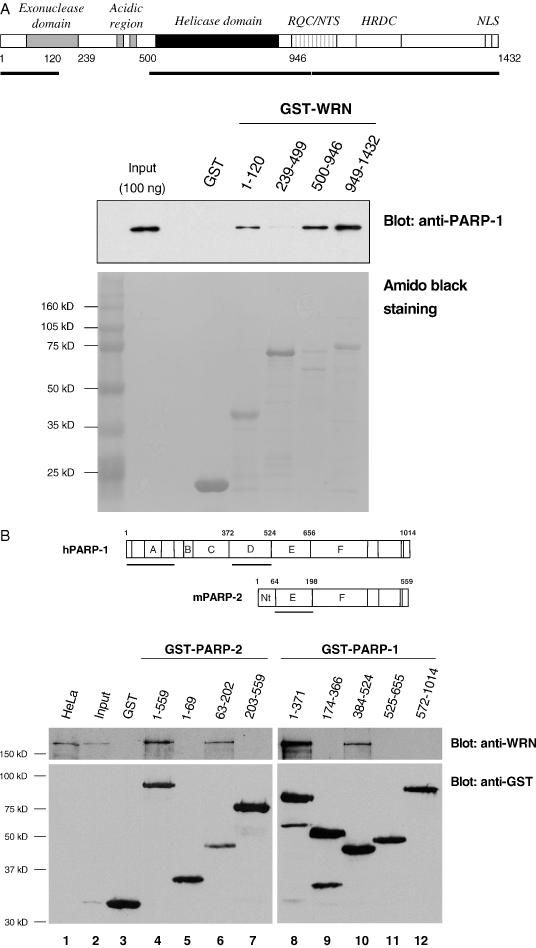
Characterization of the WRN/PARP-1 and WRN/PARP-2 physical interaction. (A) Top: schematic representation of WRN. RQC, RecQ-conserved domain; NTS, nucleolar targeting sequence; HRDC, helicase and RNase D conserved domain; NLS, nuclear localization signal. The solid line indicates PARP-1 binding sites. Bottom: GST alone and GST–WRN fragments (as indicated) were bound to glutathione beads. After washing, the beads were incubated with recombinant PARP-1 (5 μg/sample) for 2 h at 4°C. The bound PARP-1 was eluted with protein sample buffer and analyzed by western blot. The input corresponds to 2% (100 ng) of PARP-1 used in the binding reactions. The control for recombinant protein loaded is indicated by amido black staining of the membrane. (B) Top: schematic representation of PARP-1 and PARP-2 as has been described previously (23,47). A, DNA binding domain; B, nuclear localization signal; C, domain of unknown function; D, BRCT and automodification domain; E, homo- and heterodimerization domain of PARP-2; F, catalytic domain; Nt, DNA binding domain. The lines represent the WRN binding sites. Bottom: GST (lane 3) and GST-tagged deletion mutants of PARP-2 (lanes 4 to 7) or PARP-1 (lanes 8 to 12) were expressed in HeLa cells and interacting endogenous proteins were extracted by GST pull-down and analyzed by western blot using the indicated antibodies. Lane 1, crude extract from 4 × 105 HeLa cells. Lane 2, Input corresponds to 2% of the total cell extract used for the GST pull down. (C) Wells were coated with either purified recombinant PARP-1 or PARP-2 (80 nM per well). After blocking with BSA, the wells were incubated with serial dilutions of recombinant WRN (0.15, 0.3, 0.6, 1.2, 2.5, 5, 10, 20 and 40 nM per well) in the presence of 20 μg/ml of ethidium bromide. Bound WRN was detected with rabbit anti-WRN antibodies followed by colorimetric analysis. Filled squares represent the binding of WRN to PARP-1. Filled triangles represent the binding of WRN to PARP-2. Values represent the mean of two experiments performed in duplicate and were corrected for the background signal (WRN binding to BSA). The purity of recombinant PARP-1 (1 μg) and PARP-2 (1 μg) is shown by SDS–PAGE and coomassie staining (right panel).
Identification of PARP-1 and PARP-2 domains involved in the interaction with WRN
PARP-1 and PARP-2 share many of the same protein partners (23). To address whether PARP-2 also interacts with WRN, we performed in vivo pull-down experiments. As shown in Figure 1B, PARP-2 co-precipitated endogenous WRN (lane 4). To map interacting regions, GST-fusion proteins were generated, expressing truncated versions of PARP-2 and PARP-1 and over-expressed in HeLa cells (Figure 1B). Only domain E (amino acids 63–202) of PARP-2 (Figure 1B, lane 6) was able to pull down WRN as efficiently as the full-length protein. Co-precipitation of endogenous WRN was efficient with PARP-1 constructs containing either the DNA binding domain (Figure 1B, lane 8) or the BRCT domain (Figure 1B, lane 10). The DNA binding and BRCT domains of PARP-1 are involved in the homodimerization of the protein as well as in the binding to several partners (23). Domain E of PARP-2 is involved in homodimerization and in heterodimerization with PARP-1 (23). Thus, WRN binds important structural regions of PARP-1 and PARP-2, which act as interfaces for protein–protein interactions. These results also demonstrate that PARP-2 is a new WRN interacting protein.
