ABSTRACT
Staphylococcus aureus is a human pathogen, and S. aureus bacteremia can cause serious problems in humans. To identify the genes required for bacterial growth in calf serum (CS), a library of S. aureus mutants with randomly inserted transposons were analyzed for growth in CS, and the aspartate semialdehyde dehydrogenase (asd)-inactivated mutant exhibited significantly reduced growth in CS compared with the wild type (WT). The mutant also exhibited significantly reduced growth in medium, mimicking the concentrations of amino acids and glucose in CS. Asd is an essential enzyme for the biosynthesis of lysine, methionine, and threonine from aspartate. We constructed inactivated mutants of the genes for lysine (lysA), methionine (metE), and threonine (thrC) biosynthesis and found that the inactivated mutants of lysA and thrC exhibited significantly lower growth in CS than the WT, but the growth of the metE mutant was similar to that of the WT. The reduced growth of the asd mutant was recovered by addition of 100 μg/ml lysine and threonine in CS. These results suggest that S. aureus requires lysine and threonine biosynthesis to grow in CS. On the other hand, the asd-, lysA-, metE-, and thrC-inactivated mutants exhibited significantly reduced growth in mouse serum compared with the WT. In mouse bacteremia experiments, the asd-, lysA-, metE-, and thrC-inactivated mutants exhibited attenuated virulence compared with WT infection. In conclusion, our results suggest that the biosynthesis of de novo aspartate family amino acids, especially lysine and threonine, is important for staphylococcal bloodstream infection.
IMPORTANCE Studying the growth of bacteria in blood is important for understanding its pathogenicity in the host. Staphylococcus aureus sometimes causes bacteremia or sepsis. However, the factors responsible for S. aureus growth in the blood are not well understood. In this study, using a library of 2,914 transposon-insertional mutants in the S. aureus MW2 strain, we identified the factors responsible for bacterial growth in CS. We found that inactivation of the lysine and threonine biosynthesis genes led to deficient growth in CS. However, the inactivation of these genes did not affect S. aureus growth in general medium. Because the concentration of amino acids in CS is low compared to that in general bacterial medium, our results suggest that lysine and threonine biosynthesis is important for the growth of S. aureus in CS. Our findings provide new insights for S. aureus adaptation in the host and for understanding the pathogenesis of bacteremia.
INTRODUCTION
Staphylococcus aureus is one of the major pathogens that causes various suppurative diseases, food poisoning, pneumonia, and toxic shock syndrome (1, 2). S. aureus bacteremia is a serious problem in humans. In intensive care units, S. aureus bacteremia was reported to have fatality rates ranging from 20% to 50% (3). Health care-acquired infection with methicillin-resistant Staphylococcus aureus is even more difficult to treat (4).
S. aureus possesses multiple virulence factors, and some of them have been found to be associated with fitness in vivo (5–7). However, because the environmental conditions in vivo are complex and vary by site, it is sometimes difficult to identify the key factors for survival under different conditions in the host. Previously, we utilized ex vivo experimental models using calf serum (CS) to understand the virulence of S. aureus in the blood, and we demonstrated that the pattern of gene expression in CS was different than that in general bacterial medium. In particular, the expression of virulence genes and iron acquisition genes in CS was higher than that in bacterial medium (8).
Previously, several reports demonstrated that the genes encoding nutrient metabolism factors are important to adaptation in host serum. With S. aureus, Horsburgh et al. demonstrated that a putative phenylalanine permease is essential for growth on pig serum agar plates (9). With Neisseria meningitidis, Mendum et al. demonstrated that the genes for aromatic amino acids, leucine, histidine, glycine, and proline, and purine base metabolism are important for growth in human serum (HS) (10). Samant et al. reported that genes involved in purine and pyrimidine de novo biosynthesis in Escherichia coli, Salmonella enterica serovar Typhimurium, and Bacillus anthracis are critical for growth in HS (11).
