Kishi et al. find that protein C receptor (PROCR) is specifically expressed on the surface of Th17 cells and its loss exacerbates encephalitogenic Th17 cell responses.
Abstract
Th17 cells are key players in defense against pathogens and maintaining tissue homeostasis, but also act as critical drivers of autoimmune diseases. Based on single-cell RNA-seq profiling of pathogenic versus nonpathogenic Th17 cells, we identified protein C receptor (PROCR) as a cell surface molecule expressed in covariance with the regulatory module of Th17 cells. Although PROCR expression in T cells was controlled by the cooperative action of the Th17 lineage-specific transcription factors RORγt, IRF4, and STAT3, PROCR negatively regulated Th17 differentiation. CD4+ T cells from PROCR low expressor mutant mice readily differentiated into Th17 cells, whereas addition of the PROCR ligand, activated protein C, inhibited Th17 differentiation in vitro. In addition, PROCR acted as a negative regulator of Th17 pathogenicity in that it down-regulated expression of several pathogenic signature genes, including IL-1 and IL-23 receptors. Furthermore, T cell–specific deficiency of PROCR resulted in the exacerbation of experimental autoimmune encephalomyelitis (EAE) and higher frequencies of Th17 cell in vivo, indicating that PROCR also inhibits pathogenicity of Th17 cells in vivo. PROCR thus does not globally inhibit Th17 responses but could be targeted to selectively inhibit proinflammatory Th17 cells.
INTRODUCTION
Th17 cells are characterized by the production of the cytokines IL-17A, -17F, -21, and -22 and play a key role in defense against extracellular pathogens, as well as in the induction of autoimmune diseases (Bettelli et al., 2007; Korn et al., 2009). Recently, it has become clear that Th17 cells can exist in different states that have different functions and are marked by coexpression of IL-17 with other cytokines (Lee et al., 2012). In humans, coexpression of IL-17 and IFN-γ in Th17 cells is critical for defense against Candida albicans infection, whereas cells that produce IL-17 together with IL-10 are effective against Staphylococcus aureus. Expression of IFN-γ in Th17 cells is enhanced by IL-1β signaling, which suppresses IL-10 production in Th17 cells (Zielinski et al., 2012). Pathogenicity of Th17 cells to induce autoimmune diseases in mice is also associated with increased levels of IFN-γ, as well as high expression of T-bet, IL-23R, and IL-22 (Ghoreschi et al., 2011; Peters et al., 2011; Lee et al., 2012). In vitro activation of T cells in the presence of IL-1β + IL-6 results in generation of a highly pathogenic Th17 subset (Yang et al., 2008; Ghoreschi et al., 2011). Similarly, the presence of TGF-β3 rather than TGF-β1, as well as exposure to IL-1 and -23, induces a pathogenic phenotype in Th17 cells (Peters et al., 2011; Lee et al., 2012). In contrast, Th17 cells that coexpress IL-10 with -17 are nonpathogenic in murine models of autoimmunity (Lee et al., 2012), and exposure of these Th17 cells to IL-23 renders them pathogenic by decreasing IL-10 expression (McGeachy et al., 2007). Thus, IL-1β and -23 play a critical role in inducing and maintaining a pathogenic phenotype and effector functions in Th17 cells.
Co-stimulatory signals such as PD-1, CTLA-4, and ICOS have been shown to be involved in the differentiation of Th17 cells. The PD-1–PD-L1 pathway is recognized to be important for the maintenance of peripheral tolerance, and it inhibits the differentiation of Th17 cells (Francisco et al., 2010). The addition of PD-L1–neutralizing antibody completely abolishes IL-27–mediated inhibition of Th17 cell differentiation (Hirahara et al., 2012). CTLA-4 also controls the differentiation of Th17 cells. Ying et al. (2010) reported that CTLA-4–B7 interaction inhibits Th17 differentiation in vitro and in vivo and suppresses Th17-mediated autoimmunity. Although ICOS is not essential for the differentiation of Th17 cells, ICOS plays a critical role in their maintenance, as ICOS-deficient mice demonstrated a defect in the expansion of Th17 cells after IL-23 stimulation (Bauquet et al., 2009). Therefore, PD-1 and CTLA-4 are important for the regulation of Th17 differentiation, and ICOS is essential for the maintenance and expansion of Th17 cells. However, whether there are other co-stimulatory pathways that regulate the pathogenic/nonpathogenic phenotype of Th17 cells has not been addressed.
We recently performed single-cell RNA-seq profiling of pathogenic versus nonpathogenic Th17 cells to identify candidate regulators of Th17 pathogenicity (Gaublomme et al., 2015). These data suggest the presence of two anticorrelated modules influencing Th17 function—a proinflammatory module in covariance with IL17a and a regulatory module correlating with expression of Il10. Among the newly identified genes that are in covariance with the regulatory module of Th17 pathogenicity, protein C receptor (PROCR) stood out as an interesting candidate. In our studies on the transcriptional network of differentiating Th17 cells, we identified Procr mRNA to be strongly induced in Th17 (Yosef et al., 2013); however, whether PROCR plays a functional role in T cells was not investigated. Previous studies have shown that PROCR, which is also known as endothelial PROCR (EPCR), plays an important role in mediating intracellular effects of activated protein C (aPC; Esmon, 2012; Gleeson et al., 2012; Montes et al., 2012). aPC is used therapeutically to reduce mortality in patients with severe sepsis (Cao et al., 2010; Kerschen et al., 2010; Della Valle et al., 2012). Thus far, PROCR expression has been reported to be limited to a subset of CD8+ conventional DCs among immune cells, and PROCR-expressing DCs were identified as critical targets of aPC therapy (Kerschen et al., 2010). Interestingly, comparative proteomic profiles identified protein C inhibitor to be present in chronic active plaque of multiple sclerosis (MS) patients, and recombinant aPC reduced disease severity of experimental autoimmune encephalomyelitis (EAE; Han et al., 2008). In this setting, both the anticoagulant and the signaling functions of aPC contributed to the amelioration of disease (Han et al., 2008). However, whether aPC inhibits the encephalitogenic T cell response by directly engaging PROCR on T cells was not addressed.
