Abstract
Oxanine (O) is a deamination product derived from guanine with the nitrogen at the N1 position substituted by oxygen. Cytosine, thymine, adenine, guanine as well as oxanine itself can be incorporated by Klenow Fragment to pair with oxanine in a DNA template with similar efficiency, indicating that oxanine in DNA may cause various mutations. As a nucleotide, deoxyoxanosine may substitute for deoxyguanosine to complete a primer extension reaction. Endonuclease V, an enzyme known for its enzymatic activity on uridine-, inosine- and xanthosine-containing DNA, can cleave oxanosine-containing DNA at the second phosphodiester bond 3′ to the lesion. Mg2+ or Mn2+, and to a small extent Co2+ or Ni2+, support the oxanosine-containing DNA cleavage activity. All four oxanosine-containing base pairs (A/O, T/O, C/O and G/O) were cleaved with similar efficiency. The cleavage of double-stranded oxanosine-containing DNA was ∼6-fold less efficient than that of double-stranded inosine-containing DNA. Single-stranded oxanosine-containing DNA was cleaved with a lower efficiency as compared with double-stranded oxanosine-containing DNA. A metal ion enhances the binding of endonuclease V to double-stranded and single-stranded oxanosine-containing DNA 6- and 4-fold, respectively. Hypothetic models of oxanine-containing base pairs and deaminated base recognition mechanism are presented.
INTRODUCTION
DNA exposed to nitrosative stress or high temperatures experiences accelerated purine base deamination (1–3). Hypoxanthine and xanthine are two well-known deamination products of adenine and guanine, respectively. Previous studies indicate that the treatment of nucleotides and DNA with nitrous acid, nitric oxide (NO) or 1-nitrosoindole-3-acetonitrile yields another guanine deamination product, oxanine (O) (4,5). Oxanine differs from guanine at the N1 position, in which the nitrogen is substituted by oxygen, resulting in intracyclic deamination. The N-glycosidic bond in deoxyoxanosine triphosphate (dOTP) is as stable as that in deoxyguanosine triphosphate (6). The stabilities of O-containing base pairs are ∼2-fold lower than the natural G/C base pair (6). Escherichia coli Klenow Fragment of DNA polymerase I inserts dOTP to pair with both C or T in a DNA template, thus, potentially generating G/C to A/T transitions during DNA replication (7,8). Because of the O-acylisourea structure, oxanine can cross-link with glycine and proteins (9–11). Oxanosine was initially discovered as an antibiotic from Streptomyces capreolus, and as an antineoplastic agent by its ability to inhibit the growth of HeLa cells (12–15). The mutagenicity and chemical reactivity of oxanine may underlie its genotoxicity and cytotoxicity, which in turn, may account for its antibacterial and antineoplastic properties (12–15).
Endonuclease V (endo V) was initially discovered in E.coli as a DNA repair enzyme because it nicked DNA that was treated with acid, exposed to osmium tetroxide or irradiated with ultraviolet light (16,17). A series of biochemical analyses reveals that endo V is active toward several deaminated DNA bases; including uridine, inosine and xanthine (18–23). The cleavage site generated by endo V is at the second phosphodiester bond 3′ to a lesion (18,23). Endo V is also active toward AP and urea sites (18,23), base pair mismatches (23,24), Flap and pseudo Y structures (25) and small insertions/deletions (25,26). Genetic analysis in E.coli suggests that endo V is involved in the repair of inosine, xanthosine and N-6-hydroxylaminopurine (27–30).
Given the broad substrate specificities, in particular toward deaminated bases, we investigated the possible activity of endo V on O-containing DNA. Here, we report the mutagenicity of O-containing DNA and the deoxyoxanosine cleavage activity of endo V. Oxanine in DNA templates can pair with adenine, thymine, guanine, cytosine with similar efficiency; as well as oxanine, as assayed using the Klenow Fragment. Endo V nicks O-containing DNA at the second phosphodiester bond 3′ to the lesion. The metal requirement for the nicking activity can be satisfied best by Mg2+ or Mn2+, and to a much less extent by Co2+ or Ni2+. All four O-containing base pairs (A/O, T/O, G/O and C/O), as well as single-stranded O-containing DNA are substrates for endo V.
MATERIALS AND METHODS
Materials
All routine chemical reagents were purchased from Sigma Chemicals (St. Louis, MO), Fisher Scientific (Suwanee, GA) or VWR (Suwanee, GA). Klenow Fragment (3′–5′ exo-) was purchased from New England Biolabs (Beverly, MA). Deoxyribonucleoside triphosphates (dNTPs) were purchased from Sigma or Promega (Madison, WI). GeneScan Stop Buffer consisted of 80% formamide (Amresco, Solon, OH), 50 mM EDTA (pH 8.0) and 1% blue dextran. TB buffer (1×) consisted of 89 mM Tris base and 89 mM boric acid. Endo V from Thermotoga maritima (Tma) was purified as described previously (23). Recombinant Salmonella endo V was purified by Hitrap chelating and Q column chromotagraphy (H. Feng, A. M. Klutz and W. Cao, unpublished data).
Oligodeoxyribonucleotides
Oligodeoxyribonucleotides except for oxanosine-containing DNA were synthesized and purified by Integrated DNA Technologies, Inc. (Coralville, IA). dOTP was generated by treating deoxyguanosine triphosphate with acidified nitrite as described previously (8). O-containing DNA was constructed by DNA polymerase-based nucleotide extension similar to the method described previously (31,32). Briefly, dOTP was generated by treating dGTP with acidified sodium nitrite as described in (31). dOTP, which migrated ∼4 min ahead of the remaining dGTP on a reverse phase C18 column by HPLC, was pooled and freeze-dried. A single nucleotide extension reaction was performed to add an oxanine base to a 36mer. The resulting 37mer was purified by denaturing sequencing gel electrophoresis. No contaminating 36mer was detected in the purified O-containing 37mer. The 37mer was re-annealed to a DNA template (66mer) and extended to full-length (62mer) in the presence of regular dNTPs. The O-containing 62mer was purified and dried as described above, and dissolved in TE.
