Abstract
Adenovirus (Ad) precursor terminal protein (pTP) in a complex with Ad DNA polymerase (pol) serves as a primer for Ad DNA replication. During initiation, pol covalently couples the first dCTP with Ser-580 of pTP. By using an in vitro reconstituted replication system comprised of purified proteins, we demonstrate that the conserved Asp-578 and Asp-582 residues of pTP, located close to Ser-580, are important for the initiation activity of the pTP/pol complex. In particular, the negative charge of Asp-578 is essential for this process. The introduced pTP mutations do not alter the binding capacity to DNA or polymerase, suggesting that the priming mechanism is affected. The Asp-578 or Asp-582 mutations increase the Km for dCTP incorporation, and higher dCTP concentrations or Mn2+ replacing Mg2+ partially relieve the initiation defect. Moreover, the kcat/Km values are reduced as a consequence of the pTP mutations. These observations demonstrate that pTP influences the catalytic activity of pol in initiation. Since both Asp residues are situated close to the pol active site during initiation, they may contribute to correct positioning of the OH group in Ser-580. Our results indicate that specific amino acids of the protein primer influence the ability of Ad5 DNA polymerase to initiate DNA replication.
INTRODUCTION
The genome of adenovirus serotype 5 (Ad5) is a 35.935 bp double-stranded (ds) linear DNA with a terminal protein covalently attached to each DNA end. The origins of DNA replication are located at each end of the genome in 103 bp long inverted terminal repeats. Ad employs a unique protein priming mechanism to initiate its DNA replication (1,2). First, the pre-initiation complex is assembled on the origin of replication. It consists of three viral proteins, precursor terminal protein (pTP) that serves as a primer, Ad5 DNA polymerase (pol), DNA-binding protein (DBP), and two cellular transcription factors, nuclear factor I (NFI) and octamer-binding protein (Oct-1), which are able to significantly enhance the replication process. During initiation, pTP presents its Ser-580 residue to the pol active site. After binding with an incoming dCTP nucleotide, pol covalently couples dCTP with the OH group of Ser-580. This reaction depends on the affinity of pol for dCTP and pTP as well as template DNA binding, and the catalysis itself that can be kinetically described by an apparent KmKmapp (3,4). After coupling of the first dCTP, two additional nucleotides are added, forming a pTP–CAT intermediate using an internal GTA triplet at positions 4–6 as a template. Subsequently, the newly synthesized CAT trinucleotide jumps back to positions 1–3 at the beginning of the template strand (2,5). Next, pTP dissociates from pol and pol elongates DNA processively (6). Later in viral infection, pTP is cleaved by a viral protease at three specific cleavage sites (Figure 1) (7,8) resulting in the formation of a smaller terminal protein that stays attached to each DNA end for the remaining part of the viral life cycle.
Figure 1.
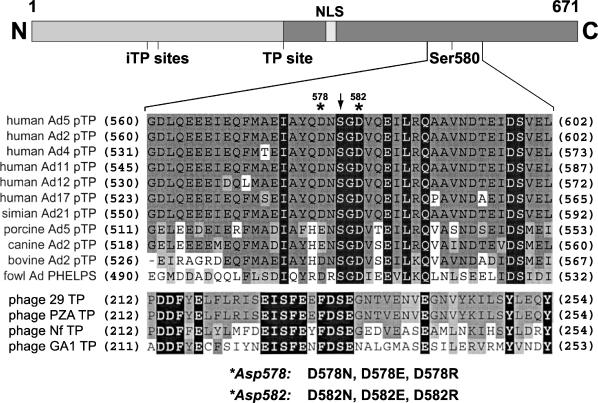
Alignment of terminal protein sequences of the region containing the priming Ser residue. The upper panel shows a schematic overview of the primary Ad5 pTP sequence (671 amino acids). Ser-580 of pTP is coupled to the first nucleotide during initiation of Ad5 DNA replication. pTP contains a nuclear localization signal (NLS) and three protease cleavage sites (two iTP sites and one TP site) used by adenoviral protease in a later infection stage (7,8). The lower panel shows the aligned sequences from adenoviruses infecting human and other species. The sequences of the TP proteins from B.subtilis phages are also shown. Numbers in parentheses indicate the amino acid position relative to the N-terminus. Residues identical among the listed viruses are shown in black squares, conserved residues are shown in dark gray squares and similar residues are shown in light gray squares. The priming Ser is marked with an arrowhead. The Asp-578 and Asp-582 of Ad5 pTP are indicated with asterisks and the introduced mutations are listed.
The priming pTP is a 75 kDa protein that can form a tight heterodimer in solution with the 140 kDa pol (pTP/pol complex) (9–12). Protease cleavage experiments and mutational studies show that a large portion of the pTP surface is involved in the interaction with pol (13–16). Moreover, studies on the molecular architecture of Ad5 pol demonstrate that pTP binds at the entrance of the primer-binding groove of pol (17). Although, the sequence specificity of DNA binding by pTP alone is very limited (18,19), the pTP/pol heterodimer binds with increased specificity to a conserved region located between 9 and 18 bp in the Ad5 origin of replication (19,20). The pTP/pol complex can be recruited to the origin by the transcription factors, Oct-1 and NFI, interacting with pTP and pol, respectively, resulting in the stimulation of replication (21–23). Furthermore, based on the dimerization properties of pTP, the proposed interaction between pTP and the terminal protein covalently bound to the viral DNA most probably stabilizes the formation of a pre-initiation complex (18).