In this study, we found that PROCR is specifically expressed on the cell surface of Th17 cells where it regulates their function. The transcription factors that are critical drivers of Th17 differentiation (STAT-3, IRF-4, and RORγt) regulate PROCR expression. PROCR expression inversely correlates with the pathogenicity of Th17 cells, and we found PROCR overexpression or engagement reduced expression of some of the key members of the proinflammatory module in Th17 cells, including IL-1R and -23R, the two key receptors that drive the pathogenic phenotype of Th17 cells. Furthermore, using active immunization and adoptive transfer models of EAE, we showed that loss or reduction in the expression of PROCR led to an increase in Th17 pathogenicity and enhanced EAE in vivo. In this study, we therefore identified PROCR as a negative regulator of Th17 pathogenicity.
RESULTS
PROCR is specifically expressed in Th17 cells
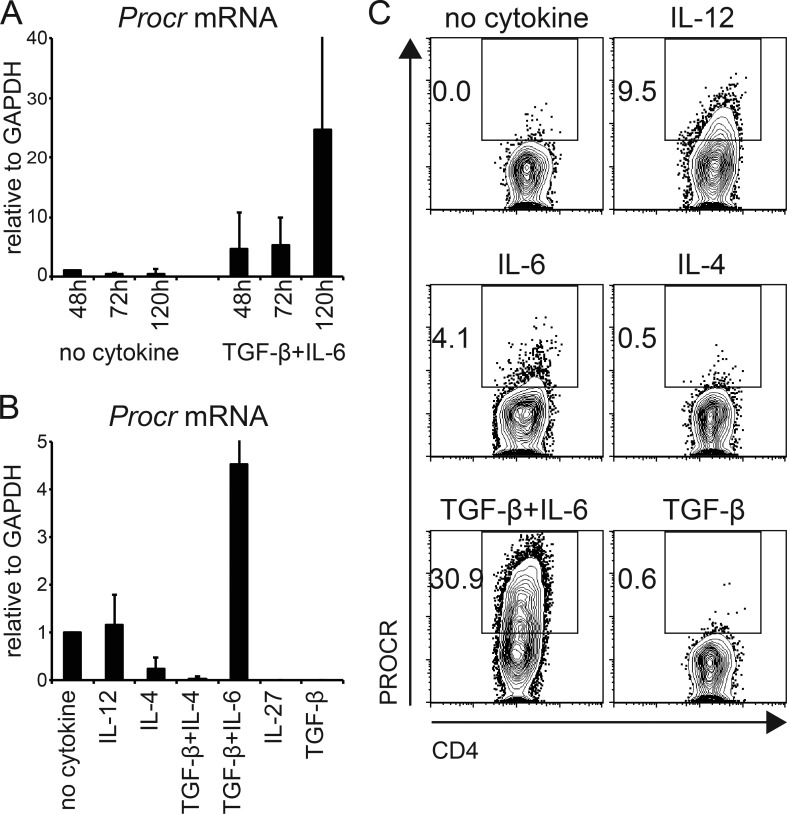
To identify candidate regulators of Th17 pathogenicity, we recently performed single-cell RNA-seq of Th17 cells differentiated under pathogenic (IL-1β + IL-6 + IL-23) versus nonpathogenic (TGF-β1 + IL-6) conditions in vitro or of Th17 cells isolated ex vivo from the lymph nodes or central nervous system of mice undergoing EAE (Gaublomme et al., 2015). We identified two distinct modules influencing Th17 pathogenicity. The proinflammatory module is in covariance with Il17a, whereas the regulatory module correlates with expression of Il10, Il9, and others. We reasoned that surface molecules that are in covariance with the regulatory module might identify novel regulators that modulate Th17 pathogenicity and therefore play a critical role in patients suffering from autoimmune diseases. We queried our dataset for potential candidates and identified PROCR as an interesting candidate. Although we had previously reported Procr mRNA expression in Th17 cells (Yosef et al., 2013), its functional role in these cells was not evaluated. We found PROCR to covary with the regulatory module in our dataset, suggesting it might play a role in inhibiting Th17 pathogenicity. First, we wanted to validate the expression of PROCR in Th17 cells differentiated under nonpathogenic (TGF-β1 + IL-6) conditions. Transcriptional expression of Procr in Th17 cells was already detectable at 48 h and reached peak after 5 d of in vitro differentiation in nonpathogenic differentiation conditions (Fig. 1 A). Procr expression was restricted to Th17 cells, but was not expressed on other in vitro–differentiated T cell subsets such as Th0 (no cytokines), Th1 (IL-12), Th2 (IL-4), Th9 (IL-4 + TGF-β1), Tr1 (IL-27), or induced regulatory T cells (TGF-β1; Fig. 1 B). Furthermore, the cell surface expression of PROCR protein on Th17 cells, but not other T cell subsets, was also confirmed by flow cytometry (Fig. 1 C).
Figure 1.