In this study, we used S. aureus MW2, isolated from a patient with fatal septicemia (12), and we identified and characterized the genes responsible for growth in CS.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. S. aureus was grown in Trypticase soy broth (TSB) (Becton Dickinson Microbiology Systems, Cockeysville, MD, USA). Tetracycline (5 μg/ml), chloramphenicol (5 μg/ml), and erythromycin (Em) (30 μg/ml) were added for maintenance of mutant strains. Eight calf sera (CS-1 to CS-8) were purchased from 5 companies: CS-1 and CS-4 with different lot numbers were purchased from Thermo Fisher Scientific (Waltham, MA, USA), CS-2 was from Sigma-Aldrich (St. Louis, MO, USA), CS-3 was from Bio West (Nuaillé, Maine-et-Loire, France), CS-5, CS-6, and CS-8 with different lot numbers were purchased from GE Healthcare Bio-Sciences (Pittsburgh, PA, USA), and CS-7 was from Medical & Biological Laboratories (Nagoya, Aichi, Japan). Mouse serum (MS) was purchased from Nippon Bio-Test Laboratories (Tokyo, Japan). HS was purchased from Bio West. Heat-inactivated serum was prepared by incubation of the serum at 56°C for 30 min. Chemically defined medium (CDM) was prepared using methods described elsewhere (13). The concentrations of amino acids and glucose were modified as necessary.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| Staphylococcus aureus | ||
| MW2 | Clinical strain, sepsis, methicillin resistant (mecA+) | 12 |
| RN4220 | Restriction-deficient transformation recipient | 20 |
| YO2082 | pMW2-cured MW2, cadmium sensitive | This study |
| YO2090 | YO2082 harboring pI258, Emr Cdr | This study |
| M3-91 | asd::Tn551 in YO2090, Emr | This study |
| M17-9 | asd::Tn551 in YO2090, Emr | This study |
| Δasd mutant | asd::pYT1 in MW2, Tcr | This study |
| ΔthrC mutant | thrC::pYT1 in MW2, Tcr | This study |
| ΔlysA mutant | lysA::pYT1 in MW2, Tcr | This study |
| ΔmetE mutant | metE::pYT1 in MW2, Tcr | This study |
| asd compl. | asd complemented in Δasd mutant by pYO18, Tcr Cpr | This study |
| Escherichia coli | ||
| XL-II | endA1 supE44 thi-1 hsdR17 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr) Amy Cmr] | Stratagene |
| Plasmids | ||
| pI258 | Thermosensitive plasmid harboring Tn551, Emr Cdr | 15 |
| pYT1 | E. coli-S. aureus shuttle vector, thermosensitive plasmid, Tcr (S. aureus) Ampr (E. coli) | 18 |
| pCL15 | E. coli-S. aureus shuttle vector, IPTG-inducible, Pspac promoter, Cpr (S. aureus) Ampr (E. coli) | 19 |
| pYO18 | pCL15 containing a PCR fragment of asd for complementation | This study |
Emr, erythromycin resistance; Cdr, cadmium resistance; Tcr, tetracycline resistance; Cpr, chloramphenicol resistance; Tetr, tetracycline resistance; Ampr, ampicillin resistance.
Construction of the transposon mutant library of S. aureus MW2.
Because S. aureus MW2 was reported to possess plasmid pMW2, which contains cadmium resistance genes (14), plasmid curing was performed. A small amount (105 CFU) of MW2 overnight culture was inoculated into 100 μl of TSB containing ethidium bromide (2 μg/ml) and grown at 37°C overnight. The diluted overnight culture was plated on Trypticase soy agar (TSA) and incubated at 37°C overnight. The colonies were replated on TSA and TSA containing cadmium nitrate (25 μg/ml), and a cadmium-sensitive colony was obtained as a parent strain (YO2082) for transposon mutagenesis. The plasmid curing was confirmed by PCR using pMW2-specific primers (Table 2). The temperature-sensitive plasmid pI258 containing transposon Tn551 (15) was transduced into YO2082 by phage 80α (16). The transformant (YO2090) was grown in TSB containing Em (30 μg/ml) and cadmium nitrate (50 μg/ml) at 30°C, and the dilution was plated on TSA containing Em (30 μg/ml) and incubated at 42°C for 48 h. Then, the colonies were replated on TSA containing cadmium nitrate (50 μg/ml). The cadmium-resistant clones were removed as the negative clones, and the sensitive clones were obtained as the transposon-insertional mutant strains.
TABLE 2.
Primers used in this study
| Plasmid type | Primer | Sequencea |
|---|---|---|
| Plasmid curing | ||
| Forward | 5′-TAAGCGGTGGTGAACAACAA | |
| Reverse | 5′-CAAATCGTTTCGCAAGCTCT | |
| TAIL-PCR | ||
| Tn551-specific primer-1 | 5′-TCTCGTCAAAATAGATCCCGAAG | |
| Tn551-specific primer-2 | 5′-AGCATATCACTTTTCTTGGAGAG | |
| Tn551-specific primer-3 | 5′-GGCCTTGAAACATTGGTTTAGTG | |
| AD primer-1 | 5′-NTCGASTWTSGWGTT | |
| AD primer-2 | 5′-NGTCGASWGANAWGAA | |
| AD primer-3 | 5′-WGTGNAGWANCANAGA | |
| asd flanking-F | 5′-CTATATGACGCATTTAAC | |
| asd flanking-R | 5′-CCCTCAAATAAATGTGTC | |
| Gene inactivation | ||
| asd KO-F | 5′-AATAAGCTTATGCTCGTGCAAGTGAAC | |
| asd KO-R | 5′-TAAGGATCCAATATCTTCTGCAGTCGC | |
| lysA KO-F | 5′-GCAAAGCTTTGGTACACCTACCATTG | |
| lysA KO-R | 5′-CTGGGATCCCAATCTGTGAACCAATATG | |
| metE KO-F | 5′-AGAAAGCTTAAGAAGAATTAGATCAAAC | |
| metE KO-R | 5′-TAGGGATCCCTGTCGTCTGTAACTAAG | |
| thrC KO-F | 5′-TGATGAAGCTTTAGAAATTG | |
| thrC KO-R | 5′-CTTGGATCCCTTGAGGTAATTTACCTTGT | |
| Complementation | ||
| asd compl-F | 5′-GGTAAGCTTCAAATATAGTTGCACATACG | |
| asd compl-R | 5′-GTGGGATCCTTGGCAGGATATATTATGAAG |
Restriction sites are underlined.