Ad5 pol can use both DNA and protein as a primer (4,24). Based on the results obtained from the study of the protein priming system in Bacillus subtilis phage 29 (φ29) (25–28), it is likely that the same Ad5 pol catalytic site is used for the interaction with both primers. We do not know the exact arrangement of pTP interaction in the pol active site, but it is remarkable that close to Ser-580 several conserved negatively charged amino acids are located that might mimic DNA (Figure 1). In this study, we raised a question on the importance of these negatively charged residues for the pTP–pol interaction and for the kinetics of polymerization. We demonstrated that the highly conserved Asp-582 and Asp-578 of Ad5 pTP are important for the optimal initiation of the Ad5 DNA replication. Furthermore, we show that the mutation of both residues influences the kinetics of initiation. Taking into consideration that Asp-578 and Asp-582 together with Ser-580 are positioned close to the pol active site, we propose that both Asp residues contribute to the arrangement of an optimally functioning active site of pol that leads to an efficient initiation.
MATERIALS AND METHODS
Construction of pTP mutants
Point mutations were introduced in the pTP gene by site-directed mutagenesis using the QuickChange method from Stratagene. As a template for the PCR-based mutagenesis, a pETpTP1S2-671 construct carrying full-length Ad5 pTP cDNA encoding amino acids 1–671 was used (22). Oligonucleotides used for the PCR-based mutagenesis were as follows. D578N: 5′-GATCGCCTATCAAAACAACTCAGGAGAC-3′, 5′-GTCTCCTGAGTTGTTTTGATAGGCGATC-3′; D578R: 5′-GATCGCCTATCAAAGAAACTCAGGAGAC-3′, 5′-GTCTCCTGAGTTTCTTTGATAGGCGATC-3′; D578E: 5′-GATCGCCTATCAAGAAAACTCAGGAGAC-3′, 5′-GTCTCCTGAGTTTTCTTGATAGGCGATC-3′; D582N: 5′-GACAACTCAGGAAACGTGCAGGAGATT-3′, 5′-AATCTCCTGCACGTTTCCTGAGTTGTC-3′; D582R: 5′-GACAACTCAGGAAGAGTGCAGGAGATT-3′,5′-AATCTCCTGCACTCTTCCTGAGTTGTC-3′; D582E: 5′-GACAACTCAGGAGAAGTGCAGGAGATT-3′, 5′-AATCTCCTGCACTTCTCCTGAGTTGTC-3′; and the mutations are represented in boldface. The presence of the mutations was confirmed using DNA sequence analysis.
Expression and purification of pTP mutants
All pTP mutants and wild-type pTP were expressed as full-length proteins with an additional C-terminal His-tag (6 × His). An aliquot of 1 liter of each BL21 expression culture was induced with 1 mM isopropyl-β-d-thiogalactopyranoside at an OD600 of 0.6 at 30°C. After 3 h of induction, cells were harvested and 9 ml of lysis buffer was added [50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 5 mM 2-mercaptoethanol, 10 mM Na2S2O5, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mg/ml lysozyme and complete protease inhibitor cocktail (Roche)]. The cells were left on ice for 10 min and freeze/thawed once. Then, the lysate was cleared by ultracentrifugation (at 2.74 × 105 g for 45 min at 4°C) and the supernatant was diluted with an equal volume of purification buffer B (20 mM HEPES-KOH, pH 8.0, 5 mM 2-mercaptoethanol, 10 mM Na2S2O5, 0.5 mM PMSF and 10% glycerol). The lysate was loaded onto a SP-sepharose column (1.8 ml) equilibrated with buffer B containing 150 mM of NaCl (B150), the column was washed with 7 ml of B150 and proteins were eluted with buffer B containing 450 mM NaCl and 20 mM imidazole (B450/20). Eluates from SP-columns were directly loaded onto Ni-nitriloacetate–agarose (1 ml) columns equilibrated with B450/20 and the columns were washed with 4 ml of B450/20 followed by additional wash with 2 ml of B150/20 in order to lower the NaCl concentration. Proteins were eluted with 500 mM imidazole in 2.5 ml of B150. Next, to stabilize purified proteins, BSA was added to a final concentration of 0.5 μg/μl and the eluates were dialyzed against buffer B150 to remove the imidazole. The purity of the proteins was estimated by Coomassie blue staining and was ∼90%.
Proteins and DNA templates
Adenovirus DNA polymerase was expressed using the baculovirus expression system and purified to near homogeneity as described previously (4). The protein dilution buffer contained 25 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 15% glycerol, 5 mM DTT and 0.5 μg/μl BSA.
As DNA templates, T30 or TD50 DNA derived from the Ad5 origin of replication was used. T30 is a single-stranded (ss) DNA representing the first 30 nt of the template strand of the Ad5 origin: T30, 5′-AATCCAAAATAAGGTATATTATTGATGATG-3′. TD50 is a dsDNA consisting of T50 and D50 that represent the first 50 nt of the template and displaced strand of the Ad5 origin, respectively.
T50: 5′-CTCATTATCATATTGGCTTCAATCCAAAATAAGGTATATTATTGATGATG-3′; D50: 5′-CATCATCAATAATATACCTTATTTTGGATTGAAGCCAATATGATAATGAG-3′. TD50 was obtained by boiling equimolar amounts of T50 and D50 oligonucleotides for 5 min in 25 mM Tris–HCl, pH 7.5, 100 mM NaCl and slow cooling to room temperature.
Protein–DNA interactions (electrophoretic mobility shift assay)
In protein–DNA binding studies T50 was used. For the preparation of the T50 probe, the T50 oligonucleotide was end-labeled using T4 polynucleotide kinase and [γ-32P]ATP (4500 Ci/mmol) in a standard kinase buffer and the labeled T50 was purified on a 10% polyacrylamide gel. In the DNA-binding assay, pTP mutants and wild-type pTP were incubated with DNA for 30 min on ice in binding buffer (25 mM HEPES-KOH, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 4 mM DTT, 0.2 μg/μl BSA and 3.2% Ficoll). Protein–DNA complexes were separated on a 7% polyacrylamide gel in TBE buffer at 4°C and the intensity of the bands was quantified using a Storm 820 PhosphorImager. Where indicated, α-pTP polyclonal antibody (Ab) (29) at a 1:100 dilution was added to the binding reaction in order to identify the pTP–DNA complexes.