PARP-1 and PARP-2 bind WRN with similar affinities
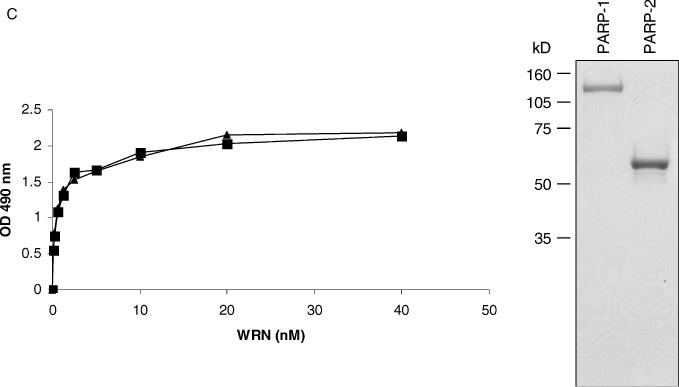
To determine direct protein–protein interactions between PARP-2 and WRN, we performed ELISA with purified recombinant proteins (Figure 1C, right panel). In vitro binding reactions contained the same molar concentration of purified recombinant PARP and increasing concentrations of WRN. To rule out that interactions were DNA-mediated, binding reactions were performed in the presence of ethidium bromide. PARP-2 bound WRN in a concentration-dependent manner (Figure 1C, left panel). Furthermore, the binding affinities between WRN and PARP-2 (Kd = 1.5 nM) were almost identical to that of WRN and PARP-1 (Kd = 1.3 nM).
PARP-1 inhibits WRN helicase activity
Based on the physical protein interactions demonstrated above, we next investigated the influence of PARP-1 and PARP-2 on WRN catalytic activities. Using a 22 bp forked duplex oligonucleotide (22Fork3/22Fork4), the WRN helicase displaced the labeled strand in a concentration-dependent manner (Figure 2A, lanes 3 and 8). The presence of increasing concentrations of PARP-1 resulted in a concentration-dependent decrease in the amount of product unwound by WRN (Figure 2A, lanes 4 to 7 and 9 to 12). At the highest concentration of PARP-1 examined (an 8-fold molar excess), WRN helicase activity was inhibited 2.4- to 3.3-fold. As PARP-1 is an abundant nuclear protein, the molar ratios of WRN:PARP-1 employed in this study are likely biologically relevant. In contrast to the findings with PARP-1, increasing concentrations of PARP-2 did not affect WRN helicase activity on a short-forked duplex (Figure 2B, lanes 4 to 7 and 9 to 12). Thus, PARP-1 inhibited the helicase activity of WRN, whereas PARP-2 did not.
Figure 2.
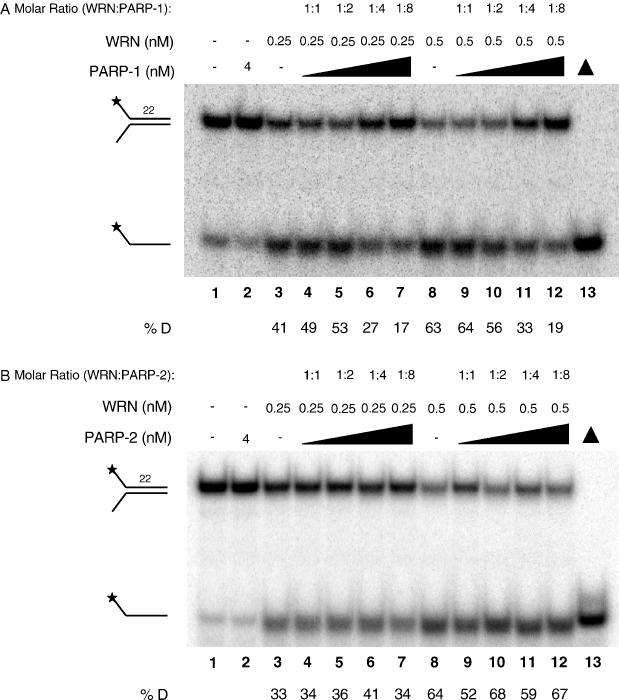
The effect of PARP-1 and PARP-2 on WRN helicase activity. (A) Reactions containing WRN (0.25 nM, lanes 3 to 7; or 0.5 nM, lanes 8 to 12) in the absence or presence of PARP-1 were incubated with a 22 bp forked substrate (0.5 nM, 22Fork3/22Fork4) for 15 min at 37°C. The concentrations of PARP-1 were 4 nM (lane 2), 0.25, 0.5, 1 and 2 nM (lanes 4 to 7, respectively), and 0.5, 1, 2 and 4 nM (lanes 9 to 12, respectively). Lane 1, substrate only. Lane 13, heat-denatured substrate. Reaction products were run on a 12% native gel and visualized using a PhosphorImager. %D, percentage of single-stranded product displaced. (B) WRN (0.25 nM, lanes 3 to 7; or 0.5 nM, lanes 8 to 12) was incubated in the absence or presence of PARP-2 for 15 min at 37°C with a 22 bp forked duplex substrate (0.5 nM, 22Fork3/22Fork4). Concentrations of PARP-2 were 4 nM (lane 2), 0.25, 0.5, 1 and 2 nM (lanes 4 to 7, respectively), and 0.5, 1, 2 and 4 nM (lanes 9 to 12, respectively). Lane 1, substrate only. Lane 13, heat-denatured substrate. Reactions were analyzed as above.