Isolation of low-growth mutants in CS from the transposon mutant library.
YO2082 and 2,914 clones of the transposon mutants (1× 107 to 4 × 107 CFU) were inoculated in 100 μl of CS-1 in 96 well microplates using sterile toothpicks and incubated at 37°C for 48 h. Bacterial growth was measured as the optical density at 660 nm (OD660) using a SpectraMax 340PC384 microplate reader (Molecular Devices, Sunnyvale, CA, USA). Mutants with 60% less growth compared to the WT were selected. The selected mutant strains were then inoculated in CS-1 again to reconfirm their growth in CS-1.
Identification of Tn551 insertion sites.
Thermal asymmetric interlaced PCR (TAIL-PCR) was performed to identify the Tn551 insertion site. TAIL-PCR was performed using the protocol from Liu et al. (17), with the following modifications. Using the chromosomal DNA of the mutant (20 ng), the DNA fragment was amplified using PCR with Tn551-specific primer-1 and 3 sets of arbitrary degenerate primers (AD primer). The thermal cycling conditions were as follows: one cycle at 94°C for 1 min and 95°C for 1 min; 5 cycles at 94°C for 1 min, 65°C for 1 min, and 72°C for 3 min; and 14 cycles at 94°C for 30 s, 68°C for 1 min, 72°C for 3 min, 94°C for 30 s, 68°C for 1 min, 72°C for 3 min, 94°C for 30 s, 44°C for 1 min, and 72°C for 3 min. After amplification, PCR products were diluted 50-fold with Tris-EDTA (TE) buffer, and 1 μl of the dilution was used as the template for the secondary PCR. The secondary PCR was performed with Tn551-specific primer-2 and 3 sets of AD primers. The thermal cycling conditions were as follows: 11 cycles at 94°C for 30 s, 64°C for 1 min, 72°C for 3 min, 94°C for 30 s, 64°C for 1 min, 72°C for 3 min, 94°C for 30 s, 44°C for 1 min, and 72°C for 3 min. After amplification, the PCR products were diluted 10-fold with TE buffer, and 1 μl of the dilution was used as the template for the third PCR. The third PCR was performed using Tn551-specific primer-3 and 3 sets of AD primers. The thermal cycling conditions were 20 cycles at 94°C for 1 min, 44°C for 1 min, and 72°C for 3 min. The PCR products were cloned into the pGEM-T easy vector (Promega, Tokyo, Japan) using E. coli XLII. After the plasmid purification, DNA sequencing was performed. On the basis of the sequence of inserted Tn551, the site of insertion was determined from the S. aureus MW2 whole-genome sequence (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCA_000011265.1_ASM1126v1/) using in silico Molecular Cloning (In Silico Biology, Yokohama, Kanagawa, Japan). To confirm the insertion, PCR was performed using primers designed for both flanking regions of the target gene. The primers are listed in Table 2.
Gene inactivation and complementation.
A method of specific gene inactivation using the thermosensitive plasmid pYT1 has been previously described (18). For gene complementation, the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible vector pCL15 was used (19). The DNA fragment for complementation was amplified by PCR, and then the DNA fragment was cloned into pCL15 using E. coli XL-II competent cells. The constructed plasmid was purified and electroporated into S. aureus RN4220, which is a recipient for foreign plasmids (20). Then, the plasmid was transduced into a mutant strain using phage 80α. To induce the cloned gene, 1 mM IPTG was added into CS-1. The primers are listed in Table 2.
Growth experiments.
Overnight cultures of S. aureus strains grown in 5 ml of TSB were pelleted by centrifugation (1,200 × g, 1 min) and resuspended in sterile water. A small amount (4 × 107 CFU) of the bacterial suspension was inoculated into 4 ml of CS-1 or CDM in a test tube (18-mm diameter by 150-mm height) and grown aerobically at 37°C with shaking at 120 rpm. Bacterial growth was monitored by measuring the OD660 for 4 to 24 h using a MiniPhoto 518R spectrophotometer (Taitec Corporation, Koshigaya, Saitama, Japan). For CFU determination, the cultures grown in 8 calf sera (CS-1 to CS-8) were diluted with phosphate-buffered saline (PBS), and the appropriate dilutions were plated on TSA plates and aerobically incubated at 37°C overnight. The growth experiment using mouse serum (MS) was conducted on a smaller scale. A small amount (106 CFU) of S. aureus strains were inoculated into 100 μl of MS in a small test tube (12-mm diameter by 105-mm height) and aerobically incubated at 37°C with shaking at 120 rpm. The cultures were diluted with PBS, and the appropriate dilutions were plated on TSA plates and incubated aerobically at 37°C overnight.