Mutagenicity of oxanine in DNA
Single nucleotide extension reactions were carried out at 37°C for 30 min in a 10 μl vol containing 50 μM dOTP or other dNTPs, 10 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 7.5 mM DTT, 10% glycerol, 0.01 U/μl of Klenow Fragment (3′ exo-), and 10 nM single nucleotide gap substrate composed of EV42O d(GGACCAATTGGACGOATCCGTCACGCGTCTGCTAGCAGGAC, O = deoxyoxanosine), O-04.B d(HEX-GTCCTGCTAGCAGACGCGTGACGGAT) and O-15.B d(CGTCCAATTGGTCC). Reactions were quenched by the addition of an equal volume of GeneScan stop buffer. The mixtures were then incubated at 94°C for 3 min and cooled on ice directly before loading. Samples (3.5 μl) were loaded onto a 7 M urea–10% denaturing polyacrylamide gel. Electrophoresis was conducted at 1500 V for 1.6 h using an ABI 377 sequencer (Applied Biosystems). Cleavage products and remaining substrates were quantified using GeneScan analysis software.
Tma endo V cleavage reaction
The cleavage reactions were performed at 65°C for 60 min unless indicated otherwise in a 10 μl reaction mixture containing 10 mM HEPES-KOH (pH 7.4), 1 mM DTT, 2% glycerol, 5 mM MgCl2 unless otherwise specified, 10 nM DNA substrate and the indicated amount of purified Tma endo V protein. In the case of E.coli and Salmonella endo V, the reactions were performed at 37°C instead of 65°C. Reactions were quenched by the addition of equal volume of GeneScan stop buffer. The mixtures were then incubated at 94°C for 3 min and cooled on ice directly before loading. Samples (3.5 μl) were loaded onto a 7 M urea–10% denaturing polyacrylamide gel. Electrophoresis was conducted at 1500 V for 1.6 h using an ABI 377 sequencer (Applied Biosystems). Cleavage products and remaining substrates were quantified using GeneScan analysis software.
Gel mobility shift assays
The binding reaction mixtures (10 μl) contained 50 nM double-stranded or single-stranded fluorescently labeled oligonucleotide DNA substrate, 5 mM CaCl2, or 2 mM EDTA, 20% glycerol, 10 mM HEPES-KOH (pH 7.4), 1 mM DTT and the indicated amount of Salmonella endo V protein. The binding reactions were carried out at 37°C for 30 min. Samples were electrophoresed on a 6% native polyacrylamide gel in 1× TB buffer supplemented with 5 mM CaCl2 or 2 mM EDTA. The bound and free DNA species were analyzed using Typhoon 9400 (Amersham Biosciences) with the following settings: PMT at 600 V, excitation at 532 nm and emission at 526 nm (filter 526 SP). Data analyses were carried out using ImageQuant 5.2 (Amersham Biosciences).
RESULTS
Mutagenicity of oxanine in DNA
The mutagenicity of dOTP has been addressed in a previous study (8). In addition to incorporating dOTP to pair with C, Klenow Fragment also misincorporates dOTP to pair with T, causing G/C to A/T transitions (8). To understand the mutagenicity of oxanine in DNA templates, we assembled a single nucleotide gap substrate with a single O in the template strand (Figure 1A). Individual dNTPs were added to the extension reaction to test which nucleotide can be incorporated to pair with the O in the template by the Klenow Fragment (3′-exo). We found that each of the four natural dNTPs was incorporated across from oxanine in the template strand with similar efficiency (∼35% incorporation) (Figure 1A). Note the primer extension product was extended slightly higher for the C/O reaction because the base 5′ adjacent to O was G, which should allow deoxycytidine triphosphate to extend one more base (Figure 1A). Surprisingly, dOTP was found to pair with the O in the template with a slightly higher efficiency (42% incorporation) (Figure 1A). These results suggested that oxanine in DNA is potentially more mutagenic than in the nucleotide form.
Figure 1.
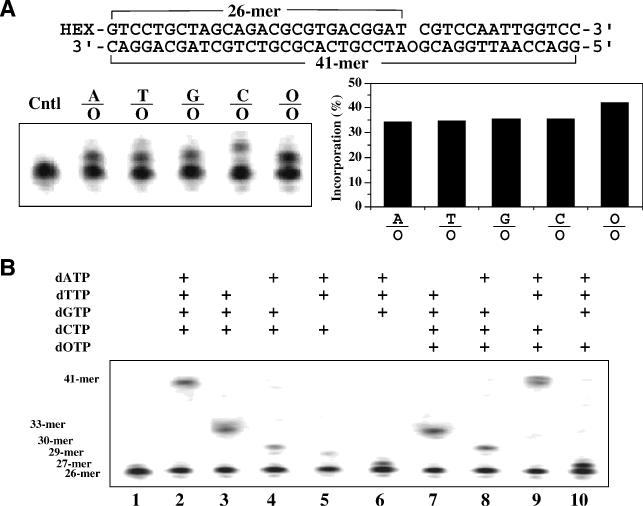
Mutagenicity of oxanine in DNA. (A) Addition of dNTP to O-containing DNA by E.coli polymerase I Klenow Fragment (3′ exo-). Addition of dOTP and individual dNTP by Klenow Fragment was assayed as detailed in Materials and Methods. HEX, fluorophore (Integrated DNA Technologies) and Cntl, substrate control. (B) Extension of primer on O-containing template. The primer extension reactions were performed as detailed in Materials and Methods with indicated species of dNTPs.
