Abstract
The gene coding for the SinI DNA-methyltransferase, a modification enzyme able to recognize and methylate the internal cytosine of the GGA/TCC sequence, was subjected to in vitro mutagenesis, DNA-shuffling and a strong selection for relaxed GGNCC recognition specificity. As a result of this in vitro evolution experiment, a mutant gene with the required phenotype was selected. The mutant SinI methyltransferase carried five amino acid substitutions. None of these was found in the ‘variable region’ that were thought to be responsible for sequence specificity. Three were located near the N-terminal end, preceding the first conserved structural motif of the enzyme; two were found between conserved motifs VI and VII. A clone engineered to carry out only the latter two replacements (L214S and Y229H) displays relaxed recognition specificity similar to that of the parental mutant, whereas the clone carrying only the N-terminal replacements showed a much weaker change in recognition specificity. The enzyme with two internal mutations was purified and characterized. Its catalytic activity (kcat/Km) was ∼5-fold lower towards GGA/TCC and 20-fold higher towards GGG/CCC than that of the wild-type enzyme.
INTRODUCTION
Type II modification enzymes are ideally suited to studying the molecular mechanisms of sequence-specific protein–DNA interactions as these enzymes are able to recognize, and act upon, 2–8 bp long DNA sequence elements with an astonishing degree of accuracy (1). One subgroup of these enzymes, the cytosine-C5 methyltransferases (C5-MTases), displays a basically similar architecture. Comparison of the amino acid sequences of 13 such enzymes showed that they share 6 strongly conserved and 4 weakly conserved sequence motifs arranged in every case in the same order [(2), Figure 1]. Motifs VIII and IX are separated by a long variable stretch of amino acids with little or no homology between different enzymes. Nearly all C5-MTases sequenced later were also shown to have the same structural organization (3). This structure suggested the intuitively attractive assumption that the common motifs might be responsible for the common enzymatic reaction, i.e. the transfer of the methyl group from S-adenosyl-l-methionine (AdoMet) to the ring C5 of the target cytosine, whereas the variable region would determine the recognition specificity that distinguishes these enzymes from each other. This notion, supported by several lines of experimental evidence, is now generally accepted in the literature.
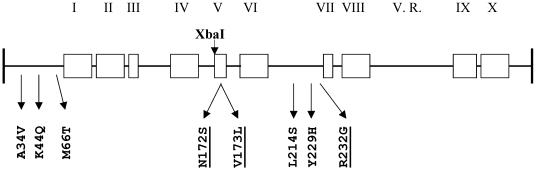
Figure 1.
Schematic representation of conserved sequence motifs in the SinI MTase. Position of an XbaI site in the M.SinI gene and positions of amino acid replacements described in this paper and in (10) (underlined) are indicated. V. R., variable region.
A subgroup of C5-MTases, the bacteriophage-coded multispecific MTases are able to methylate several different DNA sequences. These enzymes have unusually long variable regions, and segments of these regions (target recognizing domains) were shown to be responsible for the recognition of each of these sequences, as recognition specificity proved to be transferable with the transfer of target recognizing domains to the variable region of another enzyme (4,5). Although changing sequence specificity by transferring only the variable region has not been reported for any of the monospecific C5-MTases, but in two cases chimeric C5-MTases could be constructed whose methylation specificity strongly supported the role of the variable region in the DNA substrate recognition. Methylation specificity of M.HhaI–M.HpaII hybrids corresponds to that partner which contributes the variable region together with motif IX (6). In the second study, involving hybrids of M.HpaII and M.MspI, it was shown that the variable region also determined the target cytosine within the recognition sequence containing two cytosines (7).
The three-dimensional structures of the specific enzyme–DNA complexes of M.HhaI (8) and M.HaeIII (9) also confirmed the notion that the variable region determines specificity. The structures show that the enzymes consist of a larger and a smaller domain forming a cleft where the substrate DNA fits. In both X-ray models, the large domain containing most of the conserved motifs that faces the minor groove of the DNA, whereas the small domain contains the variable region and motif IX that faces the major groove. Contacts mediating sequence-specific DNA recognition were shown to occur between the small domain of the MTase and the major groove surface of the DNA (8,9).