Analysis of free amino acids in sera.
The quantitative analysis of free amino acids in CS-1, CS-3, MS, and HS was performed by a contract analysis company, Towa Environment Science Co., Ltd. (Hiroshima, Japan). Sera were spiked with norvaline and sarcosine as the internal standard and then deproteinized by adding 10 volumes of methanol. The supernatant was filtered with a Nanosep MF 0.2-μm centrifugal device (Pall Corporation, Santa Clara, NY, USA). The filtrate was then subjected to precolumn derivatization with o-phthalaldehyde for primary amino acids and 9-fluorenylmethyl chloroformate for secondary amino acids. The derivate was injected into an Agilent 1100 series high-performance liquid chromatography (HPLC) system equipped with a reversed-phase column, the Agilent Poroshell 120 EC-C18, 3.0 by 150 mm, 2.7 μm, and fluorescent detector, according to an application note by the manufacturer (Agilent Technologies, Inc., Santa Clara, CA, USA).
Mouse bacteremia experiment.
Eight-week-old female CD1 Swiss mice were purchased from Kyudo (Tosu, Saga, Japan). A small amount (108 CFU) of S. aureus strains was inoculated into 10 ml of TSB and grown aerobically at 37°C with shaking at 120 rpm. When the bacterial culture reached an OD660 of 0.8, the bacterial cells were collected and washed three times with PBS. Then, the cells were resuspended in PBS at a concentration of 1.5 × 109 CFU/ml. An aliquot of 100 μl (1.5 × 108 CFU) was injected into the tail vein of the mice. Mouse survival was monitored for 14 days. This experimental protocol was approved by the ethical committee of the Animal Research Center of Kagoshima University, Japan.
Statistical analysis.
All statistical analyses were performed with the statistical software EZR version 1.32 (http://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmedEN.html).
RESULTS
Identification of genes contributing to growth in CS.
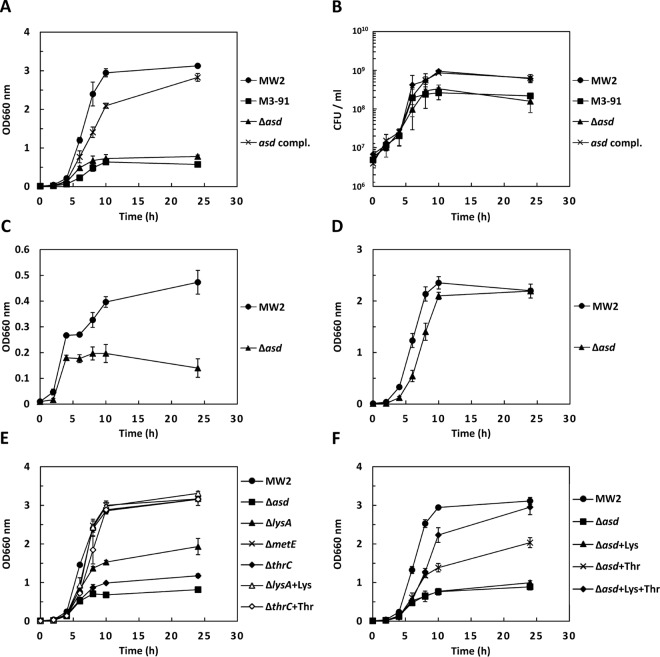
We obtained six Tn551-insertional mutants, which exhibited reduced growth in CS-1 compared with the parent strain. In two mutants (M3-91 and M17-9), we found that Tn551 was located in a gene encoding aspartate semialdehyde dehydrogenase (asd). The Tn551 insertion sites of M3-91 and M17-9 were 276 bp and 347 bp from the initiation of the asd gene, respectively. Moreover, we found the Tn551 insertion in the genes of two two-component systems (TCSs) (arlR and saeR), a sec translocase accessory protein (secDF), and an uncharacterized protein (MW0883) in mutants. In this study, we focused on the asd mutants. In the growth curve experiments, M3-91 exhibited significantly less growth than WT MW2 (Fig. 1A). We also found that the growth curve of M17-9 was similar to that of M3-91 (data not shown). To confirm the reduced growth of the mutants, we constructed an asd-inactivated mutant (Δasd mutant) using the thermosensitive plasmid pYT1 and the complemented strain using the IPTG-inducible plasmid pCL15. Similar growth was observed between the Δasd mutant and M3-91, and the complemented strain exhibited growth similar to that of the WT (Fig. 1A). Similar results were obtained using CFU determination of the strains grown in CS-1 (Fig. 1B). We also investigated the CFU of the mutant and WT strains after incubation in the additional 7 CS varieties for 24 h. The CFU of the mutant were significantly lower than in the WT in 5 of the 7 CS varieties (see Fig. S1 in the supplemental material). In CS-6, the CFU of the mutant were 5 times lower than in the WT, but no statistical significance was observed. The growth of the WT and mutant in CS-3 was much lower than that of other CS varieties. Additionally, in CS-3, the growth of the mutant was not different from that of the WT.