PROCR is specifically induced in Th17 cells. (A) Kinetic expression of Procr mRNA was measured by quantitative RT-PCR in Th17 cells differentiated with TGF-β1 + IL-6. (B) Procr mRNA expression was measured by quantitative RT-PCR analysis in different T cell subsets after 72 h of stimulation in the presence of the indicated cytokines. (C) PROCR protein expression was examined by flow cytometry in different T cell subsets generated as in B. Representative plots of three to four independent experiments are shown.
As PROCR is selectively expressed in Th17 cells, we reasoned that its expression in T cells might be regulated by Th17-specific transcription factors. RORγt is a key transcription factor that promotes development of the Th17 lineage. To determine whether RORγt also controls PROCR expression, we first analyzed its expression in Th17 cells from RORγt-deficient mice. Indeed, PROCR expression in RORγt−/− Th17 cells was strongly reduced but not completely abolished compared with WT Th17 cells (Fig. 2 A), indicating that RORγt positively regulates PROCR expression. We next queried the genomic chromatin immunoprecipitation (ChIP)-seq dataset of RORγt binding and identified two binding sites in the genomic locus of PROCR containing the TAGGTCA/TGGGTCA RORγt-binding motif (Ciofani et al., 2012; Xiao et al., 2014). Using these genomic segments, we undertook a ChIP-PCR analysis to determine whether RORγt binds to the defined genomic elements in the Procr promoter. Indeed, ChIP-PCR data confirmed that RORγt can directly bind to the Procr promoter (Fig. 2 D). In addition to RORγt, IRF4 and STAT-3 are the critical transcription factors that promote the Th17 differentiation program; therefore, we tested whether IRF4 and STAT3 could also regulate PROCR expression. Indeed, Th17 cells lacking IRF4 (CD4CreIRF4fl/fl) or STAT3 (CD4CreSTAT3fl/fl) displayed reduced PROCR protein expression under Th17-differentiating conditions (Fig. 2, B and C). By identifying the binding sites for IRF4 and STAT3 from genome wide ChIP-seq data (Kwon et al., 2009), we designed ChIP-PCR primers and again performed ChIP-PCR analysis to determine whether IRF4 and STAT-3 would physically bind to the Procr genomic region. Like RORγt, IRF4 and STAT-3 significantly bound to their respective binding motifs that were identified in the Procr promoter (AANNGAAA for IRF4 and TTCTGAGA for STAT-3; Fig. 2 D). The Il23r promoter used as a positive control showed enriched binding to all three transcription factors (Fig. 2 D). To functionally validate whether RORγt, IRF4, and STAT3 can transactivate the Procr promoter, we tested their ability to induce expression of a luciferase reporter gene under the control of the Procr reporter. Co-transfection of the Procr luciferase reporter with constructs encoding for RORγt, IRF4, or STAT3 resulted in enhanced luciferase activity (Fig. 2 E). The co-transfection of all three transcription factors resulted in additive transactivation of the construct, further supporting the notion that they act together to promote Procr expression. Collectively, our data suggest that the surface receptor PROCR is specifically expressed in Th17 cells and that its expression in T cells is controlled by the concerted action of the Th17 lineage-specific transcription factors RORγt, IRF4, and STAT3.
Figure 2.
Rorγt, IRF4, and STAT3 bind the Procr promoter and induce PROCR expression. Naive CD4+ T cells from Rorγt KO mice (A), CD4CrexIRF4fl/fl mice (B), or CD4CrexSTAT3fl/fl mice (C) and control mice were polarized under Th17 conditions (TGF-β1 and IL-6) or without cytokines (Th0) as control. On day 3, PROCR expression was determined by flow cytometry. (D) Purified WT naive CD4+ T cells were differentiated into Th17 cells in the presence of TGF-β1 and IL-6. After 4 d, binding of Rorγt, IRF4, or STAT3 to the Procr and IL23r promoters was assessed by ChIP-PCR (TF, transcription factor; mean ± SD of triplicates). (E) Procr promoter activity was measured in HEK293T cells transfected with a luciferase vector driven by 1,040 bp of the Procr promoter (pGL4-Procr), and the indicated transcription factor expression constructs (mean ± SD of triplicates). Representative plots of two to three independent experiments are shown.
