ABSTRACT
Acinetobacter baumannii is an important emerging pathogen that is capable of causing many types of severe infection, especially in immunocompromised hosts. Since A. baumannii can rapidly acquire antibiotic resistance genes, many infections are on the verge of being untreatable, and novel therapies are desperately needed. To investigate the potential utility of copper-based antibacterial strategies against Acinetobacter infections, we characterized copper resistance in a panel of recent clinical A. baumannii isolates. Exposure to increasing concentrations of copper in liquid culture and on solid surfaces resulted in dose-dependent and strain-dependent effects; levels of copper resistance varied broadly across isolates, possibly resulting from identified genotypic variation among strains. Examination of the growth-phase-dependent effect of copper on A. baumannii revealed that resistance to copper increased dramatically in stationary phase. Moreover, A. baumannii biofilms were more resistant to copper than planktonic cells but were still susceptible to copper toxicity. Exposure of bacteria to subinhibitory concentrations of copper allowed them to better adapt to and grow in high concentrations of copper; this copper tolerance response is likely achieved via increased expression of copper resistance mechanisms. Indeed, genomic analysis revealed numerous putative copper resistance proteins that share amino acid homology to known proteins in Escherichia coli and Pseudomonas aeruginosa. Transcriptional analysis revealed significant upregulation of these putative copper resistance genes following brief copper exposure. Future characterization of copper resistance mechanisms may aid in the search for novel antibiotics against Acinetobacter and other highly antibiotic-resistant pathogens.
IMPORTANCE Acinetobacter baumannii causes many types of severe nosocomial infections; unfortunately, some isolates have acquired resistance to almost every available antibiotic, and treatment options are incredibly limited. Copper is an essential nutrient but becomes toxic at high concentrations. The inherent antimicrobial properties of copper give it potential for use in novel therapeutics against drug-resistant pathogens. We show that A. baumannii clinical isolates are sensitive to copper in vitro, both in liquid and on solid metal surfaces. Since bacterial resistance to copper is mediated though mechanisms of efflux and detoxification, we identified genes encoding putative copper-related proteins in A. baumannii and showed that expression of some of these genes is regulated by the copper concentration. We propose that the antimicrobial effects of copper may be beneficial in the development of future therapeutics that target multidrug-resistant bacteria.
INTRODUCTION
Acinetobacter baumannii, a Gram-negative aerobe, is an important emerging pathogen in the United States and worldwide. Most Acinetobacter species are ubiquitous environmental bacteria, and the ability to survive in harsh, dry environments likely aided the establishment of A. baumannii as a prominent nosocomial pathogen (1). Today A. baumannii is also nearly ubiquitous in hospital environments, especially in intensive care units (ICUs). A. baumannii is capable of causing many types of infections, including pneumonia, urinary tract infections, bacteremia, skin and soft tissue infections, osteomyelitis, and even meningitis (1). Typically, A. baumannii causes serious disease only in immunocompromised individuals. However, the lack of a suitable treatment against antibiotic-resistant strains can lead to more severe disease and higher mortality (2–4). Because of the high prevalence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains, the CDC has classified A. baumannii as a “serious” threat (5).
Despite its relatively recent introduction into hospitals in the United States and countries in South America and Europe, A. baumannii is responsible for a significant proportion of hospital-acquired infections. A recent meta-analysis of studies carried out worldwide reported that A. baumannii consistently causes 5 to 10% of hospital-acquired infections and an even greater proportion of ICU-acquired infections (6). However, depending on patient demographics and the strains present, the proportion of certain infections caused by A. baumannii can be much larger than 5 to 10%. For example, a retrospective study of ICU-acquired infections in the ICU of the National Cancer Institute of Mexico reported that A. baumannii was the causative agent of 6% of total hospital-acquired infections but disproportionately represented 60% of pneumonia and 25% of bloodstream infections (7). A. baumannii has rapidly evolved into a difficult-to-treat pathogen through acquisition, mostly via integrons and transposons, of multiple drug resistance genes that supplement the already high level of intrinsic drug resistance (8). A recent study analyzing more than 1,300 globally collected bloodstream isolates included over 150 A. baumannii isolates (4). Of these clinical strains, 91.9% were MDR, 71.3% were XDR, and 1% were pandrug resistant (PDR; resistant to all antibiotic treatments). Worldwide, XDR and PDR strains limit the treatment options for physicians, which can lead to empirical therapy and complications. Not surprisingly, the mortality rate of infection correlates with the level of drug resistance of the infecting strain and whether or not the patient was given appropriate antimicrobial therapy (2–4). The extensive drug resistance in A. baumannii and other “superbugs” has prompted calls for more atypical antibacterial approaches that take into account the present resistance to most classes of antibiotics.
Most antibiotics target essential functions for bacterial growth and/or survival. Metal homeostasis is a vital process that provides an extensive list of potential new antibiotic targets. Copper is required for cellular function, e.g., for redox balance and as an enzyme cofactor. However, copper ions become toxic at high concentrations; these ions participate in Fenton chemistry to produce hydroxyl radicals that react with and damage essential biomolecules (9). Additionally, copper ions damage iron-sulfur cluster proteins by binding with sulfur and displacing iron (10). Recent studies in Escherichia coli and Salmonella spp. showed that when bacteria are placed on copper surfaces, outer membrane integrity is compromised, hydroxyl radicals are produced, respiration is inhibited, and DNA is degraded (11). Because of the need for new therapeutics to treat highly antibiotic-resistant pathogens, research into the use of copper as an antimicrobial has recently increased. Various pathogens have been assessed for their levels of copper sensitivity in vitro; unfortunately, these values are difficult to compare due to protocol differences, namely, the growth medium used. However, it is evident that among the pathogens tested, the concentration of copper required for successful inhibition or elimination in minimal medium is typically within the micromolar range (12–15). Clinical isolates of A. baumannii have been assessed for their overall levels of copper resistance; however, previous experiments used a rich medium and reported MICs in the millimolar range, similar to those for other pathogens tested in rich medium (14, 16–18). Thus, these MICs may be influenced by medium choice.
Variation in the inhibitory concentration of copper ions among pathogens and across individual strains is likely due to differences in the proteins present in the organisms that sense or modulate copper levels and/or their regulation. The details of copper homeostasis mechanisms that are critical to copper resistance have been investigated in some bacterial species. Among these, the E. coli copper resistance system is likely the most fully characterized. Generally, copper resistance in bacteria is facilitated by detoxification and export systems (19). For example, in E. coli, the copper ATPase CopA effluxes copper ions from the cytosol to the periplasm, where they are oxidized to the less toxic Cu2+ form by the copper oxidase, CueO (20, 21). Expression of both copA and cueO is induced by the transcriptional regulator CueR when it is loaded with copper (22). In addition, a two-component system, CusRS, senses copper ions in the periplasm and directly upregulates expression of an RND family efflux pump that removes copper ions from the cell (12, 17, 23). E. coli also encodes the Pco system, whose contributions to copper resistance are less clear; the system appears to help in the handling of periplasmic copper (24). Although there is extensive knowledge concerning copper homeostasis in E. coli and a few other pathogens, little is known about the overall level of copper resistance in A. baumannii. Protein sequences of E. coli copper-related genes have been used to identify putative homologous proteins in many bacteria, including A. baumannii (25), but the importance of these genes to copper resistance has not been investigated experimentally. The goal of this study was to assess the overall level of copper resistance in A. baumannii clinical isolates and to begin to investigate potential mechanisms of copper homeostasis in this important pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used in this study are listed in Table 1, and primer sequences are shown in Table 2. Bacterial strains were grown overnight at 37°C on 1.5% lysogeny broth (LB) agar plates (MoBio, Carlsbad, CA). Liquid cultures of A. baumannii were grown in M9 minimal medium (26) supplemented with 0.1% Casamino Acids (Difco, Franklin Lakes, NJ) at 37°C with shaking at 190 rpm; the amino acid-supplemented medium is referred to throughout the text as simply “M9 medium.” Overnight cultures were started from a single colony and grown for 16 to 19 h. A. baumannii strains were maintained as frozen stocks at −80°C in M9 or LB medium supplemented with 40% glycerol.
TABLE 1.
Strains used in this study
| Strain | Description, yr of isolation | Source or reference |
|---|---|---|
| A. baumannii strains | ||
| ATCC 17978 | Common laboratory strain | ATCC |
| AB0057 | Clinical isolate, 2004 | 36 |
| AB4857 | Clinical isolate, 2008 | 36 |
| AB5075 | Clinical isolate, 2008 | 36 |
| AB5256 | Clinical isolate, 2009 | 36 |
| AB5711 | Clinical isolate, 2009 | 36 |
| E. coli strain | ||
| K-12 MG1655 | Common laboratory strain | 62 |
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide use and name | Sequence (5′-3′) |
|---|---|
| Quantitative real-time PCR | |
| actP2/copA1 RT up | CCAGATATGAACTCGCTGGTC |
| actP2/copA1 RT dw | GACCGTGCCTTGTGGTAATA |
| ABUW_2708 RT up | GTGACTGCAACGATTCAAGATG |
| ABUW_2708 RT dw | ATCAACGCTTTCACTGCTTTC |
| ABUW_3322 RT up | AACTCTATTCCGTCGTAGCTTAAC |
| ABUW_3322 RT dw | GCATCAGCAGCCCAAGA |
| copR/cusR RT up | GACTGGGTTACAGATGGCTTATC |
| copR/cusR RT dw | CAGCCATCTAACTTCGGTAACA |
| actP1/copA2 RT up | AGATGGAGGCACAGTCAAATAG |
| actP1/copA2 RT dw | GGCAGTTACCGGGATGATTT |
| copC RT up | GTAGGTGCGACTATGGTTGTAG |
| copC RT dw | AGATGCATTAATAGCCGGAGTAG |
| ABUW_0265 RT up | CATGTCCGTCTTATTCTGTTTGC |
| ABUW_0265 RT dw | CATTTAAAGCTGCATGATGACCA |
| ABUW_3230 RT up | AATGCGATTGACCCAGTGTAT |
| ABUW_3230 RT dw | AGCGAGCGACTTTAGCATAAT |
| ABUW 16S RT up | GGACGGGTGAGTAATGCTTAG |
| ABUW 16S RT dw | CGTAGGACGTATGCGGTATTAG |
| Junction PCR | |
| ABUW_2705/cueR jxn up | CACCCTGTACCATCTCTAAATCG |
| ABUW_2705/cueR jxn dw | TGATTGCTGTGGTGACCAG |
| cueR/actP2 jxn up | CGCCGAAATACCCGAATGT |
| cueR/actP2 jxn dw | CAGCAGGTGCGCTATATCC |
| copB/copA jxn up | TCCATGCTTTGAACATCCGTATC |
| copB/copA jxn dw | GATGAGTTAGGTGAATGGGCTATTC |
| copA/ABUW_3322 jxn up | CTGCCATAACCCAAGTCGA |
| copA/ABUW_3322 jxn dw | CATGGACCATTCTAAGATGACGG |
| copR/copS jxn up | CGCGGAATGGGGTATGTCTTAG |
| copR/copS jxn dw | CATGATGGCCAACTAAAGCATC |
| ABUW_0265/czcC jxn up | CTTACCAGTCTCCACATCTTAATGC |
| ABUW_0265/czcC jxn dw | CATTTGCCATTTGTCCCGC |
| czcC/czcB jxn up | GTTTACGTCTAGGGCTAGAGC |
| czcC/czcB jxn dw | TCCCTCATCATGGTGTTCTTC |
| czcB/czcA jxn up | CAGCTAGGCCAACGTTCTAAAG |
| czcB/czcA jxn dw | GGAACGGCATCAATAGGGAG |
| czcA/czcD jxn up | CGATGGATGAATGAAGACAAGACG |
| czcA/czcD jxn dw | ACCTCAACAATTAAGAATGTCGTGG |
| copC/copD jxn up | CCACAATGGGTGCTGATGG |
| copC/copD jxn dw | CAGCCACGTTGCATATATCCAAG |
| ABUW_3230/3229 jxn up | CAAGAAGGTGCATTACAGGTG |
| ABUW_3230/3229 jxn dw | ATGCTGCATAGGTGTCTGAG |
| ABUW_3229/pcoA jxn up | CAGCAAGCTGTACCAGATATTG |
| ABUW_3229/pcoA jxn dw | CTTAATTGCCGCGAGAGTC |
| pcoA/pcoB jxn up | CACATGAGTGCTGGAATGATG |
| pcoA/pcoB jxn dw | CCTGTCCACCATGTTCGC |
| pcoB/ABUW_3226 jxn up | CAAACCGCTTGGCAAACTG |
| pcoB/ABUW_3226 jxn dw | CACCCACAGCTAGAACAGC |
Determination of A. baumannii sensitivity to copper in liquid culture.
