ABSTRACT
The oncoprotein MDM2 is recognized as a major negative regulator of the p53 tumor suppressor but growing evidence indicates that its oncogenic activities extend beyond p53. We show that MDM2 is recruited to chromatin independently of p53 to regulate a transcriptional program implicated in amino acid metabolism and redox homeostasis.
KEYWORDS: Cancer, chromatin, Mdm2, metabolism, redox
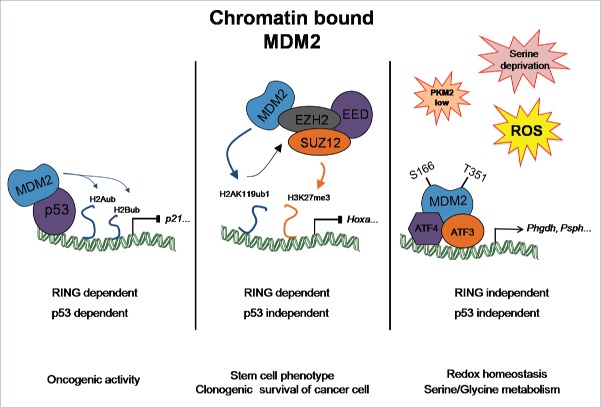
Metabolic rewiring is considered a hallmark of malignant transformation, and multiple oncogenes and tumor suppressors control various metabolic pathways.1 Among these, the p53 protein (TP53, best known as p53) is now recognized as an important regulator of metabolism and alterations of p53 functions have been proposed to directly contribute to the metabolic reprogramming of cancer cells. Beside its role in glycolysis and oxidative phosphorylation, the complexity of the metabolic network controlled by p53 is illustrated by its role in lipid and amino acid metabolism.2,3 However, the role of other components of the p53 pathway in metabolism has been poorly investigated to date. Using pan-genome approaches (ChIP-seq and gene expression arrays), we recently identified an unexpected function of the mouse double minute 2 (MDM2) oncoprotein, an essential negative regulator of p53, in the transcriptional activation of a program involved in amino acid metabolism and in the control of the redox state of cancer cells. Interestingly, we genetically demonstrated that the recruitment of MDM2 to its target genes occurs independently of p53 but through direct binding to activating transcription factors 3 and 4 (ATF3/4). Of particular note, we found that MDM2-E3 ligase activity is dispensable for its recruitment to chromatin and regulation of its target genes. As schematized in Fig. 1, these results illustrate the complex roles of chromatin-bound MDM2 that controls transcription both in a p53-dependent and independent manner. Thus, on the one hand, recent data from the Dobbelstein laboratory showed that chromatin-bound MDM2 influences transcriptional repression of genes involved in cell identity and pluripotency through its direct interaction with the polycomb repressor complex 2 (PRC2).4 As for MDM2 target genes involved in metabolism, MDM2-mediated repression of these lineage-specific genes occurred independently of p53. On the other hand, M. Oren's laboratory previously demonstrated that MDM2 is recruited to the p53 target gene p21 to repress transcription through ubiquitylation of histones H2A and H2B.5
Figure 1.
Functions of chromatin-associated MDM2. Schematic model showing that chromatin-bound MDM2 contributes to transcriptional regulation though various p53-dependent and -independent mechanisms depending on its partner proteins.
Functional characterization of these new metabolic functions of chromatin-bound MDM2 highlighted a role in amino acid metabolism and transport, and more specifically in the serine synthesis pathway. Serine/glycine metabolism supports the growth of cancer cells by contributing to their anabolic demands, their epigenome, and by regulating their redox state. Strikingly, we found that chromatin-bound MDM2 operates independently of p53 to control serine/glycine metabolism and sustain cancer growth in conditions of serine and glycine deprivation. Our results indicate that interfering with MDM2 functions when cancer cells face depleted pools of exogenous serine and glycine compromises their proliferative capacities and tumorigenic potential. These defects result, at least in part, from perturbations of their redox state, likely through several synergistic mechanisms impinging on glutathione metabolism, the NAD+/NADH ratio, and reactive oxygen species (ROS) levels. However, given the central role of serine/glycine metabolism in various anabolic pathways, the metabolic consequences of MDM2 depletion under serine and glycine deprivation are likely to be broader than the observed perturbation of the redox state of these cells. Recent findings have highlighted the essential role of serine/glycine metabolism in histone and DNA/RNA methylation through the one-carbon cycle and the generation of S-adenosyl-methionine (SAM),6 a major methyl-donor co-factor. These results raise interesting questions regarding the potential links between MDM2 deregulation and epigenetic alterations that commonly occur during cancer progression. Moreover, our pan-genome analysis of MDM2-target genes suggests that the functions of MDM2 in cancer cell metabolism extend beyond serine/glycine metabolism and may also contribute to cysteine and glutamine levels. These data underline the broad effects of MDM2 on cellular metabolism.