Recently, however, experiments from this laboratory have shown that at least in the case of two C5-MTases (M.SinI and M.EcoRII, recognition sequences are GGA/TCC and CCA/TGG, respectively) the ability of the enzyme to distinguish between A/T and G/C base pairs in the middle of the recognition sequence depends on interaction with the minor groove of the DNA (10). This observation suggested that, if these enzymes have a similar three-dimensional structure to M.HhaI and M.HaeIII, the determining elements must be in the large domain, but not in the variable region. This model was consistent with the results from random mutagenesis of a large segment (including the variable region) of the M.SinI gene. Three mutants with relaxed specificity for the central base pair of the recognition sequence were isolated. None of these mutations was found in the variable region [(10), Figure 1].
As in the previous study, only the C-terminal two-third of the M.SinI gene was mutagenized and the mutants had much lower activity than the wild-type (WT) enzyme (10), we decided to try a new approach to isolate relaxed specificity mutants of M.SinI. In this paper, we report experiments attempted to convert the specificity of M.SinI into GGNCC. Our experimental strategy involved random mutagenesis of the entire M.SinI gene, then the application of ‘DNA-shuffling’ (in vitro random recombination), followed by a strong in vitro selection for the required phenotype. The results reported here demonstrate that this strategy allowed the isolation of an M.SinI mutant displaying relaxed recognition specificity, and only a small decrease in overall catalytic acticity relative to the WT enzyme. None of the five amino acid substitutions in the mutant enzyme was located in the variable region.
METHODS
Strains, vectors and media
Plasmid pSin5 carries the M.SinI gene (10). The pTZ57R/T and pER23S(-ATG) vectors [Fermentas; (10,11)] were used for cloning and expressing the mutant genes.
Escherichia coli XL1-Blue MRF′ Kan (Stratagene) served as host in most experiments. E.coli ER1398 hsdR2 mcrB1 was used for overexpression using the vector pER23S(-ATG). In experiments using pER23S(-ATG), the host cells also contained plasmid pVH1 (12) to repress transcription (10). Cells were routinely grown in LB medium at 37°C. Enzyme production was induced at 30°C with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) or with 0.2% lactose. Ampicillin and kanamycin were used at 100 and 50 μg/ml, respectively.
Mutagenesis
Two different methods were employed to generate random point mutations in the M.SinI gene. In the ‘hypermutagenic PCR’ technique (13), the misincorporation rate is increased by the limiting concentration of one of the dNTPs (one in each of the four different reaction mixtures) and the presence of Mn2+. In the second method [‘error-prone PCR’, (14)], the misincorporation rate is increased by the presence of dITP. The PCR mutagenesis reactions contained 10 ng of pSin5 DNA template and two oligonucleotide primers (ACCGGATCCTTCAGTTTAGA and GCAGAATTCGCGACAATACG) corresponding to the ends of the 1.7 kb insert of pSin5.
Plasmid pTZS, which carried the WT M.SinI gene was generated by PCR-amplification, under non-mutagenic conditions, of the coding sequence using primers AK54 and AK55 (see below), and cloning the PCR product in pTZ57R/T.
DNA-shuffling
Two different DNA-shuffling methods were used. The steps of the method developed by Stemmer (15) are random, limited DNaseI digestion of the mutagenized PCR product to an average fragment length of 10–200 bp, random PCR without primers and final amplification by primed PCR. For this final amplification step, the primers AK54 (CCGCTCGAGATGATAATGAATGAATGACATCAATAC) and AK55 (CCGCTCGAGTTAGACCAACTCTCCAAA) were used. Both primers contained XhoI recognition sites (underlined). These primers correspond to the boundaries of the coding sequence of the gene from the translation initiation to the termination codon, thus the final PCR product was 1.4 kb, smaller than the starting mutagenized fragment. The amplified product was purified from gel, then inserted by A/T cloning into the vector pTZ57R/T and introduced by transformation into XL1-Blue MRF Kan cells.
The second shuffling protocol was described by Zhao and Arnold (16) with the modification that we employed only Taq polymerase instead of Taq/Pfu mixture.