To assess the effect of copper on A. baumannii growth, subcultures were started from overnight cultures at an optical density at 600 nm (OD600) of 0.05 in 10 ml of M9 medium containing 100 μM to 1.5 mM CuSO4 (Sigma-Aldrich, St. Louis, MO). The OD600 of the cultures was measured every hour for 6 h. At 0, 2, 4, and 6 h, an aliquot of each culture was serially diluted and plated to enumerate CFU.
Determination of A. baumannii copper sensitivity on solid metal coupons.
The metals tested were stainless steel 304, copper 110 H02 (99.9% Cu), and cartridge brass C260 (∼70% Cu) (Online Metals, Seattle, WA). Metal sheets were cut into coupons (1 × 1 × 0.04 to 0.05 cm) and degreased by vortexing in approximately 20 ml of acetone for 30 s. Coupons were stored immersed in absolute ethanol. Prior to use, coupons were flamed and placed in sterile petri dishes. Bacterial subcultures were grown as described above. At 2.5 h (log phase), 6 h (early stationary phase), and 24 h (stationary phase), 5-μl samples were taken for (i) serial dilution and plating to determine the inoculum (T0) and (ii) spotting onto metal coupons. Dishes containing coupons were incubated on the lab bench at room temperature for up to 75 min. At various times, coupons were transferred to 5-ml screw-cap tubes (Axygen, Corning, NY) containing 3 ml of phosphate-buffered saline (PBS) and mixed in a Bullet Storm 5 blender (Next Advance, Averill Park, NY) on level 1 for 1 min. The samples were serially diluted and plated, and surviving CFU were enumerated. The limit of detection was 3 CFU per coupon.
Quantification of copper ions via ICP-MS.
To measure the concentration of copper ions found in medium exposed to metal surfaces, droplets were collected and analyzed by inductively coupled plasma mass spectrometry (ICP-MS). Similar to the setup of the copper sensitivity assay described above, two 5-μl droplets of M9 medium were placed on a 1- by 1.5-cm metal coupon for 0, 15, 45, or 75 min. The droplets were collected by running an additional 100 μl of M9 medium across the surface into a 1.5-ml Eppendorf tube; the total volume was run across the surface two additional times to collect any precipitated ions. The collected sample was immediately vortexed, and 10- to 20-μl aliquots were removed to separate tubes and frozen at −20°C for later analysis. Aliquots of each preparation of M9 medium were also analyzed. Samples were thawed at room temperature and diluted 1:100 in 2% HNO3 (trace metal grade). The metal content of the samples was quantitated by ICP-MS. Copper concentrations were determined by injecting diluted samples into an Agilent 7700X ICP-MS instrument (Agilent Technologies, Santa Clara, CA). Copper levels were detected using an octopole reaction system (ORS) cell in He mode to remove interference. The ICP-MS parameters used for the analysis were as follows: a radio frequency (RF) power of 1,550 W, an argon carrier gas flow of 1.01 liters/min, an argon make-up gas flow of 0.1 liter/min, a helium gas flow of 4.3 ml/min, an octopole RF of 200 V, and an OctP bias of −18 V. Samples were directly infused using the model 7700X peristaltic pump with a speed of 0.1 rotation per s (rps) and a micromist nebulizer. Copper concentrations in samples were derived from a calibration curve generated by a series of dilutions of a copper atomic absorption standard (Fluka Analytical, St. Louis, MO) prepared in the same matrix as the samples. Data analysis was performed using Agilent's Mass Hunter software.
Assessing copper sensitivity of A. baumannii biofilms.
Biofilms of A. baumannii strain AB5711 were grown in 1 ml of LB broth in 24-well tissue culture-treated plates (Corning, Corning, NY). Cultures were started at an OD600 of 0.05 and incubated statically at 37°C for 18 h. Afterwards, the LB broth was gently removed and replaced with 1.5 ml of M9 medium containing up to 1.5 mM CuSO4. The biofilms were incubated for another 6 to 24 h. At each time point (0, 6, and 24 h), the M9 medium was transferred to a 1.5-ml Eppendorf tube and vortexed for approximately 5 s to break up any clumped planktonic bacteria in the supernatant. Additionally, the biofilm was scraped from the sides and bottom of the well by use of a pipette tip and resuspended in 1 ml of PBS. Serial dilutions of both planktonic cells (supernatant) and biofilms were plated and CFU enumerated. The wells were stained with crystal violet following processing to ensure that the biofilms were recovered equally from all wells.
Identification of putative copper-related genes in A. baumannii.
Initially, amino acid sequences from copper-related proteins in E. coli and Pseudomonas aeruginosa were used as queries for Protein BLASTp (NCBI) searches to identify putative copper-related genes in A. baumannii AB5075-UW. To determine the ultimate sequence similarity and identity between proteins in A. baumannii, E. coli, and P. aeruginosa, AB5075-UW (accession no. NZ_CP008706.1) proteins were compared to those encoded in the reference genomes of E. coli and P. aeruginosa, namely, E. coli K-12 MG1655 (accession no. NC_000913.3) and P. aeruginosa PAO1 (accession no. NC_002516.2). The identified amino acid sequences encoded by putative copper-related genes from A. baumannii AB5075-UW were entered as queries for BLASTp searches, and E. coli K-12 MG1655 (taxid 511145) and P. aeruginosa PAO1 (taxid 208964) were chosen individually as the search set. The following parameters were used: word size, 6; maximum number of matches in a query range, 0; BLOSUM62 matrix with gap costs of 11 for existence and 1 for extension; composition-based statistics; and automatic adjustments for short input sequences. To determine if each putative copper-related gene was present in the other Acinetobacter strains of the panel, the amino acid sequences from AB5075-UW were again used as queries, and each isolate was chosen as the search set. The same search parameters were used. Note that there are two locus numbers for each gene of AB5075-UW. In addition to the original annotation, there was a subsequent reannotation by GenBank that occurred very recently. Both identifiers are given in Table 4 and are available under accession no. NZ_CP008706.1. Based on the timing of the reannotation, we used the original annotation throughout this study.
TABLE 4.