Effects of PARP-1 and PARP-2 on WRN exonuclease activity
We next examined the effect of PARP-1 and PARP-2 on WRN exonuclease activity. Using a 34 bp forked duplex substrate (34ForkA/34ForkB), WRN degraded the substrate from the blunt end (Figure 3A, lanes 3 and 10). Increasing concentrations of PARP-1 inhibited WRN exonuclease activity (Figure 3A, lanes 11 to 14 and Table 2). Heat denaturation of PARP-1 before the reaction relieved the inhibition of WRN exonuclease activity (Figure 3A, lane 15). In addition to forked duplex substrates, PARP-1 strongly inhibited WRN exonuclease activity on recessed, nicked and gapped substrates (Table 2). Quantitation of undigested versus digested products revealed a marked decrease in the amount of substrate digested by WRN in the presence of increasing concentrations of PARP-1 (Figure 3B, right panel). At the highest concentration of PARP-1 employed (1:8 molar ratio of WRN:PARP-1), PARP-1 inhibited WRN exonuclease activity approximately 8-fold. Increasing concentrations of PARP-2 also inhibited the progression of the WRN exonuclease (Figure 3A, lanes 4 to 7). However, although progression of the WRN exonuclease was inhibited by PARP-2 (1:4 and 1:8 molar ratios), the amount of total digested products was similar (Figure 3B, left panel).
Figure 3.
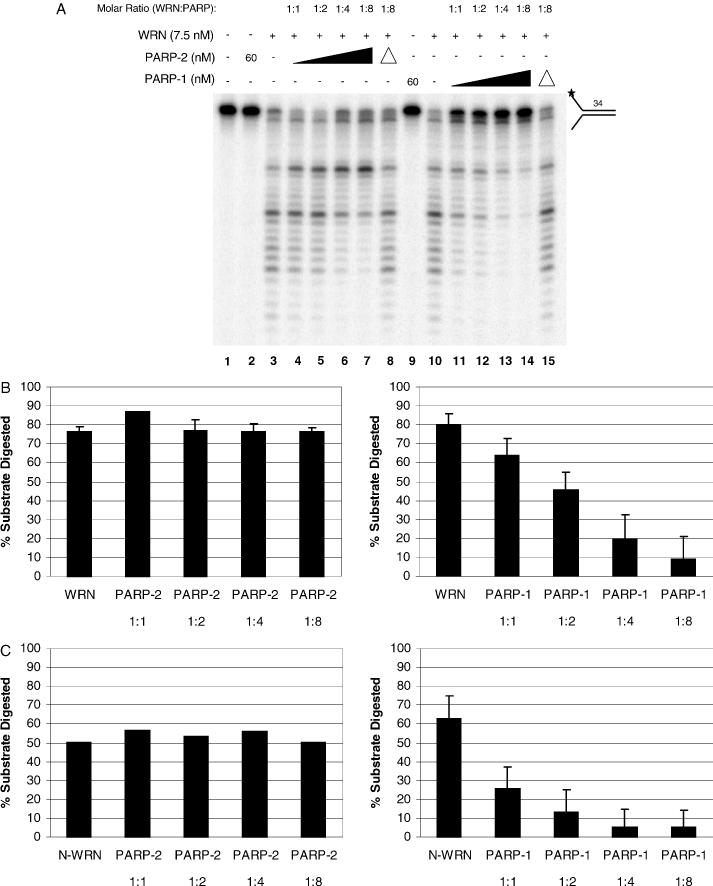
The effect of PARP-1 and PARP-2 on WRN exonuclease activity. (A) Reactions contained 7.5 nM WRN (lanes 3 to 8 and 10 to 15) and PARP-1 or PARP-2 as indicated and were incubated with a 34 bp forked substrate (0.5 nM, 34ForkA/34ForkB) for 15 min at 37°C. The concentrations of PARP-2 were 60 nM (lane 2), 7.5, 15, 30 and 60 nM (lanes 4 to 7, respectively). The concentrations of PARP-1 were 60 nM (lane 9), 7.5, 15, 30 and 60 nM (lanes 11 to 14, respectively). Lane 1, no enzyme. Lane 8, PARP-2 (60 nM) was heat-inactivated before the reaction. Lane 15, PARP-1 (60 nM) was heat-inactivated before the reaction. Products were heat-denatured for 5 min at 95°C, run on a 14% denaturing polyacrylamide gel and visualized using a PhosphorImager. (B) Quantitation of the gels shown in (A) containing PARP-2 (left) or PARP-1 (right). (C) Quantitation of gels from representative experiments containing N-WRN and PARP-2 (left) or PARP-1 (right).
Table 2. Inhibition of WRN exonuclease by PARP-1.
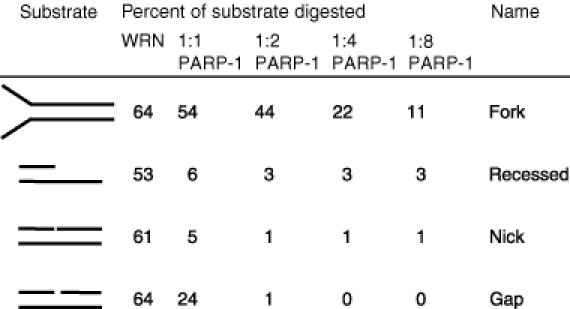
The numbers above PARP-1 refer to the molar ratio of WRN:PARP-1. Fork, oligonucleotides 34ForkA/34ForkB; Recessed, oligonucleotides 15P/34G; Nick, oligonucleotides 15P/19D/34G; Gap, oligonucleotides 15P/17D/34G.
As the exonuclease domain of WRN resides in the N-terminus of the protein (amino acids 1–368, N-WRN), we next examined the influence of PARP-1 and PARP-2 on N-WRN exonuclease activity. N-WRN has been shown to possess exonuclease activity similar to that of the full-length protein (29). Analogous to what we observed for full-length WRN, increasing concentrations of PARP-1 inhibited the exonuclease activity of N-WRN (Figure 3C, right panel). Interestingly, PARP-1 inhibited the exonuclease activity of N-WRN at lower molar ratios compared to full-length WRN (compare Figure 3C, right panel to Figure 3B, right panel). Increasing concentrations of PARP-2 did not affect the percentage of substrate digested by N-WRN (Figure 3C, left panel).
