ABSTRACT
The use of sanitizers is essential for produce safety. However, little is known about how sanitizer efficacy varies with respect to the chemical surface properties of produce. To answer this question, the disinfection efficacies of an oxidant-based sanitizer and a new surfactant-based sanitizer for porcine rotavirus (PRV) strain OSU were examined. PRV was attached to the leaf surfaces of two kale cultivars with high epicuticular wax contents and one cultivar of endive with a low epicuticular wax content and then treated with each sanitizer. The efficacy of the oxidant-based sanitizer correlated with leaf wax content as evidenced by the 1-log10 PRV disinfection on endive surfaces (low wax content) and 3-log10 disinfection of the cultivars with higher wax contents. In contrast, the surfactant-based sanitizer showed similar PRV disinfection efficacies (up to 3 log10) that were independent of leaf wax content. A statistical difference was observed with the disinfection efficacies of the oxidant-based sanitizer for suspended and attached PRV, while the surfactant-based sanitizer showed similar PRV disinfection efficacies. Significant reductions in the entry and replication of PRV were observed after treatment with either disinfectant. Moreover, the oxidant-based-sanitizer-treated PRV showed sialic acid-specific binding to the host cells, whereas the surfactant-based sanitizer increased the nonspecific binding of PRV to the host cells. These findings suggest that the surface properties of fresh produce may affect the efficacy of virus disinfection, implying that food sanitizers should be carefully selected for the different surface characteristics of fresh produce.
IMPORTANCE Food sanitizer efficacies are affected by the surface properties of vegetables. This study evaluated the disinfection efficacies of two food sanitizers, an oxidant-based sanitizer and a surfactant-based sanitizer, on porcine rotavirus strain OSU adhering to the leaf epicuticular surfaces of high- and low-wax-content cultivars. The disinfection efficacy of the oxidant-based sanitizer was affected by the surface properties of the vegetables, while the surfactant-based sanitizer was effective for both high- and low-wax leafy vegetable cultivars. This study suggests that the surface properties of vegetables may be an important factor that interacts with disinfection with food sanitizers of rotaviruses adhering to fresh produce.
INTRODUCTION
There are 48 million estimated annual incidents of foodborne illness in the United States (1–3). Of these incidents, 128,000 cases require hospitalization and 3,000 result in death (1–3). The annual cost associated with foodborne illness, including human morbidity and mortality, is estimated to range between $14.1 and $152 billion (4–6). Viruses, including norovirus and rotavirus, cause foodborne outbreaks, especially due to their persistence in the environment and their low infectious doses (10 to 100 particles) (7, 8). Murine norovirus, Tulane virus, and sapovirus have been found to attach to the surface of or within the tissues of romaine lettuce and strawberries (9–13), which suggests that preharvest viral contamination is a public health concern. To reduce the incidence of viral foodborne illnesses caused by contaminated produce, it is essential to understand the environmental and agricultural factors that control the stability and, therefore, the infectivity of foodborne viruses.
There are several conditions where fresh produce can become contaminated with viral pathogens. This may occur during the preharvest period, when plants come into contact with contaminated irrigation water or runoff (14–16). Typically, fresh produce is stored at around 4°C to maintain postharvest shelf life and to prevent bacterial growth. However, this condition is favorable for the stability of various types of viruses (7, 17, 18). In addition, the chemical composition, surface roughness, and hydrophobicity of fresh produce also play important roles in virus adhesion to produce surfaces (19–22). For example, the presence of wax crystals on the cuticular surfaces of 24 vegetable cultivars was found to reduce rotavirus adhesion (20). Moreover, rotavirus particles that attached to the surfaces of these 24 cultivars persisted even after washing with phosphate-buffered saline (PBS) (20). A similar trend was observed with hepatitis A virus adsorption to lettuce, fennel, and carrots that were washed with potable water (23). These findings emphasize the importance of disinfection practices for fresh produce.
The current sanitation treatments employed in the food industry may not effectively inactivate viruses that have adsorbed to fresh vegetables (24–27). Chlorine-based sanitizers are the most commonly used sanitizers in the food industry. However, chlorine is consumed by the organic matter that is present on produce, which may result in unstable disinfection efficacy (28). These shortcomings motivate the development of alternative sanitation methods using non-chlorine-based sanitizers (29–31), ozone (24, 32), ultrasound (33, 34), heat (35, 36), cold atmospheric gaseous plasma (26), and electron beams (37). However, it remains unclear how nonchlorine chemical sanitizers inactivate viruses on fresh produce.
To fill this research gap, this study aimed (i) to determine the efficacy of a surfactant-based and an oxidant-based food sanitizer on rotaviruses adhering to the surfaces of three fresh vegetables with different epicuticular wax compositions and (ii) to identify which stage of the rotavirus replication cycle was inhibited by the sanitizers. Two cultivars with high wax contents (Brassica napus ‘Red Russian’ and Brassica oleracea ‘Starbor’ kales) and a cultivar with a low wax content (Cichorium intybus ‘Totem’ Belgian endive), as characterized by our previous study (20), were selected. Tsunami 100, an oxidant acid-based food sanitizer authorized by the Environmental Protection Agency (EPA) and a potentially stronger disinfectant than chlorine (38, 39), and another sanitizer, a mixture of malic acid with thiamine dilauryl sulfate (TDS), were chosen because their disinfection efficacies on viruses remain unclear. We chose rotavirus as our model virus because it is a major cause of gastroenteritis worldwide, especially in children less than 5 years old (40). Although rotavirus vaccines have been used worldwide, rotavirus has been frequently detected in treated wastewater, river water, and fresh produce (8, 41–43). A better understanding of the survival of foodborne viral pathogens adhering to fresh produce will improve disinfection strategies for fresh produce and prevent foodborne illness.
MATERIALS AND METHODS
Sanitizers.
An oxidant-based sanitizer (Tsunami 100) was purchased from Ecolab (Saint Paul, MN). The ingredients for a new surfactant-based sanitizer, malic acid and thiamine dilauryl sulfate (TDS), were purchased from Sigma-Aldrich (St. Louis, MO) and Sanigen Co. Ltd. (Juam-dong, South Korea), respectively. Vibrio cholerae neuraminidase was purchased from Sigma-Aldrich (St. Louis, MO).
Greenhouse production of leafy vegetables.
In this study, ‘Red Russian’ kale (Brassica napus; total leaf wax concentration, 81.3 ± 3.7 μg/cm2) and ‘Starbor’ kale (Brassica oleracea; total leaf wax concentration, 78.4 ± 1.4 μg/cm2) were chosen as cultivars with high epicuticular wax concentrations. ‘Totem’ Belgian endive (Cichorium intybus; total leaf wax concentration, 6.3 ± 0.2 μg/cm2) was chosen as a cultivar with a low epicuticular wax content (20). All three plants were grown in the greenhouse as previously described (20). Greenhouse conditions were consistently maintained throughout the study so that replicated samples of produce could be obtained over a period of months. All seeds were purchased from Johnny's Selected Seeds (Winslow, ME). Seeds of each cultivar were germinated in 32-cell plant plug trays filled with Sunshine LC1 (Sun Gro Horticulture, Vancouver, British Columbia, Canada) professional soil mix. Seedlings were grown in a university greenhouse under a 25°C/17°C and 14 h/10 h day/night temperature regimen with supplemental lighting. The greenhouse was disinfected regularly, affording vegetable growth without substantial microbial contamination that may be detrimental to the plants. Twenty days after germination, seedlings were transferred to 4-liter pots. Leaf tissues from each type of plant were harvested 50 to 65 days after sowing seeds. Leaves from the median internodes from each leafy vegetable were harvested at market maturity for analysis.
Cell culture and rotavirus.