To further study the effect of oxanine in the DNA template, we performed primer extension reactions with different combinations of dNTPs (Figure 1B). As expected, in the presence of A, T, C and G, a completely extended product was observed (Figure 1B, lane 2). Omission of A, T, G or C resulted in limited extension to form a product at approximately 33mer, 30mer, 29mer or 27mer position, respectively (Figure 1B, lanes 3–6). When C was absent, a 1 nt extension product at about 27mer position was observed (Figure 1B, lane 6). Based on the results from Figure 1A, it is likely that other nucleotides (A, T or G) could be incorporated to pair with O in the template. Since the adjacent position 5′ upstream to O was G, the lack of C and the formation of a non-C/O base pair may make it difficult to further extend the primer strand (Figure 1B, lane 6). Similarly, the other three extension reactions in the absence of A, T or G may have formed a mismatch base pair at the 3′ end, which prevented it from further extension (Figure 1B, lanes 3–5). The next set of extension reactions examined the possibility of substituting a regular nucleotide with oxanosine (Figure 1B, lanes 7–10). Evidently, O can substitute only for G to generate a fully extended product at about 41mer location (Figure 1B, lane 9).
Endo V cleavage of O-containing DNA and its metal ion dependency
Endo V is a repair enzyme with broad substrate specificity, in particular for deaminated base damage. This biochemical property prompted us to examine its potential cleavage activity on O-containing DNA. Initially, we assayed cleavage activity using four double-stranded O-containing substrates (A/O, G/O, T/O and C/O). Cleavage products were detected with all four substrates (Figure 2). The accumulation of cleavage products was proportional to the amount of input Tma endo V (Figure 2). The primary cleavage site (38mer position) was at the second phosphodiester bond 3′ to the lesion (Figure 2 and data not shown), in keeping with previous observations with inosine-containing substrates (18,23). Minor cleavage could occur at the third phosphodiester bond 3′ to the lesion at high enzyme concentrations (Figure 2). In addition to endo V from Tma, E.coli and Salmonella endo V can also cleave O-containing DNA (data not shown).
Figure 2.
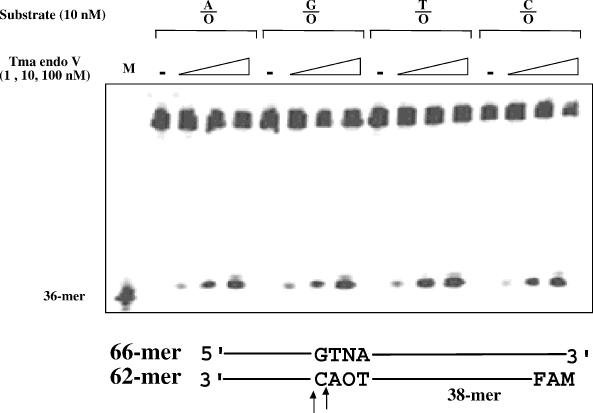
Cleavage activity of purified Tma endo V on O-containing DNA. The cleavage reactions were performed as detailed in Materials and Methods with 1, 10 and 100 nM Tma endo V protein. Cleavage products were separated from remaining substrate by electrophoresis. M, 36mer marker; N = A, T, G or C; FAM, fluorophore (Integrated DNA Technologies); and -, control reactions without addition of endo V.
Since endo V is a metal-dependent enzyme (18,23), divalent metal ions were tested for their effects on cleavage of O-containing DNA by Tma endo V. Among the seven metal ions tested, four of them supported cleavage of double-stranded T/O (Figure 3A), but the cleavage activity in the presence of Co2+ or Ni2+ was 10-fold lower than that in the presence of Mg2+ or Mn2+ (Figure 3B). These results are consistent with the previous studies using inosine-containing DNA (18).
Figure 3.
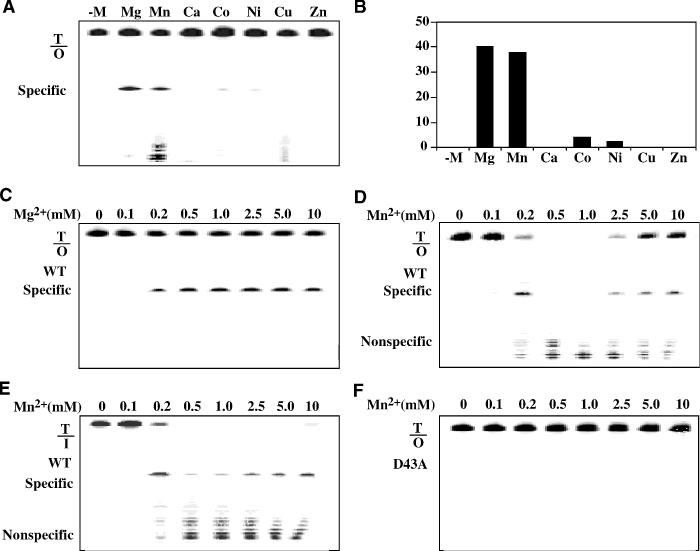
Effect of divalent metals on Tma endo V cleavage activity. The cleavage reactions containing 100 nM Tma endo V and various metal ions were incubated at 65°C for 30 min before electrophoresis. (A) Electrophoresis diagram of metal ion dependency. (B) Quantitative analysis of cleavage activity with various metal ions. (C) Titration of Mg2+ with wt Tma endo V and T/O substrate. (D) Titration of Mn2+ with wt Tma endo V and T/O substrate. (E) Titration of Mn2+ with wt Tma endo V and T/I substrate. (F) Titration of Mn2+ with D43A mutant Tma endo V and T/O substrate.