Our results demonstrate that PARP-2 had only a modest inhibitory effect on WRN exonuclease activity while PARP-1 strongly inhibited both full-length and N-WRN exonuclease activity on a variety of substrates. As shown in Figure 1A, PARP-1 binds both the N- and C-terminal regions of WRN, suggesting that protein–protein interactions may be responsible for the inhibition. However, since both PARP-1 and PARP-2 are DNA binding proteins, we cannot rule out the possibility that the inhibition of WRN catalytic activity is due to PARP-1 or PARP-2 binding the DNA and preventing WRN access to the DNA substrate.
PARP-1 and PARP-2 bind a long-forked duplex DNA substrate
The forked duplex substrates used for the helicase and exonuclease assays contain both regions of single-strand (ss) and double-strand (ds) DNA. In addition, the blunt end mimics a dsDNA break. One possible mechanism for PARP-1 inhibition of WRN helicase and exonuclease activities may be that PARP-1 binds to the DNA substrate and prevents WRN catalytic activities. To test for PARP-1 binding to the forked duplex substrate, we performed gel-shift assays (EMSA). Proteins known to bind dsDNA breaks (Ku 70/80) and ssDNA (RPA) effectively bound (shifted) the long-forked duplex substrate (Figure 4, lanes 2 to 4 and 5 to 7, respectively). In addition, PARP-1 bound the long-forked duplex (Figure 4, lanes 8 to 10) with a similar affinity as RPA. To determine whether the differential effects observed by PARP-1 and PARP-2 were due to differences in DNA binding, we also tested whether PARP-2 could bind the long forked duplex. We found that PARP-2 formed a stable complex with DNA at similar concentrations as PARP-1 (Figure 4, lanes 11-13). Thus, the differential influences of PARP-1 and PARP-2 on WRN catalytic functions do not appear to be due to differences in the affinity for the DNA substrate. However, it may be possible that PARP-1 and PARP-2 bind to different regions of the DNA substrate, as PARP-1 and PARP-2 contain distinct DNA binding domains (19).
Figure 4.
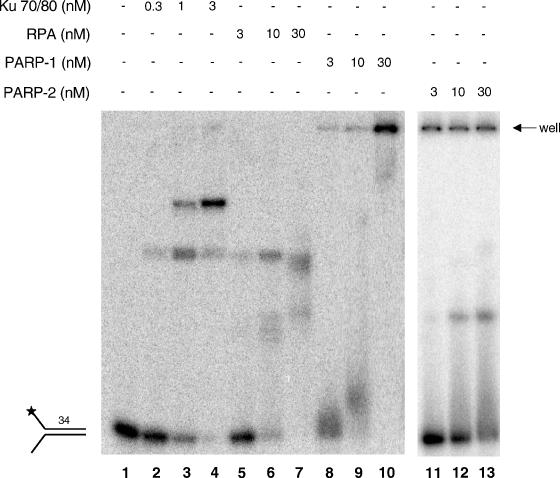
PARP-1 and PARP-2 bind a forked DNA oligonucleotide. Binding experiments were performed by incubating Ku 70/80 (lanes 2 to 4), RPA (lanes 5 to 7), PARP-1 (lanes 8 to 10), or PARP-2 (lanes 11 to 13) as indicated with a 34 bp forked duplex (3 nM, 34ForkA/34ForkB) for 30 min on ice. Lane 1, no enzyme. Reactions were run on a 5% acrylamide gel and visualized using a PhosphorImager.
Auto-poly(ADP-ribosyl)ated PARP-1 does not inhibit WRN catalytic activities
Binding of PARP-1 to DNA strand breaks results in activation and auto-poly(ADP-ribosyl)ation, which involves the addition of ADP-ribose chains using NAD+ as a substrate (13). As the above experiments were performed in the absence of NAD+, we were interested in the effects of activated and auto-poly(ADP-ribosyl)ated PARP-1 on WRN catalytic activities. PARP-1 was activated in the presence of unlabeled forked duplex oligonucleotide and NAD+ (Figure 5A, lanes 2 and 5), but not in the absence of NAD+ (Figure 5A, lanes 1 and 4). As seen previously, unmodified PARP-1 inhibited WRN exonuclease activity (Figure 5B, lane 2). ADP-ribosylation of PARP-1 restored WRN exonuclease activity (Figure 5B, lane 3). Similarly, unmodified PARP-1 inhibited WRN helicase activity (Figure 5C, lane 6), whereas modified PARP-1 did not (Figure 5C, lane 7). These results demonstrate that the poly(ADP-ribosyl)ation status of PARP-1 regulates WRN catalytic activities.
Figure 5.