Co-immunoprecipitation assay of pTP with pol
An aliquot of 30 μl of pre-swollen 50% protein A–agarose beads was washed twice with IP buffer (25 mM HEPES-KOH, pH 8.0, 0.5 mM 2-mercaptoethanol, 1 mM Na2S2O5, 0.5 mM PMSF, 0.1% NP-40 and 10% glycerol) containing 500 mM NaCl and 2% BSA (IP500/2%). Next, 10 μl of α-pol Ab (rabbit, polyclonal) (4,29) was added to the beads in 400 μl of IP500/2%. The beads were tumbled for 30 min at 4°C and washed twice with IP500/0.02% and twice with IP300/0.02%. In a separate tube, 600 ng Ad5 pol and 800 ng wild-type pTP or pTP mutants (a 2-fold molar excess of pTP over pol) were preincubated on ice for 15 min in 70 μl of IP300/0.02%. Then, the protein mixtures were added to the beads and incubation continued for 1 h at 4°C in 400 μl of IP300/0.02%. Non-bound pTP was removed by washing three times with IP300 without BSA. After the last wash the supernatant was carefully removed, 17 μl of Laemmli sample buffer was added, the samples were boiled for 10 min and loaded onto a 7.5% SDS–PAGE. Proteins were detected by western blotting using α-His Ab (mouse, monoclonal) (Qiagen) and α-pol Ab (rabbit, polyclonal) for pTP and pol detection, respectively.
In vitro initiation and partial elongation assays
In vitro initiation and partial DNA elongation was assayed using 60 ng of pol and 50 ng of wild-type pTP or pTP mutants in a 25 μl reaction mixture containing initiation buffer (20 mM HEPES-KOH, pH 7.5, 50 mM NaCl, 1 mM MgCl2, 1 mM DTT and 40 ng/μl BSA) and 4 μCi of [α-32P]dCTP (3000 Ci/mmol) at the final concentration of 53 nM. When indicated, 3 mM MnCl2 was used in the initiation buffer instead of MgCl2. As a template, 10 ng of TD50 was used for the initiation reaction and 100 ng of T30 for the partial elongation reaction. In addition, for partial elongation assays, 0.7 μM dCTP, 40 μM dTTP and 40 μM dATP were added to allow DNA elongation. The reactions were incubated for 45 min at 37°C and stopped by the addition of EDTA to a final concentration of 80 mM. The reaction products were precipitated with 20% trichloroacetic acid (TCA), washed with 5% TCA, dissolved in sample buffer and analyzed on a 7.5% SDS–PAGE. The intensity of the initiation and partial elongation bands was quantified using a Storm 820 PhosphorImager at the linear range of the signal.
The kinetic analysis of dCTP incorporation by pol for wild-type and pTP mutants was determined by performing initiation assays as described above with the addition of increasing concentrations of unlabeled dCTP (0.6–20 μM). Kmapp and Vmaxapp were determined from Lineweaver–Burk plots. Kcatapp was calculated from kcat = Vmaxapp/Et, where [Et] is the total amount of enzyme (pol). The average values were calculated from at least two independent experiments.
RESULTS
Conservation of negatively charged residues in the priming region of Ad5 pTP
The crucial amino acid of Ad5 pTP is Ser-580, which is used to prime Ad5 DNA replication (30). There are several negatively charged amino acids located in the neighborhood of Ser-580 in adenoviruses and also in other protein priming systems (Figure 1). In this study, we analyzed Asp-578 and Asp-582 of Ad5 pTP located near Ser-580 (Figure 1). The amino acid residue at position 578 exists as Asp or Glu in different serotypes and Asp-582 is completely conserved in all adenovirus serotypes that are known to date. To analyze the role of these two residues in initiation, they were mutated to Asn (D to N) in order to remove a negative charge, or mutated to Arg (D to R) to replace a negative charge by a positive charge. In addition, to verify the significance of the negative charge, Asp was changed to Glu (D to E), keeping the negative charge unchanged. All mutants were expressed and purified as His-tagged full-length pTP proteins as described in Materials and methods.
Mutation of Asp-578 or Asp-582 of pTP affects the initiation activity
During initiation covalent coupling of the first C-residue to the Ser-580 OH group of pTP by pol results in the formation of the pTP–dCMP product (pTP–C), which can be detected by SDS–PAGE. The ability of the pTP mutants to serve as a primer was tested in an initiation assay (Figure 2 and Table 2). The origin-dependent formation of pTP–C was studied using a dsDNA fragment (TD50) consisting of the first 50 nt of the Ad5 core origin. Substitution of Asp-578 to Asn or Arg decreased pTP initiation activity to 29 and 7% of wild-type activity for D578N and D578R, respectively. Changing Asp-578 to Glu (D578E) did not affect the initiation activity at all. Mutation of Asp-582 resulted in more severe effects on initiation. D582N showed only 15% activity, whereas the activity of D582R was completely abolished. Furthermore, the initiation activity of D582E decreased to 35%. When higher amounts of pTP up to 2-fold were used, the relative initiation activity of the pTP mutants compared with wild-type did not change (data not shown). These results demonstrate the importance of Asp-578 and Asp-582 in initiation.
Figure 2.