Noticing the significant non-specific cleavage in the presence of Mn2+ (Figure 3A), we performed a metal titration with the two most active metal ions, Mg2+ or Mn2+. Consistent with the results shown in Figure 3A, we did not observe non-specific cleavage across the 10 mM Mg2+ range (Figure 3C). However, non-specific cleavage was detected from 0.2 to 10 mM Mn2+ range (Figure 3D). In the range of 0.5–1.0 mM Mn2+, all the T/O substrate was digested to non-specific product. These results indicated that the Mn2+-dependent non-specific nuclease activity was most active with 0.2–1.0 mM Mn2+ (Figure 3D). To determine whether the non-specific cleavage activity was unique to O-containing substrate, we titrate Mn2+-dependent cleavage of a T/I substrate. Non-specific cleavage was also observed in the range of 0.2–10 mM Mn2+ (Figure 3E). The Mn2+-dependent non-specific nuclease activity on the T/I substrate was most active with 0.5–1.0 mM Mn2+ (Figure 3E). To confirm the Mn2+-dependent non-specific nuclease activity was intrinsic to endo V, Tma endo V D43A mutant, which is unable to undergo catalysis due to an amino acid substitution within the active site of the enzyme (33), was used to perform the assay. No signs of specific or non-specific cleavage were observed, indicating that both the lesion-specific endonuclease activity and Mn2+-dependent non-specific nuclease activity were intrinsic to endo V (Figure 3F).
Cleavage and binding kinetics of endo V on double-stranded O-containing DNA
To determine the effects bases opposite the oxanine lesion may have on cleavage, we studied the cleavage kinetics using substrate in excess [enzyme:substrate (E:S) ratio of 1:10], E:S in equal molar ratio (E:S = 1:1), and enzyme in excess (E:S = 10:1) (Figure 4A). When the substrate was in excess, cleavage of the T/O substrate was ∼2% (Figure 4B). When the E:S ratio was raised to 1:1, the cleavage reached above 10–15%. There was no significant difference in cleavage efficiency of the four O-containing base pairs (data not shown). However, under the same assay conditions, cleavage of the T/I substrate approached completion (Figure 4C and D), which suggests that the cleavage efficiency of T/O was at least 6-fold less efficient than that of T/I. Excess of enzyme increased the cleavage of the T/O substrates to ∼35% completion, with an apparent rate constant of 0.23 min−1 (Figure 4B). Similar levels of cleavage were observed with A/O, G/O and C/O under enzyme in excess conditions (data not shown).
Figure 4.
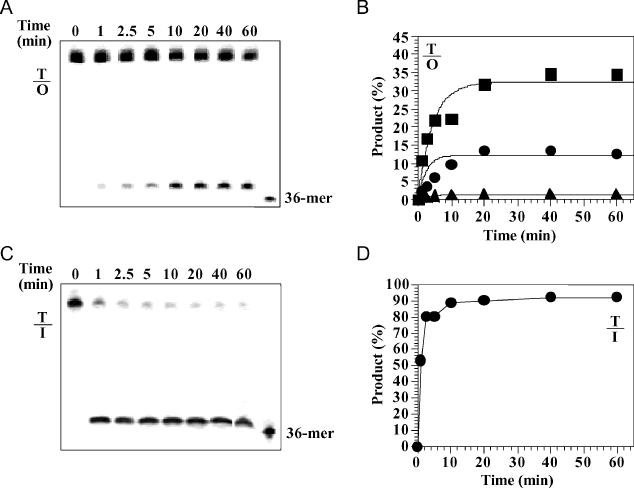
Time course analysis of T/O and T/I cleavage by Tma endo V. Cleavage reactions were performed as described in Materials and Methods with 1 nM Tma endo V (triangles), 10 nM (circles) and 100 nM (squares). Reactions were stopped on ice at indicated time points and followed by adding equal volume GeneScan stop buffer. (A) Representative GeneScan gel analysis of T/O cleavage (E:S = 10:1). (B) Plots of T/O cleavage. (C) Representative GeneScan gel analysis of T/I cleavage (E:S = 1:1). (D) Plot of T/I cleavage (E:S = 1:1).
To study the binding affinity of endo V to O-containing DNA, we initially used Tma endo V protein. However, no retarded bands were observed (data not shown). We subsequently switched to the Salmonella endo V, which binds tightly to deaminated DNA (H. Feng, A. M. Klutz and W. Cao, unpublished data). Distinct retarded bands were detected with Salmonella endo V (Figure 5A and B). Like E.coli endo V on inosine-containing DNA (34), Salmonella endo V forms multiple retarded bands on O-containing DNA (Figure 5A and B). The binding in the absence of metal ion was weak, with an apparent Kd of 485 nM for the primary ES complex (Figure 5C). The effect of metal ion on binding was determined using Ca2+, which promotes binding without triggering DNA cleavage [Figure 3A and (23)]. The apparent Kd value was 77 nM for the ES complex, indicating that a metal ion can enhance the binding to double-stranded O-containing DNA by 6-fold. To compare with non-specific binding, gel mobility shift analysis was also performed on identical double-stranded DNA except that the T/O base pair was changed to a T/A base pair. Only a small amount of diffused shifted band was detectable at very high enzyme concentrations in the presence of Ca2+ (data not shown). Since the highest enzyme concentration used was 500 nM, the apparent Kd value for undamaged DNA should be >500 nM.
Figure 5.