FIG 1.
Inactivation of lysine and/or threonine biosynthesis genes cause growth deficiency in CS. (A and B) Bacterial growth of S. aureus MW2 WT, M3-91, Δasd mutant and asd complement strain (compl.) grown in CS-1 were analyzed by measurement of bacterial density (A) and CFU (B). (C and D) Bacterial growth (bacterial density) of the WT and Δasd mutant grown in CDM mimicking the concentrations of amino acids and glucose in CS-1 (C) or standard CDM (D). (E) Bacterial growth (bacterial density) of the WT and the Δasd, ΔlysA, ΔmetE, and ΔthrC mutants grown in CS-1 or CS-1 containing additional lysine or threonine. (F) Bacterial growth (bacterial density) of the WT and Δasd mutant grown in CS-1 or CS-1 containing additional lysine and/or threonine. + Lys indicates l-lysine addition in CS-1 (100 μg/ml). + Thr indicates l-threonine addition in CS-1 (100 μg/ml). + Lys + Thr indicates l-lysine and l-threonine addition in CS-1 (100 μg/ml, respectively). The data shown represent the means and standard deviations (SD) of measurements performed in triplicate.
Lysine and threonine biosynthesis via Asd are important for growth in CS.
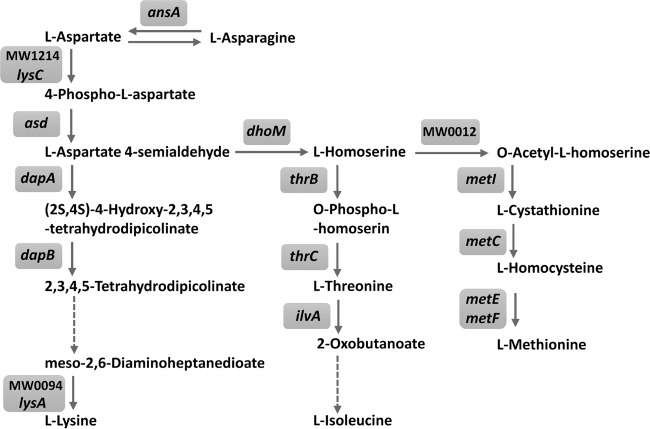
Because Asd is known as the enzyme for the biosynthesis of the aspartate family of amino acids in prokaryotes, fungi, and some higher plants (21), we hypothesized that the concentrations of amino acids in the CS affected the growth of the asd-inactivated mutant. Analysis of free amino acids in CS-1, except for asparagine and glutamine, showed that the concentrations of amino acids were less than those in CDM (Table 3). We constructed modified CDM to mimic the concentrations of free amino acids (Table 3) and glucose (0.5 g/liter) in CS-1, and we analyzed the growth of the asd-inactivated mutant and WT. The mutant exhibited reduced growth compared with the WT in CDM mimicking CS-1 (Fig. 1C), unlike in standard CDM (Fig. 1D). Asd catalyzes 4-phospho-l-aspartate to l-aspartate 4-semialdehyde, which is a precursor for the biosynthesis of lysine, methionine, threonine, and isoleucine (Fig. 2). The asd-inactivated mutant could not grow in the CDM lacking either lysine, methionine, or threonine, but it grew in the CDM lacking isoleucine (Fig. S2 in the supplemental material). Next, we constructed inactivated mutants of genes for the biosynthesis of lysine (lysA), threonine (thrC), and methionine (metE), and we found that these mutants could not grow in the CDM lacking the respective amino acid (see Fig. S3 in the supplemental material). The lysA- or thrC-inactivated mutant exhibited reduced growth compared to the WT in CS-1, but the growth of the two mutants was greater than that of the asd-inactivated mutant. However, the growth of the metE-inactivated mutant was similar to the WT in CS-1 (Fig. 1E). The growth of the lysA- or thrC-inactivated mutant was completely recovered by the addition of 100 μg/ml lysine or threonine, respectively (Fig. 1E). In the asd-inactivated mutant, the growth was not recovered by the addition of lysine, but it was partially recovered by the addition of threonine. The growth of the asd-inactivated mutant was recovered by the addition of 100 μg/ml lysine and threonine. (Fig. 1F).
TABLE 3.
Concentrations of free amino acids in CS-1 and CDM
| Amino acid | Concn (μM) |
|
|---|---|---|
| CS-1a | CDM | |
| Alanine | 353.6 ± 32.9 | 1,122 |
| Arginine | 266.4 ± 45.5 | 574 |
| Asparagine | 16.3 ± 3.3 | 0 |
| Aspartic acid | 9.0 ± 2.1 | 751 |
| Cystine | 5.4 ± 1.0 | 416 |
| Glutamine | 253.8 ± 30.8 | 0 |
| Glutamic acid | 135.5 ± 27.6 | 680 |
| Glycine | 61.3 ± 9.1 | 1,332 |
| Histidine | 33.9 ± 0.6 | 645 |
| Isoleucine | 141.1 ± 24.1 | 762 |
| Leucine | 255.7 ± 41.1 | 762 |
| Lysine | 43.3 ± 8.7 | 684 |
| Methionine | 17.0 ± 4.2 | 670 |
| Phenylalanine | 70.5 ± 12.8 | 605 |
| Proline | 240.3 ± 27.0 | 869 |
| Serine | 136.9 ± 12.2 | 952 |
| Threonine | 78.0 ± 5.3 | 839 |
| Tryptophan | 45.4 ± 16.4 | 490 |
| Tyrosine | 46.0 ± 10.5 | 552 |
| Valine | 297.6 ± 38.8 | 854 |
Values are means ± SD.