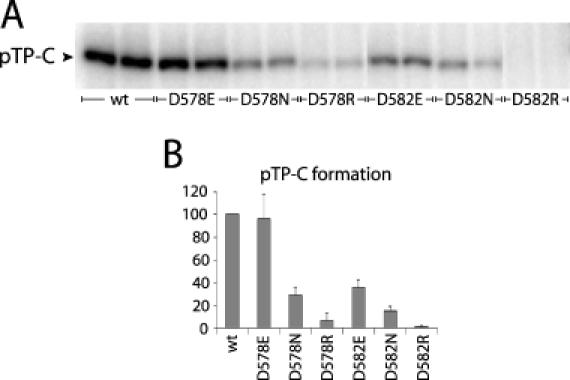
Initiation activity of the pTP mutants is affected. (A) The initiation activity of Asp-578 and Asp-582 mutants was measured by the formation of the [α-32P]dCMP-pTP product (pTP–C). A total of 60 ng of polymerase and 50 ng of wild-type pTP or the pTP mutants were incubated with 10 ng of TD50 template. (B) Percentage of initiation activity of the pTP mutants compared with the wild-type activity (set at 100%). The error bars represent the standard deviation from three independent experiments.
Table 2. Analysis of the initiation activity of the pTP mutants.
| pTP | Initiation activity (%) | ||
|---|---|---|---|
| 1 mM Mg2+ | 1 mM Mg2+ + 20 μM dCTP | 3 mM Mn2+ | |
| Wild-type | 100 | 100 | 100 |
| D578E | 97 | 118 | 145 |
| D578N | 29 | 48 | 90 |
| D578R | 7 | 26 | 65 |
| D582E | 35 | 65 | 69 |
| D582N | 15 | 46 | 45 |
| D582R | <1 | <1 | <1 |
aNumbers represent the percentage of initiation activity of the pTP mutants compared with wild-type pTP. Wild-type activity was set to 100% for each condition. [α-32P]dCTP was present at the standard 53 nM concentration.
bThe values represent the activity in the presence of an additional 20 μM of unlabeled dCTP.
cThe initiation activity of wild-type pTP in the presence of 3 mM Mn2+ was 8-fold higher than in the presence of 1 mM Mg2+.
Mutation of Asp-578 or Asp-582 of pTP does not decrease the DNA elongation
When all nucleotides are present, the pTP–C complex can be elongated leading first to a pTP–CAT initiation intermediate, which jumps back and is subsequently extended generating longer products. In the absence of dGTP elongation proceeds only to position 25, since at position 26 the first G residue should be incorporated. This leads to the formation of a pTP-25 product that migrates ∼90 kDa in SDS–PAGE (Figure 3A). At a low dCTP concentration only part of the pTP–CAT intermediate elongates, enabling the study of the pTP–CAT formation and DNA elongation simultaneously. As shown in Figure 3A and B, the formation of pTP–CAT decreased to 25 and 14% of wild-type activity for D578N and D578R, respectively, whereas D578E showed 80% activity. The activity of D582N decreased to 27% and D582R showed no detectable activity. D582E showed 30% activity compared with wild-type pTP. The ability of the pTP mutants to form pTP–CAT (Figure 3A and B) correlated with their ability to form the initiation pTP–C product (Figure 2).
Figure 3.
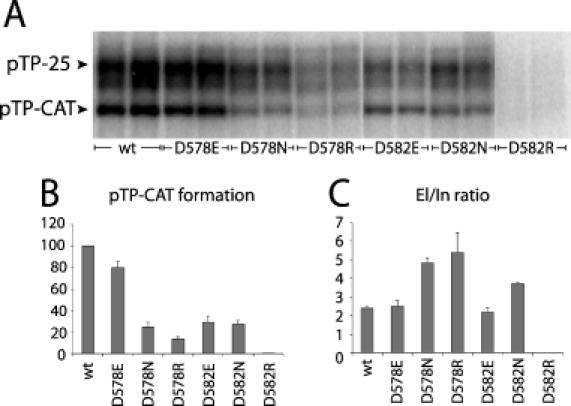
Elongation activity of the pTP mutants is not decreased. (A) Partial elongation was measured in the absence of dGTP resulting in DNA elongation only up to 25 nt (pTP-25). At low-dCTP concentration only part of the pTP–CAT intermediate jumps back and elongates, allowing simultaneous analysis of both products. A total of 60 ng of polymerase and 50 ng of wild-type pTP or the pTP mutants were incubated with 100 ng of T30 template. The positions of the pTP–CAT and pTP-25 product are indicated with arrowheads. The band directly below pTP-25 represents a faster migrating pTP-25 product due to secondary structure formation of the 25 nt long ssDNA. For our measurements of DNA elongation, we have added these two signals. (B) Amount of pTP–CAT formation for the pTP mutants compared with the wild-type. Wild-type pTP activity was set to 100%. (C) El/In values calculated as the ratio between the amount of pTP-25 and pTP–C product. The error bars in both graphs represent the standard deviation from three independent experiments.
The level of DNA elongation was calculated from the pTP-25/pTP–CAT ratio (elongation/initiation ratio, El/In; Figure 3C). Under the experimental conditions used, the El/In ratio for wild-type pTP was 2.4 ± 0.12. Mutants D578E and D582E also showed a wild-type El/In ratio of 2.5 ± 0.26 and 2.2 ± 0.21, respectively. This indicates that the substitution of Asp-578 and Asp-582 with Glu does not disturb the jumping back process or the subsequent DNA elongation. For the D578N, D578R and D582N, the El/In ratio was even higher than that of wild-type pTP, 4.9 ± 0.25, 5.4 ± 1.07 and 3.7 ± 0.06, respectively, showing that more elongating product (pTP-25) was formed after initiation. No elongation was observed for the D582R mutant.