Putative copper-related genes/proteins in AB5075
| Annotated A. baumannii protein name/new namea | Locus no. (old/new)b | Putative function |
E. coli K-12 MG1655 (taxid 511145) homologue |
P. aeruginosa PAO1 (taxid 208964) homologue |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein name, locus | % identity/% similarity | Bit score | Expect value | % query coverage | Protein name, locus | % identity/% similarity | Bit score | Expect value | % query coverage | |||
| ABUW_0265c,e | ABUW_0265/ABUW_RS01300 | None | None | |||||||||
| CzcCe | ABUW_0266/ABUW_RS01305 | RND family efflux pump | None | None | ||||||||
| CzcB | ABUW_0267/ABUW_RS01310 | RND family efflux pump | CusB, b0574 | 25/44 | 69.3 | 1E−13 | 80 | CzcB, PA2521 | 34/53 | 182 | 5E−52 | 85 |
| CzcA | ABUW_0268/ABUW_RS01315 | RND family efflux pump | CusA, b0575 | 34/56 | 582 | 0E+00 | 97 | CzcA, PA2520 | 62/78 | 1,227 | 0E+00 | 97 |
| CzcD | ABUW_0269/ABUW_RS01320 | RND family efflux pump | ZitB (zinc efflux system), b0752 | 40/59 | 155 | 9E−45 | 86 | Cation efflux system protein, PA0397 | 56/73 | 286 | 2E−95 | 90 |
| ABUW_1334 | ABUW_1334/ABUW_RS06510 | Cation efflux | CusC, b0572 | 31/50 | 181 | 2E−51 | 96 | Hypothetical protein, PA2525 | 37/56 | 283 | 5E−89 | 95 |
| ABUW_2705c | ABUW_2705/ABUW_RS13125 | Hypothetical proteind | 28/45 | 57.8 | 8E−10 | 85 | Hypothetical protein, PA2706 | 34/51 | 87.8 | 9E−23 | 91 | |
| CueR | ABUW_2706/ABUW_RS13130 | Transcriptional regulator | CueR, b0487 | 41/66 | 106 | 3E−30 | 94 | CueR, PA4778 | 49/68 | 127 | 6E−38 | 96 |
| ActP2/CopA1 | ABUW_2707/ABUW_RS13135 | Copper-transporting P-type ATPase | CopA, b0484 | 41/61 | 561 | 0E+00 | 98 | Metal transporting P-type ATPase, PA3920 | 46/64 | 577 | 0E+00 | 98 |
| ABUW_2708 | ABUW_2708/ABUW_RS13140 | Copper chaperone | None | Hypothetical protein, PA3520 | 46/58 | 54.3 | 1E−11 | 87 | ||||
| ABUW_3226c | ABUW_3226/ABUW_RS15660 | Cation efflux | Sodium/proton antiporterd | 51/69 | 286 | 3E−94 | 90 | Sodium/proton antiporterd | 52/69 | 290 | 2E−96 | 90 |
| PcoB | ABUW_3227/ABUW_RS15665 | CopBd | 30/50 | 83.6 | 3E−17 | 79 | Copper resistance protein B, PA2064 | 27/52 | 77.4 | 5E−17 | 65 | |
| PcoA | ABUW_3228/ABUW_RS15670 | Copper oxidase | CueO, b0123 | 23/35 | 65.1 | 7E−12 | 91 | Copper resistance protein A, PA2065 | 46/60 | 473 | 8E−159 | 92 |
| ABUW_3229c,e | ABUW_3229/ABUW_RS15675 | None | None | |||||||||
| ABUW_3230c | ABUW_3230/ABUW_RS15680 | Sugar phosphatase YbiV, b0822 | 41/62 | 214 | 1E−68 | 99 | None | |||||
| CopB | ABUW_3320/ABUW_RS16130 | Copper resistance protein Bd | 27/46 | 84.7 | 3E−17 | 67 | Copper resistance protein B, PA2064 | 31/46 | 94 | 6E−26 | 65 | |
| CopA/CueO | ABUW_3321/ABUW_RS16135 | Copper oxidase | CueO, b0123 | 24/38 | 53.5 | 4E−08 | 71 | Copper resistance protein A, PA2065 | 45/59 | 479 | 3E−160 | 93 |
| ABUW_3322c | ABUW_3322/ABUW_RS16140 | Copper oxidased | 52/59 | 44.7 | 1E−04 | 80 | Copper oxidased | 62/68 | 60.1 | 2E−10 | 53 | |
| CopR/CusR | ABUW_3323/ABUW_RS16145 | Response regulator | CusR, b0571 | 62/81 | 286 | 8E−98 | 99 | CopR, PA2809 | 63/80 | 279 | 7E−95 | 99 |
| CopS/CusS | ABUW_3324/ABUW_RS16150 | Sensor kinase | CusS, b0570 | 34/54 | 265 | 6E−83 | 94 | CopS, PA2810 | 34/53 | 256 | 1E−79 | 98 |
| ActP1/CopA2 | ABUW_3325/ABUW_RS16155 | Copper-transporting P-type ATPase | CopA, b0484 | 46/65 | 507 | 8E−168 | 83 | Metal transporting P-type ATPase, PA3920 | 43/63 | 471 | 6E−154 | 79 |
| CopC | ABUW_3326/ABUW_RS16160 | Copper chaperone | CopC family protein YobA, b1841 | 33/43 | 57.4 | 4E−12 | 90 | CopC precursord | 39/53 | 58.9 | 7E−10 | 82 |
| CopDe | ABUW_3327/ABUW_RS16165 | None | None | |||||||||
Some of the proteins identified in AB5075 were annotated with names commonly used for different copper-related proteins. To avoid confusion in discussing A. baumannii systems alongside those of other bacteria, we referred to these proteins by new names that match those used elsewhere. In these cases, we first provide the current annotated name of the protein followed by the new name used in this publication.
Two locus numbers are given for each protein in AB5075-UW: one from the original annotation and one from the subsequent reannotation by GenBank. Both are available under accession no. NZ_CP008706.1.
Genes located in operons with identified copper-related genes were included even if they were not also identified as being copper related.
No homologue was found in the selected E. coli or P. aeruginosa strain, but significant homology was found with a different strain when the species E. coli (taxid 562) or P. aeruginosa (taxid 287) was chosen as the search set. The ABUW_2705 homologue was found in E. coli KTE51, with the locus tag A1SA_00101. The ABUW_3226 homologues were identified in multiple E. coli strains (accession no. WP_004364974.1) and multiple P. aeruginosa strains (accession no. WP_031693306.1). The PcoB homologue was identified in multiple E. coli strains (accession no. WP_020996127.1). The CopB homologue was found in E. coli strain HM46, with the locus tag HM46_RS23285. The ABUW_3322 homologue was found in E. coli HM46, with the locus tag HM46_RS23290, and was identified in multiple P. aeruginosa strains (accession no. WP_043096474.1). The CopC homologue was found in P. aeruginosa strain E15_London_28_01_14, with the locus tag PAERUG_E15_London_28_01_14_00104.
Four proteins included here do not have homologues in E. coli or P. aeruginosa, but there is homology to proteins found in other organisms, e.g., Bacillus spp., Moraxella spp., and Shewanella spp., when BLASTp searches are used to search all protein sequences.
RNA extraction, cDNA synthesis, and qRT-PCR.
A large subculture of A. baumannii cells was grown to exponential phase (2.5 h) as described above and then aliquoted in 10-ml volumes into 25-ml flasks. Various concentrations of CuSO4 (2.5 to 500 μM) were added directly to the cultures, and the cells were incubated with shaking for an additional 30 min. Cells were harvested by centrifugation at room temperature, and RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) as described previously (27), with the exceptions that the aqueous phase was used directly in an RNeasy cleanup protocol (Qiagen, Hilden, Germany) and a 60-min DNase (Qiagen) digestion was performed during the cleanup. cDNA was synthesized using a Quantitect reverse transcription kit (Qiagen) according to the manufacturer's instructions and as previously described (28). Briefly, quantitative reverse transcriptase PCR (qRT-PCR) was performed using a Roto-gene Q instrument (Qiagen), 1× SYBR green RT-PCR master mix (Qiagen), 3 pmol each of the forward and reverse primers, and 1 μl of cDNA. A reaction mixture in which no reverse transcriptase enzyme was added was included as a no-RT control. Cycling conditions were as follows: an initial activation step for 5 min at 95°C and 35 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 20 s, and extension at 72°C for 10 s. SYBR green fluorescence was measured during each extension step. Relative gene expression was calculated by the 2−ΔΔCT method, using the 16S rRNA gene as the internal reference.
Confirmation of operonic structure by junction PCR.
To detect each of the operon gene junctions, qualitative RT-PCR was conducted using GoTaq master mix (Promega, Madison, WI) per the manufacturer's instructions. Junctional PCRs were performed using 0.5 μl of cDNA, synthesized as described above, as the template. Chromosomal DNA was used as a positive control (0.5 μl; ∼100 ng), and the original RNA sample was used as a negative control (0.5 μl; 150 to 200 ng). Each 25-μl reaction mixture was incubated for 5 min at 95°C, followed by 25 cycles of denaturation at 95°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 30 s. PCR products were separated in 1% agarose gels and visualized by staining with ethidium bromide. Because the genomic DNA (gDNA) template produced more PCR product, a smaller volume was loaded on the gel in order to visualize all bands well. The gel was loaded with the following amounts of PCR products: gDNA, 1 μl; RNA, 20 μl; and cDNA, 1 to 20 μl.
Data analysis and statistics.
All graphs and statistical analyses were carried out using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA). Brown-Forsythe and Bartlett's tests were used to test for equality of variances, and graphical methods were used to assess normality. Welch's correction was used in the case of unequal variances, and nonparametric methods were used in the case of nonnormal data. Two-way analysis of variance (ANOVA) with Tukey's correction for multiple comparisons was used to evaluate differences in growth for various copper treatments, using log-transformed data. Data are presented as geometric means plus standard errors of the means (SEM). The Kruskal-Wallis test with Dunn's correction for multiple comparisons was used to compare surviving CFU on the coupons at each time point; data show numbers of CFU per coupon or percent survival and are presented as arithmetic means and interquartile ranges. One-way ANOVA with Welch's correction followed by linear contrasts with Bonferroni adjustment was used to compare biofilm and planktonic cell percentages of survival at each concentration and time point. Data are presented as arithmetic means and ranges. For all tests, a two-sided α level set at 0.05 determined significance. Copper concentrations measured by ICP-MS of droplets on all steel coupons (0 to 75 min) are presented as arithmetic means ± standard deviations.
RESULTS
Copper sensitivity of A. baumannii strains in liquid culture.
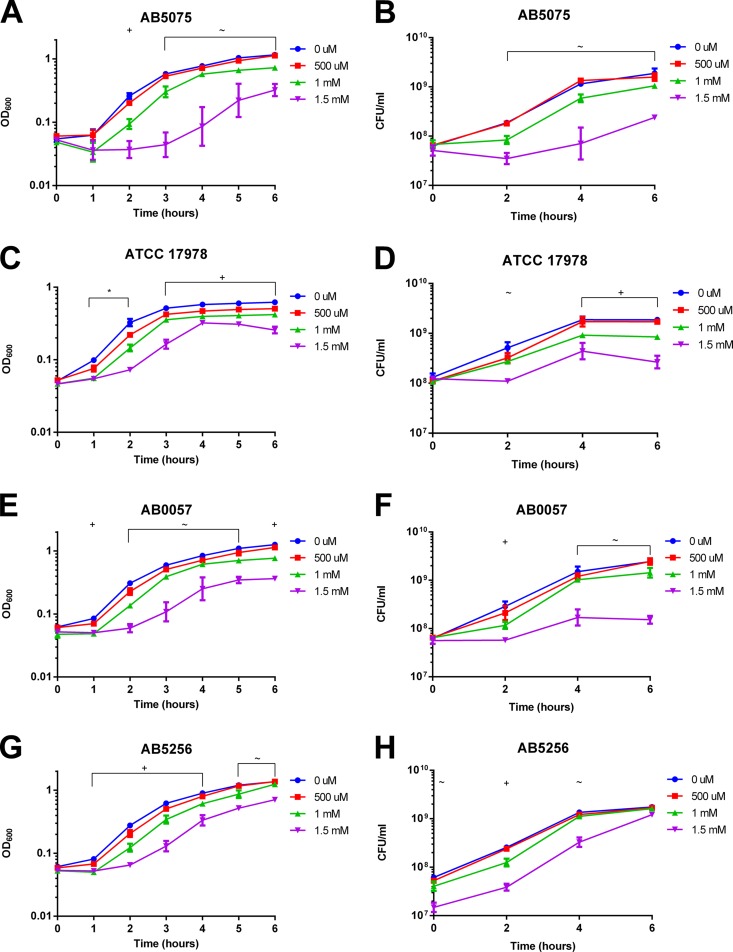
Few published studies have investigated copper resistance levels in A. baumannii, and most prior studies examined environmental isolates of Acinetobacter spp. (29–33). A single study examined clinical isolates of A. baumannii, and it found that MICs for copper in rich medium were in the millimolar range (16). Thus, we first sought to determine the resistance levels of various A. baumannii isolates in minimal medium for a panel of recent clinical and laboratory strains (Table 1). We supplemented liquid cultures of A. baumannii with 100 μM to 1.5 mM CuSO4 and measured bacterial growth (Fig. 1). We found that the A. baumannii isolates grouped into the following two levels of copper resistance: highly resistant and sensitive. The more resistant A. baumannii strains (AB5075, ATCC 17978, AB0057, and AB5256) grew similarly to control cultures in up to 500 μM CuSO4 (Fig. 1A to H). For these strains, 1 mM copper minimally inhibited growth, while 1.5 mM copper significantly delayed growth. However, no cell death was observed, and each of these strains was able to grow in 1.5 mM CuSO4 following a 1- to 3-h delay. Unfortunately, we were unable to test the effects of higher copper concentrations in this assay, since the addition of higher concentrations of copper resulted in visible precipitation of copper from the solution.
