Abstract
Autophagy controls cell homeostasis and provides a rapid response to a variety of stresses. Although many steps of the autophagy process have been elucidated, how they are temporally regulated is less well characterized. Recently, we reported that dynamic interaction of the pro-autophagic factor AMBRA1 with CULLIN E3 ubiquitin ligases ensures the timely onset and termination of the autophagy response.
Keywords: autophagy, AMBRA1, ubiquitin proteasome system, DEPTOR, ELONGIN B, DDB1
Autophagy is a rapid response that is activated by cells to cope with a variety of stresses. By engulfing damaged or supernumerary components within autophagosomal vesicles and delivering them to the lysosome for degradation, autophagy attempts to remove damage caused by the stress, in addition to providing substrates for new synthesis and energy production.1
Stress signaling pathways activate the autophagy machinery through post-translational modifications of the upstream autophagy complexes of the serine/threonine-protein kinase ULK1 and BECLIN-1, including phosphorylation, ubiquitination, and acetylation.2 An important role in conveying signals to the autophagy machinery is played by activating molecule in BECN1-regulated autophagy protein 1 (AMBRA1), a WD40 protein that interacts with both BECLIN-1 and ULK1 and is required for activation of these autophagy complexes by mediating K63-chain ubiquitination.3
To be cell protective, the autophagy response should be highly selective and temporally controlled to avoid unwanted and excessive digestion. Selectivity is ensured by families of receptor proteins that bridge the autophagosomal protein microtubule-associated proteins 1A/1B light chain 3 (LC3) to substrates, which are usually labeled by specific modifications such as ubiquitination or glycosylation.4
Less well characterized are the mechanisms governing the temporal dynamics of autophagy. We recently showed that the timing of the autophagy response is controlled by dynamic interaction between the pro-autophagic protein AMBRA1 and two CULLIN E3 ubiquitin ligases.5
CULLINs are involved in the regulation of numerous cellular processes, such as proliferation, differentiation, cell metabolism, and stress response.6 Seven canonical CULLIN complexes have been identified in mammals.6 These E3 ligase complexes contain: (i) a RING finger protein, RBX1 (E3 ubiquitin-protein ligase RBX1) or RBX2, which mediates the interaction with the E2 enzyme, (ii) a scaffold protein CULLIN, (iii) a receptor-adaptor specific for each CULLIN (with the exception of CULLIN 3): S-phase kinase-associated protein 1 (SKP1) for CULLIN 1 and 7; transcription elongation factor B polypeptide 2 B/C (ELONGIN B/C) for CULLIN 2 and 5; and DNA damage-binding protein 1 (DDB1) for CULLIN 4A and 4B; and (iii) a member of a large family of substrate receptors, which confer specificity to the E3 ligase.
In this study, we showed that AMBRA1 interacts with different adaptors of the CULLIN complexes and that these interactions play a role in the temporal regulation of the onset and termination of the autophagy response.
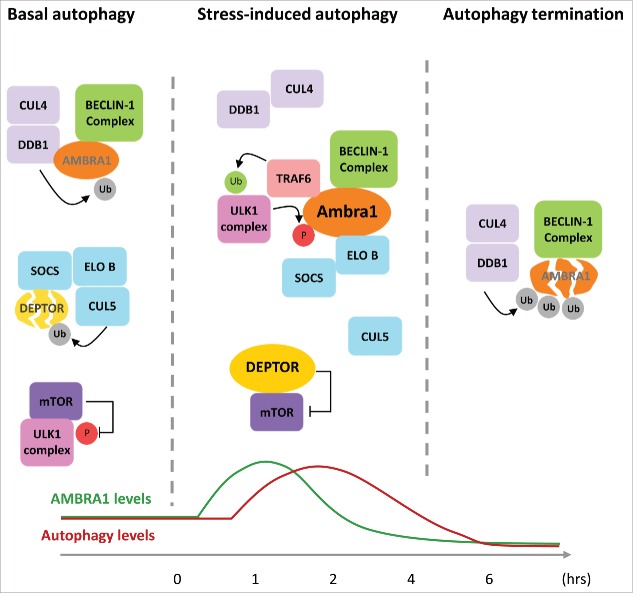
In unstressed conditions, AMBRA1 is maintained at a low level, with a large fraction of the protein being ubiquitinated by the CULLIN 4 complex and degraded by the proteasome (Fig. 1, left side). Recruitment to CULLIN 4 is mediated by the amino-terminal WD40 domain of AMBRA1, which directly interacts with the CULLIN 4 adaptor DDB1. This interaction is relevant to limit basal autophagy, since downregulation of DDB1 leads to an increase in the rate of autophagosome formation.
Figure 1.
Crosstalk between AMBRA1 and CULLIN E3 ubiquitin ligases regulates autophagy. In unstressed conditions, AMBRA1 protein levels are kept low by the DDB1/CULLIN 4 E3 ubiquitin ligase (CUL4) complex, ULK1 activity is repressed by mTOR, and DEPTOR protein is degraded by a CULLIN 5 (CUL5) complex containing ELONGIN B (ELO B) and a substrate receptor of the suppressor of cytokine signaling (SOCS) family. Autophagy stimuli rapidly lead to AMBRA1 protein stabilization by inducing AMBRA1-DDB1 dissociation in an ULK1-dependent manner. AMBRA1 is then able to interact with TRAF6 and activate ULK1 and BECLIN-1 complexes. Moreover, once free from CULLIN 4, AMBRA1 is able to inhibit the activity of the distinct E3 ubiquitin ligase complex, CULLIN 5, by interacting with its adaptor protein ELONGIN B. This leads to stabilization of the mTor inhibitor DEPTOR, which allows timely activation of ULK1 by decreasing the inhibitory phosphorylation mediated by mTOR. In prolonged stress conditions, CULLIN 4 re-associates with AMBRA1, causing rapid degradation and ending the autophagy response. Ub: Ubiquitin, P: Phosphorylation.