Porcine rotavirus (PRV) strain OSU was used in this study because the structure of its outer protein is similar to that of human rotavirus strain Wa and because of its stability in the environment (44). PRV was propagated in the monkey MA104 cell line, which was purchased from ATCC, and maintained at 37°C in a 5% CO2 incubator with minimal essential medium (MEM) with 10% fetal bovine serum (FBS). PRV was propagated using confluent cells in a 150-cm2 flask, and cells were washed three times with prewarmed serum-free MEM as recommended (45). PRV was activated with trypsin at a final concentration of 10 μg/ml for 30 min at 37°C. Following dilution with serum-free MEM, the trypsin-activated rotavirus solution was added to these confluent cells. After incubation at 37°C for 60 min in a 5% CO2 incubator, the viral solution was aspirated and washed twice with serum-free MEM. Then, serum-free MEM was added to the flask and incubated for 4 to 5 days at 37°C without the presence of trypsin until most of the cells were detached. After this propagation step, rotavirus solution was sequentially frozen at −80°C and thawed three times. The rotavirus solution was centrifuged at 1,000 × g for 10 min at room temperature and filtered through a 0.45-μm-pore-size filter to remove cell debris. After this step, MEM was removed from the virus stock by filtering the virus suspension through a 100-kDa ultrafiltration membrane as described previously (44). Rotavirus was resuspended in 1 mM NaCl plus 0.1 mM CaCl2 and stored at 4°C for up to 4 weeks. Virus titers were quantified by using the focus-forming unit (FFU) assay as previously described (46) (Table 1).
TABLE 1.
Original titer, inactivated virus titer, and recovered virus titer for the PRV disinfection experiments with sanitizers
| Figure | Cultivar or sanitizer | Virus inoculum (FFU/ml) | Inactivated viruses (FFU/ml) | Recovered viruses (FFU/ml) |
|---|---|---|---|---|
| 1 | ‘Totem’ Belgian endive | 1,000,000 | 755,014 ± 124,485 | 244,986 ± 124,485 |
| ‘Starbor’ kale | 1,000,000 | 994,500 ± 10,370 | 5,500 ± 10,370 | |
| ‘Red Russian’ kale | 1,000,000 | 998,700 ± 733 | 1,300 ± 733 | |
| 2 | ‘Totem’ Belgian endive | 1,000,000 | 997,168 ± 1,180 | 2,832 ± 1,180 |
| ‘Starbor’ kale | 1,000,000 | 991,257 ± 5,493 | 8,743 ± 5,293 | |
| ‘Red Russian’ kale | 1,000,000 | 987,500 ± 13,640 | 12,501 ± 13,640 | |
| 3 | Oxidant-based sanitizer | 1,000,000 | 99,977 ± 14 | 23 ± 14 |
| Surfactant-based sanitizer | 1,000,000 | 99,867 ± 55 | 132 ± 55 |
FFU assay.
Trypsin-activated PRV stock was serially diluted with serum-free MEM. Next, PRV aliquots were applied to MA104 cellular monolayers in a 96-well plate and incubated at 37°C for 30 min in a 5% CO2 incubator. Virus was aspirated, and each well was washed twice using serum-free MEM. A final 50 μl of serum-free MEM was added to each well, and the plates were incubated at 37°C for 18 h in a 5% CO2 incubator to allow viruses to replicate.
Next, the medium was removed from each well, and cells were fixed by adding 100 μl of 9:1 methanol/glacial acetic acid per well and incubated for 2 min. One hundred microliters of 70% ethanol was added to each well and incubated for 5 min to rehydrate cells before adding 100 μl of 50% ethanol and incubating for another 5 min. Endogenous peroxidase activity was quenched by adding 50 μl of 3% H2O2 in wash buffer (96 mM Tris-HCl, 350 mM NaCl, 29 mM Tris base, and 0.25% Triton X-100) per well and incubating for 10 min at room temperature. After washing fixed cells with wash buffer, 50 μl of wash buffer containing 5% normal goat serum was added to each well and incubated for 20 min at room temperature. Then, 50 μl of primary antibody (rabbit anti-rotavirus group A; AbD Serotec, Raleigh, NC; 1:100) in wash buffer was added to each well and incubated at 37°C for 1 h. Wells were rinsed twice with wash buffer. Then, 50 μl of wash buffer containing secondary antibody (biotinylated goat anti-rabbit IgG; Vector Laboratories, CA; 1:200) and 1.5% normal goat serum was added to each well and incubated for 20 min at room temperature. Wells were rinsed twice with wash buffer. Then, 50 μl of Vectastain ABC reagent, containing 2% reagent A and 2% reagent B (Vectastain ABC kit; Vector Laboratories, CA) and first incubated in wash buffer for 30 min, was added to each well and incubated for 20 min. After washing wells twice with wash buffer, 50 μl of diaminobenzidine (DAB) solution that was diluted in distilled water following the manufacturer's recommendations (DAB substrate kit; Vector Laboratories, CA) was added to each well and incubated for 2 min. DAB was solution aspirated, and Milli-Q water was replaced into each well. Focus-forming units were enumerated using a microscope.
Disinfection experiments for PRV adhering to leaves.
Each set of leaves in this study, consisting of three biological replicates for each cultivar, was gently washed with Milli-Q water, and the water on the leaves was then lightly wiped off with a Kimwipe (Kimberly-Clark, Irving, TX). Two disks were excised from each leaf with a 15.6-mm-diameter cork borer. One disk was sampled on the adaxial surface, and the other was sampled on the abaxial surface. Two drops of 20 μl of viral solution (PRV) were applied onto each disk surface and incubated for 1 h to allow virus adsorption. Next, each disk was washed with 4 ml of each kind of prechilled sanitizer solution at 4°C in a well of a 12-well plate as a function of time. Each sanitizer concentration was as follows: 50 ppm oxidant-based sanitizer (Tsunami 100) at pH 3.7 and 0.25% malic acid with 0.025% TDS for the surfactant-based sanitizer at pH 2.7. Next, 10 μl of 1 M NaOH was immediately added into the sanitizer-containing solution to raise the pH to 7. The leaf was removed from the well, and the remaining PRV on the leaf was eluted with 500 μl of serum-free MEM in a 1.5-ml centrifuge tube by vortexing for 30 s. Four hundred microliters from the tube was transferred into a new 1.5-ml centrifuge tube for trypsin activation of both sanitizer-treated samples and rotavirus stock with known FFU. Integrated cell culture-quantitative PCR (ICC-qPCR) was then conducted.
Rotavirus decay experiment.
Following PRV adsorption to leaves, decay experiments were conducted to clarify the effect of each sanitizer on PRV suspensions without vegetable leaf tissue. PRV suspensions were treated with the same concentration of each sanitizer described above as a function of time at 4°C. As a control, PRV suspensions were treated with distilled water at 4°C. As above, the solutions were adjusted to pH 7. Following trypsin activation, the PRV solution was immediately diluted with serum-free MEM. ICC-qPCR was then conducted.
ICC-qPCR.
ICC-qPCR was used to quantify the infectious viruses remaining after the disinfection experiments for PRV adhering to leaves and the rotavirus decay experiment. This method was employed instead of the focus-forming unit (FFU) assay because this methodology allows for the detection of infectious viruses more rapidly and sensitively than the FFU assay (47). In addition, this method allows for the quantification of the RNA genomes of the viruses replicated inside the host cells. The quantitative principle of this method is based on a calibration curve for the number of copies of NSP3 genes from the replicated viruses inside MA104 cells infected by either the virus solutions with known FFUs or the infectious viruses, which remained after exposure to the sanitizers. The x axis of this calibration curve consists of log10 copy numbers of NSP3 genes from infectious rotaviruses. The y axis of this calibration curve consists of log10 FFUs obtained from the virus solution with a known FFU. A monolayer of confluent cells on a 24-well plate was washed twice with prewarmed serum-free MEM, and then trypsin-treated rotavirus from the disinfection experiments for PRV adhering to leaves, the rotavirus decay experiment, or serially diluted rotavirus stock with a known FFU was added onto the cells and incubated at 37°C for 30 min in a 5% CO2 incubator. After the infection step, cells were washed twice with serum-free MEM and incubated with 500 μl of serum-free MEM at 37°C for 18 h in a 5% CO2 incubator. During this 18 h of incubation, only infectious viruses can enter cells and replicate. This method enables us to quantify the infectivity of viruses remaining after the sanitizer treatment, using the known infectivity of a viral stock. After the incubation, 350 μl of lysis buffer from an RNA extraction kit (E.Z.N.A. total RNA kit I; Omega Bio-Tek) was added to each well and incubated for 30 min at room temperature. RNA extraction was conducted according to the manufacturer's recommendations. After the extraction, reverse transcription-quantitative PCR (RT-qPCR) was performed using the rotavirus gene-specific primers and cellular gene-specific primers.