FIG 1.
Copper sensitivity of A. baumannii strains in liquid culture. Strains were grown with shaking at 37°C for 6 h in M9 medium supplemented with the indicated concentrations of CuSO4. Bacterial growth was measured by OD600 measurement (A, C, E, G, I, K, and M) and colony enumeration (CFU per milliliter) (B, D, F, H, J, L, and N). The data are presented as geometric means and SEM for three biologically independent experiments. E. coli K-12 MG1655 was included as a positive control (M and N). At each time point, growth in the presence of each copper concentration was compared to that of the 0 μM control by two-way ANOVA with Tukey's adjustment for multiple comparisons. The symbols above the graphs indicate which copper treatments were statistically significantly different (P < 0.05) from the 0 μM control, as follows: ∼, growth in 1.5 mM CuSO4 was different from the control; +, growth in 1 mM and 1.5 mM CuSO4 was different from the control; *, growth in 500 μM, 1 mM, and 1.5 mM CuSO4 was different from the control; #, growth in 250 μM, 500 μM, 1 mM, and 1.5 mM CuSO4 was different from the control; and ∧, growth in 100 μM, 250 μM, 500 μM, 1 mM, and 1.5 mM CuSO4 was different from the control.
The panel of tested strains included two A. baumannii isolates that were drastically more copper sensitive than the others: AB5711 and AB4857 (Fig. 1I to L). These strains were able to grow normally in 100 μM copper, but growth was delayed by as little as 250 μM copper. Furthermore, these strains were partially killed by copper concentrations of 500 μM or higher. A common lab strain of E. coli, K-12 substrain MG1655, was tested as a positive control for copper sensitivity (Fig. 1M and N); K-12-derived strains of E. coli have been shown to be inhibited significantly by 250 μM CuSO4 in modified M9 broth as well as on copper coupons (12, 34). Overall, we observed that the A. baumannii strains tested had various levels of copper resistance; some strains were significantly affected by low micromolar concentrations of copper, while others continued to grow in media with millimolar concentrations of copper.
Survival of A. baumannii isolates on metal surfaces.
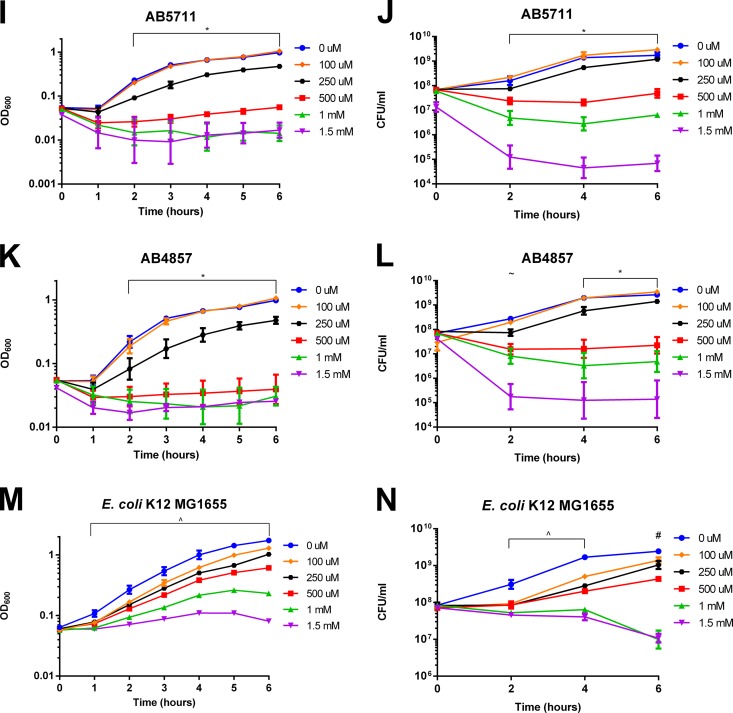
Given the recent deployment of copper as an environmental decontamination strategy (35), we additionally investigated the survival of A. baumannii isolates on metal surfaces. First, we assessed the effects of various metals on survival of strain AB5075. We chose to focus on this strain because it is a recent clinical isolate that is both highly copper resistant (Fig. 1A and B) and virulent in animal models of lung and wound infections (36, 37). To test survival on copper-containing surfaces, coupons of copper, cartridge brass, and stainless steel (negative control) were inoculated with AB5075 cells grown to log phase (2.5 h), and bacterial survival was determined at 15-min increments. As expected, AB5075 showed no loss of viability on stainless steel over the 75-min test period (Fig. 2A). Conversely, AB5075 was rapidly killed on the copper surface; the number of CFU dropped approximately 6 log and reached the limit of detection (3 CFU per coupon) in 30 min. On cartridge brass, which is approximately 70% copper, AB5075 was killed more slowly, but the limit of detection was still reached within 60 min.
FIG 2.
Survival of A. baumannii isolates on metal surfaces. (A) Log-phase (2.5 h) bacterial cultures of AB5075 were spotted onto stainless steel 304, copper 110 H02 (99.9% copper), and cartridge brass C260 (∼70% copper) coupons. Bacteria were recovered after the indicated times and plated for enumeration of surviving CFU. The data are presented as numbers of CFU per coupon for five biologically independent experiments; medians and interquartile ranges are shown. The dashed line indicates the limit of detection (3 CFU/coupon). At each time point, numbers of CFU per milliliter were compared among all three metals by the Kruskal-Wallis test with Dunn's correction for multiple comparisons. The symbols indicate statistically significant comparisons (P < 0.05), as follows: +, brass and copper different from steel; and *, copper different from steel. (B to D) Similar to the experiments in panel A, droplets of AB5075 were spotted onto steel, brass, and copper coupons. Bacteria were taken from log-phase (2.5 h), early-stationary-phase (6 h), and stationary-phase (24 h) cultures. Due to differences in the concentration of cells over the time course, data are presented as percent survival compared to that of the inoculum; open symbols indicate values at or below the limit of detection for all replicates. Data represent five biologically independent experiments; medians and interquartile ranges are shown. *, log- and stationary-phase cultures were significantly different from each other (P < 0.05) at the indicated time point. (E and F) Early-stationary-phase (6 h) cultures of each indicated strain were spotted onto steel and brass coupons. The results for the panel of strains were separated into two graphs for clarity, with AB5075 shown in both panels for comparison. The data are presented as numbers of CFU per coupon for 3 to 5 biologically independent experiments; medians and ranges are shown.
Numerous studies have suggested that bacterial stress responses are affected by the growth phase, including in A. baumannii (38). Thus, we next investigated whether the growth phase of the culture affected survival on copper-containing surfaces. This was accomplished by testing temporal samples from an AB5075 culture, i.e., samples taken during log phase (2.5 h), early stationary phase (6 h), and stationary phase (24 h). As in Fig. 2A, stainless steel caused no change in viability; this was true regardless of the growth phase of the cells (Fig. 2B). However, copper resistance positively correlated with the age of the culture on both brass and copper coupons; AB5075 was most copper sensitive in log phase and most resistant in stationary phase (Fig. 2C and D). Indeed, the percentages of survival of stationary- and log-phase cells on brass and copper were significantly different from each other at 15, 30, and 45 min (P < 0.05).
Since the panel of A. baumannii isolates showed various levels of copper resistance in the liquid culture assay, we also individually assessed the full panel by using the coupon assay. In order to best discern differences among strains, we chose to focus on early-stationary-phase cultures and cartridge brass coupons; under these conditions, AB5075 died evenly and gradually, but its level still reached the limit of detection within 75 min. Stainless steel coupons and the E. coli strain were included as controls. While the differences were not statistically significant, ATCC 17978, AB5256, and AB0057 appeared to survive better on the brass surface than did AB5075, AB5711, AB4857, and E. coli K-12 MG1655 (Fig. 2E and F). Despite these differences in sensitivity, all strains were reduced approximately 7 log, to the limit of detection, during the 75-min test period. This finding highlights the ability of copper-containing surfaces to effectively eliminate A. baumannii.