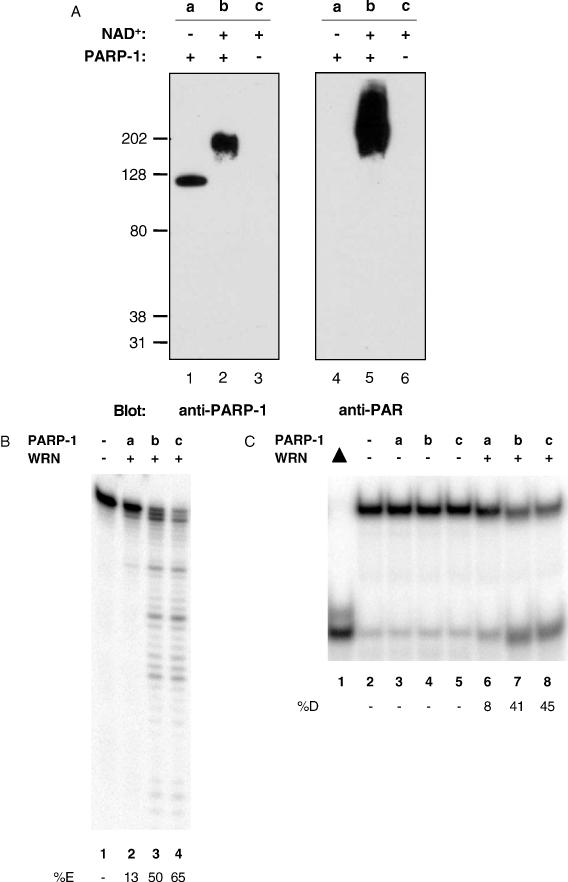
Activated and poly(ADP-ribosyl)ated PARP-1 does not inhibit WRN catalytic activities. (A) PARP-1 (500 ng) and NAD+ (100 μM) as indicated were incubated with an unlabeled forked duplex for 10 min at room temperature. Western blot analysis was performed using anti-PARP-1 (left panel) or anti-PAR (right panel) antibodies. (B) WRN (7.5 nM, lanes 2 to 4) and a radiolabeled 34 bp forked duplex (10 fmol, 34ForkA/34ForkB) were incubated with aliquots from the reactions performed in (A) containing PARP-1 (lane 2), PARP-1 and NAD+ (lane 3), or NAD+ (lane 4) for 15 min at 37°C. Lane 1, no enzyme. Products were heat-denatured for 5 min at 95°C, run on a 14% denaturing polyacrylamide gel, and visualized using a PhosphorImager. (C) Aliquots from the reactions performed in (A) containing PARP-1 (lanes 3 and 6), PARP-1 and NAD+ (lanes 4 and 7), or NAD+ (lanes 5 and 8) were incubated with a radiolabeled 22 bp forked duplex (20 fmol, 22Fork3/22Fork4) in the absence (lanes 3 to 5) or presence (lanes 6 to 8) of WRN (1 nM) for 15 min at 37°C. Lane 1, heat-denatured substrate. Lane 2, no enzyme. Products were run on a 12% native polyacrylamide gel and visualized using a PhosphorImager.
DISCUSSION
We and others have recently identified a physical interaction between WRN and PARP-1 (11,30,31), and we have shown that the WRN/PARP-1 interaction is important for the poly(ADP-ribosyl)ation of nuclear proteins after DNA damage (11). In this paper, we further mapped the interacting domains between WRN and PARP-1 and identified PARP-2 as a new WRN interacting protein. We also demonstrated that unmodified PARP-1 inhibited both the exonuclease and helicase activities of WRN. With an estimated PARP-1 concentration of 2 × 105 to 5 × 105 molecules per cell (32,33) and a WRN concentration of 3.4 × 104 to 8.4 × 104 molecules per cell (34), the strong inhibition of WRN catalytic activities with an 8-fold molar excess of PARP-1 may likely be observed in vivo. This inhibition of WRN by unmodified PARP-1 appears to be specific, as PARP-2 failed to inhibit WRN helicase activity and only modestly inhibited WRN exonuclease activity, despite showing binding affinities to WRN and the DNA substrate similar to that of PARP-1. Activation and auto-poly(ADP-ribosyl)ation of PARP-1 in the presence of DNA strand breaks relieved the inhibition of WRN catalytic activities. Consistent with our results, Li et al. (31) found that poly(ADP-ribosyl)ated PARP-1 did not inhibit WRN helicase or exonuclease activities. In contrast, they did not observe inhibition of WRN catalytic activities by unmodified PARP-1 (31).
Other WRN protein partners including p53 and BLM have been shown to inhibit WRN exonuclease activity. p53 strongly inhibits full-length WRN but not N-WRN exonuclease activity and does not influence WRN helicase activity (35). BLM inhibits both full-length and N-WRN exonuclease activity and does not effect WRN helicase activity (8). To our knowledge, PARP-1 is the only known WRN protein partner that strongly inhibits both WRN exonuclease and helicase activites. As PARP-1 binds to both WRN and DNA, it is not clear which is responsible for the inhibitory effect. However, Ku is also an abundant nuclear protein with a high affinity for DNA ends and which binds WRN. As Ku strongly stimulates WRN exonuclease activity (36), the inhibition of WRN catalytic activities by other proteins cannot simply be due to their binding to DNA and blocking WRN access, and suggests that the mechanism of PARP-1 inhibition also involves protein–protein interactions.
The in vivo regulation of WRN catalytic activities is likely mediated by protein–protein interactions and/or post-translational modifications. The regulation of WRN exonuclease activity by DNA-PK (Ku 70/80 and DNA-PKcs) suggests a direct role for WRN in non-homologous end joining. Interestingly, the two components of DNA-PK differentially affect the exonuclease activity. Ku 70/80 strongly stimulates WRN exonuclease activity (36). This stimulation is due to the direct binding of the Ku 70 subunit to the N-terminal region of WRN (27). In contrast, DNA-PKcs inhibits both the exonuclease and helicase activities of WRN either by Ser/Thr phosphorylation (28) or by physical association (37). Interestingly, the in vitro inhibition by DNA-PKcs is dominant over the Ku 70/80 stimulation (28). Although the precise role of these functional interactions has still not been elucidated, it is possible that release of one of WRN's modulators from the DNA end would allow another to regulate WRN helicase and/or exonuclease activities.