Selection
Transformant colonies obtained after mutagenesis and shuffling were pooled and used to inoculate a liquid culture. At mid-log phase, 1 mM IPTG was added and shaking at 30°C was continued for 4 h. Induction of MTase production by IPTG was thought to be necessary because the M.SinI gene cloned in pTZ57R lacked its own promoter. A sample (500 ng) of the plasmid DNA isolated from the culture was digested with 15 U of Cfr13I. Cfr13I does not cut GGNCC sites C5-methylated at the internal cytosine (17). Progress of the digestion was followed by determining the number of transformants obtained from aliquots taken at intervals from the digestion mixture. Plasmid pTZS containing the non-mutagenized M.SinI gene was used as a control.
Protein purification
The mutant MTases enzyme was purified using a modified version of the method described for the WT M.SinI (10). Briefly, after induction for 4 h at 30°C cells were harvested, resuspended and disrupted by sonication. Crude extracts were centrifuged and the supernatant was loaded onto a phosphocellulose column equilibrated with PC buffer (20 mM K-phosphate, pH 7.5, 10 mM 2-mercaptoethanol, 1 mM EDTA and 5% glycerol). Proteins were eluted by a salt gradient (0.2–1 M NaCl in PC buffer). Active fractions (eluted between 0.46 and 0.5 M) were dialyzed against PC buffer containing 0.2 M NaCl and loaded onto a heparin–agarose column. Elution conditions were the same as in the previous step and active fractions were recovered between 0.5 and 0.57 M NaCl. These fractions were concentrated by dialysis against a storage solution (0.1 M Tris–HCl, pH 7.5, 50 mM KCl, 0.1 mM EDTA, 1 mM DTT and 50% glycerol) and stored at −20°C.
MTase assay
MTase activity measurements using [methyl-3H]AdoMet and 19 bp double-stranded oligonucleotide substrates containing GGA/TCC or GGG/CCC sites (A/T and G/C oligo, respectively), and the calculation of steady-state kinetic parameters were carried out as described previously (10).
For a qualitative check for the presence of MTase activity during the purification procedure, a non-radioactive ‘protection’ assay was used. Samples of the fractions were incubated with 500 ng plasmid DNA substrate, 2.5 μM AdoMet, 50 mM Tris–HCl, pH 8, 50 mM NaCl, 10 mM EDTA and 1 mM DTT in 25 μl volume for 20 min at 37°C. After the MTase reaction, 4 μl of 100 mM MgCl2 and 1 U of Cfr13I was added, the sample was further incubated for 30 min at 37°C, and subsequently electrophoresed in a 1% agarose gel.
Methylation of GGA/TCC sites in plasmid DNA preparations was tested by digestion with Eco47I and methylation of GGG/CCC sites was tested by digestion with Cfr13I.
Other methods
Cloning, transformation, plasmid isolation, agarose electrophoresis and PAGE, and fragment isolation were done by standard methods as described previously (18). For cloning PCR products, the InsT/Aclone cloning kit of Fermentas was used.
Enzymes were purchased from Fermentas or New England Biolabs. [methyl-3H]AdoMet was from New England Nuclear.
RESULTS
Selection of the mutant SinI MTase
The gene encoding M.SinI was mutagenized using two different methods. The PCR-products obtained in the two reactions were pooled, then split in two portions and subjected to two different shuffling procedures as described in Methods. Approximately 7000 AmpR clones were obtained. Plasmid DNA isolated from the culture was digested with an excess of Cfr13I and the digested DNA was used to transform E.coli. Plasmid pTZ57R, the vector used in this experiment has 2 GGA/TCC and 6 GGG/CCC sites. Selection for the mutant MTase with the required relaxed recognition specificity was based on the assumption that expression of such a mutant MTase would render the plasmid encoding it resistant to digestion with the Cfr13I restriction endonuclease (recognition specificity GGNCC), while expression of the WT M.SinI enzyme would leave the GGG/CCC sites on the plasmid sensitive to digestion. A single transformant was obtained.