Reverse transcription-quantitative PCR.
Reverse transcription-quantitative PCR (RT-qPCR) was conducted to quantify the PRV NSP3 and cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcripts in infected MA104 cells using an iTaq universal SYBR green one-step kit (Bio-Rad Laboratories, Hercules, CA) in MicroAmp optical 384-well reaction plates with a 7900HT Fast real-time PCR system (Applied Biosystems, Foster, CA). Plasmids containing the rotavirus NSP3 gene (Integrated DNA Technologies, Coralville, IA) were used to develop a standard curve for the amount of cDNA present (x axis, expressed as log10 genome copies per microliter) versus threshold cycle (CT) values obtained from qPCR on the y axis. For the quantification of cells that were exposed to PRV, genomic RNAs extracted from cells were used as a standard. A total reaction mixture of 10 μl for quantification of rotavirus consisted of 3 μl of RNA sample, 5 μl of 2× iTaq universal SYBR green reaction mix, 0.125 μl of iScript reverse transcriptase, 0.3 μl of 10 μM JVK forward primer (5′-CAGTGGTTGATGCTGAAGAT-3′), 0.3 μl of 10 μM JVK reverse primer (5′-TCATTGTAATCATATTGAATACCCA-3′) (48, 49), and 1.275 μl of nuclease-free water. For the quantification of cells, a total reaction mixture of 10 μl for quantification of rotavirus consisted of 3 μl of RNA sample, 5 μl of 2× iTaq universal SYBR green reaction mix, 0.125 μl of iScript reverse transcriptase, 0.6 μl of 10 μM GAPDH forward primer (5′-AATCCCATCACCATCTTCCAG-3′), 0.6 μl of 10 μM GAPDH reverse primer (5′-AAATGAGCCCCAGCCTTC-3′) (50), and 0.675 μl of nuclease-free water. The thermal cycling conditions for both the NSP3 gene and GAPDH gene quantification were as follows: reverse transcription reaction at 50°C for 10 min and polymerase activation and DNA denaturation at 95°C for 1 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. After these steps, a dissociation stage for the dissociation curve analysis was conducted at 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s. Data were obtained from the software package SDS 2.4.2 (Applied Biosystems, Foster, CA) after RT-qPCRs. In this study, the amount of rotavirus RNA was normalized by the number of housekeeping gene copies of GAPDH in cellular RNA.
The qPCR specificity was checked by gel electrophoresis using RT-qPCR products, with 2% agarose containing SYBR Safe DNA gel stain (Thermo Fisher Scientific, Waltham, MA). Only one band between 100 and 200 bp was observed for both products amplified with JVK primers and GAPDH primers.
Virus entry and replication assay.
PRVs were treated with each sanitizer at 4°C as a function of time. After this sanitation step, the viral solution was adjusted to pH 7 and activated with trypsin. This solution of activated rotaviruses was diluted with serum-free MEM, 300 μl of which was added to the monolayer of confluent MA104 cells and incubated for 30 min at 37°C in a 5% CO2 incubator. Following this incubation, cells were washed twice with serum-free MEM and incubated for 18 h at 37°C in a 5% CO2 incubator. RNA extraction and RT-qPCR were conducted as described above.
Virus binding assay.
The assays for detection of PRV binding to MA104 cells were conducted similarly to the virus entry and replication assay. The only difference from the entry and replication assays was that after the sanitizer treatment of PRV at 4°C for 5 min, the sanitizer-treated PRV was incubated with MA104 cells at 4°C for 1 h, followed by aspiration of the viral solution and two washes of MA104 cells with serum-free MEM. RNA extraction and RT-qPCR were conducted as described above. For the control experiment to check adhesion of intact PRV to MA104 cells, viruses were treated with distilled water without sanitizer at 4°C for 5 min. To check for nonspecific binding of rotaviruses to cells, A549 cells without the receptors for PRV (GM3 receptors) (51, 52) were used for the binding control experiment.
Binding assay with neuraminidase.
Confluent monolayers of MA104 cells in a 6-well plate were washed twice with prewarmed serum-free MEM. One milliliter of serum-free MEM containing neuraminidase V. cholerae at a final concentration of 40.3 mU/ml was added to the cells in each well and incubated for 1 h at 37°C in a 5% CO2 incubator. As a positive control for each sample, 1 ml of prewarmed serum-free MEM without neuraminidase was added to cells and incubated similarly as described above. Intact PRV as a control experiment and sanitizer-treated PRV were adjusted to pH 7 and activated with trypsin before they were incubated with cells and washed in the same manner as the virus binding assay, followed by RNA extraction and RT-qPCR.
Plaque assay with neuraminidase.
The plaque assay for the detection of PRV-infected MA104 cells was conducted similarly to the binding assay with neuraminidase. The only difference from the binding assay with neuraminidase was that the incubation of the sanitizer-treated PRV with MA104 cells was conducted at 37°C for 1 h in a 5% CO2 incubator, followed by washing cells twice with serum-free MEM. The cells were overlaid with MEM, containing 1% agarose, 2.2 μg/ml trypsin at the final concentration, 7.5% sodium bicarbonate, 15 mM HEPES, 0.1 mg of kanamycin/ml, 0.05 mg of gentamicin/ml, and incubated at 37°C for 72 h in a 5% CO2 incubator. Following incubation, the cells were fixed with 10% formaldehyde in 1× PBS and stained with 0.05% crystal violet in 10% ethanol. PRV-infected MA104 cells were visualized and counted.
Statistical analyses.
Statistical analyses were conducted with OriginPro 2016 (OriginLab Corporation, MA, USA). For the disinfection experiments of PRV adhering to leaves, significant differences in disinfection efficacies between cultivars and sanitizer treatments were determined using a three-way analysis of variance (ANOVA). Also, for the rotavirus decay experiment, disinfection efficacies were analyzed for sanitizer treatments using a two-way ANOVA. For other assays, t tests were conducted. Relationships were considered significant when the P value was <0.05.
RESULTS
Disinfection of PRV adhering to leaf surfaces.
The ratio of the number of infectious PRVs remaining after the sanitizer treatment (FFU) over the initial number of infectious PRVs (FFU0) was determined to identify the effectiveness of each sanitizer in inactivating PRV when PRV was adsorbed to the tested plant surfaces. The disinfection of PRV was expressed as a disinfection ratio (FFU/FFU0) obtained by the ICC-RT-qPCR method. Figures 1 and 2 show comparisons of log10 reductions of PRV treated with an oxidant-based sanitizer at 50 ppm and a surfactant-based sanitizer (0.25% malic acid with 0.025% TDS) on leaves of the three vegetable cultivars, respectively.
FIG 1.
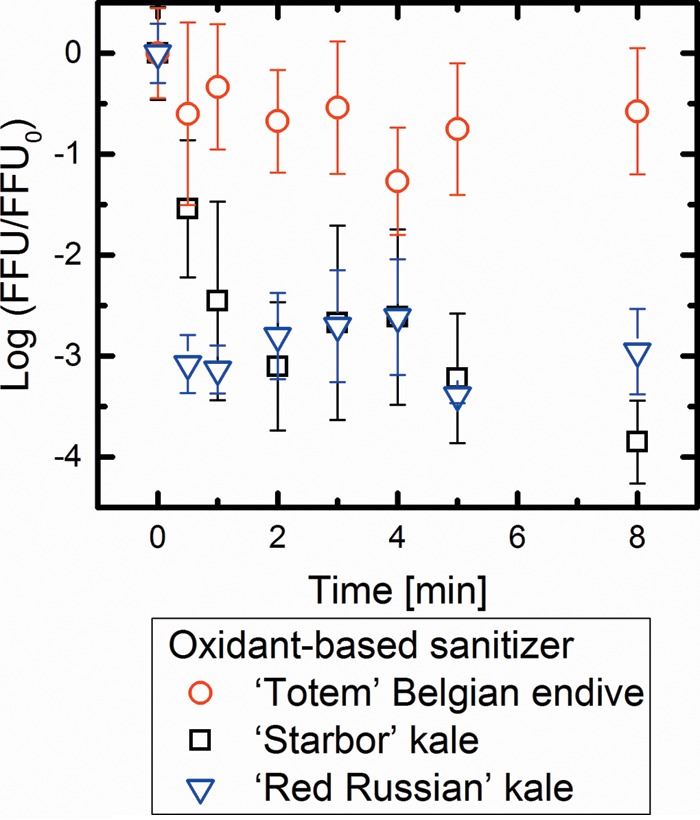
Comparisons of log10 disinfection of PRV on ‘Totem’ Belgian endive, ‘Starbor’ kale, and ‘Red Russian’ kale using the oxidant-based sanitizer. Values in the figure are the averages of results from biological replicates (n = 6) at each contact time (min), with standard deviations shown as vertical error bars. There was a significant difference in the disinfection ratios of PRV between ‘Totem’ Belgian endive and ‘Starbor’ and ‘Red Russian’ kale (P < 0.05).