PROCR regulates Th17 differentiation
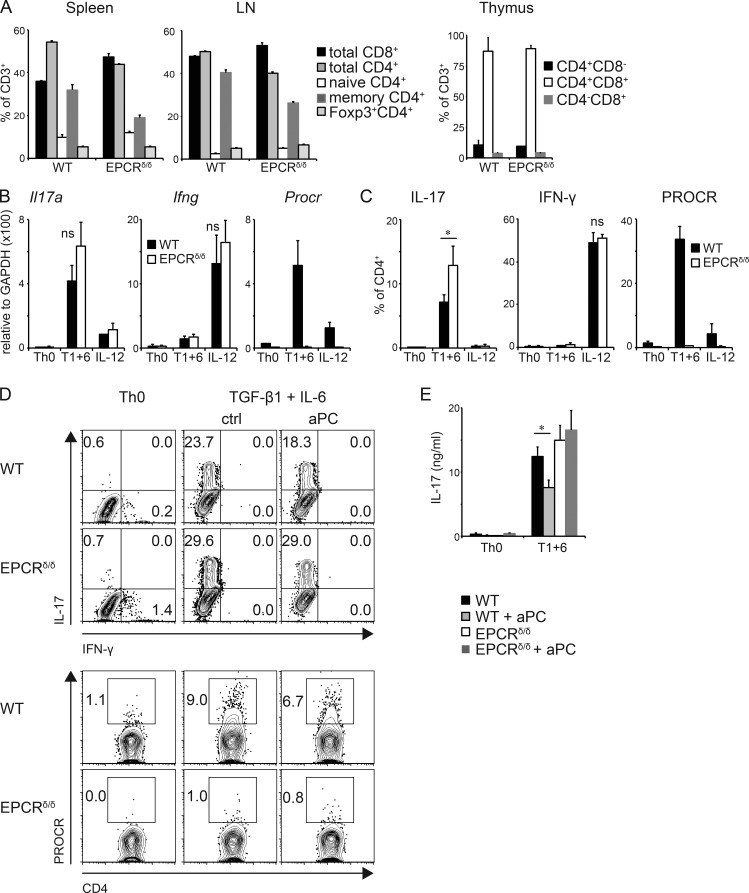
As PROCR is specifically expressed in Th17 cells, we next analyzed whether it also plays a role in Th17 differentiation by using cells that are deficient in PROCR. Because the total “knock-out” mice are embryonically lethal, we used PROCR low expressor (EPCRδ/δ) mice. EPCRδ/δ mice have an altered 5′-untranslated region of the Procr gene, which results in severely reduced expression of PROCR (Castellino et al., 2002). The mice are viable and do not have a phenotype in their immune cells or T cell homeostasis (Fig. 3 A), as exemplified by the expression of equal numbers of CD4+, CD8+, and Foxp3+ regulatory T cells in generative and peripheral lymphoid organs. We stimulated naive CD4+ T cells from WT and EPCRδ/δ mice under Th17 differentiation conditions and analyzed their cytokine profile. Loss of PROCR resulted in a slight but not significant difference in Il17a mRNA expression (Fig. 3 B). However, under Th17 differentiation conditions, cell from EPCRδ/δ mice generated significantly more IL-17A producing Th17 cells at the protein level as determined by intracellular cytokine staining (Fig. 3 C). IFN-γ production by Th1 cells was not altered in EPCRδ/δ cells. To further extend these observations, we tested whether PROCR engagement by its ligand aPC could affect expression of IL-17 in differentiating Th17 cells. Indeed, the frequency of IL-17+ CD4+ T cells was slightly but significantly reduced when WT Th17 cells were activated in the presence of aPC (Fig. 3, D and E). In contrast, aPC had no effect on IL-17 production in EPCRδ/δ cells (Fig. 3, D and E), confirming that aPC inhibits Th17 differentiation through engagement of PROCR and this effect is lost in the absence of PROCR. These results indicate that engagement of PROCR by its ligand aPC leads to a modest inhibition of Th17 differentiation.
Figure 3.
Role of PROCR in Th17 differentiation. (A) Cells from spleen, peripheral LNs, and thymus were isolated from WT and EPCRδ/δ mice, and frequencies of CD8+, CD4+, memory CD62L−CD44+CD4+, naive CD62L+CD44−CD4+, and regulatory Foxp3+CD4+ T cells were determined by flow cytometry (four mice/group). (B–E) Naive CD4+ T cells from WT and EPCRδ/δ mice were differentiated into Th17 cells by anti-CD3/anti-CD28 stimulation in the presence of TGF-β1 + IL-6 (T1+6) or no cytokines (Th0) or IL-12 as control. Il17a, Ifng, and Procr mRNA levels were determined after 96 h using quantitative PCR (B), and IL-17A and IFN-γ protein secretion, as well as PROCR expression levels, were examined by flow cytometry (C). (D and E) Cells were differentiated by anti-CD3/anti-CD28 stimulation with or without 300 nM aPC and cytokine production was analyzed 96 h later using flow cytometry (D) and ELISA (E). (B–D) Bar graphs represent mean ± SD of three independent experiments; Student’s t test; *, P < 0.05; ns, not significant.
PROCR controls the pathogenic signature genes of Th17 cells and inhibits expression of IL-23R and IL-1R
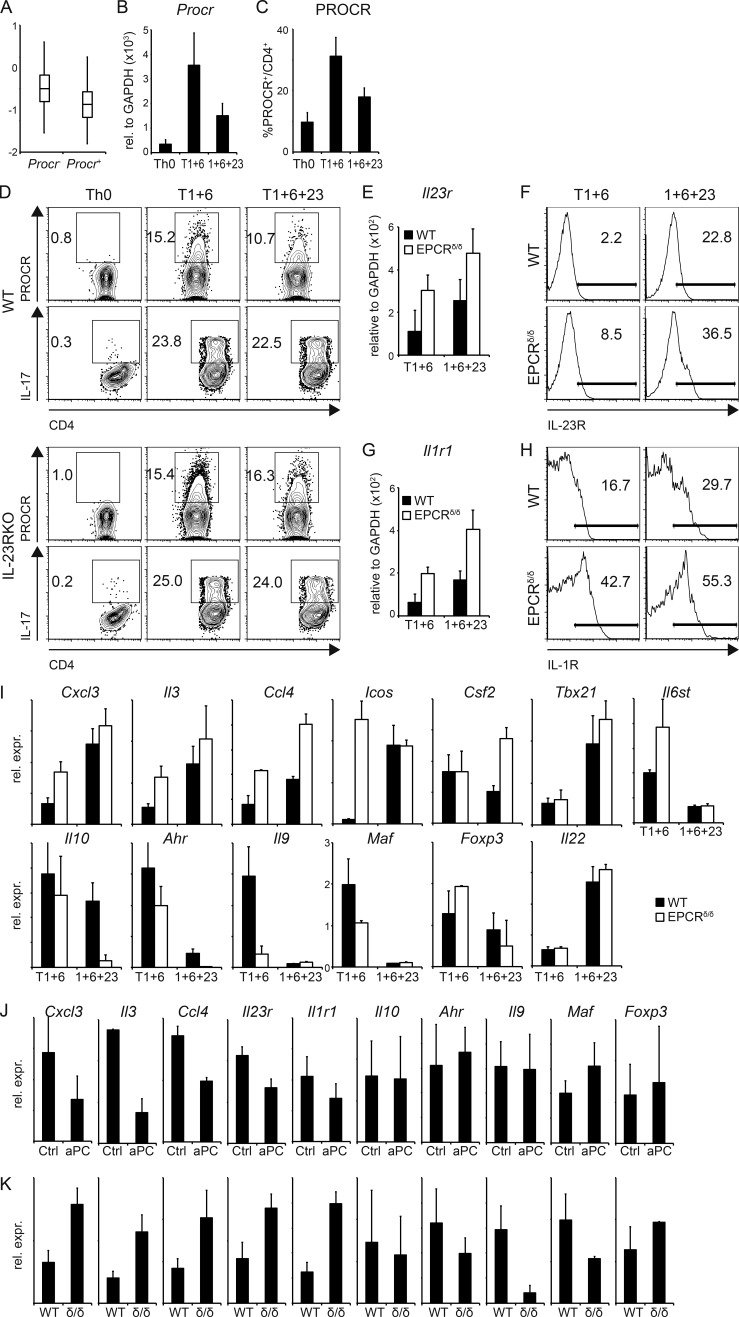
As PROCR expression covaries with the regulatory module in our single-cell RNA-seq analysis of Th17 cells, we speculated that PROCR might affect pathogenicity of Th17 cells. We previously defined a transcriptional gene signature of 23 genes, which discriminates pathogenic from nonpathogenic Th17 cells (Lee et al., 2012) and used this to test our hypothesis. We analyzed expression of the pathogenic Th17 signature in PROCR+ vs. PROCR− Th17 cells in the single-cell RNA-seq Th17 dataset and found a significant enrichment of the pathogenic signature in PROCR− Th17 cells (Fig. 4 A). We then tested PROCR expression on in vitro differentiated Th17 subsets generated under different conditions. Naive CD4+ T cells were stimulated under