Characterization of the mutant gene
The plasmid (named pTZSmut) isolated from an IPTG-induced culture of the single transformant clone was completely resistant to Eco47I and highly resistant to Cfr13I (Figure 2) indicating that the MTase encoded by pTZSmut, in addition to the WT target site GGA/TCC also methylates GGG/CCC sites. As expected, orientation of the MTase gene corresponds to that of the transcription starting from the lac promoter on pTZ57R. To check the specificity for methylation, the pTZSmut plasmid was digested with a few other restriction endonucleases (MspI, HhaI, Sau3AI and BspRI) known to be sensitive to C5-methylation. All these digestions generated patterns identical to that of the WT plasmid, except in the case of the BspRI-digestion (data not shown). This was to be expected, because methylation of the GGG/CCC sites would render overlapping BspRI sites (GGCC) resistant against BspRI digestion.
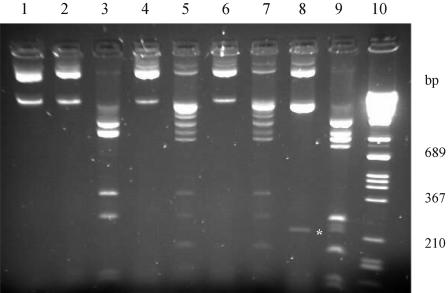
Figure 2.
Modification in vivo of plasmid GGA/TCC and GGG/CCC sites by the WT and mutant SinI MTase. Methylation status of the plasmids was tested by digestion with Eco47I (specificity GGA/TCC) or Cfr13I (GGNCC). Lane 1, pTZS (undigested); lane 2, pTZS–Eco47I; lane 3, pTZS–Cfr13I; lane 4, pTZSmut–Eco47I; lane 5, pTZSmut–Cfr13I; lane 6, pTZSmut45–Eco47I; lane 7, pTZSmut45–Cfr13I; lane 8, pTZSmut123–Eco47I; lane 9, pTZSmut123–Cfr13I; and lane 10, DNA size marker. All plasmids tested have two GGA/TCC sites. The 222 bp fragment generated by digestion at the GGA/TCC sites is indicated by asterisk. pTZS and pTZSmut123 carry nine GGG/CCC sites (six in the vector, three in the M.SinI gene), whereas pTZSmut and pTZSmut45 have an additional GGG/CCC site created by the mutagenesis process. Electrophoresis carried out in a 1.5% agarose gel.
MTase activity measurements with crude extract of the mutant clone and synthetic oligonucleotides (A/T-oligo and G/C-oligo) also indicated methylation of GGG/CCC sites.
DNA sequencing revealed that the mutant gene contained nine point mutations, seven transitions and two transversions. Four of these (nucleotide positions T96C, C169T, C242T and G618C) were ‘silent’ [numbering refers to the sequence in (19) starting with the A of the translation-initiation codon]. Five mutations (C101T, A130C, T197C, T641C and T685C) resulted in amino acid replacements (A34V, K44Q, M66T, L214S and Y229H). Three of these replacements were located in the N-terminal part, preceding the first conserved structural motif; two replacements in the middle of the molecule between conserved blocks VI and VII (Figure 1). No substitution was found in the variable region (between blocks VIII and IX). One of the ‘silent’ mutations (the G–C transversion at nucleotide position 618) created a new GGG/CCC site in the gene.
Separation of the internal and the N-terminal mutations
The fact that three out of five amino acid substitutions in the mutant gene resided in the region coding for the N-terminal end of the protein seemed to be somewhat surprising, because this region is missing from most C5-MTases. To study their contribution to the altered phenotype, the two groups of mutations were separated. An XbaI site located between the gene segments coding for motifs IV and V (Figure 1) allowed the exchange of a 898 bp long XbaI fragment between the pTZS and pTZSmut to yield pTZSmut123 (A34V + K44Q + M66T) and pTZSmut45 (L214S + Y229H). As shown in Figure 2, methylation status of pTZSmut and pTZSmut45, as revealed by Eco47I and Cfr13I digestion, was similar, whereas in pTZSmut123 the GGG/CCC sites were only very partially modified and methylation of even the GGA/TCC sites was not complete. This observation indicates that altered sequence specificity of the mutant enzyme is mainly caused by one or both of the two replacements between motifs VI and VII, and contribution to the phenotype of the three N-terminal mutations was minimal.