PARP-1 has also been shown to interact with Ku 70/80 and DNA-PKcs (38,39). DNA-PK phosphorylates PARP-1 and PARP-1 poly(ADP-ribosyl)ates each of the three subunits of DNA-PK (38,39). Poly(ADP-ribosyl)ation of Ku 70/80 by PARP-1 results in decreased DNA binding of Ku 70/80 and a decreased ability of Ku 70/80 to stimulate WRN exonuclease activity (31). As PARP-1 is believed to be the first protein that binds a DNA break to protect the ends from nuclease degradation and aberrant DNA exchanges (40), regulation and coordination between PARP-1, DNA-PK, and WRN are likely important to ensure correct processing of the DNA double-strand break.
PARP-1 and PARP-2 also play important roles in SSBR/BER. Mouse embryonic fibroblasts lacking either PARP-1 (12) or PARP-2 (23) display defects in alkylation-induced DNA strand break repair. In addition, parp-1−/− and parp-2−/− mice are sensitive to ionizing radiation (12,21). PARP-1 interacts with many proteins involved in SSBR/BER including XRCCI (15,41), lig III (17,23), and pol β (14). PARP-1 recruits XRCCI to the damage site and is required for the formation of XRCCI foci following oxidative DNA damage (42). Interestingly, XRCCI (15) and lig III (17) preferentially interact with poly(ADP-ribosyl)ated PARP-1.
WRN also interacts physically and functionally with many BER components, including pol β (10), pol δ (43), and FEN-1 (7). WRN helicase activity stimulates strand displacement DNA synthesis of pol β (10) and nucleotide incorporation by pol δ (44). In addition, WRN stimulates FEN-1 flap cleavage (7). A WRN/PARP-1 complex may act in DNA damage recognition and in recruitment of DNA repair proteins to the damage site. Binding of PARP-1 to DNA breaks results in PARP-1 activation and the poly(ADP-ribosyl)ation of nuclear proteins, a process involved in the opening of chromatin around the damage site as well as in the recruitment of DNA repair proteins (i.e. XRCC1) mediated through binding to poly(ADP-ribose) (45) (Figure 6). Poly(ADP-ribosyl)ated PARP-1 binds not only less efficiently to DNA, but also to WRN (11), leaving the damaged site and relieving the inhibition of WRN catalytic activities (Figure 6). WRN may then act by recruiting other DNA repair proteins, as we have previously seen that cells lacking WRN are deficient in the poly(ADP-ribosyl)ation of other proteins following oxidative stress (11). Furthermore, WRN could play an active role in the repair process, possibly allowing for subsequent stimulation of pol β strand displacement and/or FEN-1 flap cleavage by WRN during long-patch BER (Figure 6).
Figure 6.
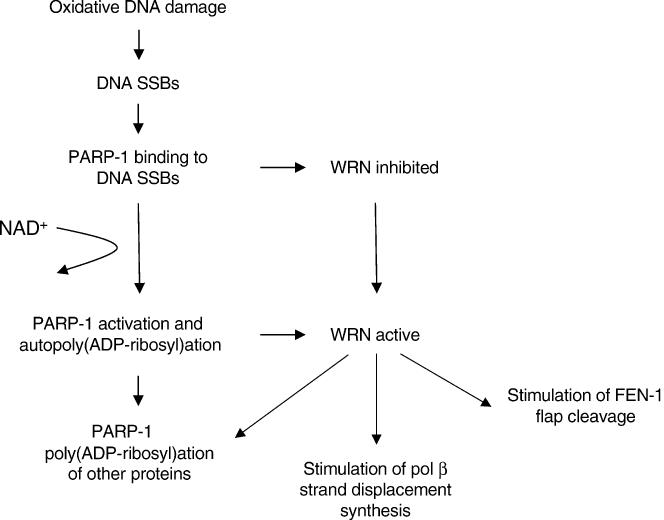
Model representing the regulation of WRN by PARP-1. Reactive oxygen species (ROS) generate DNA single-strand breaks (SSBs). PARP-1 binds to SSBs which prevents WRN catalytic activities. Binding of PARP-1 to SSBs also results in activation and auto-poly(ADP-ribosyl)ation, causing release of PARP-1 from the DNA and PARP-1 poly(ADP-ribosyl)ation of other proteins. Poly(ADP-ribosyl)ated PARP-1 binds less efficiently to WRN and subsequently relieves the inhibition of WRN catalytic activities.
PARP-2 also interacts with XRCCI, lig III and pol β (23). It has been proposed that PARP-2 may not be involved in the DNA strand break recognition and signaling steps of SSBR/BER like PARP-1, but rather in a subsequent step of the repair process, which when absent, leads to a repair defect observed in PARP-2−/− cells (23). The apparently distinct roles of PARP-1 and PARP-2 in SSBR/BER may explain their differential effect on WRN catalytic activities.
Neither PARP-1 nor WRN are essential components of the basic process of DNA strand break repair (either DSBR or SSBR/BER). However, their presence may be necessary to ensure correct and efficient DNA repair, and in their absence, improper (i.e. inappropriate recombination) and/or inefficient repair may lead to various pathologies with age. Consistently, PARP-1−/−/WRNΔhel/Δhel mice have an increased frequency of cancer and fibroblasts derived from those animals display elevated levels of chromosomal breaks, fragments and rearrangements (46).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Tina Thorslund, Dr Nadja de Souza-Pinto, Dr Patricia Opresko and Dr Sudha Sharma for comments and critical reading of the manuscript.