The DNA-binding capacity of pTP mutants is not affected
To verify that the mutations introduced in pTP did not affect DNA binding, we performed electrophoretic mobility shift assays (EMSAs) using ssDNA representing the first 50 nt of the Ad5 origin template strand (T50). Addition of pTP to the DNA gave rise to multiple protein–DNA complexes (Figure 4A). Two of these were specific pTP–DNA complexes containing one (pTP1) or two pTP molecules (pTP2), which is consistent with a previous study of DNA-binding properties of pTP (18). Two different concentrations of pTP were tested for each mutant (Figure 4A). The binding affinity of the pTP mutants was not significantly changed compared to wild-type binding, demonstrating that mutations of Asp-578 and Asp-582 do not affect the pTP–DNA interaction.
Figure 4.
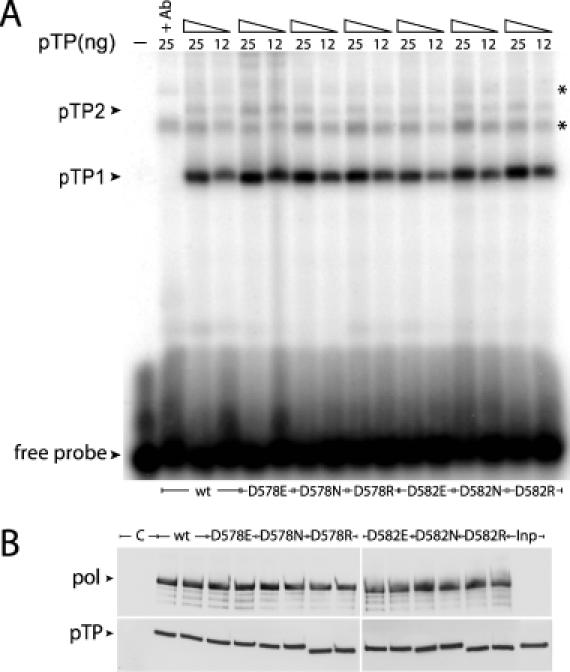
Binding of the Ad5 pTP mutants to DNA or to pol is not altered. (A) EMSA experiments with single-stranded T50 probes representing the first 50 nt of the Ad5 template strand. The positions of the pTP–DNA complexes (pTP1 monomer and pTP2 dimer) and free DNA are marked with arrowheads. The DNA probe was incubated with 12 ng or 25 ng of wild-type pTP or the pTP mutants. In the second lane (+Ab) α-pTP polyclonal antibody was added to the binding reaction with wild-type pTP at a 1:100 dilution in order to identify the protein–DNA complexes containing pTP molecules. Two asterisks point to non-specific protein–DNA complexes that do not contain pTP, possibly as a result of some impurity in the pTP samples. The first lane represents free probe. (B) Analysis of pTP/pol complex formation in co-immunoprecipitations. A mixture of 800 ng of wild-type pTP or the pTP mutants with 600 ng of pol was incubated with protA-αpolAb beads. The ability of pTP to bind pol was analyzed by western blotting using specific antibodies against pTP or pol. Positions of pol and pTP are marked with arrowheads. The first lane is a control lane (C) where pol was omitted in the incubation mixture and the last lane represents 10% of the pTP added to the beads (Inp). The smaller size of D578R and D582R pTP is most likely caused by the fact that an Arg substitution affects the protein migration in the gel. Careful restriction analysis and sequencing of the expression vectors carrying the D578R and D582R pTP mutants did not show any sequence deletions. The weaker signals below the actual pol bands are caused by pol degradation.
The interaction of pTP mutants with pol is not disturbed
It is likely that during initiation the priming region of pTP interacts with the pol active site. Therefore, it is possible that the defects in initiation are caused by a reduced interaction between pTP and pol. The contribution of the Asp-578 and Asp-582 residues to pTP–pol interaction was analyzed in a co-immunoprecipitation assay. The control experiment without pol showed no non-specific pTP binding to the protA beads or to the α-pol antibody (Figure 4B). When pol was added, ∼10% of pTP was co-immunoprecipitated at the conditions used. All mutants showed unaffected pol binding (Figure 4B). We also performed experiments testing the stability of the pTP–pol interaction at 37°C for up to 60 min, but no differences in half-lives were observed for the pTP/pol complexes containing pTP mutants compared to the wild-type (data not shown). Based on these results, we conclude that binding of the pTP mutants to pol is not disturbed.
The kinetics of the initiation reaction is affected by Asp-578 or Asp-582 mutations of pTP
Since a decreased activity of the pTP mutants was noticeable already in the protein-priming step, we performed a kinetic analysis of the initiation reaction. Unlabeled dCTP ranging from 0.6 to 20 μM was added to the reaction mixture containing wild-type or mutated pTP and wild-type polymerase. The reactions were incubated for 45 min, which is still in the linear time range. Based on the Lineweaver–Burk plots, the apparent Kmapp for dCTP incorporation was determined (Table 1). Interestingly, the Kmapp values increased for the D578N, D578R, D582E and D582N pTP mutants. When initiation was performed in the presence of 20 μM dCTP, the D578N, D578R, D582E and D582N mutants gained activities up to 48, 26, 65 and 46% of wild-type activity, respectively (Table 2). However, the severe initiation defect of D582R could not be rescued. The increased values of the Kmapp of the pTP/pol complexes containing pTP mutants and their increased activities at higher dCTP concentrations suggest a reduced affinity of pol for dCTP when it is in complex with the pTP mutants.
Table 1. Analysis of the kinetics of the initiation of Ad5 DNA replicationa.
| pTP | Kmapp (μM dCTP) | kcatapp/Kmappb (M−1 s−1) |
|---|---|---|
| Wild-type | 2.06 ± 0.33 | 2.20 |
| D578E | 2.12 ± 0.11 | 2.01 |
| D578N | 4.16 ± 0.16 | 1.16 |
| D578R | 5.68 ± 1.48 | 0.31 |
| D582E | 3.45 ± 0.62 | 1.13 |
| D582N | 4.12 ± 0.30 | 0.98 |
| D582R | — | — |
aThe Kmapp and Vmaxapp values of pTP deoxycytidylation by polymerase in the initiation reaction were calculated from Lineweaver–Burk plots (0.6–20 μM dCTP) based on at least two independent experiments for each pTP. The Kmapp and Vmaxapp of D582R pTP could not be determined since the initiation activity of this mutant was below the detection limit.
bkcatapp/Kmapp ratio describes the catalytic efficiency of initiation. kcatapp was calculated from kcatapp = Vmaxapp/Et, where [Et] is the total amount of enzyme (pol).