Purification and characterization of the Leu214Ser, Tyr229His double mutant enzyme
As the MTase level in the cell extracts of clones carrying the pTZ57R/T-based plasmids was low, all three mutant genes were transferred (by using XhoI digestion) to the SalI site of the expression vector plasmid pER23S(-ATG). In the case of the mutants carrying the three N-terminal mutations alone, or all five mutations, this transfer did not help, because majority of the mutant enzymes was found in insoluble inclusion bodies after induction and the enzymes could not be purified. However, the enzyme carrying only the two internal mutations was proved to be soluble and could be purified to near homogeneity (Figure 3).
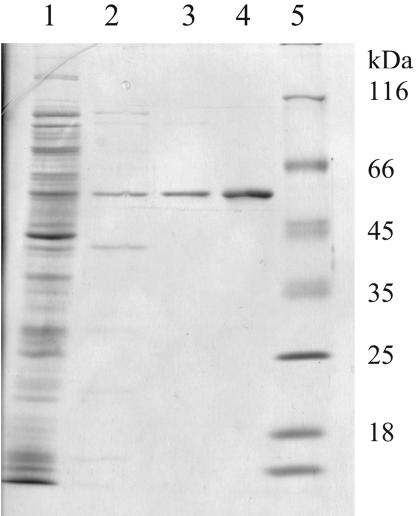
Figure 3.
SDS–PAGE of the purified mutant enzyme coded by pERSmut45. Lane 1, crude extract; lane 2, after phosphocellulose chromatography; lane 3, after heparin–agarose chromatography; lane 4, purified WT M.SinI enzyme; and lane 5, protein size markers.
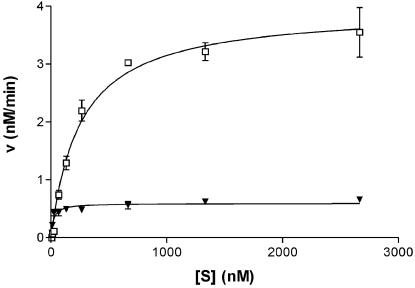
Steady-state kinetic parameters of the double-mutant SinI MTase were determined (Figure 4 and Table 1). M.SinI(L214S + Y229H) has a 4.5-fold lower kcat/Km value for the canonical substrate GGA/TCC than the WT enzyme. This change is mainly due to the decrease in kcat. The mutant has 2-fold higher Km than the WT enzyme for the GGG/CCC substrate, however, this loss is more than compensated by the large (40-fold) increase in kcat. As a consequence, ∼150-fold preference of the WT enzyme for GGA/TCC sites over GGG/CCC sites has changed to 1.7-fold preference in the mutant.
Figure 4.
Steady-state kinetic analysis of DNA methylation by the M.SinI(L214S + Y229H) mutant MTase. Triangles, GGA/TCC and open squares, GGG/CCC.
Table 1. Steady-state kinetic parameters of the WT M.SinI and the L214S + Y229H double-mutant enzyme.
| DNA substrate | M.SinI (WT) | M.SinI (L214S + Y229H) | ||
|---|---|---|---|---|
| GGA/TCC | GGG/CCC | GGA/TCC | GGG/CCC | |
| Km (nM) | 17 | 116 | 20.7 ± 0.34 | 255 ± 15 |
| kcat (min−1) | 0.3 | 0.014 | 0.08 ± 0.0048 | 0.56 ± 0.06 |
| kcat/Km (105 M−1 s−1) | 2.94 | 0.02 | 0.65 | 0.37 |
DISCUSSION
This work was started with two aims. First, we wanted to test the hypothesis put forward in a previous paper (10) that the discrimination by M.SinI between GGA/TCC and GGG/CCC sites is mediated by the large domain of the enzymes. Second, we wanted to test whether in vitro evolution techniques can be applied to alter the recognition specificity of C5-MTases, which have been proved to be resistant to more conventional approaches.