either pathogenic (IL-1β + IL-6 + IL-23) or nonpathogenic (TGF-β1 + IL-6) differentiation conditions, and Procr mRNA expression was determined by quantitative RT-PCR. Whereas Procr mRNA was detected in both subsets, expression was significantly higher in the nonpathogenic Th17 cells induced by TGF-β1 + IL-6 (Fig. 4 B). The qPCR data were confirmed by flow cytometry, where a higher frequency of PROCR+ cells was detected in the nonpathogenic Th17 cells (Fig. 4 C). These results are in line with the data obtained from the single-cell RNA-seq analysis and suggest that PROCR expression on Th17 cells inversely correlates with their expression of pathogenic signature genes.
Figure 4.
PROCR regulates genes of the proinflammatory module in Th17 cells. (A) Th17 cells differentiated in vitro with TGF-β1 + IL-6 were analyzed using single-cell RNA-seq (Gaublomme et al., 2015), and Procr expression in these cells was correlated with the pathogenic Th17 signature (Lee et al., 2012). P = 1.18 × 10−3 (Wilcoxon ranks test comparing signature scores of Procr+ vs. Procr – cells). (B and C) Naive CD4+ T cells from WT mice were differentiated into Th17 cells by anti-CD3/anti-CD28 stimulation in the presence of TGF-β1 + IL-6 (T1+6), IL-1β + IL-6 + IL-23 (1+6+23), or no cytokines as control (Th0). PROCR expression was determined by quantitative RT-PCR (B) and flow cytometry (C) 72 h later. Representative plots of three independent experiments are shown. (D) WT or IL-23R KO Th17 cells generated by anti-CD3/anti-CD28 stimulation in the presence of TGF-β1 + IL-6 (T1+6), TGF-β1 + IL-6 + IL-23 (T1+6+23), or no cytokines as control were analyzed for PROCR expression and IL-17 secretion by flow cytometry 72 h later. Representative plots of three independent experiments are shown. (E–H) Naive CD4+ T cells from WT and EPCR δ/δ mice were differentiated into Th17 cells as in B. IL-23R (E and F) and IL-1R1 (G and H) expression was determined by quantitative RT-PCR (E and G; mean ± SD of three independent experiments) and flow cytometry (F and H) 96 h later. (I) Quantitative RT-PCR analysis of pathogenic and nonpathogenic signature genes in WT (filled bars) and EPCR δ/δ (open bars) Th17 cells at 96 h. (J and K) Quantitative RT-PCR analysis of pathogenic and nonpathogenic signature genes (J) in Th17 cells differentiated with TGF-β1 + IL-6 with or without (Ctl) aPC and (K) in Th17 cells from WT or EPCRδ/δ (δ/δ) mice after 3 d of culture. (I–K) Bar graphs represent mean ± SEM of at least three independent experiments.
IL-23 plays a critical role in the differentiation of pathogenic versus nonpathogenic Th17 cells (Awasthi et al., 2009; Ghoreschi et al., 2011; Lee et al., 2012). Activation of naive T cells in the presence of TGF-β + IL-6 induces Th17 cells that are generally nonpathogenic in models of autoimmunity, but exposure of these cells to IL-23 results in the acquisition of a pathogenic Th17 phenotype (McGeachy et al., 2007; Peters et al., 2011; Lee et al., 2012). In addition, IL-23R expression on T cells is essential for Th17 stability and pathogenicity (Cua et al., 2003; Awasthi et al., 2009; Lee et al., 2012). Based on the inverse correlation between PROCR expression and Th17 pathogenicity, we hypothesized that IL-23 signaling might affect PROCR expression and vice versa. To test this hypothesis, we determined PROCR levels on nonpathogenic (TGF-β1 + IL-6) and pathogenic (TGF-β1 + IL-6 + IL-23) Th17 cells from WT and IL-23R–deficient mice. Addition of IL-23 reduced the expression of PROCR in Th17 cells from WT mice, when added to TGF-β1 + IL-6 differentiation conditions (Fig. 4 D). However, loss of IL-23R in IL-23R KO mice increased the expression of PROCR and brought it back to the levels observed in WT T cells activated under nonpathogenic Th17 conditions (Fig. 4 D).