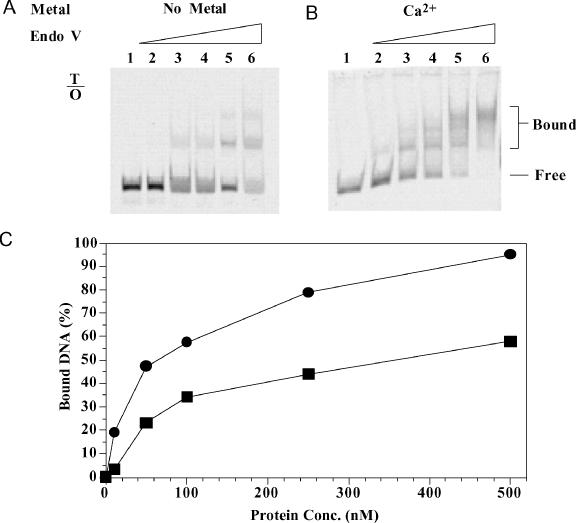
Gel mobility shift analysis of Salmonella endo V with double-stranded O-containing substrate. Lanes: 1, 0 nM endo V; 2, 10 nM endo V; 3, 50 nM endo V; 4, 100 nM endo V; 5, 250 nM endo V; and 6, 500 nM endo V. (A) Binding of T/O substrate without metal. (B) Binding of T/O substrate with Ca2+. (C) Quantitative analysis of endo V binding to T/O substrate without metal (squares) or with Ca2+ (circles).
Cleavage and binding kinetics of endo V on single-strandedO-containing DNA
Endo V is known to cleave single-stranded inosine-containing DNA (18,23). We therefore tested the cleavage of single-stranded O-containing DNA. Indeed, single-stranded O-containing DNA was a substrate for endo V (Figure 6A). Cleavage was detected in all three E:S ratios, albeit the cleavage efficiency was lower than that for double-stranded O-containing DNA (Figures 4B and 6B). At the E:S ratio of 1:1, the cleavage of single-stranded O-containing DNA reached ∼7%. However, under the same assay conditions, cleavage of the single-stranded I substrate approached ∼90% completion (Figure 6C and D), which suggests that the cleavage efficiency of single-stranded O was ∼13-fold less efficient than that of single-stranded I. Excess of enzyme increased the cleavage of the single-stranded O to ∼20% completion, with an apparent rate constant of 0.15 min−1 (Figure 6A and B).
Figure 6.
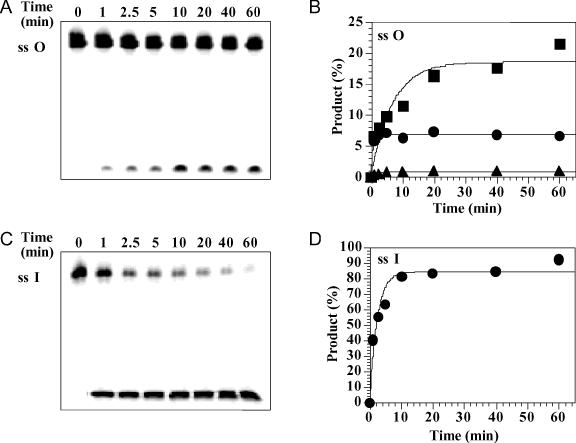
Time course analysis of single-stranded O and I cleavage by Tma endo V. Cleavage reactions were performed as described in Materials and Methods with 1 nM Tma Endo V (triangles), 10 nM (circles) and 100 nM (squares). Reactions were stopped on ice at indicated time points and followed by adding equal volume of GeneScan stop buffer. (A) Representative GeneScan gel analysis of single-stranded O cleavage (E:S = 10:1). (B) Plots of single-stranded O cleavage. (C) Representative GeneScan gel analysis of single-stranded I cleavage (E:S = 1:1). (D) Plot of single-stranded I cleavage (E:S = 1:1).
Distinct retarded bands were observed with single-stranded O-containing DNA (Figure 7A and B). The apparent Kd of the ES complex in the absence of metal ion was ∼384 nM for the single-stranded O-containing DNA (Figure 7C). In the presence of Ca2+, the apparent Kd was reduced to 88 nM (Figure 7C), indicating a more than 4-fold tighter binding. Binding to undamaged single-stranded DNA was very weak even in the presence of Ca2+, since no distinct shifted band was detected at all enzyme concentrations (data not shown).
Figure 7.
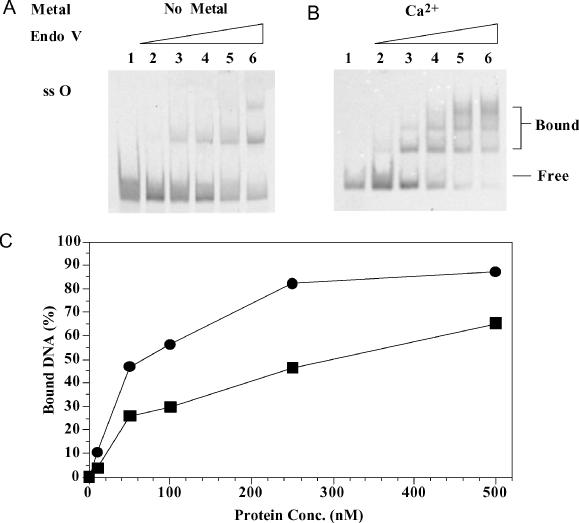
Gel mobility shift analysis of Salmonella endo V with single-stranded O-containing substrate. Lanes: 1, 0 nM endo V; 2, 10 nM endo V; 3, 50 nM endo V; 4, 100 nM endo V; 5, 250 nM endo V; and 6, 500 nM endo V. (A) Binding of single-stranded O-containing substrate without metal. (B) Binding of single-stranded O-containing substrate with Ca2+. (C) Quantitative analysis of endo V binding to single-stranded O-containing substrate without metal (squares) or with Ca2+ (circles).