FIG 2.
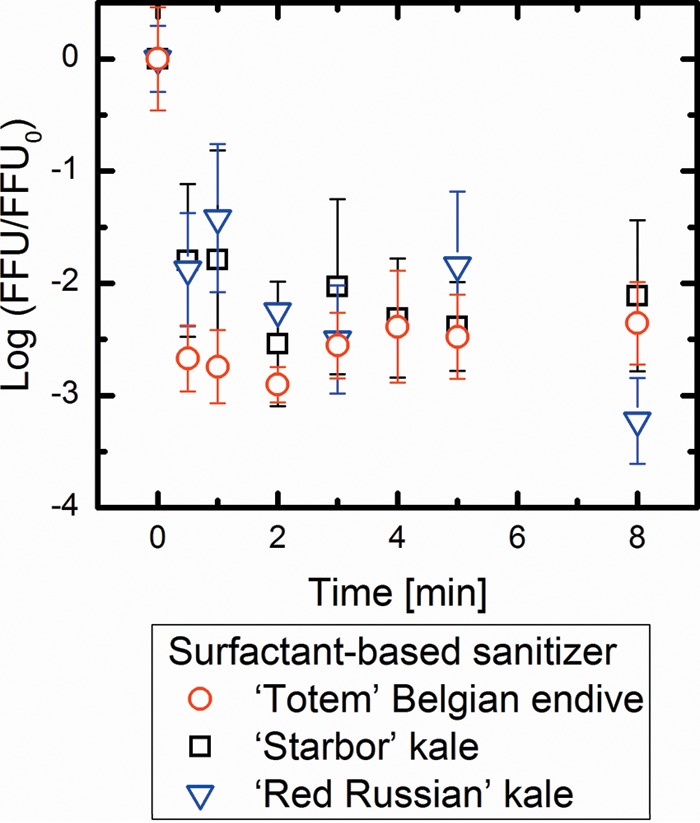
Comparisons of log10 disinfection of PRV on ‘Totem’ Belgian endive, ‘Starbor’ kale, and ‘Red Russian’ kale treated with the surfactant-based sanitizer. Values in the figure are the averages of results from biological replicates (n = 6) at each contact time (min), with standard deviations shown as vertical error bars. There were no significant differences in the disinfection ratios of PRV between all of the cultivars (P > 0.05).
As shown in Fig. 1, the oxidant-based sanitizer caused statistically similar disinfection rates of PRV that was adsorbed to the ‘Starbor’ kale and ‘Red Russian’ kale surfaces (P > 0.05). In contrast, only an approximately 1-log10 PRV disinfection was observed when PRV was adsorbed to the ‘Totem’ Belgian endive surfaces. In contrast, as shown in Fig. 2, the surfactant-based sanitizer exhibited similar disinfection of PRVs on all three cultivars (P > 0.05). The disinfection of PRV adhering to the ‘Totem’ Belgian endive surfaces achieved approximately 3-log10 inactivation when the surfactant-based sanitizer was used, while the oxidant-based sanitizer showed only 1-log10 inactivation. These results suggest that the surfactant-based sanitizer is more effective than the oxidant-based sanitizer in inactivating PRV adhering to the leaf surface of ‘Totem’ Belgian endive. Taken together, these results imply that the different sanitizer efficacies for leafy vegetables vary with different epicuticular chemical properties.
Disinfection of suspended PRV without vegetables.
To determine how each sanitizer interacted with PRV, we explored how PRV in suspension, and in the absence of plants, was disinfected. Figure 3 shows the comparisons of the log10 disinfection of suspended PRVs treated with distilled water (control), the oxidant-based sanitizer, or the surfactant-based sanitizer. Similar to Fig. 1 and 2, the disinfection of PRV was expressed as the disinfection ratio of FFU/FFU0 obtained by the ICC-RT-qPCR method. Compared to the control experiment, both sanitizers had statistically different disinfection ratios (P < 0.05). For the disinfection efficacies of the surfactant-based sanitizer, it was observed that the efficacies were statistically similar when comparing the disinfection of PRV in solution versus PRV adsorbed to plant leaves (Fig. 3 compared to Fig. 2). However, the oxidant-based sanitizer disinfected suspended PRV to a statistically significant greater degree than it did when PRV was associated with the plant leaves (P < 0.05). Thus, the efficacy of the oxidant-based sanitizer was different when PRV was in suspension compared to when it was attached to vegetable surfaces, whereas the surfactant-based sanitizer showed a similar disinfection efficacy on PRV with or without vegetable leaf tissue. These data imply that PRV interaction with the leaf surface may reduce the efficacy of the oxidant-based sanitizer.
FIG 3.

Comparisons of log10 disinfection of suspended PRV (in the absence of leafy vegetable tissues) with distilled water (control), the oxidant-based sanitizer, or the surfactant-based sanitizer. Values in the figure are the averages of results from biological replicates (n = 4) at each contact time [min], with standard deviations shown as vertical error bars. There were no significant differences in the disinfection of PRV between the oxidant-based sanitizer and the surfactant-based sanitizer (P > 0.05), while there was a significant difference in the disinfection between each sanitizer treatment and the control (P < 0.05).
Influence of sanitizers on the PRV replication cycle.
It is unknown how sanitizers inactivate PRV on a molecular level. As a means to understand this process, we examined how each sanitizer impacted virus entry and replication and binding steps. For these assays, the suspensions of PRV that were exposed to the sanitizers (as opposed to the PRV adhering to vegetable surfaces) were examined. Figure 4 shows the comparison of the number of PRV RNA copies (NSP3 gene) replicated in MA104 cells after the sanitizer treatments of PRV 18 h postinfection. PRV treated with the oxidant-based sanitizer and PRV treated with the surfactant-based sanitizer had significant reductions in the numbers of PRV RNA copies replicated in MA104 cells 18 h postinfection compared to the control, where PRV was exposed to distilled water instead of a sanitizer. Treatment with the oxidant-based sanitizer or the surfactant-based sanitizer led to significantly less PRV RNA copy numbers compared to treatment with the control (P < 0.05). Thus, each sanitizer altered PRV particles in a manner that prevented PRV entry and replication in MA104 cells.
FIG 4.
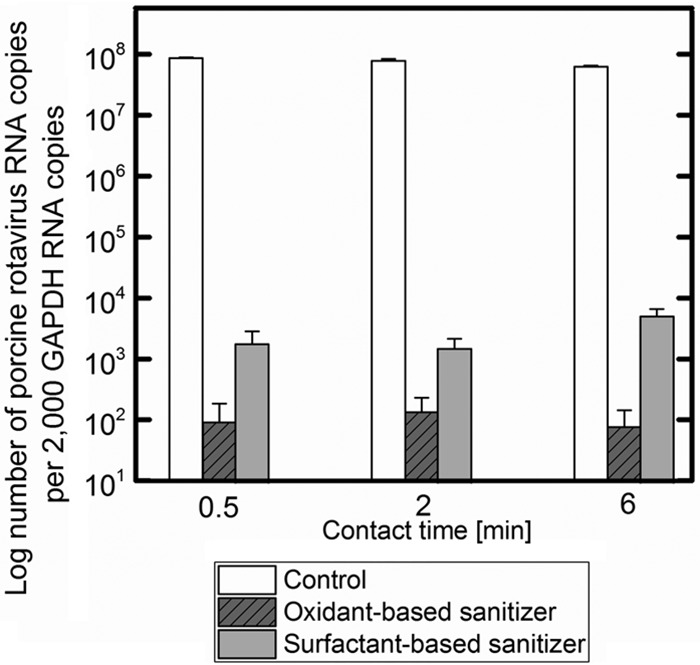
Comparisons of rotavirus NSP3 RNA transcript copy numbers replicated in MA104 cells after different sanitizer treatments. Values in the figure are the averages of the results from biological replicates (n = 6), with standard deviations shown as vertical error bars. There was a significant reduction in copy numbers in treatments with the oxidant-based sanitizer and the surfactant-based sanitizer compared to the control (P < 0.05).