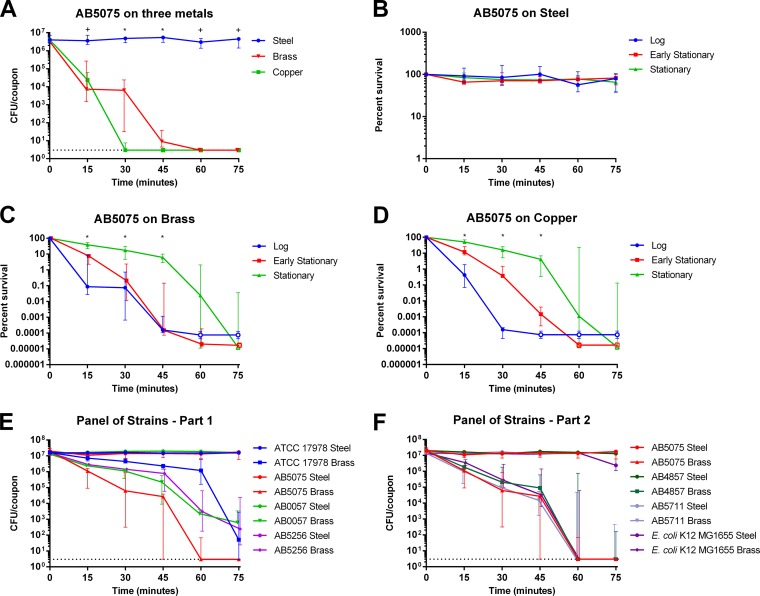
Because we saw very limited killing in the liquid culture experiments (Fig. 1A to L) but saw dramatic killing of A. baumannii on the brass and copper surfaces, we hypothesized that the concentration of copper experienced by bacteria in the droplets of medium on the metal coupons must be much higher than the maximum copper concentration (1.5 mM) achievable in growth assays. To test this hypothesis, we used ICP-MS to measure the concentration of copper that accumulated in the droplets (Fig. 3). As a control, we also assessed the copper content of the M9 growth medium. The M9 growth medium routinely contained approximately 1 to 2 μM copper. The droplets exposed to stainless steel accumulated relatively low concentrations of copper, which were insufficient to kill any of the bacterial strains (131 ± 86 μM). The observed increase in copper ions following exposure to steel is presumably due to trace metal contamination in the stainless steel sheet preparation; the company specification sheet listed 0.54% Cu for the product, and the presence of copper in the steel coupons was confirmed by ICP-MS analysis of a steel sample treated with concentrated nitric acid (data not shown). While the concentrations of copper from steel were not sufficient to kill A. baumannii, the concentrations of copper in the droplets incubated on brass or copper surfaces were extremely high. The copper concentration in the droplets incubated on brass temporally increased to as high as 2 mM over the 75-min test period. Similarly, the copper concentration in the droplets incubated on pure copper temporally increased from >2 mM at 15 min to approximately 8 mM at 75 min. Thus, on these two metal surfaces, substantial concentrations of copper are released into the liquid environment, likely explaining why the bacteria were killed so rapidly under these conditions.
FIG 3.
Concentrations of copper that accumulated in droplets on metal surfaces. Five-microliter droplets of M9 minimal medium were placed onto stainless steel, cartridge brass (∼70% Cu), or 99.9% copper coupons for the indicated times. Droplets were recovered by washing an additional 100 μl of M9 medium across the surface. Copper content was then measured by ICP-MS. Six biologically independent replicates were completed with six individual preparations of medium. Values for each replicate are shown as individual points, and the geometric means are represented by bars.
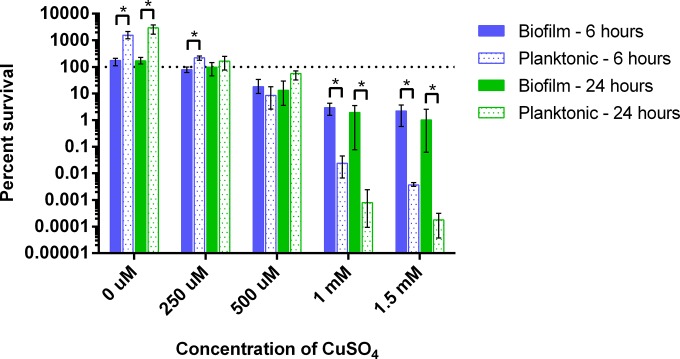
Killing of Acinetobacter biofilms by copper.
Since A. baumannii forms biofilms on biotic and abiotic surfaces (39), and because bacteria found in biofilms are often more resistant to environmental stress (40, 41), we next sought to determine whether copper was efficacious against A. baumannii cells found within a biofilm. We tested the effects of copper on preformed biofilms of AB5711 because this strain formed substantial biofilms in vitro and was relatively sensitive to the copper concentration that is achievable in M9 medium (Fig. 1I and J). We observed dose-dependent killing of both planktonic and biofilm cells (Fig. 4). Bacteria within biofilms were reduced approximately 2 log over the 24-h period when the biofilms were treated with 1 or 1.5 mM copper. The planktonic cells were significantly more sensitive and were reduced approximately 5.5 log over the 24-h test period. Therefore, although biofilms did show increased copper resistance relative to that of planktonic cells, copper was still able to kill AB5711 within a biofilm.
FIG 4.
Killing of Acinetobacter biofilms and planktonic cells by copper. AB5711 cultures were incubated statically overnight at 37°C in a 24-well tissue culture-treated plate to establish biofilms. The LB broth was removed and replaced with M9 medium containing the indicated concentration of CuSO4. After 0, 6, or 24 h of additional incubation, bacteria from both the supernatant (planktonic) and the biofilm were serially diluted and plated for enumeration of CFU. Percent survival was calculated as the percentage of the bacteria present at time zero under each condition. Data are presented as mean and ranges for four biologically independent replicates. One-way ANOVA with Welch's correction followed by linear contrasts with Bonferroni adjustment was used to compare biofilm and planktonic cell percentages of survival at each concentration and time point. Statistically significant comparisons (P < 0.01) are indicated with asterisks.
Copper tolerance response.
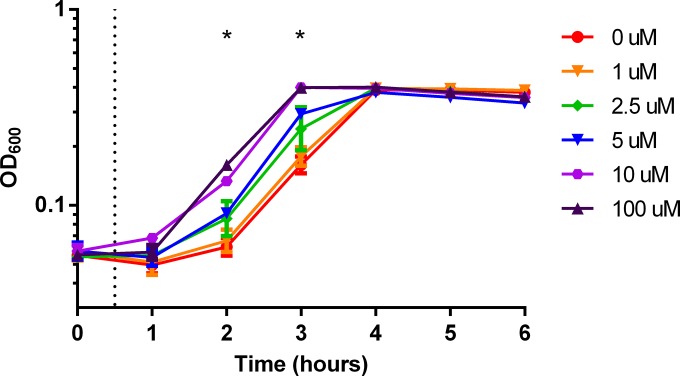
In the liquid assay, we observed that growth of strain AB5075 was delayed by the addition of 1.5 mM copper but later continued at an ample rate (Fig. 1A). This pattern suggested that AB5075 was able to adapt to the copper stress. Tolerance responses following priming have been shown for other environmental stresses, e.g., the acid tolerance response (42). Therefore, we tested if copper sensitivity was altered by preexposure of the bacteria to a subinhibitory concentration of copper. To this end, we preexposed AB5075 to increasing subinhibitory concentrations of copper (1 to 100 μM) before adding copper to a final inhibitory concentration of 1.5 mM. Preexposure of cells to very small amounts of copper allowed the bacteria to initiate growth in the presence of 1.5 mM copper much sooner than if they were not preexposed (Fig. 5; Table 3). Furthermore, the pattern showed a classic dose-response relationship. As little as 2.5 μM copper significantly reduced the lag in growth seen in the presence of 1.5 mM copper (P < 0.05), and preexposure to higher concentrations of copper further decreased the lag in growth; this trend was represented by a decrease in P values (Table 3). However, priming reached the maximum effect at approximately 10 μM during the preexposure stage. Overall, preexposure of AB5075 cells to very low concentrations of copper shortened the time required to initiate growth with a high concentration of copper, which further suggests that AB5075 is able to mount a tolerance response to copper stress.
FIG 5.
Preexposure to a low concentration of copper induces a copper tolerance response. AB5075 was subcultured at an OD600 of 0.05 in M9 medium containing the indicated concentration of CuSO4. After 30 min of incubation (dashed line), copper was added to a final concentration of 1.5 mM. Culture density was then measured by determining the OD600 at the indicated time points. The data are presented as geometric means and standard errors of the means for three biologically independent experiments. For each time point, growth under each priming condition was compared by two-way ANOVA with Tukey's adjustment for multiple comparisons. Significance (P < 0.05) was found at hours 2 and 3; P values are shown in Table 2.
TABLE 3.
Adjusted P values for AB5075 growth following copper priminga
| Copper priming concn (μM) comparison |
P value |
|
|---|---|---|
| 2 h | 3 h | |
| 0 vs 1 | 0.9884 | 0.9481 |
| 0 vs 2.5 | 0.0452 | 0.0038 |
| 0 vs 5 | 0.0096 | <0.0001 |
| 0 vs 10 | <0.0001 | <0.0001 |
| 0 vs 100 | <0.0001 | <0.0001 |
| 1 vs 2.5 | 0.1961 | 0.0527 |
| 1 vs 5 | 0.0571 | 0.0004 |
| 1 vs 10 | <0.0001 | <0.0001 |
| 1 vs 100 | <0.0001 | <0.0001 |
| 2.5 vs 5 | 0.9943 | 0.6264 |
| 2.5 vs 10 | 0.0023 | 0.0006 |
| 2.5 vs 100 | <0.0001 | 0.0006 |
| 5 vs 10 | 0.0131 | 0.0737 |
| 5 vs 100 | <0.0001 | 0.0784 |
| 10 vs 100 | 0.5352 | >0.9999 |
Differences in growth were evaluated among priming conditions for each time point by using two-way ANOVA with Tukey's correction for multiple comparisons. No significance was found at 0, 4, 5, or 6 h. P values of <0.05 are highlighted in bold.
Putative copper-related genes.
Copper resistance in bacteria is mediated by a variety of copper homeostasis mechanisms, mainly copper detoxification and efflux (19). The ability of AB5075 to adapt to growth in a high concentration of copper suggests that this bacterium likely encodes copper resistance mechanisms that are induced by copper exposure. To identify putative copper resistance mechanisms in AB5075, we used the amino acid sequences encoded by known copper-related genes from E. coli and P. aeruginosa to search the AB5075 genome (19, 25); many of the identified targets were already annotated with copper-related functions. The levels of homology of each of the identified AB5075 proteins to E. coli K-12 MG1655 and P. aeruginosa PAO1 proteins are presented in Table 4. Given the closer relationship of the two, most A. baumannii proteins were more similar to those of P. aeruginosa than to those of E. coli. Overall, the level of homology to copper-related proteins in both organisms was high, suggesting that these copper resistance mechanisms are well conserved.
In total, 23 putative proteins of AB5075 were analyzed via comparison to E. coli and P. aeruginosa (Table 4). The most well-known copper resistance systems were present and well conserved in AB5075. The CopA (copper ATPase), CueO (copper oxidase), and CueR (regulator) system of detoxification was complete (ActP2/CopA1, CopA/CueO, and CueR). Additionally, AB5075 appeared to encode a second copper ATPase (ActP2/CopA1) and copper oxidase (PcoA). The copper-sensing two-component system, CopRS/CusRS, was also present and highly conserved. However, the presumed target of CopRS/CusRS regulation, the RND family efflux pump encoded by czcABCD, appeared to be less well conserved. Homologues of CzcA, CzcB, and CzcD were clearly present, but the CzcC protein did not match any proteins in E. coli or P. aeruginosa. However, a distally located gene, ABUW_1334, showed clear homology to the genes encoding CusC/CzcC of E. coli and an outer membrane efflux protein of P. aeruginosa. In addition, many homologues of copper-related genes with less-well-characterized functions were also conserved in AB5075: PcoB, CopB, and CopC.