DISCUSSION
Base deamination is a common DNA lesion caused by nitrosative stress. Oxanine is an intracyclic deamination product derived from guanine. This work investigates the mutagenicity of oxanine in DNA and cleavage activity of endo V on O-containing DNA. A previous study indicates that dOTP can be incorporated by Klenow Fragment in DNA to pair with both C and T (8). In this work, we have extended the previous study by determining the mutagenicity of oxanine once it is incorporated in the DNA templates. One significant finding is that oxanine in DNA is more mutagenic than in nucleotide form, as evidenced by the incorporation of all four regular dNTPs as well as dOTP to pair with O in the DNA template by Klenow Fragment (Figure 1A). This broad misincorporation does not seem to be limited to Klenow Fragment. Human DNA polymerase β can also misincorporate A, T, G to pair with O in DNA, albeit with different efficiencies (31). This suggests that the mutagenicity of oxanine largely increases once it makes its way into DNA, allowing it to pair with any of the four natural bases, dOTP and possibly other damaged bases as well. dOTP appears to be able to substitute for dGTP but not other nucleotides in primer extension reactions [Figure 1B and (8)], suggesting that a C/O base pair may adopt a Watson–Crick base pair symmetry to allow efficient extension. Other O-containing base pairs, although formed, may be difficult to extend by a DNA polymerase. The low efficiency of incorporation of G to pair with O in DNA by human DNA polymerase β is an indication that G/O base pair deviates more substantially from Watson–Crick base pair symmetry (31).
Taken together, we propose a hypothetic model for the formation of several O-containing base pairs (Figure 8A). O can pair with C in essentially the same way G pairs with C, retaining conventional Watson–Crick base pair symmetry. Because of the deamination that substitutes the N1 nitrogen with oxygen, the middle hydrogen bond is eliminated (Figure 8A). O can pair with T or A by forming two hydrogen bonds, but the base pair symmetries are slightly deviated from standard Watson–Crick base pair. The proposed geometry of the C/O and T/O base pairs is identical to a previous model (8). The formation of G/O base pair is likely more difficult. Based on the previous studies with 8-oxo-guanine (8-oxoG), which is flipped to a syn conformation to form an anti:syn oxo-G/A base pair (35,36), we surmise that oxanine may also switch to a syn conformation to pair with G (Figure 8A).
Figure 8.
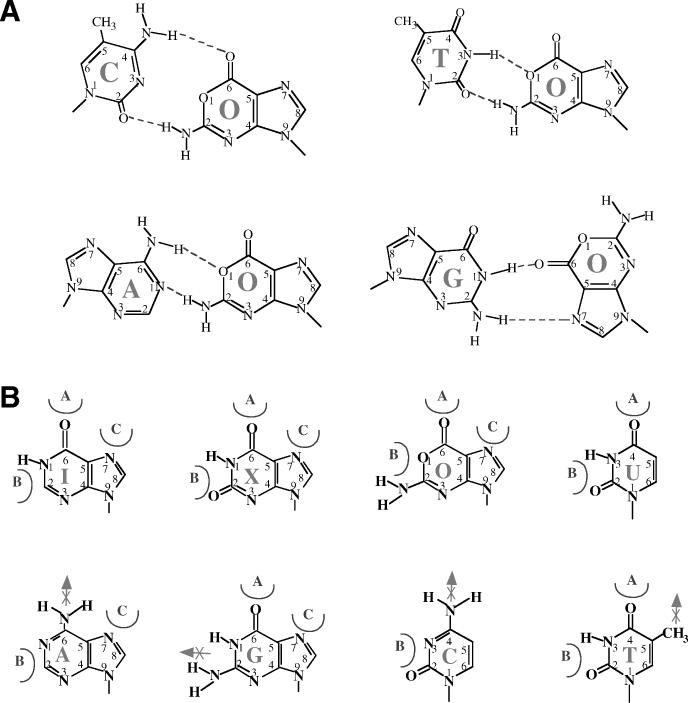
Models of oxanine base pairs and deamianted base recognition mechanism. (A) Proposed O-containing base pairs. (B) A hypothetic model of deamianted base recognition mechanism.
Non-specific nuclease activity in endo V has been reported previously (16,23,25). It is known that Mn2+ stimulates non-specific cleavage activity (23). The interesting finding as a result of this work is that the non-specific nuclease activity is very sensitive to Mn2+ concentration (Figure 3D and E). It is most active in the range of 0.2–1.0 mM Mn2+. The non-specific nuclease activity of endo V is abated at Mn2+ concentrations >5 mM, while the lesion-specific cleavage started to peak. The biochemical basis of this phenomenon is not known. We speculate that endo V may possess two Mn2+-binding sites. At low Mn2+ concentrations, the catalytic site is occupied, triggering non-specific activity. With increased Mn2+ concentrations, another Mn2+-binding site (low-affinity site) is occupied. The binding of the second Mn2+ may cause a subtle conformational change in the enzyme. As a result, the non-specific nuclease activity is reduced, while the lesion-specific cleavage activity may be stimulated.
Oxanine DNA glycosylase activities from E.coli AlkA and endo VIII have been reported recently (32). Compared with endo V, the ODG activities in AlkA and endo VIII appear low. It was reported that 30-fold excess of enzyme was needed to detect excision products (32). In contrast, we were able to observe cleavage products even when the E:S ratio was 1:10 (Figure 2). When the enzyme was in 10-fold excess, we observed over 35% cleavage products, which suggests that endo V is a more robust enzyme than AlkA or endo VIII for oxanine lesions (Figure 4). Under the conditions where endo V was in 30-fold excess, the cleavage of T/O was increased to 70% (data not shown). Taken together, these results indicate that the amount of cleavage product is proportional to the input of enzyme. However, the cleavage efficiency for oxanosine is still lower than that for inosine (Figure 4). Interestingly, E.coli endo VIII can serve as a backup system for the removal of 8-oxoG in the absence of MutM (Fpg) and MutY, despite its rather low in vitro 8-oxoG DNA glycosylase activity (37). Another distinct feature revealed from this study is that all four O-containing base pairs are cleaved with similar efficiencies, suggesting that cleavage of O-containing DNA by endo V is not dependent on base opposite oxanine. It was reported that the ODG activities of AlkA and endo VIII were dependent on the opposite base, with A/O, G/O, C/O but not T/O base pairs preferentially removed (32).