FIG 2.
Aspartate amino acid biosynthesis pathway. The biosynthesis pathway of aspartate family amino acids in S. aureus strain MW2 was adapted from the KEGG pathway database (http://www.genome.jp/kegg-bin/show_pathway?sam01230). Dashed arrows indicate multiple reactions.
Growth of the deficient mutants of lysine, methionine, and/or threonine biosynthesis in MS and CDM mimicking HS.
We performed the growth experiment for asd-, lysA-, metE-, and thrC-inactivated mutants in MS and found that these mutants exhibited significantly lower growth than the WT (see Fig. S4 in the supplemental material). In HS, the exact growth of S. aureus strains could not be analyzed because S. aureus showed extensive aggregation in HS. Concentrations of free amino acids in HS are shown in Table S1 in the supplemental material. We constructed modified CDM to mimic the concentrations of free amino acids and glucose (0.5 g/liter) in HS and found that the asd and metE mutants exhibited significantly lower growth than the WT (see Fig. S5 in the supplemental material).
Aspartate family amino acid biosynthesis is critical for virulence in mice.
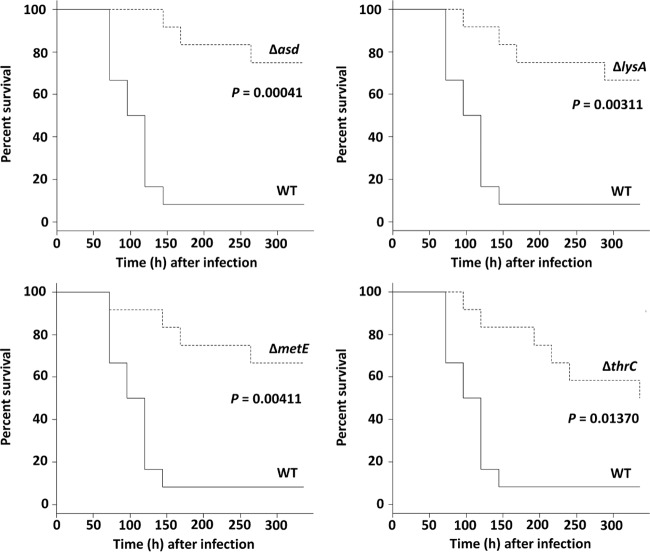
The mouse survival experiment was performed using the bacteremia model with WT S. aureus and the asd-, lysA-, metE-, and thrC-inactivated mutants. The majority of WT-infected mice (11 of 12 mice) died within 150 h. All mutant strains displayed a significant reduction in virulence compared to the WT. The mouse survival ratio among the mutants was not significantly different (Fig. 3).
FIG 3.
Mouse survival experiment. Survival percentage of CD-1 Swiss mice (n = 12) after challenge with an intravenous injection of 1.5 × 108 CFU of S. aureus MW2 WT (solid lines) and mutant strains (dashed lines). Mouse survival was plotted at 24-h intervals for 336 h. The P value from a log rank test is denoted in each graph.