One possibility is that the inhibition of PRV entry and replication was due to transformed PRV particles that could no longer bind to the host cells. This question was answered by conducting a virus-binding assay as shown in Fig. 5. In this experiment, sanitizer-treated PRV was incubated with MA104 cells, cellular monolayers were washed, and PRV particles that remained attached to cells were quantified by RT-qPCR. As shown in Fig. 5, PRV genes were more abundant when PRV particles were treated with either disinfectant compared to when they were treated with water. This finding indicated that sanitizers increased the binding of PRV to the MA104 cells. This same assay was performed in parallel using A549 cells, cells that lack GM3 receptors (52), which allows PRV to initialize penetration into the host cells through an entry receptor integrin. When using the A549 cell line, the highest number of bound PRVs was observed when PRV was treated with the surfactant-based sanitizer, followed by the intact PRV and then the PRV treated with the oxidant-based sanitizer. The increased binding of PRV after treatment with sanitizers suggests that sanitizers themselves may alter the capsid such that there is an increase in the nonspecific binding of PRV to both A549 cells and MA104 cells.
FIG 5.
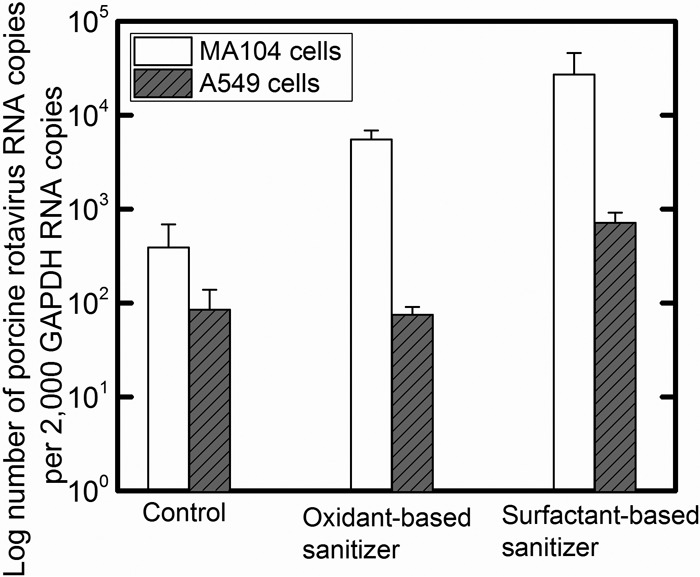
Comparisons of NSP3 gene copy numbers bound to MA104 and A549 cells after different sanitizer treatments. Values in the figure are the averages of results from biological replicates (n = 6), with standard deviations shown as vertical error bars. Viral binding to MA104 cells was significantly different between the control, the oxidant-based sanitizer, and the surfactant-based sanitizer treatments (P < 0.05). Virus binding to A549 cells was significantly different between the control and the treatment with the surfactant-based sanitizer (P < 0.05), whereas there was no significant difference in the viral binding to A549 cells between the control and the treatment with the oxidant-based sanitizer (P > 0.05).
Although A549 cells do not express the GM3 receptor (52), both A549 and MA104 cells express sialic acid on the cellular surface (53), a molecule that can serve as an attachment receptor for PRV. Therefore, the effect of the sanitizers on the disruption of PRV-sialic acid interactions cannot be evaluated using this assay. To determine whether a sanitizer treatment influences PRV binding to sialic acid on the MA104 cellular surface, binding assays were conducted in which MA104 cells were incubated with V. cholerae neuraminidase to digest sialic acid moieties on the cellular surface (Fig. 6). The removal of sialic acid from the cellular surface significantly reduced the number of untreated and oxidant-based-sanitizer-treated PRVs that bound to MA104 cells as determined by RT-qPCR. This observation suggests that sialic acid plays an important role in PRV attachment to MA104 cells, where receptor-specific binding of the control sample (intact PRV) and oxidant-based-sanitizer-treated PRV were reduced by the removal of sialic acid (P < 0.05). However, no statistical difference was observed when using surfactant-based-sanitizer-treated PRV (P > 0.05), which indicates that the nonspecific binding of PRV to the cellular surface occurred regardless of the presence of sialic acid. These data imply that each sanitizer may alter PRV in distinct manners.
FIG 6.
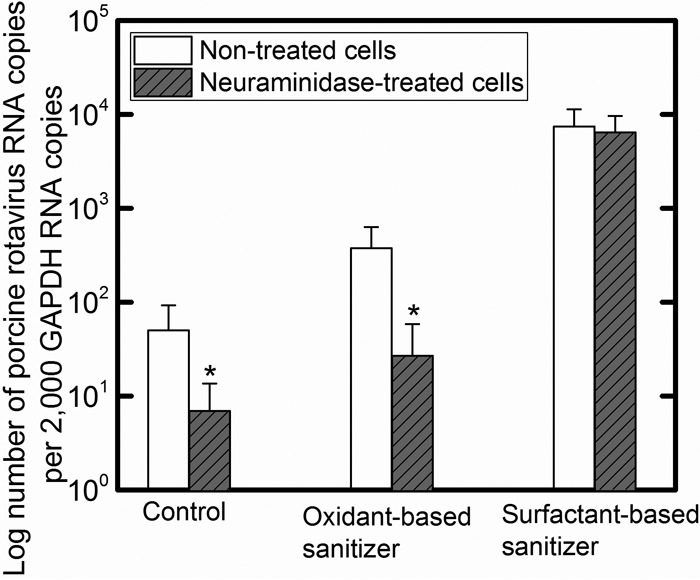
Comparisons of PRV attachment to MA104 cells pretreated with 0 and 40.3 mU/ml Vibrio cholerae neuraminidase. PRV was treated with distilled water (control), the oxidant acid-based sanitizer, or the surfactant-based sanitizer prior to exposure to MA104 cells. Values in the figure are the averages of results from biological replicates (n = 6) at each contact time (min), with standard deviations shown as vertical error bars. Statistical analyses were performed with t test. *, P value of <0.05.
To identify whether the sialic acid digestion by neuraminidase affects PRV infection, a plaque assay was conducted. As shown in Fig. 7, a significant difference in plaque formation (PRV infection) was observed with the control sample (intact PRV) between nontreated infected cells and neuraminidase-treated infected cells (P < 0.01). A similar trend was observed for PRV treated with the oxidant-based sanitizer and the surfactant-based sanitizer (P < 0.001 and 0.05, respectively). The removal of sialic acid from the ganglioside on the cellular surface reduced PRV infection as well as the attachment of intact PRV and PRV treated with the oxidant-based sanitizer to MA104 cells. This indicates that PRV nonspecifically bound to MA104 cells could not replicate effectively in the cells.
FIG 7.
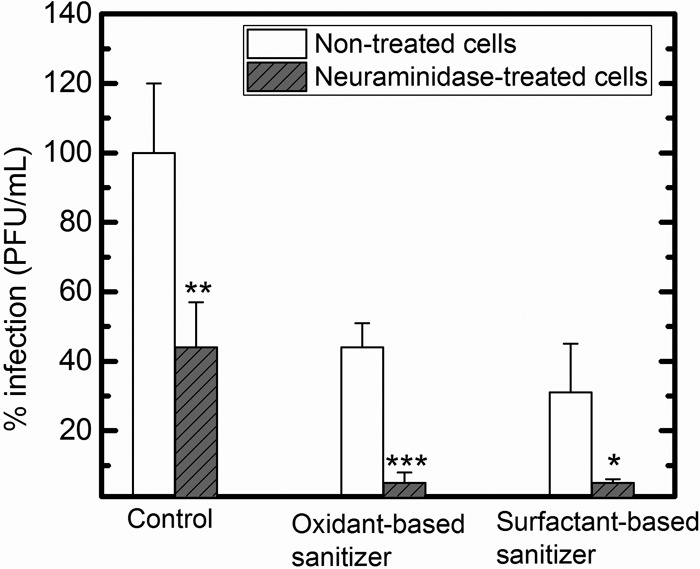
Comparisons of PRV infections in MA104 cells pretreated with 0 and 40.3 mU/ml Vibrio cholerae neuraminidase. PRV was treated with distilled water (control), the oxidant-based sanitizer, or the surfactant-based sanitizer prior to exposure to MA104 cells. Values in the figure are the averages of results from biological replicates (n = 3) at each contact time (min), with standard deviations shown as vertical error bars. Statistical analyses were performed with t test. *, P value of <0.05; **, P value of <0.01; ***, P value of <0.001.