In addition to the Kmapp, the kcatapp/Kmapp ratios of pTP/pol complexes in initiation were determined (Table 1). The kcatapp/Kmapp ratios were 7-fold lower for D578R, and 2-fold lower for D578N, D582E and D582N pTP, demonstrating that the catalytic efficiency of the initiation reaction is reduced by these pTP mutations. These results correlate with the decreased initiation activity of the mutants (Figure 2).
The pTP/pol complex containing D578E showed a wild-type Kmapp and kcatapp/Kmapp (Table 1), which is in agreement with the wild-type activity found in initiation. The initiation activity of D582R was below the detection limit, so the specific kinetic properties could not be determined.
The presence of Mn2+ partially relieves the initiation defect of the pTP mutants
Ad5 pol uses two Mg2+ ions in the active site for efficient catalysis. However, it can also make use of other divalent cations with different efficiencies depending on the cation (31). Mn2+ increases the activity of DNA polymerase not only of Ad5 pol but also of other polymerases (31,32). This suggests that Mn2+ induces subtle alterations in the active site of pol that might result in a more efficient catalysis. To analyze the influence of Mn2+ on the activity of the pTP mutants, initiation assays were performed in the presence of 3 mM Mn2+, the optimal conditions for Mn2+, instead of the standard 1 mM Mg2+ (Table 2). Indeed, Mn2+ increased the initiation activity of the pTP mutants resulting in 145, 90, 65, 69 and 45% of the wild-type activity for D578E, D578N, D578R, D582E and D582N, respectively. The abolished initiation activity of the D582R pTP mutant could not be rescued by Mn2+, confirming that the D582R mutation affects pTP/pol function severely. These data demonstrate that in the presence of Mn2+ pol tolerates the mutations present in pTP and that Mn2+ can lead to a more efficient initiation for the pTP mutants compared with the initiation in the presence of Mg2+ (Figure 2 and Table 2).
DISCUSSION
Functional importance of Asp-578 and Asp-582 of pTP for Ad5 DNA replication
We show that two conserved amino acids, Asp-578 and Asp-582, of Ad5 pTP are essential for the initiation of replication. Substitution of these residues with Asn or Arg greatly reduced the ability of pTP to form the pTP–C and pTP–CAT products (Figures 2, 3A and B). The strongest effect was observed when Asp was changed into a positively charged residue. We demonstrate that specifically the negative charge of Asp-578 is essential for the initiation, since its change to Glu did not affect the initiation. Some Ad serotypes have a Glu at this position (Figure 1), confirming that this residue is already functional when a negative charge is present. In contrast, Asp-582 is completely conserved, and cannot be substituted by a Glu residue without loss of initiation activity. Since Glu has a larger side chain than Asp, it is likely that such a mutation is not tolerated by a functionally optimal protein structure.
During initiation, Ser-580 of pTP is located in the active site of pol. Asp-578 and Asp-582 must be in or near the active site as well, and may contact pol or even template DNA. However, with the assays used in this study we cannot find any effect on the interaction with DNA or pol (Figure 4). Since multiple regions of pTP are involved in pol binding (14–16), it is likely that single point mutations do not significantly disrupt this interaction. Our assay conditions do not rule out that the mutations locally affect the pTP/pol interaction within the active site. This could affect the optimal positioning of Ser-580 of pTP in the pol active site leading to an impaired initiation.
After initiation, formation of pTP–CAT and jumping back is followed by dissociation of pTP from pol, which is almost complete by the time 7 nt are synthesized (5,6). The El/In ratio (Figure 3C) shows that the substitution of Asp-578 and Asp-582 with Glu does not impair the jumping back event and the subsequent DNA elongation. Interestingly, in the case of D578N, D578R and D582N mutants the elongation was more efficient than the wild-type pTP. It is possible that these mutations locally weaken the interaction of pTP with pol, thereby facilitating the dissociation of pol from pTP after jumping back, leading to an increased amount of pTP-25. An alternative explanation might be that the D578N, D578R and D582N mutants need a lower dNTP concentration to elongate efficiently and that the increased El/In ratio is a result of changed kinetics of the DNA elongation step. Kinetic studies also showed that a higher dCTP concentration is required for optimal activity of pol during elongation compared with initiation (3,6).
Contribution of pTP to the kinetics of the initiation of Ad5 DNA replication
We show that mutations of Asp-578 and Asp-582 change the kinetics of the initiation reaction. The Kmapp for incorporation of dCTP was increased for the pTP/pol complexes containing pTP mutants shown to be defective in initiation. Moreover, at high dCTP concentrations the mutants gained some initiation activity suggesting a reduced affinity of the pTP/pol complex for dCTP. These observations demonstrate that pTP contributes to the affinity of pol for dCTP in the initiating pTP/pol complex. We also observed that the presence of Mn2+ instead of Mg2+ partially relieved the initiation defect of the pTP mutants. Reduced kcatapp/Kmapp values show that also the overall catalytic efficiency of the reaction is decreased. These results demonstrate that the decrease in initiation activity caused by the Asp-578 and Asp-582 mutations is a direct consequence of changed kinetics of initiation. So far, polymerase mutations were only reported to affect the kinetics of initiation for Ad5 or φ29 (4,32,33). In this study, we show that the protein primer pTP, interacting with polymerase directly influences the kinetics of the initiation. Such an effect was already suggested by our previous study that revealed a change in the active site of pol after dissociation from pTP upon transition from initiation to elongation (6). Also, the elongation and exonuclease activities of pol are inhibited by pTP (24), which show that pTP indeed influences the pol active site. The kinetic properties of the active site of pol are described by Kmapp for dCTP incorporation and might depend on several parameters like affinity of pol for dCTP, pTP and template DNA binding and positioning of the OH group of Ser-580. The increase in the Kmapp caused by the Asp-578 and Asp-582 mutations indicates that pTP contributes to the correct arrangement of the active site of pol resulting in the optimal activity of pol.