DISCUSSION
In this study, we identified six mutants, including two asd mutants, that showed significantly reduced growth in CS compared with that of WT. Then, we focused on asd and its related genes to demonstrate the importance of amino acids for growth in CS. Asd is an enzyme at the first branch point in the following pathways for the synthesis of four amino acids from aspartate (Fig. 2): (i) lysine biosynthesis: l-aspartate 4-semialdehyde, which is a resulting metabolite by the catalysis of Asd and a common precursor of the threonine and methionine pathways, is converted into lysine via multiple steps containing the synthesis of diaminopimelic acid using the lysine biosynthesis genes (dapABD and lysA) and multiple unidentified genes. (ii) Threonine and isoleucine biosynthesis: l-aspartate 4-semialdehyde is converted into homoserine by homoserine dehydrogenase (DhoM), and homoserine is converted into threonine by homoserine kinase (ThrB) and threonine synthase (ThrC). Isoleucine biosynthesis is processed using ilvABCDE from threonine. (iii) Methionine biosynthesis; homoserine is catalyzed by homoserine O-acetyltransferase (MW0012), the metabolite is converted into cystathionine to react with cysteine by cystathionine γ-synthase (MetI), and the cystathionine is converted into methionine via 2 steps using the genes metC, metE, and metF. Based on these pathways, the inactivation of asd is likely to impair lysine, threonine, isoleucine, and methionine biosynthesis in S. aureus. As shown in Fig. S2 in the supplemental material, the asd-inactivated mutant did not have the auxotrophy for isoleucine in the CDM, while the mutant had auxotrophy for lysine, threonine, and methionine. In most bacteria, isoleucine biosynthesis is initiated in the conversion from threonine to 2-oxobutanoate by threonine dehydratase (IlvA) (22). Previous studies reported that an alternative pathway for isoleucine biosynthesis was observed in Leptospira interrogans, E. coli, and Geobacter sulfurreducens (23–25). Therefore, alternative pathways for isoleucine biosynthesis might also exist in S. aureus. We investigated the growth of the inactivated mutants of lysA, thrC, and metE to elucidate which pathway is responsible for growth in CS-1, and we found that the inactivated mutants of lysA and thrC exhibited reduced growth compared with the WT (Fig. 1E). These results suggested that the concentrations of lysine and threonine in CS-1 were not sufficient for S. aureus growth without the biosynthesis of these amino acids. However, the inactivated mutant of metE exhibited growth almost similar to that of the WT in CS-1 (Fig. 1E). Therefore, de novo biosynthesis of methionine may not be necessary for growth in CS-1. In this study, we also isolated arlR and saeR mutants, which exhibited significantly reduced growth in CS-1. Because TCSs are known to regulate several genes (26), we investigated the expression of asd and its related genes in these mutants and found no alterations in these mutants grown in CS-1 (data not shown).
Analysis of free amino acids revealed that the concentration of each in CS-1 was lower than that in CDM. The concentrations of lysine and threonine were 43.3 μM (standard deviation [SD], 8.7 μM) and 78.0 μM (SD, 5.3 μM), respectively (Table 3). We investigated the growth of the asd-inactivated mutant in CDM containing only the concentration of lysine or threonine in CS-1. Contrary to expectations, we found that the growth was similar to that of the WT (data not shown). However, in CDM containing the concentrations of all amino acids and glucose in CS-1, we found that the asd-inactivated mutant showed reduced growth compared with the WT (Fig. 1C). These results suggest that the de novo synthesis of lysine and threonine is important for the growth of S. aureus under restricted conditions, such as in CS, which has low concentrations of amino acids and glucose compared to CDM or TSB.
In mouse bacteremia experiments, the metE-inactivated mutant exhibited attenuated virulence at the same level as the asd-, lysA-, and thrC-inactivated mutants (Fig. 3). This result was not in accordance with the results of in vitro experiments using CS-1. However, the growth of the mutants in MS was different compared to their growth in CS-1, especially the metE mutant (see Fig. S4 in the supplemental material). The metE mutant also showed lower growth in MS, as did other mutants. Therefore, in bacteremia experiments, the mortality rate among the S. aureus strains corresponded with the result of the growth assay using MS. However, comparing the amino acid concentrations between CS-1 and MS, the concentration of methionine was not significantly different (Table 3; see also Table S1 in the supplemental material). In addition, when CS-3 was used for the growth experiment, the asd mutant did not exhibit significant reduced growth compared to the WT (see Fig. S1 in the supplemental material). The total concentration of amino acids in CS-3 was approximately 40% less than in CS-1, but the concentrations of lysine and methionine were similar to those in CS-1 (Table 3; see also Table S1). Also, in modified CDM mimicking HS, the growth of thrC and lysA mutants was similar to that of WT, while asd and metE mutants showed reduced growth. The concentration of threonine in HS was similar to that in CS-1 or MS (Table 3; see also Table S1), while the concentration of lysine in HS was higher than that of CS-1. These results suggest that the asd mutant showed reduced growth in human, calf, and mouse serum, but other mutants showed different growth patterns. These results indicate that the composition of total amino acid or other factors is associated with growth in the sera. Further studies will be required to elucidate these findings. Because we found a correlation between mouse mortality and the bacterial growth in serum, it is thought that suppressed growth may cause suppressed virulence in vivo. In S. aureus, the toxins and exoenzymes are tightly regulated by several transcriptional regulators, mainly the Agr system (27). In the late-exponential and stationary phases, S. aureus produces significant quantities of several virulence factors (28). Therefore, the suppressed growth causes a reduction in the virulence, affecting mouse mortality.
In this study, we demonstrated that the biosynthesis of aspartate family amino acids, especially lysine and threonine, was important for the growth of S. aureus in serum. Our data strongly suggest that the genes for lysine and threonine biosynthesis are critical factors for the multiplication of S. aureus in the blood.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01399-16.