REFERENCES
- 1.Hickson I.D. (2003) RecQ helicases: caretakers of the genome. Nat. Rev. Cancer, 3, 169–178. [DOI] [PubMed] [Google Scholar]
- 2.Harrigan J.A. and Bohr,V.A. (2003) Human diseases deficient in RecQ helicases. Biochimie, 85, 1185–1193. [DOI] [PubMed] [Google Scholar]
- 3.Opresko P.L., Cheng,W.H., von Kobbe,C., Harrigan,J.A. and Bohr,V.A. (2003) Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis, 24, 791–802. [DOI] [PubMed] [Google Scholar]
- 4.Shen J.C., Gray,M.D., Oshima,J., Kamath-Loeb,A.S., Fry,M. and Loeb,L.A. (1998) Werner syndrome protein. I. dna helicase and dna exonuclease reside on the same polypeptide. J. Biol. Chem., 273, 34139–34144. [DOI] [PubMed] [Google Scholar]
- 5.von Kobbe C. and Bohr,V.A. (2002) A nucleolar targeting sequence in the Werner syndrome protein resides within residues 949-1092. J. Cell Sci., 115, 3901–3907. [DOI] [PubMed] [Google Scholar]
- 6.von Kobbe C., Thoma,N.H., Czyzewski,B.K., Pavletich,N.P. and Bohr,V.A. (2003) Werner syndrome protein contains three structure-specific DNA binding domains. J. Biol. Chem., 278, 52997–53006. [DOI] [PubMed] [Google Scholar]
- 7.Brosh R.M. Jr, von Kobbe,C., Sommers,J.A., Karmakar,P., Opresko,P.L., Piotrowski,J., Dianova,I., Dianov,G.L. and Bohr,V.A. (2001) Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J., 20, 5791–5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Kobbe C., Karmakar,P., Dawut,L., Opresko,P., Zeng,X., Brosh,R.M.,Jr, Hickson,I.D. and Bohr,V.A. (2002) Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem., 277, 22035–22044. [DOI] [PubMed] [Google Scholar]
- 9.Opresko P.L., von Kobbe,C., Laine,J.P., Harrigan,J., Hickson,I.D. and Bohr,V.A. (2002) Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem., 277, 41110–41119. [DOI] [PubMed] [Google Scholar]
- 10.Harrigan J.A., Opresko,P.L., von Kobbe,C., Kedar,P.S., Prasad,R., Wilson,S.H. and Bohr,V.A. (2003) The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J. Biol. Chem., 278, 22686–22695. [DOI] [PubMed] [Google Scholar]
- 11.von Kobbe C., Harrigan,J.A., May,A., Opresko,P.L., Dawut,L., Cheng,W.H. and Bohr,V.A. (2003) Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol. Cell. Biol., 23, 8601–8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shall S. and de Murcia,G. (2000) Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res., 460, 1–15. [DOI] [PubMed] [Google Scholar]
- 13.Smith S. (2001) The world according to PARP. Trends Biochem. Sci., 26, 174–179. [DOI] [PubMed] [Google Scholar]
- 14.Dantzer F., De La,R.G., Menissier-de Murcia,J., Hostomsky,Z., de Murcia,G. and Schreiber,V. (2000) Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry, 39, 7559–7569. [DOI] [PubMed] [Google Scholar]
- 15.Masson M., Niedergang,C., Schreiber,V., Muller,S., Menissier-de Murcia,J. and de Murcia,G. (1998) XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol., 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pleschke J.M., Kleczkowska,H.E., Strohm,M. and Althaus,F.R. (2000) Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem., 275, 40974–40980. [DOI] [PubMed] [Google Scholar]
- 17.Leppard J.B., Dong,Z., Mackey,Z.B. and Tomkinson,A.E. (2003) Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell. Biol., 23, 5919–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shieh W.M., Ame,J.C., Wilson,M.V., Wang,Z.Q., Koh,D.W., Jacobson,M.K. and Jacobson,E.L. (1998) Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem., 273, 30069–30072. [DOI] [PubMed] [Google Scholar]
- 19.Ame J.C., Rolli,V., Schreiber,V., Niedergang,C., Apiou,F., Decker,P., Muller,S., Hoger,T., Menissier-de Murcia,J. and de Murcia,G. (1999) PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem., 274, 17860–17868. [DOI] [PubMed] [Google Scholar]
- 20.Tong W.M., Hande,M.P., Lansdorp,P.M. and Wang,Z.Q. (2001) DNA strand break-sensing molecule poly(ADP-Ribose) polymerase cooperates with p53 in telomere function, chromosome stability, and tumor suppression. Mol. Cell. Biol., 21, 4046–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menissier d.M., Ricoul,M., Tartier,L., Niedergang,C., Huber,A., Dantzer,F., Schreiber,V., Ame,J.C., Dierich,A., LeMeur,M. et al. (2003) Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J., 22, 2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ame J.C., Schreiber,V., Fraulob,V., Dolle,P., de Murcia,G. and Niedergang,C.P. (2001) A bidirectional promoter connects the poly(ADP-ribose) polymerase 2 (PARP-2) gene to the gene for RNase P RNA. Structure and expression of the mouse PARP-2 gene. J. Biol. Chem., 276, 11092–11099. [DOI] [PubMed] [Google Scholar]
- 23.Schreiber V., Ame,J.C., Dolle,P., Schultz,I., Rinaldi,B., Fraulob,V., Menissier-de Murcia,J. and de Murcia,G. (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem., 277, 23028–23036. [DOI] [PubMed] [Google Scholar]