Our data also revealed an important role of MDM2 in the metabolic network regulated by pyruvate kinase M2 (PKM2), a key glycolytic enzyme that converts phopshoenolpyruvate (PEP) into pyruvate and is commonly deregulated in cancer cells. We found that MDM2 is efficiently recruited to chromatin in cells exhibiting low PKM2 activity, as well as in 2 experimental conditions that alter PKM2 activity, oxidative stress and serine deprivation. PKM2 inhibits MDM2 recruitment to chromatin through its phosphorylation on Ser166 and Thr351. Nevertheless, our results obtained with recombinant proteins, as well as those from a recent study showing that PKM2 lacks protein kinase activity,7 do not support the notion that MDM2 is a direct target of PKM2 but rather a target of a yet unidentified kinase that associates with PKM2. Together, our data highlight a previously unsuspected network involving PKM2, MDM2, and ATFs in the regulation of a transcriptional program involved in serine metabolism and redox homeostasis.
A previous study highlighted that p53-mutated cells are hypersensitive to serine and glycine deprivation.3 The potential connections of these observations with our findings showing that MDM2 regulates serine metabolism may reflect the well-described role of p53 in Mdm2 transcription and the documented downregulation of MDM2 protein level in many p53 null cells. However, other reports showing that p53 directly represses several MDM2 target genes identified in our study, including phosphoserine phosphatase (Phgdh) and neutral amino-acid transporter (Slc7a11), illustrate the complex roles of the p53 pathway in these metabolic pathways.2,8 Further studies are warranted to study how these metabolic functions of MDM2 synergize with its well-described antagonistic role on p53. Finally, the clinical implications of our findings are underlined by bioinformatic analyses showing the significance of the expression level of genes involved in serine metabolism to predict breast and lung cancer patient survival.9,10
Together, our data showing that MDM2 controls cancer cell metabolism pave the way for therapeutic strategies targeting these unexpected functions of this commonly deregulated oncogene, in particular in cancers harboring MDM2 overexpression.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by grants from the Institut National du Cancer (INCa), the Ligue contre le Cancer (Equipe labelisée 2016), and the INSERM-Avenir program. R.R was supported by a fellowship from the French ministry of research and the ARC foundation.
References
- 1.Boroughs LK, Deberardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 2015; 17:351-9; PMID:25774832; http://dx.doi.org/ 10.1038/ncb3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le J, Kon N, Li T, Wang S-J, Su T, Hibshoosh H, Baer R, Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015; 520:1-6; PMID:25799988; http://dx.doi.org/10.103823242140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maddocks ODK, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 2013; 493:542-6; PMID:23242140; http://dx.doi.org/ 10.1038/nature11743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wienken M, Dickmanns A, Nemajerova A, Kramer D, Najafova Z, Weiss M, Karpiuk O, Kassem M, Zhang Y, Lozano G, et al.. MDM2 Associates with Polycomb Repressor Complex 2 and Enhances Stemness-Promoting Chromatin Modifications Independent of p53. Mol Cell 2016; 61:68-83; PMID:26748827; http://dx.doi.org/ 10.1016/j.molcel.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell 2004; 16:631-9; PMID:15546622; http://dx.doi.org/ 10.1016/j.molcel.2004.10.016 [DOI] [PubMed] [Google Scholar]
- 6.Maddocks ODK, Labuschagne CF, Adams PD, Vousden KH. Serine metabolism supports the methionine cycle and DNA/RNA methylation through de novo ATP synthesis in cancer cells. Mol Cell 2016; 61(2):1-1; PMID:26748607; http://dx.doi.org/10.1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosios AM, Fiske BP, Gui DY, Vander Heiden MG. Lack of evidence for PKM2 protein kinase activity. Mol Cell 2015; 59:850-7; PMID:26300261; http://dx.doi.org/ 10.1016/j.molcel.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ou Y, Wang S-J, Jiang L, Zheng B, Gu W. p53 Protein-mediated regulation of phosphoglycerate dehydrogenase (PHGDH) is crucial for the apoptotic response upon serine starvation. J Biol Chem 2015; 290:457-66; PMID:25404730; http://dx.doi.org/ 10.1074/jbc.M114.616359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeNicola GM, Chen P-H, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, et al.. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 2015; 12:1475-81; PMID:26482881; http://dx.doi.org/ 10.1038/ng.3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonov A, Agostini M, Morello M, Minieri M, Melino G, Amelio I. Bioinformatics analysis of the serine and glycine pathway in cancer cells. Oncotarget 2014; 22:11004-13; PMID:25436979; http://dx.doi.org/ 10.18632/oncotarget.2668 [DOI] [PMC free article] [PubMed] [Google Scholar]