A structural model of protein priming
Studies on Ad5 pol and B.subtilis φ29 polymerase demonstrated that the protein primer as well as the DNA primer bind to a common site of polymerase (17,25,26). Therefore, it is likely that protein primers mimic DNA in order to interact with pol in the primer-binding groove. Interestingly, the priming region of pTP surrounding Ser-580 contains 13 out of 43 conserved acidic residues (Figure 1). They cluster at one side of two predicted α-helices. We observed that two of these residues (Glu-585 and Asp-594) are also important for the initiation of replication (data not shown). Similarly, the priming region of φ29 TP is also rich with negatively charged residues, conserved among other B.subtilis phages (11 amino acids out of 43 amino acids; Figure 1). The secondary structure prediction of the Ad5 pTP and the φ29 TP priming region revealed that in both proteins the priming Ser residue is located in a β-turn surrounded from both sides by α-helices (34). Asp-578 and Asp-582 of Ad5 pTP could similarly be positioned in a β-turn and surface exposed. Such a location could facilitate the optimal charge–charge interactions between pTP and the active site of pol leading to the optimal positioning of the Ser-580 OH group. Similarly, TP of φ29 also contains conserved acidic residues located next to the priming Ser-232 (Asp-231 and Glu-233; Figure 1), but their role in initiation has not been studied so far.
Binding of pTP to pol might structurally resemble interaction of the regulatory factor GreB to bacterial RNA polymerase (RNAP) (35,36). The globular C-terminal domain of GreB binds at the edge of the RNAP active site channel, while the N-terminal coiled-coil domain extends into a channel leading to the RNAP active site with its tip making contacts in the active site. Two conserved acidic residues in the coiled-coil tip (Asp-41 and Glu-44) are located very close to the RNAP active site and are proposed to influence its function. Mutations of these residues cause severe functional defects without affecting the interaction with RNAP, while substitution of Asp-41 with Glu had little or no effect on GreB function (36). Similar roles are proposed for Asp-290 and Glu-291 of the eukaryotic elongation factor TFIIS (37).
Our results demonstrate that the negatively charged residues Asp-578 and Asp-582 of Ad5 pTP play an important role in the arrangement of an optimally functioning polymerase active site, leading to an efficient initiation of Ad5 DNA replication. This shows that pTP is an integral component of the pol active site contributing to the catalytic activity of pol. It is likely that the contribution of the acidic residues to protein priming is a general mechanism among other organisms that use protein as a primer to start the DNA replication.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Lars Meijer, Richard Heideman and Marjoleine Bleijenberg for helpful discussions.
REFERENCES
- 1.de Jong R.N. and van der Vliet,P.C. (1999) Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene, 236, 1–12. [DOI] [PubMed] [Google Scholar]
- 2.de Jong R.N., van der Vliet,P.C. and Brenkman,A.B. (2003) Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP. Curr. Top. Microbiol. Immunol., 272, 187–211. [DOI] [PubMed] [Google Scholar]
- 3.Mul Y.M. and van der Vliet,P.C. (1993) The adenovirus DNA binding protein effects the kinetics of DNA replication by a mechanism distinct from NFI or Oct-1. Nucleic Acids Res., 21, 641–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenkman A.B., Heideman,M.R., Truniger,V., Salas,M. and van der Vliet,P.C. (2001) The (I/Y)XGG motif of adenovirus DNA polymerase affects template DNA binding and the transition from initiation to elongation. J. Biol. Chem., 276, 29846–29853. [DOI] [PubMed] [Google Scholar]
- 5.King A.J. and van der Vliet,P.C. (1994) A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J., 13, 5786–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King A.J., Teertstra,W.R. and van der Vliet,P.C. (1997) Dissociation of the protein primer and DNA polymerase after initiation of adenovirus DNA replication. J. Biol. Chem., 272, 24617–24623. [DOI] [PubMed] [Google Scholar]
- 7.Webster A., Leith,I.R. and Hay,R.T. (1994) Activation of adenovirus-coded protease and processing of preterminal protein. J. Virol., 68, 7292–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webster A., Leith,I.R., Nicholson,J., Hounsell,J. and Hay,R.T. (1997) Role of preterminal protein processing in adenovirus replication. J. Virol., 71, 6381–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smart J.E. and Stillman,B.W. (1982) Adenovirus terminal protein precursor. Partial amino acid sequence and the site of covalent linkage to virus DNA. J. Biol. Chem., 257, 13499–13506. [PubMed] [Google Scholar]
- 10.Chen M., Mermod,N. and Horwitz,M.S. (1990) Protein–protein interactions between adenovirus DNA polymerase and nuclear factor I mediate formation of the DNA replication preinitiation complex. J. Biol. Chem., 265, 18634–18642. [PubMed] [Google Scholar]
- 11.Enomoto T., Lichy,J.H., Ikeda,J.E. and Hurwitz,J. (1981) Adenovirus DNA replication in vitro: purification of the terminal protein in a functional form. Proc. Natl Acad. Sci. USA, 78, 6779–6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stillman B.W., Tamanoi,F. and Mathews,M.B. (1982) Purification of an adenovirus-coded DNA polymerase that is required for initiation of DNA replication. Cell, 31, 613–623. [DOI] [PubMed] [Google Scholar]