- 24.Orren D.K., Brosh,R.M.,Jr, Nehlin,J.O., Machwe,A., Gray,M.D. and Bohr,V.A. (1999) Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res., 27, 3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machwe A., Ganunis,R., Bohr,V.A. and Orren,D.K. (2000) Selective blockage of the 3′→5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res., 28, 2762–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giner H., Simonin,F., de Murcia,G. and Menissier-de Murcia,J. (1992) Overproduction and large-scale purification of the human poly(ADP-ribose) polymerase using a baculovirus expression system. Gene, 114, 279–283. [DOI] [PubMed] [Google Scholar]
- 27.Karmakar P., Snowden,C.M., Ramsden,D.A. and Bohr,V.A. (2002) Ku heterodimer binds to both ends of the Werner protein and functional interaction occurs at the Werner N-terminus. Nucleic Acids Res., 30, 3583–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karmakar P., Piotrowski,J., Brosh,R.M.,Jr, Sommers,J.A., Miller,S.P., Cheng,W.H., Snowden,C.M., Ramsden,D.A. and Bohr,V.A. (2002) Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J. Biol. Chem., 277, 18291–18302. [DOI] [PubMed] [Google Scholar]
- 29.Huang S., Li,B., Gray,M.D., Oshima,J., Mian,I.S. and Campisi,J. (1998) The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease. Nature Genet., 20, 114–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adelfalk C., Kontou,M., Hirsch-Kauffmann,M. and Schweiger,M. (2003) Physical and functional interaction of the Werner syndrome protein with poly-ADP ribosyl transferase. FEBS Lett., 554, 55–58. [DOI] [PubMed] [Google Scholar]
- 31.Li B., Navarro,S., Kasahara,N. and Comai,L. (2004) Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J. Biol. Chem., 279, 13659–13667. [DOI] [PubMed] [Google Scholar]
- 32.Ludwig A., Behnke,B., Holtlund,J. and Hilz,H. (1988) Immunoquantitation and size determination of intrinsic poly(ADP-ribose) polymerase from acid precipitates. An analysis of the in vivo status in mammalian species and in lower eukaryotes. J. Biol. Chem., 263, 6993–6999. [PubMed] [Google Scholar]
- 33.Yamanaka H., Penning,C.A., Willis,E.H., Wasson,D.B. and Carson,D.A. (1988) Characterization of human poly(ADP-ribose) polymerase with autoantibodies. J. Biol. Chem., 263, 3879–3883. [PubMed] [Google Scholar]
- 34.Moser M.J., Kamath-Loeb,A.S., Jacob,J.E., Bennett,S.E., Oshima,J. and Monnat Jr,R.J. (2000) WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res., 28, 648–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brosh J., Karmakar,P., Sommers,J.A., Yang,Q., Wang,X.W., Spillare,E.A., Harris,C.C. and Bohr,V.A. (2001) p53 modulates the exonuclease activity of Werner syndrome protein. J.Biol.Chem., 276, 35093–35102. [DOI] [PubMed] [Google Scholar]
- 36.Cooper M.P., Machwe,A., Orren,D.K., Brosh,R.M., Ramsden,D. and Bohr,V.A. (2000) Ku complex interacts with and stimulates the Werner protein. Genes Dev., 14, 907–912. [PMC free article] [PubMed] [Google Scholar]
- 37.Yannone S.M., Roy,S., Chan,D.W., Murphy,M.B., Huang,S., Campisi,J. and Chen,D.J. (2001) Werner syndrome protein is regulated and phosphorylated by dna- dependent protein kinase. J. Biol. Chem., 276, 38242–38248. [DOI] [PubMed] [Google Scholar]
- 38.Ruscetti T., Lehnert,B.E., Halbrook,J., Le Trong,H., Hoekstra,M.F., Chen,D.J. and Peterson,S.R. (1998) Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem., 273, 14461–14467. [DOI] [PubMed] [Google Scholar]
- 39.Ariumi Y., Masutani,M., Copeland,T.D., Mimori,T., Sugimura,T., Shimotohno,K., Ueda,K., Hatanaka,M. and Noda,M. (1999) Suppression of the poly(ADP-ribose) polymerase activity by DNA-dependent protein kinase in vitro. Oncogene, 18, 4616–4625. [DOI] [PubMed] [Google Scholar]
- 40.Lindahl T., Satoh,M.S., Poirier,G.G. and Klungland,A. (1995) Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci., 20, 405–411. [DOI] [PubMed] [Google Scholar]
- 41.Caldecott K.W., Aoufouchi,S., Johnson,P. and Shall,S. (1996) XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El Khamisy S.F., Masutani,M., Suzuki,H. and Caldecott,K.W. (2003) A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res., 31, 5526–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szekely A.M., Chen,Y.H., Zhang,C., Oshima,J. and Weissman,S.M. (2000) Werner protein recruits DNA polymerase delta to the nucleolus. Proc. Natl Acad. Sci. USA, 97, 11365–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamath-Loeb A.S., Johansson,E., Burgers,P.M. and Loeb,L.A. (2000) Functional interaction between the Werner Syndrome protein and DNA polymerase delta. Proc. Natl Acad. Sci. USA, 97, 4603–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muiras M.L. (2003) Mammalian longevity under the protection of PARP-1's multi-facets. Ageing Res. Rev., 2, 129–148. [DOI] [PubMed] [Google Scholar]
- 46.Lebel M., Lavoie,J., Gaudreault,I., Bronsard,M. and Drouin,R. (2003) Genetic cooperation between the Werner syndrome protein and poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice. Am. J. Pathol., 162, 1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Murcia G. and Menissier,d.M. (1994) Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci., 19, 172–176. [DOI] [PubMed] [Google Scholar]