To test whether PROCR might in turn regulate IL-23R expression, we analyzed Il23r expression in EPCRδ/δ T cells differentiated in nonpathogenic versus pathogenic Th17 conditions. Indeed, Th17 cells from EPCRδ/δ mice differentiated in both pathogenic and nonpathogenic Th17 conditions showed an increased Il23r mRNA expression in the absence of PROCR (Fig. 4 E). Furthermore, increased expression of IL-23R in the absence of PROCR could be confirmed on the protein level (Fig. 4 F), indicating that PROCR regulates IL-23R expression. In addition to IL-23R signaling, IL-1R plays an important role in inducing pathogenic Th17 cells and in promoting Th17 pathogenicity in the context of EAE (Sutton et al., 2006; Chung et al., 2009). We therefore sought to determine whether PROCR also regulated expression of IL-1R. Th17 cells generated from EPCRδ/δ mice showed increased expression of Il1r1 mRNA under both pathogenic and nonpathogenic Th17 conditions (Fig. 4 G). To further confirm this observation, we tested the effect of loss of PROCR signaling on the expression of IL1-R expression at the protein level by flow cytometry. WT Th17 cells differentiated under nonpathogenic Th17 differentiation conditions (TGF-β1 + IL-6) showed very little expression of IL-1R but loss of PROCR led to a profound increase in the expression of IL-1R, even under nonpathogenic Th17 differentiation conditions. Similarly under pathogenic Th17 differentiation conditions, T cells derived from EPCRδ/δ mice showed a substantial increase in the expression of IL-1R (Fig. 4 H). These data suggest that PROCR signaling negatively regulates expression of IL-23R and IL-1R, both of which are important drivers of pathogenic Th17 cells.
We next tested whether PROCR could affect any of the genes that comprise the proinflammatory and regulatory modules in Th17 cells by comparing their expression in WT and EPCRδ/δ Th17 cells (Fig. 4 I). Loss of PROCR in EPCRδ/δ Th17 cells led to an increase in the expression of approximately half of the proinflammatory signature genes tested when compared with WT Th17 cells. Conversely, expression of some nonpathogenic genes was decreased in EPCRδ/δ Th17 cells. The most strongly affected genes included Il3, Cxcl3, Ccl4, Csf2, and Il9. Engagement of PROCR by its ligand aPC, as well as overexpression of PROCR by retroviral transduction (not depicted), resulted in a decrease in the expression of members of the proinflammatory module (Cxcl3, Il3, Ccl4, Il23r, and Il1r; Fig. 4 J). In line with these results, in the absence of PROCR expression of Cxcl3, Il3, Ccl4, Il23r, and Il1r1 was increased in Th17 cells (Fig. 4 K). PROCR therefore acts as a negative regulator of some of the key members of the proinflammatory module. Most importantly, this includes regulation of expression of both IL-23R and -1R, both of which play a critical role in promoting pathogenicity of Th17 cells.
PROCR dampens Th17 pathogenicity in vivo
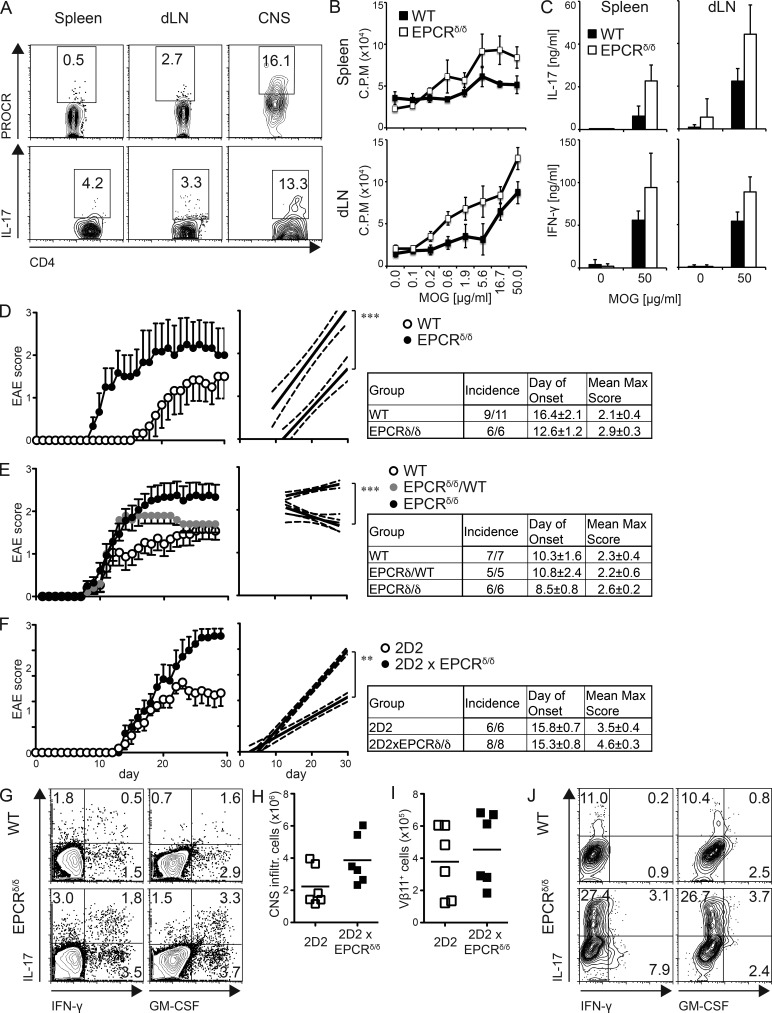
Th17 cells are the main drivers of several autoimmune diseases, including EAE, the animal model of MS. Interestingly, comparative proteomic profiles of chronic active plaques in MS patients identified protein C inhibitor, which limits the expression of the PROCR ligand aPC, to be present within these plaques (Han et al., 2008). Furthermore, in vivo administration of recombinant aPC was able to reduce the severity in EAE (Han et al., 2008). However, it is not clear how administration of aPC inhibited development of EAE. We hypothesized that these effects might be a result of the action of aPC on its receptor PROCR on Th17 cells. First, we wanted to determine whether PROCR was expressed on T cells during EAE in vivo. To this end, we induced EAE by immunizing mice subcutaneously with MOG35-55 peptide in CFA, and then analyzed PROCR expression in T cells isolated from spleen, lymph node, and CNS by flow cytometry at the peak of disease. Although PROCR was not detected on T cells in spleen or draining lymph nodes, it was highly expressed in CNS-infiltrating T cells, where Th17 cells were observed at the highest frequency (Fig. 5 A). To test whether PROCR plays a functional role in EAE, we immunized WT and EPCRδ/δ mice for EAE induction, analyzed the resulting T cell response, and monitored the mice for the development of clinical disease. To determine the magnitude of the MOG35-55-specific T cell response, we isolated cells from spleen