REFERENCES
- 1.Foster TJ. 2004. The Staphylococcus aureus “superbug.” J Clin Invest 114:1693–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Blot SI, Vandewoude KH, Hoste EA, Colardyn FA. 2002. Outcome and attributable mortality in critically Ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Arch Intern Med 162:2229–2235. doi: 10.1001/archinte.162.19.2229. [DOI] [PubMed] [Google Scholar]
- 4.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. 2006. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 5.Bunce C, Wheeler L, Reed G, Musser J, Barg N. 1992. Murine model of cutaneous infection with gram-positive cocci. Infect Immun 60:2636–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Y, Onodera Y, Lee JC, Hooper DC. 2008. NorB, an efflux pump in Staphylococcus aureus strain MW2, contributes to bacterial fitness in abscesses. J Bacteriol 190:7123–7129. doi: 10.1128/JB.00655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valentino MD, Foulston L, Sadaka A, Kos VN, Villet RA, Santa Maria J Jr, Lazinski DW, Camilli A, Walker S, Hooper DC, Gilmore MS. 2012. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. mBio 5(5):e01729-14. doi: 10.1128/mBio.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oogai Y, Matsuo M, Hashimoto M, Kato F, Sugai M, Komatsuzawa H. 2011. Expression of virulence factors by Staphylococcus aureus grown in serum. Appl Environ Microbiol 77:8097–8105. doi: 10.1128/AEM.05316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horsburgh MJ, Wiltshire MD, Crossley H, Ingham E, Foster SJ. 2004. PheP, a putative amino acid permease of Staphylococcus aureus, contributes to survival in vivo and during starvation. Infect Immun 72:3073–3076. doi: 10.1128/IAI.72.5.3073-3076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendum TA, Newcombe J, Mannan AA, Kierzek AM, McFadden J. 2011. Interrogation of global mutagenesis data with a genome scale model of Neisseria meningitidis to assess gene fitness in vitro and in sera. Genome Biol 12:R127. doi: 10.1186/gb-2011-12-12-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samant S, Lee H, Ghassemi M, Chen J, Cook JL, Mankin AS, Neyfakh AA. 2008. Nucleotide biosynthesis is critical for growth of bacteria in human blood. PLoS Pathog 4:e37. doi: 10.1371/journal.ppat.0040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 13.Hussain M, Hastings JG, White PJ. 1991. A chemically defined medium for slime production by coagulase-negative staphylococci. J Med Microbiol 34:143–147. doi: 10.1099/00222615-34-3-143. [DOI] [PubMed] [Google Scholar]
- 14.O'Brien FG, Yui Eto K, Murphy RJ, Fairhurst HM, Coombs GW, Grubb WB, Ramsay JP. 2015. Origin-of-transfer sequences facilitate mobilisation of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res 43:7971–7983. doi: 10.1093/nar/gkv755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattee PA. 1981. Distribution of Tn551 insertion sites responsible for auxotrophy on the Staphylococcus aureus chromosome. J Bacteriol 145:479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novick RP. 1963. Analysis by transduction of mutations affecting penicillinase formation in Staphylococcus aureus. J Gen Microbiol 33:121–136. doi: 10.1099/00221287-33-1-121. [DOI] [PubMed] [Google Scholar]
- 17.Liu YG, Mitsukawa N, Oosumi T, Whittier RF. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8:457–463. doi: 10.1046/j.1365-313X.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 18.Kajimura J, Fujiwara T, Yamada S, Suzawa Y, Nishida T, Oyamada Y, Hayashi I, Yamagishi J, Komatsuzawa H, Sugai M. 2005. Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol Microbiol 58:1087–1101. doi: 10.1111/j.1365-2958.2005.04881.x. [DOI] [PubMed] [Google Scholar]
- 19.Luong TT, Lee CY. 2006. The arl locus regulates Staphylococcus aureus type 5 capsule via mgr dependent and independent pathways. Microbiology 152:3123–3131. doi: 10.1099/mic.0.29177-0. [DOI] [PubMed] [Google Scholar]
- 20.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 21.Hadfield A, Kryger G, Ouyang J, Petsko GA, Ringe D, Viola R. 1999. Structure of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis. J Mol Biol 289:991–1002. doi: 10.1006/jmbi.1999.2828. [DOI] [PubMed] [Google Scholar]
- 22.Umbarger HE. 1978. Amino acid biosynthesis and its regulation. Annu Rev Biochem 47:533–606. doi: 10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, Zhang Y, Guo X, Ren S, Staempfli AA, Chiao J, Jiang W, Zhao G. 2004. Isoleucine biosynthesis in Leptospira interrogans serotype lai strain 56601 proceeds via a threonine-independent pathway. J Bacteriol 186:5400–5409. doi: 10.1128/JB.186.16.5400-5409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips AT, Nuss JI, Moosic J, Foshay C. 1972. Alternate pathway for isoleucine biosynthesis in Escherichia coli. J Bacteriol 109:714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risso C, Van Dien SJ, Orloff A, Lovley DR, Coppi MV. Elucidation of an alternate isoleucine biosynthesis pathway in Geobacter sulfurreducens. J Bacteriol 190:2266–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3:165–170. doi: 10.1016/S1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 27.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 28.Ziebandt AK, Becher D, Ohlsen K, Hacker J, Hecker M, Engelmann S. 2004. The influence of agr and sigmaB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics 4:3034–3047. doi: 10.1002/pmic.200400937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.