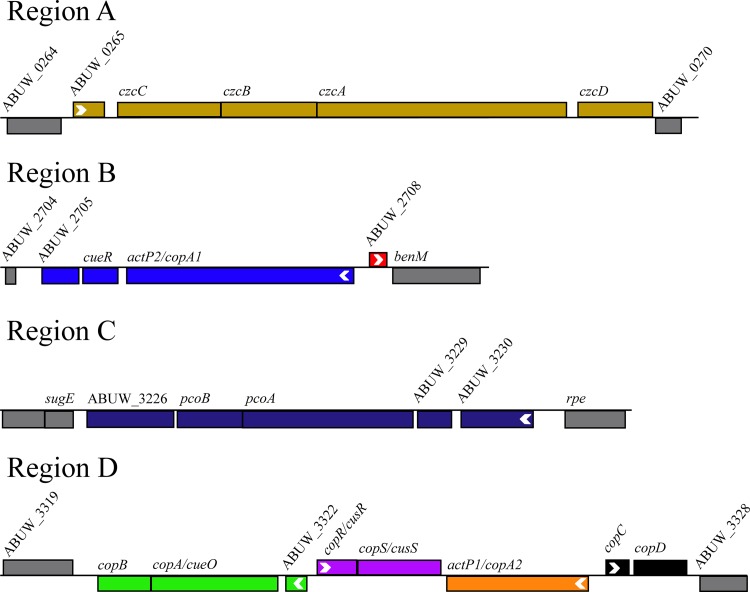
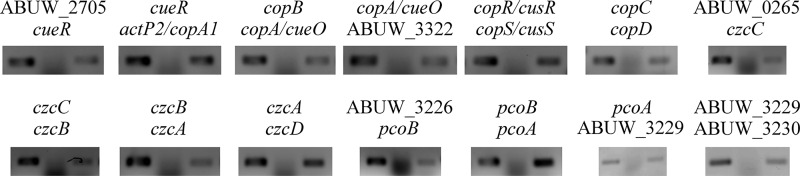
We noted that the putative copper-related proteins were encoded by genes that were located in only four areas of the AB5075 chromosome; we arbitrarily named these regions A through D (Fig. 6). The genes were arranged as 6 apparent operons with 2 adjacent, monocistronic genes. The organization and operonic structure were confirmed experimentally using junctional PCR analysis of cDNA (Fig. 7).
FIG 6.
Genomic organization of putative copper-related genes. Putative copper-related genes that were identified in AB5075 are located in four regions of the chromosome, arbitrarily named regions A, B, C, and D. Colors represent contiguous mRNA molecules and match the colors used in Fig. 8. The direction of transcription is indicated by a white arrowhead at the N terminus of each individual gene or initial gene of an operon. Locus identification numbers are given for genes without annotated names. The annotation of five genes does not match the names used for other organisms, and thus the new gene names are included following the original name, as follows: actP2/copA1, actP1/copA2, copA/cueO, copR/cusR, and copS/cusS. This image was adapted from the AB5075-UW transposon browser (http://tools.uwgenomics.org/tn_mutants).
FIG 7.
Junctional PCR confirms the operonic structure of putative copper-related genes. To detect each of the gene junctions between genes of a putative operon, qualitative reverse transcription-PCR was conducted. PCRs were performed using genomic DNA, RNA, or cDNA as the template. Each reaction mixture was incubated for 5 min at 95°C, followed by 25 cycles of denaturation at 95°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 30 s. PCR products were separated in 1% agarose gels and visualized by staining with ethidium bromide. Each PCR was performed three times using biologically independent cDNA samples.
To determine if the other A. baumannii isolates in the panel also carry the identified putative copper-related genes, we next used the amino acid sequences of AB5075 proteins to search the other genomes. Genomes were searchable by BLASTp searches for all strains except AB4857. In most of the A. baumannii strains, we identified all of the putative copper-related genes identified in AB5075. However, AB5711 did not appear to carry all of the genes found in the other strains. AB5711 lacked all of the genes found in region D in AB5075, i.e., copB, copA/cueO, ABUW_3322, copR/cusR, copS, actP1/copA2, copC, and copD.
Expression of putative copper-related genes.
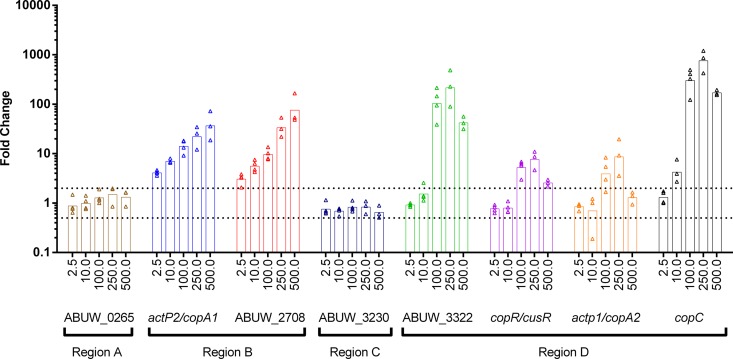
Finally, to determine if expression of the putative copper-related genes in AB5075 was regulated by copper, we used qRT-PCR to measure changes in transcript levels following 30 min of exposure to subinhibitory concentrations of copper (Fig. 8). For genes located in regions B and D, we saw significant increases (≥2-fold) in expression in response to copper. In many cases, a dose-dependent increase in expression was observed as the copper concentration increased. Note that the observed expression patterns were most similar for genes located in close proximity on the chromosome. For example, genes located in region B were upregulated by as little as 2.5 μM CuSO4 and showed a steady increase in expression as more copper was added. Conversely, all four gene sets in region D required addition of ≥10 μM CuSO4 to induce increased expression, which then peaked at 250 μM CuSO4. Expression of the operons in regions A and C did not increase in response to copper. Overall, we observed that many of the putative copper-related genes were induced by exposure to copper, suggesting that the corresponding proteins may indeed have functions related to copper resistance.
FIG 8.
Expression of putative copper-related genes in response to copper exposure. Expression of putative copper-related genes was measured by qRT-PCR, using primers specific to the initial gene of each operon. RNA was isolated from AB5075 cells exposed for 30 min to the indicated increasing concentrations (micromolar) of CuSO4. Generated cDNA was used as template DNA for qRT-PCR. Relative gene expression was calculated by the 2−ΔΔCT method, using the 16S rRNA gene as the internal reference. Three or four biologically independent replicates of each experiment were performed. Values for each replicate are shown as individual points, and the geometric means are represented by bars. Dashed lines represent significant (2-fold) up- and downregulation.
DISCUSSION
A. baumannii causes multiple types of severe nosocomial infections, for which treatment options are limited by antibiotic resistance found in MDR, XDR, and PDR strains. The intrinsic antimicrobial properties of copper give it unique potential for use in novel therapeutics. Thus, we sought to perform an initial characterization of copper resistance in recent clinical isolates of A. baumannii. In this study, we demonstrated that copper sensitivities varied among the A. baumannii isolates (Fig. 1). However, all of the tested A. baumannii strains were killed rapidly on copper-containing surfaces (Fig. 2). This was likely due in part to the high concentration of copper ions that accumulated in the droplets of medium (Fig. 3). While A. baumannii found in a biofilm was more resistant to copper toxicity than planktonic cells, biofilm cells were also susceptible to copper (Fig. 4). Additionally, we identified many putative proteins that may be involved in copper resistance and showed that expression of many of the genes that encode these proteins is upregulated following exposure to copper (Table 4; Fig. 8). Increased expression of these genes in response to copper likely mediates improved adaptation of AB5075 to copper shock when the culture is pretreated with subinhibitory concentrations of copper (Fig. 5). To our knowledge, this is the first comprehensive study characterizing copper resistance in A. baumannii.
Due to the increase in antibiotic resistance seen across an extensive array of bacterial pathogens, there is a pressing need to identify new means to treat infections caused by these organisms. The potential to use copper as a novel therapeutic to treat infections has led to a number of studies that have sought to determine the sensitivity of various pathogens to this metal. Unfortunately, it becomes very difficult to compare these experiments to one another because the chosen growth medium can drastically affect the amount of copper required to inhibit bacterial growth. The MIC of soluble copper ions is dramatically increased in rich medium; this is presumably due to binding of copper ions by proteins in the medium, which effectively reduces the concentration of free copper ions able to impose damage. For example, the MIC of copper for P. aeruginosa PAO1 increases from 100 μM in minimal medium to 6 mM in rich medium (14). While we recognize the potential limitations of comparing our results to those of experiments with different protocols, we note that the concentration of copper required to inhibit bacterial growth of A. baumannii appears to be strikingly different from that for other pathogens. Similar experiments conducted in minimal media found that the MIC of copper is 500 μM or less for Mycobacterium tuberculosis strain H37Rv, E. coli strain BW25113, P. aeruginosa strain PAO1, and Salmonella enterica serovar Typhimurium strain SL1344 (12–15). Conversely, we found that growth of most of the A. baumannii isolates was only slightly delayed by 1.5 mM CuSO4 in minimal medium, and we were unable to kill these strains with soluble copper. Thus, it appears that A. baumannii may be more copper resistant than other bacterial pathogens that have been tested. The high levels of copper resistance may be related to the fact that the progenitor A. baumannii strain may have been a soil organism, as the levels of copper in soil may be significantly higher than those typically encountered by other bacterial species (43). Since copper resistance may also play a role in survival in the human host or hospital environment, the genes were retained. Additionally, A. baumannii is an exceptional example of a pathogen that rapidly obtains genetic information from other organisms via horizontal gene transfer; indeed, copper resistance gene clusters have been identified in mobile genetic elements in A. baumannii, alongside its many antibiotic resistance genes (44).