and lymph nodes 10 d after immunization and restimulated them with MOG35-55 peptide in vitro. T cells derived from EPCRδ/δ mice that lack PROCR displayed slightly but not significantly increased proliferative responses (Fig. 5 B). Furthermore, IL-17 and IFN-γ production of T cells isolated from spleen and draining lymph node of EPCRδ/δ mice was significantly increased upon restimulation with MOG35-55 peptide (Fig. 5 C). Most importantly, EPCRδ/δ mice showed an earlier EAE disease onset and compared with WT control mice when immunized with a suboptimal dose of MOG35-55 peptide (Fig. 5 D). In addition, EPCRδ/δ mice showed enhanced EAE severity compared with WT littermate control or heterozygous mice from the same cross (Fig. 5 E), indicating a functional role for PROCR in disease pathogenicity. This was also reflected in increased levels of the proinflammatory cytokines IL-17A, IFN-γ, and GM-CSF in the CNS-infiltrating lymphocytes in EPCRδ/δ mice (Fig. 5 G). To analyze whether the observed difference in EAE susceptibility was caused by the effects of PROCR on Th17 pathogenicity, we compared the ability of WT myelin-specific 2D2 Th17 cells and EPCRδ/δ 2D2 Th17 cells to induce disease. As Th17 cells differentiated from EPCRδ/δ 2D2 mice yielded a higher frequency of IL-17+ T cells, we normalized the amount of transferred cells to the IL-17+ cells to be able to compare their pathogenicity per Th17 cell. PROCR-deficient 2D2 Th17 cells displayed markedly enhanced pathogenicity, as reflected in increased clinical score and lethality (Fig. 5 F). Differences in number and function of WT vs. EPCRδ/δ 2D2 Th17 cells in the CNS could account for the increased pathogenicity. The total number of CNS-infiltrating cells (Fig. 5 H), and the number of infiltrating 2D2 cells (Fig. 5 I) was comparable in both groups. IFN-γ production in CNS-infiltrating T cells was low because disease was induced by Th17 cells. Nevertheless, IFN-γ, as well as IL-17 and GM-CSF, production were dramatically increased in CNS-infiltrating EPCRδ/δ 2D2 T cells (Fig. 5 J). Our results therefore suggest that PROCR decreases pathogenicity of Th17 cells in vivo by regulating cytokines of the proinflammatory module, affecting pathogenicity and not necessarily the differentiation of Th17 cells.
Figure 5.
PROCR ameliorates Th17 pathogenicity in vivo. (A) Frequency of CD4+ T cells expressing IL-17A and PROCR isolated from EAE mice 21 d after immunization with MOG35-55 in CFA. (B) 10 d after immunization, spleen and lymph node cells were isolated and restimulated with MOG35-55 peptide in vitro and proliferation was determined by 3H-Thymidine incorporation 2 d later. (C) IL-17 secretion of T cells in supernatants from B was determined using ELISA. (A–C) Representative plots from one of three independent experiments are depicted (5–11 mice/group). (D) WT and EPCRδ/δ mice were immunized for EAE with a suboptimal dose of MOG35-55 (20 µg/mouse) and observed for clinical signs of disease. Mean clinical score ± SD (left) and linear regression curves of the disease for each group are shown (right; the 95% confidence intervals are represented with dashed lines). (E) WT, EPCRδ/WT, and EPCRδ/δ littermates were immunized for EAE (100 µg MOG35-55/mouse) and observed for clinical signs of disease. Mean clinical score ± SD (left) and linear regression curves of the disease for each group are shown (right; the 95% confidence intervals are represented with dashed lines). (F–I) WT recipients were transferred with WT 2D2 or EPCRδ/δ x 2D2 TCR transgenic Th17 cells differentiated with TGF-β1 + IL-6 and monitored for EAE. The data represents one of three transfers (n = 6). (F) Mean clinical score ± SD (left) is shown, and linear regression curves of the disease for each group are depicted on the right (95% confidence intervals are represented with dashed lines). (G) IFN-γ, IL-17A, and GM-CSF secretion among CNS infiltrating CD4+ cells on day 16 after immunization. The number of total (H) and Vβ11+ 2D2 (I) CNS infiltrating cells was determined 20 d after transfer. (J) IFN-γ, IL-17A, and GM-CSF secretion among Vβ11+CD4+ cells was determined by flow cytometry. Representative plots of at least three independent experiments are depicted. EAE experiments were performed with 5–11 mice/group as indicated, and plots of representative experiments of two to three repeats are depicted. **, P < 0.01; ***, P < 0.001 (Student's t test).