- 13.Parker E.J., Botting,C.H., Webster,A. and Hay,R.T. (1998) Adenovirus DNA polymerase: domain organization and interaction with preterminal protein. Nucleic Acids Res., 26, 1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webster A., Leith,I.R. and Hay,R.T. (1997) Domain organization of the adenovirus preterminal protein. J. Virol., 71, 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roovers D.J., van der Lee,F.M., van der Wees,J. and Sussenbach,J.S. (1993) Analysis of the adenovirus type 5 terminal protein precursor and DNA polymerase by linker insertion mutagenesis. J. Virol., 67, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botting C.H. and Hay,R.T. (2001) Role of conserved residues in the activity of adenovirus preterminal protein. J. Gen. Virol., 82, 1917–1927. [DOI] [PubMed] [Google Scholar]
- 17.Brenkman A.B., Breure,E.C. and van der Vliet,P.C. (2002) Molecular architecture of adenovirus DNA polymerase and location of the protein primer. J. Virol., 76, 8200–8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong R.N., Meijer,L.A.T. and van der Vliet,P.C. (2003) DNA binding properties of the adenovirus DNA replication priming protein pTP. Nucleic Acids Res., 31, 3274–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Temperley S.M. and Hay,R.T. (1992) Recognition of the adenovirus type 2 origin of DNA replication by the virally encoded DNA polymerase and preterminal proteins. EMBO J., 11, 761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rijnders A.W., van Bergen,B.G., van der Vliet,P.C. and Sussenbach,J.S. (1983) Specific binding of the adenovirus terminal protein precursor–DNA polymerase complex to the origin of DNA replication. Nucleic Acids Res., 11, 8777–8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botting C.H. and Hay,R.T. (1999) Characterization of the adenovirus preterminal protein and its interaction with the POU homeodomain of NFIII (Oct-1). Nucleic Acids Res., 27, 2799–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jong R.N., Mysiak,M.E., Meijer,L.A., van der Linden,M. and van der Vliet,P.C. (2002) Recruitment of the priming protein pTP and DNA binding occur by overlapping Oct-1 POU homeodomain surfaces. EMBO J., 21, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Leeuwen H.C., Rensen,M. and van der Vliet,P.C. (1997) The Oct-1 POU homeodomain stabilizes the adenovirus preinitiation complex via a direct interaction with the priming protein and is displaced when the replication fork passes. J. Biol. Chem., 272, 3398–3405. [DOI] [PubMed] [Google Scholar]
- 24.King A.J., Teertstra,W.R., Blanco,L., Salas,M. and van der Vliet,P.C. (1997) Processive proofreading by the adenovirus DNA polymerase. Association with the priming protein reduces exonucleolytic degradation. Nucleic Acids Res., 25, 1745–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truniger V., Blanco,L. and Salas,M. (2000) Analysis of φ29 DNA polymerase by partial proteolysis: binding of terminal protein in the double-stranded DNA channel. J. Mol. Biol., 295, 441–453. [DOI] [PubMed] [Google Scholar]
- 26.de Vega M., Blanco,L. and Salas,M. (1998) phi29 DNA polymerase residue Ser122, a single-stranded DNA ligand for 3′–5′ exonucleolysis, is required to interact with the terminal protein. J. Biol. Chem., 273, 28966–28977. [DOI] [PubMed] [Google Scholar]
- 27.Salas M., Miller,J.T., Leis,J. and DePamphilis,M.L. (1996) Mechanisms for priming DNA synthesis. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 131–176. [Google Scholar]
- 28.Meijer W.J., Horcajadas,J.A. and Salas,M. (2001) Phi29 family of phages. Microbiol. Mol. Biol. Rev., 65, 261–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coenjaerts F.E., van Oosterhout,J.A. and van der Vliet,P.C. (1994) The Oct-1 POU domain stimulates adenovirus DNA replication by a direct interaction between the viral precursor terminal protein–DNA polymerase complex and the POU homeodomain. EMBO J., 13, 5401–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettit S.C., Horwitz,M.S. and Engler,J.A. (1989) Mutations of the precursor to the terminal protein of adenovirus serotypes 2 and 5. J. Virol., 63, 5244–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pronk R., Van Driel,W. and van der Vliet,P.C. (1994) Replication of adenovirus DNA in vitro is ATP-independent. FEBS Lett., 337, 33–38. [DOI] [PubMed] [Google Scholar]
- 32.Dufour E., Mendez,J., Lazaro,J.M., de Vega,M., Blanco,L. and Salas,M. (2000) An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Mol. Biol., 304, 289–300. [DOI] [PubMed] [Google Scholar]
- 33.Truniger V., Lazaro,J.M. and Salas,M. (2004) Two positively charged residues of phi29 DNA polymerase, conserved in protein-primed DNA polymerases, are involved in stabilisation of the incoming nucleotide. J. Mol. Biol., 335, 481–494. [DOI] [PubMed] [Google Scholar]
- 34.Hermoso J.M., Mendez,E., Soriano,F. and Salas,M. (1985) Location of the serine residue involved in the linkage between the terminal protein and the DNA of phage phi 29. Nucleic Acids Res., 13, 7715–7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opalka N., Chlenov,M., Chacon,P., Rice,W.J., Wriggers,W. and Darst,S.A. (2003) Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell, 114, 335–345. [DOI] [PubMed] [Google Scholar]
- 36.Laptenko O., Lee,J., Lomakin,I. and Borukhov,S. (2003) Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J., 22, 6322–6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kettenberger H., Armache,K.J. and Cramer,P. (2003) Architecture of the RNA polymerase II–TFIIS complex and implications for mRNA cleavage. Cell, 114, 347–357. [DOI] [PubMed] [Google Scholar]