Ultimately, the levels of copper resistance are no doubt directly influenced by the repertoire of copper-related genes carried by the organism. Thus, we identified a large set of putative copper-related genes in strain AB5075 (Table 4). While the functions of the putative proteins encoded by these genes remain to be elucidated for A. baumannii, the functions of some of the orthologous proteins have been well characterized for E. coli. For example, CopA, CueO, and CueR make up a canonical copper detoxification system, consisting of a copper ATPase to efflux copper ions to the periplasm, a periplasmic copper oxidase to detoxify the copper ions, and a transcriptional regulator to activate expression of the system in response to cytoplasmic copper ions. While homologues of all three of these proteins are present in AB5075, they are not encoded together (ActP2/CopA1, CopA/CueO, and CueR). Additionally, AB5075 appears to encode a second copper ATPase and copper oxidase (AcP1/CopA2 and PcoA, respectively) in its genome. In addition to the detoxification system, homologues of a copper efflux pump (CzcCBAD) and a potential regulator of this system (CopRS/CusRS) were also identified in AB5075. We note that while expression of the two-component system encoded by copRS/cusRS was increased upon copper exposure, expression of the RND family efflux pump encoded by czcCBAD did not increase (Fig. 8). Thus, it is possible that the pump is regulated differently in A. baumannii than in E. coli, that the pump is constitutively active, or that this pump is not actually used for copper. AB5075 encodes many putative efflux pumps, and thus czcCBAD may have been misannotated as a copper pump. Indeed, future functional analysis will be required to conclusively determine the function of any of the putative proteins encoded by these copper-related genes.
Two of the A. baumannii clinical isolates tested, AB4857 and AB5711, were significantly more sensitive to copper than the rest of the strains of the panel (Fig. 1). Unfortunately, the genome of AB4857 is available only as a draft and is not searchable by BLASTp for comparison. However, AB5711 lacks a large group of copper-related genes that are present in AB5075 region D. These include copB, copA/cueO, ABUW_3322, copR/cusR, copS, actP1/copA2, copC, and copD. Thus, since the level of copper resistance of AB5711 is significantly lower than that of the other A. baumannii strains, it is tempting to speculate that the proteins encoded by the genes carried in region D are crucial to the higher level of copper resistance seen in the other A. baumannii isolates. Future mutational analysis will be required to define the role of each of these proteins in copper resistance.
Our analysis showed copper-dependent regulation of genes found in regions B and D of the chromosome; genes in region B were exquisitely sensitive to the copper concentration (Fig. 8). Indeed, expression from the two divergently transcribed genes/operons in region B increased significantly in response to the addition of as little as 2.5 μM copper sulfate, and it continued to increase with additional copper. Conversely, increased expression from region D, which carries adjacent pairs of divergently transcribed genes/operons, required the addition of ≥10 μM copper. We note that the threshold for upregulation of copper-related genes fits well with the copper tolerance response we observed (Fig. 5). Indeed, pretreatment with as little as 2.5 μM CuSO4 significantly reduced the lag in growth seen with extreme copper shock, and pretreatment with up to 10 μM CuSO4 further shortened the lag in growth. Thus, perhaps the increased expression of copper-related genes found in regions B and D is responsible for the identified copper tolerance response. Future analysis of strains containing mutations in genes located in these regions will help to define the importance of these genes to the adaptation response.
In addition to the possible utilization of copper in novel therapeutics, copper is also being tested for use in environmental decontamination. Recent clinical trials showed that covering high-touch surfaces in hospital rooms with copper can significantly decrease the bacterial burden and can reduce the number of hospital-acquired infections by more than 50% (35, 45–48). Because A. baumannii fomite-associated hospital outbreaks have been documented, specifically from a contaminated hospital bed and burn theater, evidence that copper effectively eliminates environmental A. baumannii is important (49). Indeed, we found that all tested strains of A. baumannii were reduced to near or below the limit of detection in only 60 to 75 min when exposed to brass and copper surfaces (Fig. 2). A. baumannii clinical isolates have previously been shown to be sensitive to copper-containing metal coupons (50–52). The data cannot be compared directly due to protocol differences, but similar trends were observed, with full survival on control surfaces, significant reductions on copper-containing surfaces, and more efficient killing by alloys with a higher copper content. In this study, we additionally showed the growth-phase-dependent effects on survival on copper-containing surfaces and the variation in copper sensitivity among isolates of A. baumannii. Overall, these results demonstrate that copper has great potential to reduce A. baumannii in the environment.
Because we saw dramatic killing of A. baumannii on the copper coupons but only limited killing of some strains in the liquid assay, we used ICP-MS to measure the concentration of copper that accumulated over time in the 5-μl droplets of medium on the metal surface. On the copper coupons, we saw a rapid accumulation of millimolar concentrations of copper ions, which aids in explaining the dramatic killing of bacteria under these conditions. A similar experiment was performed previously, in which millimolar concentrations of copper were reported to accumulate in a 25-μl droplet of Tris-Cl on a copper coupon (53). However, the rates at which ions were taken up into the droplets were very different between the experiments. This is likely due to the difference in droplet size between the two experiments; this minor protocol difference likely dramatically affects the resulting data.
Previously, biofilm formation by environmental Acinetobacter spp. was shown to be inhibited by increasing concentrations of copper (31). In the same vein, we found that copper was effective at killing modern clinical isolates of A. baumannii found within a preformed biofilm structure. Not surprisingly, the biofilm was partially protective, and cells in the biofilm were significantly more resilient than planktonic cells; however, even the biofilm was effectively reduced by high concentrations of copper (Fig. 4). We speculate that A. baumannii biofilms may be able to be eliminated completely by copper concentrations higher than those achievable in M9 medium. Our results demonstrate that copper-based therapies have the potential to be effective treatments, even against A. baumannii strains that have formed biofilms.
In summary, due to its intrinsic antimicrobial properties, copper has the potential to be used in new therapeutics for A. baumannii infection. Furthermore, if particular protein functions can be identified that are critical for copper resistance, novel antibiotics may be able to be designed to target those functions as a means to enhance the inherent antibacterial properties of copper. Such a strategy may make a very limited amount of copper very effective against infection. The human immune system utilizes copper in various ways to fight infection, and concentrations upwards of 500 μM have been measured in macrophage phagosomes (54–56). Furthermore, methods of increasing the concentration of copper in the body to enhance its antimicrobial effects are being investigated (57–60). Additional targeting and blocking of copper resistance systems may make these strategies increasingly efficacious; indeed, many copper resistance systems have been shown to be critical for full virulence in various pathogens (61). We hope that future identification of mechanisms that are critical for copper resistance and virulence in A. baumannii and other bacterial pathogens will open doors for development of novel copper-based therapeutics.
ACKNOWLEDGMENTS
C.L.W., J.J.G., S.L.J.M., D.V.Z., and D.S.M. conceived and designed the experiments. C.L.W. and H.M.N. performed the experiments. D.V.Z. contributed reagents/materials. C.L.W. and D.S.M. wrote the paper.
We thank Cara Olsen for help with statistics. We thank G. W. Liechti and A. T. Maurelli for providing E. coli K-12 MG1655. C.L.W. is grateful to the members of the Merrell and Zurawski labs for their microbiology expertise.
The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense, the Uniformed Services University of the Health Sciences, the Department of the Army, or any other agency of the U.S. Government. This material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication.
Funding Statement
Research in the laboratory of D.S.M. is supported by funding from the National Institutes of Health (NIH) and the Department of Defense (DoD). S.L.J.M. thanks the National Science Foundation (NSF) (grant CHE1306208). Research in the laboratory of D.V.Z. is supported by grants from the Military Infectious Diseases Research Program.
REFERENCES
- 1.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metan G, Sariguzel F, Sumerkan B. 2009. Factors influencing survival in patients with multi-drug-resistant Acinetobacter bacteraemia. Eur J Intern Med 20:540–544. doi: 10.1016/j.ejim.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Prata-Rocha ML, Gontijo-Filho PP, Melo GB. 2012. Factors influencing survival in patients with multidrug-resistant Acinetobacter baumannii infection. Braz J Infect Dis 16:237–241. [PubMed] [Google Scholar]
- 4.Tabah A, Koulenti D, Laupland K, Misset B, Valles J, Bruzzi de Carvalho F, Paiva JA, Cakar N, Ma X, Eggimann P, Antonelli M, Bonten MJ, Csomos A, Krueger WA, Mikstacki A, Lipman J, Depuydt P, Vesin A, Garrouste-Orgeas M, Zahar JR, Blot S, Carlet J, Brun-Buisson C, Martin C, Rello J, Dimopoulos G, Timsit JF. 2012. Characteristics and determinants of outcome of hospital-acquired bloodstream infections in intensive care units: the EUROBACT International Cohort Study. Intensive Care Med 38:1930–1945. doi: 10.1007/s00134-012-2695-9. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. CDC, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf#page=59. [Google Scholar]
- 6.Timsit JF, Soubirou JF, Voiriot G, Chemam S, Neuville M, Mourvillier B, Sonneville R, Mariotte E, Bouadma L, Wolff M. 2014. Treatment of bloodstream infections in ICUs. BMC Infect Dis 14:489. doi: 10.1186/1471-2334-14-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornejo-Juarez P, Vilar-Compte D, Perez-Jimenez C, Namendys-Silva SA, Sandoval-Hernandez S, Volkow-Fernandez P. 2015. The impact of hospital-acquired infections with multidrug-resistant bacteria in an oncology intensive care unit. Int J Infect Dis 31:31–34. doi: 10.1016/j.ijid.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Poirel L, Bonnin RA, Nordmann P. 2011. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 63:1061–1067. doi: 10.1002/iub.532. [DOI] [PubMed] [Google Scholar]
- 9.Liochev SI, Fridovich I. 2002. The Haber-Weiss cycle—70 years later: an alternative view. Redox Rep 7:55–57. doi: 10.1179/135100002125000190. [DOI] [PubMed] [Google Scholar]