DISCUSSION
The oxidant-based sanitizer, containing 15.2% peroxyacetic acid combined with 11.2% hydrogen peroxide, has been used in the food industry because of its low reactivity with organics (38) and because it is less dependent on pH than chlorine (54). In the food industry, the concentration range allowed for food sanitation is from 30 to 80 ppm, with a 1.5-min mixing time (55). In this study, we employed a concentration at 50 ppm with an exposure time of 30 s to 8 min. Oxidants, such as hydrogen peroxide, have been shown to inactivate viruses (44, 56). For example, a 0.32-log10 disinfection of human rotavirus strain Wa was observed with 0.6 mM hydrogen peroxide after 8 min of exposure at 25°C (44). Furthermore, 0.2 mM hydrogen peroxide achieved a 2-log10 inactivation on MS2 coliphage after a 30-min incubation (56). The hydrogen peroxide concentration in the studied sanitizer is 2 mM, which is much higher than the concentration used in the previous studies. The high concentrations of hydrogen peroxide and another oxidant, peroxyacetic acid, used in our study allowed a 3-log10 inactivation of PRV adhering to vegetables' leaf surfaces with high epicuticular wax concentrations (‘Starbor' kale and ‘Red Russian’ kale) and a 1-log10 inactivation of PRV adhering to a vegetable cultivar with a low wax concentration (‘Totem' Belgian endive). The 100-fold difference in PRV disinfection efficacy using this sanitizer suggests that a strong interaction between PRV and the more hydrophilic surfaces of endive protected the PRVs that were adhering to the leaf surface from disinfection. Rotaviruses, including PRV, are negatively charged and hydrophilic (57, 58). A weak interaction between hydrophilic PRV and hydrophobic kale surfaces may allow the oxidants to inactivate the adsorbed PRV. However, for PRVs attached strongly to hydrophilic leaf surfaces, a sanitizer that can denature or oxidize the attached capsid is more desirable.
The surfactant-based sanitizer consists of 0.25% malic acid, an organic acid that has been used as an antimicrobial (29, 54), and 0.025% thiamine dilauryl sulfate (TDS), a vitamin B1 derivative and also a negatively charged surfactant. This combination of malic acid and TDS has been recently proposed and is not yet in use in the food industry (29). The disinfection efficacy of 10% malic acid and 1% TDS on Escherichia coli O157:H7 on alfalfa seeds was as effective as chlorine at 20,000 ppm (29). TDS was found to have synergistic effects for the inactivation of total mesophilic bacteria and coliforms when it was combined with a commercially available sanitizer, such as ethanol, chlorine, or hydrogen peroxide (59). Similarly, chlorine at 200 ppm in combination with one of the following surfactants, sodium dodecyl sulfate (SDS), Triton X-100, or NP-40, had a higher sanitation efficacy than chlorine alone for murine norovirus adhering to the surfaces of strawberries, raspberries, cabbage, and romaine lettuce (25). Moreover, each of these surfactants had virucidal effects against murine norovirus when the viruses were incubated with one of the surfactants at 37°C for 72 h, disrupting the outer protein of murine norovirus (25). Charged surfactants have been found to have the ability to bind to and denature proteins (60). In our study, despite the strong interaction between PRV and the hydrophilic surface of ‘Totem’ Belgian endive, the surfactant-based sanitizer was still effective, suggesting that TDS may denature the protein capsid of PRV adhering to the leaf surface. Moreover, the surfactant-based sanitizer also had similar inactivation efficacies on PRVs adhering to the leaves of the two kale cultivars (‘Starbor' kale and ‘Red Russian’ kale). One possible reason behind these similar inactivation efficacies may be the ability of TDS and malic acid with both hydrophilic and hydrophobic groups to reach PRVs adhering to both kale and endive leaf surfaces. Taken together, the findings presented in this study demonstrate that the surfactant-based sanitizer effectively inactivated PRVs adhering to leafy surfaces with both high and low wax concentrations.
The observed effectiveness of the oxidant-based sanitizer for PRVs adhering to the leaf surfaces suggests that the surfaces of PRVs were oxidized by the sanitizer. The PRV capsid may also be denatured by surfactant-based sanitizers. We hypothesize that the oxidized or denatured capsids could nonspecifically bind to MA104 cells; however, the nonspecifically bound PRVs cannot replicate in MA104 cells effectively. This hypothesis was tested with the binding assays to determine the effects of the sanitizers on PRV attachment to MA104 cells and replication inside the cells. We found that sanitizer treatment of PRV led to an increase in the PRVs attached to MA104 cells, indicating nonspecific binding of PRV particles to MA104 cells or specific binding to the cellular receptors followed by ineffective penetration or replication in the cells, given that PRV treated with the sanitizers could not effectively replicate in MA104 cells as observed in the virus entry and replication assay. Furthermore, we conducted binding experiments with neuraminidase to digest sialic acid on the cellular surface, which is an important factor to initialize the PRV penetration step into the host cell. Rotaviruses attach to terminal sialic acid on their receptors (GM3 receptor for PRV strain OSU) and utilize sialic acid binding to penetrate into the cell via integrin, which is an entry receptor for rotaviruses (61, 62). Therefore, by digesting sialic acid moieties on the cellular surface, specific binding of PRV to sialic acid moieties was reduced as was found with intact PRV and PRV treated with the oxidant-based sanitizer. However, the surfactant-based sanitizer treatment did not cause a reduction in the PRV bound to the cellular surface with or without sialic acid digestion. This observation can be attributed to the nonspecific binding of PRV to the cellular surface caused by the surfactant-based sanitizer treatment. In the plaque assay with neuraminidase, PRV infection (plaque formation) was reduced by digesting sialic acid from MA104 cells using neuraminidase in all of the samples. Based on the fact that binding of the surfactant-based-sanitizer-treated PRV to MA104 cells was not affected by the removal of sialic acid in the binding assay with neuraminidase, the plaque assay to analyze PRV binding was needed to evaluate nonspecific binding as well as the binding assay quantified by RT-qPCR.
In summary, our mechanistic studies show that the effectiveness of the food sanitizer depends on the sanitizer properties and the epicuticular leaf surface properties. We found that the oxidant-based sanitizer was less effective for the inactivation of PRV adhering to hydrophilic leaf surfaces due to stronger interactions with PRV. This knowledge will facilitate the selection of effective food sanitizers for virus disinfection. Based on our results, nonspecific binding of PRV was increased by the surfactant-based sanitizer treatment. Future studies will identify which factors provided by the sanitizers are contributing to the nonspecific binding of PRV to MA104 cells. A potential factor may be capsid damage on rotavirus particles after the exposure to sanitizers. To identify capsid damage caused by oxidative stress, carbonyl groups on oxidatively damaged viral particles can be marked using biotin (63). Alternative methods can be applied in this study, and the sanitizer effect on PRV particles can be further analyzed.
ACKNOWLEDGMENTS
We thank Lu Lu for helping with the design of the sanitizer experiment. We also appreciate Andrew Page Storm for helping to conduct the disinfection experiments for PRV adhering to leaves.
This project was supported by the USDA National Institute of Food and Agriculture under grants ILLU-000-615 and RD83582201-0 from the U.S. Environmental Protection Agency (EPA).
Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the EPA or NIFA. Further, the EPA and NIFA do not endorse the purchase of any commercial products or services mentioned in the publication.