By a combination of in vitro mutagenesis, DNA shuffling and strong selection for the required phenotype, we have isolated a mutant SinI MTase, which in addition to its canonical substrate sequence GGA/TCC efficiently methylates also GGG/CCC sites. Of the five mutations leading to amino acid replacements, three were shown to have only minor contribution to the altered phenotype. Future experiments will determine whether both remaining mutations (L214S and Y229H) are needed for the relaxed recognition specificity phenotype. The replacements L214S and Y229H are between motifs VI and VII. Two other mutations isolated previously, and shown to lead to relaxed sequence specificity (N172S and V173L) are located in the conserved motif V, and a third mutation (R232G) characterized by a smaller change in phenotype is located between motifs VI and VII [(10), Figure 1]. Although in both studies [(10) and this work], the mutagenized region includes the variable region; no relaxed specificity mutant with a substitution in the variable region was obtained. Thus, the location of all mutations so far isolated is consistent with the hypothesis that the A/T base pair is recognized by the large domain of M.SinI. The common architecture suggests that also other C5-MTases characterized by an A/T ambiguity in the recognition sequence use large domain—minor groove contacts to exclude sites containing G/C base pairs. Owing to the lack of crystallographic data, it would obviously be premature to draw any firm conclusion or generalization concerning the structural determinants of specificity from these results. For instance, positions 214 and 229 are in a region, which is fairly long in M.SinI (19,3), thus the functional importance of this segment might be a unique feature of this enzyme.
The M.SinI mutant described here looked more interesting from the beginning than the mutants isolated previously (10) because the total MTase activity measured in crude extracts with [methyl-3H]AdoMet and both DNA substrates (A/T plus G/C oligo) was similar to that of the WT clone. Moreover, under the high DNA concentrations used in the assay, the G/C oligo seemed to be a much better substrate than the A/T oligo (data not shown). We have, therefore, decided to purify the mutant MTase and determine its basic catalytic constants. Steady-state kinetic analysis revealed that for the mutant enzyme, GGG/CCC sites are almost as good substrates as GGA/TCC sites (Table 1) qualifying M.SinI(L214S + Y229H) a bona fide GGNCC-specific MTase. Interestingly, this change was mainly caused by a large increase in kcat.
Methylation status of the plasmids isolated from cells producing M.SinI(L214S + Y229H) is consistent with the catalytic constants. GGA/TCC sites were fully protected, whereas protection of GGG/CCC sites was not complete (Figure 2). This difference can be explained by the larger number of GGG/CCC sites in the E.coli genome (4218 sites versus 2874 GGA/TCC sites; (20) and J.Pósfai, personal communication) and in the plasmid pTZSmut (10 versus 2).
Changing recognition specificity of modification methylases by various techniques of protein engineering has long been a desirable goal of several groups working in this field. In one successful attempt, rational protein design was used to change the target base preference of M.EcoRV from adenine to cytosine. However, this change in specificity was accompanied by a large decrease in overall catalytic activity (21).
While this work was in progress, other authors successfully employed a directed in vitro evolution technique to achieve a change in the specificity of the HaeIII MTase. The catalytic efficiency of the mutant enzyme was better than that of the WT enzyme with the principal substrate (22).
To the best of our knowledge, our results represent the second example of a change in recognition specificity without significant impairment of activity. The success of achieving a change in substrate specificity using in vitro evolution provides new ammunition for those arguing for the superiority of random, over ‘designed’ approaches.
The strategy used previously (10) and in this work to selecting relaxed specificity mutants of an MTase is the extension of a method developed for cloning MTase genes (23). This method can, in principle, be applied to all cases where the aim is selection of a mutant with lower specificity. A necessary precondition of the method is the availability of a restriction enzyme with the aimed specificity. The second precondition is that the endonuclease should be sensitive to the type of modification produced by the MTase (position and type of the methylated base). Potential applications include selection of mutants, which have lost the capacity to distinguish between certain base pairs [e.g. conversion of GTCGm6AC (SalI) specificity into GTYRm6AC (HincII) specificity], and the selection of mutants that recognize a shorter sequence than the parental enzyme [e.g. conversion of SalI specificity to TaqI (TCGm6A) specificity].
Acknowledgments
ACKNOWLEDGEMENTS
Thanks are due to E. Magyaródi for expert technical assistance and T. Raskó for valuable advice in enzyme purification. This work was supported by OTKA grants T 034264 and T 038343 to P.V. and A.K., respectively.