- 10.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warnes SL, Caves V, Keevil CW. 2012. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ Microbiol 14:1730–1743. doi: 10.1111/j.1462-2920.2011.02677.x. [DOI] [PubMed] [Google Scholar]
- 12.Gudipaty SA, Larsen AS, Rensing C, McEvoy MM. 2012. Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol Lett 330:30–37. doi: 10.1111/j.1574-6968.2012.02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osman D, Waldron KJ, Denton H, Taylor CM, Grant AJ, Mastroeni P, Robinson NJ, Cavet JS. 2010. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J Biol Chem 285:25259–25268. doi: 10.1074/jbc.M110.145953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, Parsek MR. 2006. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J Bacteriol 188:7242–7256. doi: 10.1128/JB.00837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. 2010. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol Microbiol 77:1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deshpande LM, Kapadnis BP, Chopade BA. 1993. Metal resistance in Acinetobacter and its relation to beta-lactamase production. Biometals 6:55–59. [DOI] [PubMed] [Google Scholar]
- 17.Franke S, Grass G, Rensing C, Nies DH. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol 185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Achard ME, Tree JJ, Holden JA, Simpfendorfer KR, Wijburg OL, Strugnell RA, Schembri MA, Sweet MJ, Jennings MP, McEwan AG. 2010. The multi-copper-ion oxidase CueO of Salmonella enterica serovar Typhimurium is required for systemic virulence. Infect Immun 78:2312–2319. doi: 10.1128/IAI.01208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arguello JM, Raimunda D, Padilla-Benavides T. 2013. Mechanisms of copper homeostasis in bacteria. Front Cell Infect Microbiol 3:73. doi: 10.3389/fcimb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grass G, Rensing C. 2001. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys Res Commun 286:902–908. doi: 10.1006/bbrc.2001.5474. [DOI] [PubMed] [Google Scholar]
- 21.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci U S A 97:652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Outten FW, Outten CE, Hale J, O'Halloran TV. 2000. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J Biol Chem 275:31024–31029. doi: 10.1074/jbc.M006508200. [DOI] [PubMed] [Google Scholar]
- 23.Kim EH, Nies DH, McEvoy MM, Rensing C. 2011. Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol 193:2381–2387. doi: 10.1128/JB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SM, Grass G, Rensing C, Barrett SR, Yates CJ, Stoyanov JV, Brown NL. 2002. The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem Biophys Res Commun 295:616–620. doi: 10.1016/S0006-291X(02)00726-X. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez-Montes G, Arguello JM, Valderrama B. 2012. Evolution and diversity of periplasmic proteins involved in copper homeostasis in gamma proteobacteria. BMC Microbiol 12:249. doi: 10.1186/1471-2180-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 27.Schoolnik GK, Voskuil MI, Schnappinger D, Yildiz FH, Meibom K, Dolganov NA, Wilson MA, Chong KH. 2001. Whole genome DNA microarray expression analysis of biofilm development by Vibrio cholerae O1 E1 Tor. Methods Enzymol 336:3–18. doi: 10.1016/S0076-6879(01)36573-4. [DOI] [PubMed] [Google Scholar]
- 28.Gilbreath JJ, West AL, Pich OQ, Carpenter BM, Michel S, Merrell DS. 2012. Fur activates expression of the 2-oxoglutarate oxidoreductase genes (oorDABC) in Helicobacter pylori. J Bacteriol 194:6490–6497. doi: 10.1128/JB.01226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhakephalkar PK, Chopade BA. 1994. High levels of multiple metal resistance and its correlation to antibiotic resistance in environmental isolates of Acinetobacter. Biometals 7:67–74. [DOI] [PubMed] [Google Scholar]
- 30.Huang HI, Shih HY, Lee CM, Yang TC, Lay JJ, Lin YE. 2008. In vitro efficacy of copper and silver ions in eradicating Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Acinetobacter baumannii: implications for on-site disinfection for hospital infection control. Water Res 42:73–80. doi: 10.1016/j.watres.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Mogilnaya OA, Lobova TI, Kargatova TV, Popova LY. 2005. Biofilm formation by bacterial associations under various salinities and copper ion stress. Biofouling 21:247–255. doi: 10.1080/08927010500445848. [DOI] [PubMed] [Google Scholar]
- 32.Shih HY, Lin YE. 2010. Efficacy of copper-silver ionization in controlling biofilm- and plankton-associated waterborne pathogens. Appl Environ Microbiol 76:2032–2035. doi: 10.1128/AEM.02174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yadav KK, Mandal AK, Chakraborty R. 2013. Copper susceptibility in Acinetobacter junii BB1A is related to the production of extracellular polymeric substances. Antonie Van Leeuwenhoek 104:261–269. doi: 10.1007/s10482-013-9946-9. [DOI] [PubMed] [Google Scholar]
- 34.Espirito Santo C, Taudte N, Nies DH, Grass G. 2008. Contribution of copper ion resistance to survival of Escherichia coli on metallic copper surfaces. Appl Environ Microbiol 74:977–986. doi: 10.1128/AEM.01938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt MG, Attaway HH, Sharpe PA, John J Jr, Sepkowitz KA, Morgan A, Fairey SE, Singh S, Steed LL, Cantey JR, Freeman KD, Michels HT, Salgado CD. 2012. Sustained reduction of microbial burden on common hospital surfaces through introduction of copper. J Clin Microbiol 50:2217–2223. doi: 10.1128/JCM.01032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. 2014. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5:e01076-14. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson MG, Black CC, Pavlicek RL, Honnold CL, Wise MC, Alamneh YA, Moon JK, Kessler JL, Si Y, Williams R, Yildirim S, Kirkup BC Jr, Green RK, Hall ER, Palys TJ, Zurawski DV. 2014. Validation of a novel murine wound model of Acinetobacter baumannii infection. Antimicrob Agents Chemother 58:1332–1342. doi: 10.1128/AAC.01944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiester SE, Actis LA. 2013. Stress responses in the opportunistic pathogen Acinetobacter baumannii. Future Microbiol 8:353–365. doi: 10.2217/fmb.12.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longo F, Vuotto C, Donelli G. 2014. Biofilm formation in Acinetobacter baumannii. New Microbiol 37:119–127. [PubMed] [Google Scholar]
- 40.Harrison JJ, Ceri H, Turner RJ. 2007. Multimetal resistance and tolerance in microbial biofilms. Nat Rev Microbiol 5:928–938. doi: 10.1038/nrmicro1774. [DOI] [PubMed] [Google Scholar]
- 41.Olsen I. 2015. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis 34:877–886. doi: 10.1007/s10096-015-2323-z. [DOI] [PubMed] [Google Scholar]
- 42.Merrell DS, Camilli A. 2002. Acid tolerance of gastrointestinal pathogens. Curr Opin Microbiol 5:51–55. doi: 10.1016/S1369-5274(02)00285-0. [DOI] [PubMed] [Google Scholar]
- 43.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie JM, Raoult D, Medigue C, Weissenbach J, Cruveiller S. 2008. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt MG, Attaway HH III, Fairey SE, Steed LL, Michels HT, Salgado CD. 2013. Copper continuously limits the concentration of bacteria resident on bed rails within the intensive care unit. Infect Control Hosp Epidemiol 34:530–533. doi: 10.1086/670224. [DOI] [PubMed] [Google Scholar]
- 46.Salgado CD, Sepkowitz KA, John JF, Cantey JR, Attaway HH, Freeman KD, Sharpe PA, Michels HT, Schmidt MG. 2013. Copper surfaces reduce the rate of healthcare-acquired infections in the intensive care unit. Infect Control Hosp Epidemiol 34:479–486. doi: 10.1086/670207. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt MG, von Dessauer B, Benavente C, Benadof D, Cifuentes P, Elgueta A, Duran C, Navarrete MS. 2016. Copper surfaces are associated with significantly lower concentrations of bacteria on selected surfaces within a pediatric intensive care unit. Am J Infect Control 44:203–209. doi: 10.1016/j.ajic.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 48.von Dessauer B, Navarrete MS, Benadof D, Benavente C, Schmidt MG. 2016. Potential effectiveness of copper surfaces in reducing health care-associated infection rates in a pediatric intensive and intermediate care unit: a nonrandomized controlled trial. Am J Infect Control 44:e133–e139. doi: 10.1016/j.ajic.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 49.Halachev MR, Chan JZ, Constantinidou CI, Cumley N, Bradley C, Smith-Banks M, Oppenheim B, Pallen MJ. 2014. Genomic epidemiology of a protracted hospital outbreak caused by multidrug-resistant Acinetobacter baumannii in Birmingham, England. Genome Med 6:70. doi: 10.1186/s13073-014-0070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eser OK, Ergin A, Hascelik G. 2015. Antimicrobial activity of copper alloys against invasive multidrug-resistant nosocomial pathogens. Curr Microbiol 71:291–295. doi: 10.1007/s00284-015-0840-8. [DOI] [PubMed] [Google Scholar]
- 51.Mehtar S, Wiid I, Todorov SD. 2008. The antimicrobial activity of copper and copper alloys against nosocomial pathogens and Mycobacterium tuberculosis isolated from healthcare facilities in the Western Cape: an in-vitro study. J Hosp Infect 68:45–51. doi: 10.1016/j.jhin.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 52.Souli M, Galani I, Plachouras D, Panagea T, Armaganidis A, Petrikkos G, Giamarellou H. 2013. Antimicrobial activity of copper surfaces against carbapenemase-producing contemporary Gram-negative clinical isolates. J Antimicrob Chemother 68:852–857. doi: 10.1093/jac/dks473. [DOI] [PubMed] [Google Scholar]
- 53.Mathews S, Hans M, Mucklich F, Solioz M. 2013. Contact killing of bacteria on copper is suppressed if bacterial-metal contact is prevented and is induced on iron by copper ions. Appl Environ Microbiol 79:2605–2611. doi: 10.1128/AEM.03608-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodgkinson V, Petris MJ. 2012. Copper homeostasis at the host-pathogen interface. J Biol Chem 287:13549–13555. doi: 10.1074/jbc.R111.316406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner D, Maser J, Lai B, Cai Z, Barry CE III, Honer Zu Bentrup K, Russell DG, Bermudez LE. 2005. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol 174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- 56.Stafford SL, Bokil NJ, Achard ME, Kapetanovic R, Schembri MA, McEwan AG, Sweet MJ. 2013. Metal ions in macrophage antimicrobial pathways: emerging roles for zinc and copper. Biosci Rep 33:e00049. doi: 10.1042/BSR20130014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashfaq M, Verma N, Khan S. 2016. Copper/zinc bimetal nanoparticles-dispersed carbon nanofibers: a novel potential antibiotic material. Mater Sci Eng C Mater Biol Appl 59:938–947. doi: 10.1016/j.msec.2015.10.079. [DOI] [PubMed] [Google Scholar]
- 58.Borkow G, Okon-Levy N, Gabbay J. 2010. Copper oxide impregnated wound dressing: biocidal and safety studies. Wounds 22:301–310. [PubMed] [Google Scholar]
- 59.Klinkajon W, Supaphol P. 2014. Novel copper (II) alginate hydrogels and their potential for use as anti-bacterial wound dressings. Biomed Mater 9:045008. doi: 10.1088/1748-6041/9/4/045008. [DOI] [PubMed] [Google Scholar]
- 60.Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. 2011. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Djoko KY, Ong CL, Walker MJ, McEwan AG. 2015. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem 290:18954–18961. doi: 10.1074/jbc.R115.647099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guyer MS, Reed RR, Steitz JA, Low KB. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp Quant Biol 45:135–140. doi: 10.1101/SQB.1981.045.01.022. [DOI] [PubMed] [Google Scholar]