REFERENCES
- 1.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States—unspecified agents. Emerg Infect Dis 17:16–22. doi: 10.3201/eid1701.P21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2011. Estimates of foodborne illness in the United States. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 4.Hoffmann S, Batz MB, Morris JG Jr. 2012. Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J Food Prot 75:1292–1302. doi: 10.4315/0362-028X.JFP-11-417. [DOI] [PubMed] [Google Scholar]
- 5.Scharff RL. 2010. Health-related costs from foodborne illness in the United States. The Pew Charitable Trusts, Philadelphia, PA. [Google Scholar]
- 6.Scharff RL. 2012. Economic burden from health losses due to foodborne illness in the United States. J Food Prot 75:123–131. doi: 10.4315/0362-028X.JFP-11-058. [DOI] [PubMed] [Google Scholar]
- 7.Seymour I, Appleton H. 2001. Foodborne viruses and fresh produce. J Appl Microbiol 91:759–773. doi: 10.1046/j.1365-2672.2001.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei J, Kniel KE. 2010. Pre-harvest viral contamination of crops originating from fecal matter. Food Environ Virol 2:195–206. doi: 10.1007/s12560-010-9050-5. [DOI] [Google Scholar]
- 9.DiCaprio E, Culbertson D, Li JR. 2015. Evidence of the internalization of animal caliciviruses via the roots of growing strawberry plants and dissemination to the fruit. Appl Environ Microbiol 81:2727–2734. doi: 10.1128/AEM.03867-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiCaprio E, Purgianto A, Li JR. 2015. Effects of abiotic and biotic stresses on the internalization and dissemination of human norovirus surrogates in growing romaine lettuce. Appl Environ Microbiol 81:4791–4800. doi: 10.1128/AEM.00650-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei J, Jin Y, Sims T, Kniel KE. 2011. Internalization of murine norovirus 1 by Lactuca sativa during irrigation. Appl Environ Microbiol 77:2508–2512. doi: 10.1128/AEM.02701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esseili MA, Wang Q, Zhang Z, Saif LJ. 2012. Internalization of sapovirus, a surrogate for norovirus, in romaine lettuce and the effect of lettuce latex on virus infectivity. Appl Environ Microbiol 78:6271–6279. doi: 10.1128/AEM.01295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei J, Jin Y, Sims T, Kniel KE. 2010. Manure- and biosolids-resident murine norovirus 1 attachment to and internalization by romaine lettuce. Appl Environ Microbiol 76:578–583. doi: 10.1128/AEM.02088-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choffnes ER, Relman DA, Olsen L, Hutton R, Mack RA. 2012. Improving food safety through a one health approach. The National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 15.Garcia BCB, Dimasupil MAZ, Vital PG, Widmer KW, Rivera WL. 2015. Fecal contamination in irrigation water and microbial quality of vegetable primary production in urban farms of Metro Manila, Philippines. J Environ Sci Health B 50:734–743. doi: 10.1080/03601234.2015.1048107. [DOI] [PubMed] [Google Scholar]
- 16.Erickson MC. 2010. Microbial risks associated with cabbage, carrots, celery, onions, and deli salads made with these produce items. Compr Rev Food Sci Food Safety 9:602–619. doi: 10.1111/j.1541-4337.2010.00129.x. [DOI] [PubMed] [Google Scholar]
- 17.Dawson DJ, Paish A, Staffell LM, Seymour IJ, Appleton H. 2005. Survival of viruses on fresh produce, using MS2 as a surrogate for norovirus. J Appl Microbiol 98:203–209. doi: 10.1111/j.1365-2672.2004.02439.x. [DOI] [PubMed] [Google Scholar]
- 18.Stine SW, Song I, Choi CY, Gerba CP. 2005. Effect of relative humidity on preharvest survival of bacterial and viral pathogens on the surface of cantaloupe, lettuce, and bell peppers. J Food Prot 68:1352–1358. [DOI] [PubMed] [Google Scholar]
- 19.Kukavica-Ibrulj I, Darveau A, Jean J, Fliss I. 2004. Hepatitis A virus attachment to agri-food surfaces using immunological, virological and thermodynamic assays. J Appl Microbiol 97:923–934. doi: 10.1111/j.1365-2672.2004.02366.x. [DOI] [PubMed] [Google Scholar]
- 20.Lu L, Ku KM, Palma-Salgado SP, Storm AP, Feng H, Juvik JA, Nguyen TH. 2015. Influence of epicuticular physicochemical properties on porcine rotavirus adsorption to 24 leafy green vegetables and tomatoes. PLoS One 10(7):e0132841. doi: 10.1371/journal.pone.0132841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deboosere N, Pinon A, Caudrelier Y, Delobel A, Merle G, Perelle S, Temmam S, Loutreul J, Morin T, Estienney M, Belliot G, Pothier P, Gantzer C, Vialette M. 2012. Adhesion of human pathogenic enteric viruses and surrogate viruses to inert and vegetal food surfaces. Food Microbiol 32:48–56. doi: 10.1016/j.fm.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Vega E, Garland J, Pillai SD. 2008. Electrostatic forces control nonspecific virus attachment to lettuce. J Food Prot 71:522–529. [DOI] [PubMed] [Google Scholar]
- 23.Croci L, De Medici D, Scalfaro C, Fiore A, Toti L. 2002. The survival of hepatitis A virus in fresh produce. Int J Food Microbiol 73:29–34. doi: 10.1016/S0168-1605(01)00689-4. [DOI] [PubMed] [Google Scholar]
- 24.Predmore A, Sanglay G, Li JR, Lee K. 2015. Control of human norovirus surrogates in fresh foods by gaseous ozone and a proposed mechanism of inactivation. Food Microbiol 50:118–125. doi: 10.1016/j.fm.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Predmore A, Li J. 2011. Enhanced removal of a human norovirus surrogate from fresh vegetables and fruits by a combination of surfactants and sanitizers. Appl Environ Microbiol 77:4829–4838. doi: 10.1128/AEM.00174-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aboubakr HA, Williams P, Gangal U, Youssef MM, El-Sohaimy SAA, Bruggeman PJ, Goyal SM. 2015. Virucidal effect of cold atmospheric gaseous plasma on feline calicivirus, a surrogate for human norovirus. Appl Environ Microbiol 81:3612–3622. doi: 10.1128/AEM.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiCaprio E, Purgianto A, Ma YM, Hughes J, Dai XJ, Li JR. 2015. Attachment and localization of human norovirus and animal caliciviruses in fresh produce. Int J Food Microbiol 211:101–108. doi: 10.1016/j.ijfoodmicro.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez G, Elizaquivel P, Aznar R, Selma MV. 2015. Virucidal effect of high power ultrasound combined with a chemical sanitizer containing peroxyacetic acid for water reconditioning in the fresh-cut industry. Food Control 52:126–131. doi: 10.1016/j.foodcont.2014.12.021. [DOI] [Google Scholar]
- 29.Fransisca L, Park HK, Feng H. 2012. E. coli O157:H7 population reduction from alfalfa seeds with malic acid and thiamine dilauryl sulfate and quality evaluation of the resulting sprouts. J Food Sci 77:M121–M126. doi: 10.1111/j.1750-3841.2011.02553.x. [DOI] [PubMed] [Google Scholar]
- 30.Ölmez H, Kretzschmar U. 2009. Potential alternative disinfection methods for organic fresh-cut industry for minimizing water consumption and environmental impact. LWT Food Sci Technol 42:686–693. doi: 10.1016/j.lwt.2008.08.001. [DOI] [Google Scholar]
- 31.Beuchat LR, Adler BB, Lang MM. 2004. Efficacy of chlorine and a peroxyacetic acid sanitizer in killing Listeria monocytogenes on iceberg and romaine lettuce using simulated commercial processing conditions. J Food Prot 67:1238–1242. [DOI] [PubMed] [Google Scholar]
- 32.Hudson JB, Sharma M, Vimalanathan S. 2009. Development of a practical method for using ozone gas as a virus decontaminating agent. Ozone Sci Eng 31:216–223. doi: 10.1080/01919510902747969. [DOI] [Google Scholar]