REFERENCES
- 1.Cheng X. (1995) Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct., 24, 293–318. [DOI] [PubMed] [Google Scholar]
- 2.Pósfai J., Bhagwat,A., Pósfai,G. and Roberts,R. (1989) Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res., 17, 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar S., Cheng,X., Klimasauskas,S., Mi,S., Pósfai,J., Roberts,R.J. and Wilson,G.G. (1994) The DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 22, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilke K., Rauhut,E., Noyer-Weidner,M., Lauster,R., Pawlek,B., Behrens,B. and Trautner,T.A. (1988) Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J., 7, 2601–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balganesh T.S., Reiners,L., Lauster,R., Noyer-Weidner,M., Wilke,K. and Trautner,T.A. (1987) Construction and use of chimeric SPR/F3T DNA methyltransferases in the definition of sequence recognizing enzyme regions. EMBO J., 6, 3543–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klimasauskas S., Nelson,J.L. and Roberts,R.J. (1991) The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res., 19, 6183–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mi S. and Roberts,R.J. (1992) How M.MspI and M.HpaII decide which base to methylate. Nucleic Acids Res., 20, 4811–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng X., Kumar,S., Pósfai,J., Pflugrath,J.W. and Roberts,R.J. (1993) Crystal structure of the HhaI methyltransferase complexed with S-adenosylmethionine. Cell, 74, 299–307. [DOI] [PubMed] [Google Scholar]
- 9.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 10.Kiss A., Pósfai,G., Zsurka,G., Raskó,T. and Venetianer,P. (2001) Role of DNA minor groove interactions in substrate recognition by the M.SinI and M.EcoRII DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 29, 3188–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukacsovich T., Orosz,A., Balikó,G. and Venetianer,P. (1990) A family of expression vectors based on the rrnB P2 promoter of Escherichia coli. J. Biotechnol., 16, 49–56. [DOI] [PubMed] [Google Scholar]
- 12.Haring V., Scholz,P., Scherzinger,E., Frey,J., Derbyshire,K., Hatfull,G., Willetts,N.S. and Bagdasarian,M. (1985) Protein RepC is involved in copy number control of the broad host range plasmid RSF1010. Proc. Natl Acad. Sci. USA, 82, 6090–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchholz F., Angrand,P.-O. and Stewart,F. (1998) Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol., 16, 657–662. [DOI] [PubMed] [Google Scholar]
- 14.Spee J.H., de Vos,W.M. and Kuipers,O.P. (1993) Efficient random mutagenesis method with adjustable mutation frequency by use of PCR and dITP. Nucleic Acids Res., 21, 777–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stemmer W.P.C. (1994) DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl Acad. Sci. USA, 91, 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao H. and Arnold,F.H. (1997) Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res., 25, 1307–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bitinaite J.B., Klimasauskas,S.J., Butkus,V.V. and Janulaitis,A.A. (1985) Characterization of restriction-modification enzymes Cfr13 I from Citrobacter freundii RFL13. FEBS Lett., 182, 509–513. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J., Fritsch,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 19.Karreman C. and de Waard,A. (1988) Cloning and complete nucleotide sequences of the type II restriction-modification genes of Salmonella infantis. J. Bacteriol., 170, 2527–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T, Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F., Gregor,J., Davis,N.W., Kirkpatrick,H.A., Goeden,M.A., Rose,D.J., Mau,B. and Shao,Y. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 21.Roth M. and Jeltsch,A. (2001) Changing the target base specificity of the EcoRV DNA methyltransferase by rational de novo protein design. Nucleic Acids Res., 29, 3137–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen H.M., Tawfik,D.S. and Griffiths,A.D. (2004) Altering the specificity of HaeIII methyltransferase by directed evolution using in vitro compartmentalization. Prot. Eng. Des. Sel., 17, 3–11. [DOI] [PubMed] [Google Scholar]
- 23.Szomolányi É., Kiss,A. and Venetianer,P. (1980) Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene, 10, 219–225. [DOI] [PubMed] [Google Scholar]