This work broadens the substrate specificity of endo V to include all known deaminated bases, uracil from cytosine, hypoxanthine from adenine, and xanthine and oxanine from guanine. How an enzyme like endo V can recognize all pyrimidine and purine deaminated bases is an unanswered question, although some aspects of recognition mechanisms have been addressed (20,23). Based on the observed broad substrate specificities and previous models, we propose that the enzyme may utilize three recognition elements to distinguish a deaminated base from a normal base (Figure 8B). The 6-keto (or 4-keto in uracil) and N7 position of a deaminated purine base may be recognized by elements A and C, as proposed previously (20,23). Element A may interact favorably with the 4-keto and element B may be snug between the 2-keto and N3 in uracil (Figure 8B). Adenine and cytosine can be excluded by the unfavorable interaction by the 6- or 4-amino group on element A. For guanine, the N1 hydrogen in combination with the 2-amino group could prevent the insertion of element B. Thymine may be excluded by the steric hindrance caused by the 5-methyl group, as proposed previously (20). In summation, the enzyme may discriminate a normal base by the occurrence of unfavorable interaction(s) and lack of favorable interaction(s). The speculative nature of these discussions should be noted. A better understanding of the endo V recognition mechanism awaits the solution of endo V–DNA complex structures.
Acknowledgments
ACKNOWLEDGEMENTS
We thank members of the Cao laboratory for stimulating discussions and Dr James Morris for critically reading the manuscript. We also thank Dr Hong Feng for the purification of Salmonella endonuclease V protein. This work was supported in part by CSREES/USDA (SC-1700153, technical contribution No. 4993), NIH (GM 067744) and Concern Foundation.
REFERENCES
- 1.Caulfield J.L., Wishnok,J.S. and Tannenbaum,S.R. (1998) Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J. Biol. Chem., 273, 12689–12695. [DOI] [PubMed] [Google Scholar]
- 2.Karran P. and Lindahl,T. (1980) Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry, 19, 6005–6011. [DOI] [PubMed] [Google Scholar]
- 3.Spencer J.P., Whiteman,M., Jenner,A. and Halliwell,B. (2000) Nitrite-induced deamination and hypochlorite-induced oxidation of DNA in intact human respiratory tract epithelial cells. Free Radic. Biol. Med., 28, 1039–1050. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki T., Yamaoka,R., Nishi,M., Ide,H. and Makino,K. (1996) Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligodeoxynucleotide and calf thymus DNA treated by nitrous-acid and nitric-oxide. J. Am. Chem. Soc., 118, 2515–2516. [Google Scholar]
- 5.Lucas L.T., Gatehouse,D. and Shuker,D.E. (1999) Efficient nitroso group transfer from N-nitrosoindoles to nucleotides and 2′-deoxyguanosine at physiological pH. A new pathway for N-nitrosocompounds to exert genotoxicity. J. Biol. Chem., 274, 18319–18326. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki T., Matsumura,Y., Ide,H., Kanaori,K., Tajima,K. and Makino,K. (1997) Deglycosylation susceptibility and base-pairing stability of 2′-deoxyoxanosine in oligodeoxynucleotide. Biochemistry, 36, 8013–8019. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez B., Soliva,R., Luque,F.J. and Orozco,M. (2000) Misincorporation of 2′-deoxyoxanosine into DNA: a molecular basis for NO-induced mutagenesis derived from theoretical calculations. Nucleic Acids Res., 28, 4873–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki T., Yoshida,M., Yamada,M., Ide,H., Kobayashi,M., Kanaori,K., Tajima,K. and Makino,K. (1998) Misincorporation of 2′-deoxyoxanosine 5′-triphosphate by DNA polymerases and its implication for mutagenesis. Biochemistry, 37, 11592–11598. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki T., Yamada,M., Ide,H., Kanaori,K., Tajima,K., Morii,T. and Makino,K. (2000) Identification and characterization of a reaction product of 2′-deoxyoxanosine with glycine. Chem. Res. Toxicol, 13, 227–230. [DOI] [PubMed] [Google Scholar]
- 10.Nakano T., Terato,H., Asagoshi,K., Masaoka,A., Mukuta,M., Ohyama,Y., Suzuki,T., Makino,K. and Ide,H. (2003) DNA–protein cross-link formation mediated by oxanine. A novel genotoxic mechanism of nitric oxide-induced DNA damage. J. Biol. Chem., 278, 25264–25272. [DOI] [PubMed] [Google Scholar]
- 11.Katafuchi A., Nakano,T., Masaoka,A., Terato,H., Iwai,S., Hanaoka,F. and Ide,H. (2004) Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem., 279, 14464–14471. [DOI] [PubMed] [Google Scholar]
- 12.Yagisawa N., Shimada,N., Naganawa,H., Takita,T., Hamada,M., Takeuchi,T., Umezawa,H., Nakamura,H. and Iitaka,Y. (1981) Oxanosine, a novel nucleoside from actinomycetes. Nucleic Acids Symp. Ser., 55–58. [PubMed] [Google Scholar]
- 13.Shimada N., Yagisawa,N., Naganawa,H., Takita,T., Hamada,M., Takeuchi,T. and Umezawa,H. (1981) Oxanosine, a novel nucleoside from actinomycetes. J. Antibiot. (Tokyo), 34, 1216–1218. [DOI] [PubMed] [Google Scholar]