- 33.Chrysikopoulos CV, Manariotis ID, Syngouna VI. 2013. Virus inactivation by high frequency ultrasound in combination with visible light. Colloids Surf B Biointerfaces 107:174–179. doi: 10.1016/j.colsurfb.2013.01.038. [DOI] [PubMed] [Google Scholar]
- 34.Zhou B, Feng H, Pearlstein AJ. 2012. Continuous-flow ultrasonic washing system for fresh produce surface decontamination. Innov Food Sci Emerg Technol 16:427–435. doi: 10.1016/j.ifset.2012.09.007. [DOI] [Google Scholar]
- 35.Bozkurt H, D'Souza HD, Davidson PM. 2014. Thermal inactivation of human norovirus surrogates in spinach and measurement of its uncertainty. J Food Prot 77:276–283. doi: 10.4315/0362-028X.JFP-13-289. [DOI] [PubMed] [Google Scholar]
- 36.Bozkurt H, Ye XF, Harte F, D'Souza DH, Davidson PM. 2015. Thermal inactivation kinetics of hepatitis A virus in spinach. Int J Food Microbiol 193:147–151. doi: 10.1016/j.ijfoodmicro.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 37.Predmore A, Sanglay GC, DiCaprio E, Li J, Uribe RM, Lee K. 2015. Electron beam inactivation of Tulane virus on fresh produce, and mechanism of inactivation of human norovirus surrogates by electron beam irradiation. Int J Food Microbiol 198:28–36. doi: 10.1016/j.ijfoodmicro.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 38.Alvaro JE, Moreno S, Dianez F, Santos M, Carrasco G, Urrestarazu M. 2009. Effects of peracetic acid disinfectant on the postharvest of some fresh vegetables. J Food Eng 95:11–15. doi: 10.1016/j.jfoodeng.2009.05.003. [DOI] [Google Scholar]
- 39.Kitis M. 2004. Disinfection of wastewater with peracetic acid: a review. Environ Int 30:47–55. doi: 10.1016/S0160-4120(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 40.Marshall GS. 2009. Rotavirus disease and prevention through vaccination. Pediatr Infect Dis J 28:355–364. [DOI] [PubMed] [Google Scholar]
- 41.Lodder WJ, de Roda Husman AM. 2005. Presence of noroviruses and other enteric viruses in sewage and surface waters in The Netherlands. Appl Environ Microbiol 71:1453–1461. doi: 10.1128/AEM.71.3.1453-1461.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aw TG, Wengert S, Rose JB. 2016. Metagenomic analysis of viruses associated with field-grown and retail lettuce identifies human and animal viruses. Int J Food Microbiol 223:50–56. doi: 10.1016/j.ijfoodmicro.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Parada-Fabian JC, Juarez-Garcia P, Natividad-Bonifacio I, Vazquez-Salinas C, Quinones-Ramirez EI. 2016. Identification of enteric viruses in foods from Mexico City. Food Environ Virol 8:215–220. doi: 10.1007/s12560-016-9244-6. [DOI] [PubMed] [Google Scholar]
- 44.Romero-Maraccini OC, Sadik NJ, Rosado-Lausell SL, Pugh CR, Niu X-Z, Croue J-P, Nguyen TH. 2013. Sunlight-induced inactivation of human Wa and porcine OSU rotaviruses in the presence of exogenous photosensitizers. Environ Sci Technol 47:11004–11012. doi: 10.1021/es402285u. [DOI] [PubMed] [Google Scholar]
- 45.Arnold M, Patton JT, McDonald SM. 2009. Culturing, storage, and quantification of rotaviruses. Curr Protoc Microbiol Chapter 15:Unit 15C.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romero OC, Straub AP, Kohn T, Nguyen TH. 2011. Role of temperature and Suwannee River natural organic matter on inactivation kinetics of rotavirus and bacteriophage MS2 by solar irradiation. Environ Sci Technol 45:10385–10393. doi: 10.1021/es202067f. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Li M, Brockman K, Nguyen TH. 2016. Reduction of MS2 bacteriophage and rotavirus in biosand filters. Environ Sci Water Res Technol 2:483–491. doi: 10.1039/C5EW00297D. [DOI] [Google Scholar]
- 48.Mattioli MC, Pickering AJ, Gilsdorf RJ, Davis J, Boehm AB. 2013. Hands and water as vectors of diarrheal pathogens in Bagamoyo, Tanzania. Environ Sci Technol 47:355–363. doi: 10.1021/es303878d. [DOI] [PubMed] [Google Scholar]
- 49.Jothikumar N, Kang G, Hill VR. 2009. Broadly reactive TaqMan assay for real-time RT-PCR detection of rotavirus in clinical and environmental samples. J Virol Methods 155:126–131. doi: 10.1016/j.jviromet.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 50.Bhowmick R, Halder UC, Chattopadhyay S, Nayak MK, Chawla-Sarkar M. 2013. Rotavirus-encoded nonstructural protein 1 modulates cellular apoptotic machinery by targeting tumor suppressor protein p53. J Virol 87:6840–6850. doi: 10.1128/JVI.00734-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rolsma MD, Kuhlenschmidt TB, Gelberg HB, Kuhlenschmidt MS. 1998. Structure and function of a ganglioside receptor for porcine rotavirus. J Virol 72:9079–9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Azuma Y, Ishikawa Y, Kawai S, Tsunenari T, Tsunoda H, Igawa T, Iida S, Nanami M, Suzuki M, Irie RF, Tsuchiya M, Yamada-Okabe H. 2007. Recombinant human hexamer-dominant IgM monoclonal antibody to ganglioside GM3 for treatment of melanoma. Clin Cancer Res 13:2745–2750. doi: 10.1158/1078-0432.CCR-06-2919. [DOI] [PubMed] [Google Scholar]
- 53.Nilsson EC, Jamshidi F, Johansson SM, Oberste MS, Arnberg N. 2008. Sialic acid is a cellular receptor for coxsackievirus A24 variant, an emerging virus with pandemic potential. J Virol 82:3061–3068. doi: 10.1128/JVI.02470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herdt J, Feng H. 2009. Aqueous antimicrobial treatments to improve fresh and fresh-cut produce safety. In Fan X, Niemira BA, Doona CJ, Feeherry FE, Gravani RB (ed). Microbial safety of fresh produce. Wiley-Blackwell, Oxford, United Kingdom. [Google Scholar]
- 55.US Environmental Protection Agency. 2015. Tsunami 100. US Environmental Protection Agency, Office of Chemical Safety and Pollution Prevention, Washington, DC. [Google Scholar]
- 56.Kohn T, Nelson KL. 2007. Sunlight-mediated inactivation of MS2 coliphage via exogenous singlet oxygen produced by sensitizers in natural waters. Environ Sci Technol 41:192–197. doi: 10.1021/es061716i. [DOI] [PubMed] [Google Scholar]
- 57.Gutierrez L, Li X, Wang J, Nangmenyi G, Economy J, Kuhlenschmidt TB, Kuhlenschmidt MS, Nguyen TH. 2009. Adsorption of rotavirus and bacteriophage MS2 using glass fiber coated with hematite nanoparticles. Water Res 43:5198–5208. doi: 10.1016/j.watres.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 58.Farkas K, Varsani A, Pang L. 2015. Adsorption of rotavirus, MS2 bacteriophage and surface-modified silica nanoparticles to hydrophobic matter. Food Environ Virol 7:261–268. doi: 10.1007/s12560-014-9171-3. [DOI] [PubMed] [Google Scholar]
- 59.Oh SR, Park SY, Ha SD. 2014. Combined effects of chlorine and thiamine dilauryl sulfate on reduction of Listeria monocytogenes in chicken breast and development of predictive growth models. Poult Sci 93:1503–1510. doi: 10.3382/ps.2013-03427. [DOI] [PubMed] [Google Scholar]
- 60.Otzen D. 2011. Protein-surfactant interactions: a tale of many states. Biochim Biophys Acta 1814:562–591. doi: 10.1016/j.bbapap.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Baker M, Prasad BVV. 2010. Rotavirus cell entry, p 121–148. In Johnson EJ. (ed), Cell entry by non-enveloped viruses. Springer, Berlin, Germany. [Google Scholar]
- 62.Coulson BS. 2015. Expanding diversity of glycan receptor usage by rotaviruses. Curr Opin Virol 15:90–96. doi: 10.1016/j.coviro.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 63.Sano D, Ohta T, Nakamura A, Nakagomi T, Nakagomi O, Okabe S. 2015. Culture-independent evaluation of nonenveloped-virus infectivity reduced by free-chlorine disinfection. Appl Environ Microbiol 81:2819–2826. doi: 10.1128/AEM.03802-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