Autophagy stimuli lead to a rapid accumulation of AMBRA1 as a result of its release from DDB1 upon phosphorylation by ULK1 (Fig. 1, center).
Once stabilized, AMBRA1 contributes to autophagy induction not only by stimulating BECLIN-1, TRAF6 (TNF receptor-associated factor 6), and ULK1 activity as previously characterized,7,8 but also by interacting with ELONGIN B, the adaptor protein of distinct CULLIN complexes CULLIN 2 and 5. Interestingly, competition experiments showed that the interactions of AMBRA1 with DDB1 and ELONGIN B are mutually exclusive.
While searching for a role of this interaction in the autophagy response, we found that Ambra1 binds ELONGIN B to inhibit CULLIN 5 activity and allow the stabilization of DEP domain-containing mTOR-interacting protein (DEPTOR), an inhibitor of the serine/threonine-protein kinase mTOR.9 The stabilization of DEPTOR mediated by AMBRA1 has been proved to be important for rapid onset of autophagy, as indicated by its role in the sharp decrease in the inhibitory phosphorylation of ULK1 mediated by mTOR.
The interaction of AMBRA1 with ELONGIN B is transient. Its dissociation is followed by re-binding of DDB1 and CULLIN 4-mediated degradation, which leads to timely termination of the autophagy response, even when stress conditions persist (Fig. 1, right side). In fact, we observed that silencing DDB1 extends the duration of the autophagy process, which results in a higher rate of cell death.
CULLIN 4 is known to play an important role in the response to DNA damage.10 Interestingly, Cullin 4A mutant cells show an increased resistance to UV-mediated DNA damage associated with a prolonged DNA repair response as a result of reduced degradation of DNA damage sensors, such as DDB2 and DNA repair protein complementing XP-C (XPC).10 In light of our results, it is tempting to speculate that CULLIN 4 function may have evolved to temporally limit the induction of different stress pathways.
In conclusion, our data show that interaction of the pro-autophagic protein AMBRA1 with 2 CULLIN complexes plays an important role in temporal control of the autophagic response. Identification of this interplay between autophagy and the ubiquitin-proteasome system may provide novel insights into how autophagy is altered in human diseases. For example, as CULLIN 4 activity is upregulated in a variety of tumors,11 it is conceivable that this alteration leads to autophagy deregulation and contributes to tumor development.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from AIRC (IG2012–13529), the Telethon Foundation (GEP12072), the Italian Ministry of University and Research (FIRB Accordi di Programma 2011) and the Italian Ministry of Health (Ricerca Finalizzata and Ricerca Corrente).
References
- 1.Mizushima N. Autophagy in protein and organelle turnover. Cold Spring Harb Symp Quant Biol 2011; 76:397-402; PMID: 21813637; http://dx.doi.org/ 10.1101/sqb.2011.76.011023 [DOI] [PubMed] [Google Scholar]
- 2.Wirth M, Joachim J, Tooze SA. Autophagosome formation–the role of ULK1 and beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol 2013; 23:301-9; PMID: 23727157; http://dx.doi.org/ 10.1016/j.semcancer.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 3.Fimia GM, Corazzari M, Antonioli M, Piacentini M. Ambra1 at the crossroad between autophagy and cell death. Oncogene 2012; 32:3311-8; PMID: 23069654; http://dx.doi.org/ 10.1038/onc.2012.455 [DOI] [PubMed] [Google Scholar]
- 4.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 2014; 16:495-501; PMID: 24875736; http://dx.doi.org/ 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- 5.Antonioli M, Albiero F, Nazio F, Vescovo T, Perdomo AB, Corazzari M, Marsella C, Piselli P, Gretzmeier C, Dengjel J, et al.. AMBRA1 interplay with CULLIN E3 ubiquitin ligases regulates autophagy dynamics. Dev Cell 2014; 31:734-46; PMID: 25499913; http://dx.doi.org/ 10.1016/j.devcel.2014.11.013 [DOI] [PubMed] [Google Scholar]
- 6.Lydeard JR, Schulman BA, Harper JW. Building and remodelling CULLIN-RING E3 ubiquitin ligases. EMBO Rep 2013; 14:1050-61; PMID: 24232186; http://dx.doi.org/ 10.1038/embor.2013.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al.. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 2010; 191:155-68; PMID: 20921139; http://dx.doi.org/ 10.1083/jcb.201002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al.. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15:406-16; PMID: 23524951; http://dx.doi.org/ 10.1038/ncb2708 [DOI] [PubMed] [Google Scholar]
- 9.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009; 137:873-86; PMID: 19446321; http://dx.doi.org/ 10.1016/j.cell.2009.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, Yin Y, Koff A, Ma L, Zhou P. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell 2009; 34:451-60; PMID: 19481525; http://dx.doi.org/ 10.1016/j.molcel.2009.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]