- 14.Yagisawa N., Shimada,N., Takita,T., Ishizuka,M., Takeuchi,T. and Umezawa,H. (1982) Mode of action of oxanosine, a novel nucleoside antibiotic. J. Antibiot. (Tokyo), 35, 755–759. [DOI] [PubMed] [Google Scholar]
- 15.Itoh O., Kuroiwa,S., Atsumi,S., Umezawa,K., Takeuchi,T. and Hori,M. (1989) Induction by the guanosine analogue oxanosine of reversion toward the normal phenotype of K-ras-transformed rat kidney cells. Cancer Res., 49, 996–1000. [PubMed] [Google Scholar]
- 16.Gates F.T. III and Linn,S. (1977) Endonuclease V of Escherichia coli. J. Biol. Chem., 252, 1647–1653. [PubMed] [Google Scholar]
- 17.Demple B. and Linn,S. (1982) On the recognition and cleavage mechanism of Escherichia coli endodeoxyribonuclease V, a possible DNA repair enzyme. J. Biol. Chem., 257, 2848–2855. [PubMed] [Google Scholar]
- 18.Yao M., Hatahet,Z., Melamede,R.J. and Kow,Y.W. (1994) Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′ endonuclease, from Escherichia coli. J. Biol. Chem., 269, 16260–16268. [PubMed] [Google Scholar]
- 19.Yao M., Hatahet,Z., Melamede,R.J. and Kow,Y.W. (1994) Deoxyinosine 3′ endonuclease, a novel deoxyinosine-specific endonuclease from Escherichia coli. Ann. NY Acad. Sci., 726, 315–316. [DOI] [PubMed] [Google Scholar]
- 20.Yao M. and Kow,Y.W. (1997) Further characterization of Escherichia coli endonuclease V. J. Biol. Chem., 272, 30774–30779. [DOI] [PubMed] [Google Scholar]
- 21.He B., Qing,H. and Kow,Y.W. (2000) Deoxyxanthosine in DNA is repaired by Escherichia coli endonuclease V. Mutat. Res., 459, 109–114. [DOI] [PubMed] [Google Scholar]
- 22.Moe A., Ringvoll,J., Nordstrand,L.M., Eide,L., Bjoras,M., Seeberg,E., Rognes,T. and Klungland,A. (2003) Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res., 31, 3893–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J., Lu,J., Barany,F. and Cao,W. (2001) Multiple cleavage activities of endonuclease V from Thermotoga maritima: recognition and strand nicking mechanism. Biochemistry, 40, 8738–8748. [DOI] [PubMed] [Google Scholar]
- 24.Yao M. and Kow,Y.W. (1994) Strand-specific cleavage of mismatch-containing DNA by deoxyinosine 3′-endonuclease from Escherichia coli. J. Biol. Chem., 269, 31390–31396. [PubMed] [Google Scholar]
- 25.Yao M. and Kow,Y.W. (1996) Cleavage of insertion/deletion mismatches, flap and pseudo-Y DNA structures by deoxyinosine 3′-endonuclease from Escherichia coli. J. Biol. Chem., 271, 30672–30676. [DOI] [PubMed] [Google Scholar]
- 26.Huang J., Kirk,B., Favis,R., Soussi,T., Paty,P., Cao,W. and Barany,F. (2002) An endonuclease/ligase based mutation scanning method especially suited for analysis of neoplastic tissue. Oncogene, 21, 1909–1921. [DOI] [PubMed] [Google Scholar]
- 27.Schouten K.A. and Weiss,B. (1999) Endonuclease V protects Escherichia coli against specific mutations caused by nitrous acid. Mutat. Res., 435, 245–254. [DOI] [PubMed] [Google Scholar]
- 28.Weiss B. (2001) Endonuclease V of Escherichia coli prevents mutations from nitrosative deamination during nitrate/nitrite respiration. Mutat. Res., 461, 301–309. [DOI] [PubMed] [Google Scholar]
- 29.Burgis N.E., Brucker,J.J. and Cunningham,R.P. (2003) Repair system for noncanonical purines in Escherichia coli. J. Bacteriol., 185, 3101–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradshaw J.S. and Kuzminov,A. (2003) RdgB acts to avoid chromosome fragmentation in Escherichia coli. Mol. Microbiol., 48, 1711–1725. [DOI] [PubMed] [Google Scholar]
- 31.Hitchcock T.M., Dong,L., Connor,E.E., Meira,L.B., Samson,L.D., Wyatt,M.D. and Cao,W. (2004) Oxanine DNA glycosylase activity from mammalian alkyladenine glycosylase. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 32.Terato H., Masaoka,A., Asagoshi,K., Honsho,A., Ohyama,Y., Suzuki,T., Yamada,M., Makino,K., Yamamoto,K. and Ide,H. (2002) Novel repair activities of AlkA (3-methyladenine DNA glycosylase II) and endonuclease VIII for xanthine and oxanine, guanine lesions induced by nitric oxide and nitrous acid. Nucleic Acids Res., 30, 4975–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J., Lu,J., Barany,F. and Cao,W. (2002) Mutational analysis of endonuclease V from Thermotoga maritima. Biochemistry, 41, 8342–8350. [DOI] [PubMed] [Google Scholar]
- 34.Yao M. and Kow,Y.W. (1995) Interaction of deoxyinosine 3′-endonuclease from Escherichia coli with DNA containing deoxyinosine. J. Biol. Chem., 270, 28609–28616. [DOI] [PubMed] [Google Scholar]
- 35.McAuley-Hecht K.E., Leonard,G.A., Gibson,N.J., Thomson,J.B., Watson,W.P., Hunter,W.N. and Brown,T. (1994) Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry, 33, 10266–10270. [DOI] [PubMed] [Google Scholar]
- 36.Kouchakdjian M., Bodepudi,V., Shibutani,S., Eisenberg,M., Johnson,F., Grollman,A.P. and Patel,D.J. (1991) NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry, 30, 1403–1412. [DOI] [PubMed] [Google Scholar]
- 37.Blaisdell J.O., Hatahet,Z. and Wallace,S.S. (1999) A novel role for Escherichia coli endonuclease VIII in prevention of spontaneous G→T transversions. J. Bacteriol., 181, 6396–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]