DISCUSSION
The Th17 lineage includes a diverse spectrum of functional subtypes that are adapted and specialized to fulfill different functions. These range from being important mediators of host defense and homeostasis at barrier sites to acting as the effector cell type in several inflammatory and autoimmune diseases. Although several different cytokines have been identified that favor the differentiation of Th17 cells with different functional phenotypes, the cellular receptors that mediate the functional transition between proinflammatory and regulatory Th17 subtypes have not been described. In this study, we identified PROCR as such a receptor that acts as a negative regulator of Th17-mediated pathogenicity.
Single-cell genomic analysis of pathogenic and nonpathogenic Th17 cells revealed the presence of two gene modules that influence Th17 function (Gaublomme et al., 2015). PROCR is preferentially expressed in nonpathogenic Th17 cells and covaries with the regulatory module, suggesting that it could potentially act as a negative regulator of Th17 pathogenicity. Analyzing PROCR function, we found that it only has a minor dampening effect on Th17 differentiation. However, PROCR regulates several key molecules of the proinflammatory module in Th17 cells that determine their pathogenicity, including expression of IL-1R and -23R, both of which are critical for the generation of pathogenic Th17 cells (Cua et al., 2003; Sutton et al., 2006; Awasthi et al., 2009; Chung et al., 2009). Interestingly, there appears to be some sort of antagonism between expression of IL-1 and -23 receptors on one hand and PROCR on the other hand. Consistent with this observation, we detected an increase in the expression of PROCR in the absence of IL-23R signaling in IL-23R−/− Th17 cells and loss of PROCR signaling in the PROCR mutant mice resulted in an increase in the expression of both IL-23R and -1R. This suggests that one possible mechanism by which IL-23 promotes Th17 pathogenicity might be by reducing the expression of PROCR, which regulates pathogenicity by inhibiting expression of multiple members of the proinflammatory module. Although the molecular mechanism for this cross-regulation between PROCR and IL-23R/IL-1R is still being worked out, this study shows that PROCR and IL-1R/IL-23R functionally antagonize each other and support conversion from pathogenic to nonpathogenic Th17 cells or vice versa. This observation is further supported by independent observations in humans, where Th17 cells that coproduce IL-17 with IFN-γ require IL-1β signaling for their induction (Zielinski et al., 2012). In addition, the nonpathogenic Th17 cells that coproduce IL-10 and have lower expression of IL-23R, also produce IL-1RN, which actively inhibits IL-1 signaling.
In healthy individuals, the majority of Th17 cells are nonpathogenic and found at mucosal sites where they promote tissue repair and help maintain barrier functions (McAleer and Kolls, 2011). It would therefore make sense that the default Th17 differentiation program would favor a nonpathogenic phenotype that can then be converted into pathogenic Th17 cells in the presence of additional proinflammatory signals, such as IL-1β or -23. Interestingly, PROCR expression is induced by RORγt, STAT3, and IRF4, three transcription factors that drive differentiation of Th17 cells. This suggests that the Th17 differentiation program includes the induction of surface molecules that act as intrinsic negative regulators (like PROCR) to control pathogenicity of the Th17 lineage and to skew Th17 cells toward a nonpathogenic phenotype under steady-state conditions. In addition to tipping the balance to favor nonpathogenic Th17 cells under steady-state conditions, PROCR might also be acting as an active brake during an ongoing proinflammatory Th17 response. The proinflammatory cytokines IL-1β and -23 actively down-regulate this brake, promoting a proinflammatory phenotype in Th17 cells. Although IL-1β and -23 can induce pathogenic Th17 cells, PROCR engagement through its ligand aPC allows for a functional conversion of pathogenic into nonpathogenic Th17 cells. The fact that the PROCR ligand aPC has a known antiinflammatory function and is used clinically as a treatment in severe sepsis would support this hypothesis. An active regulatory role through aPC ligation is further supported by the fact, that protein C inhibitor is contained in the highly inflammatory environment of chronic active plaques of MS patients (Han et al., 2008). Exogenous administration of aPCs was shown to ameliorate EAE and affect cytokine production by macrophages, astrocytes, and T cells (Han et al., 2008). Our data suggests that aPCs may directly engage PROCR on Th17 cells and limit their pathogenicity by inhibiting genes of the proinflammatory module in Th17 cells. In line with these results, we observed increased EAE severity in EPCRδ/δ mice. This increase in EAE severity could also be achieved by simply deleting PROCR expression on encephalitogenic Th17 cells, suggesting that the difference in disease severity observed after aPC administration must have been caused in part by its direct effects on encephalitogenic Th17 cells, supporting the idea that PROCR acts as a negative regulator of Th17 pathogenicity in vivo.
A follow up study showed that aPC not only has a beneficial effect in EAE when given at disease induction but is also effective when given therapeutically (Han et al., 2008). Our data would suggest that administration of aPC results in increased binding of PROCR on Th17 cells and promotes a functional shift from a pathogenic toward a nonpathogenic phenotype in these cells, partly by inhibiting IL-1R and -23R signaling by controlling the receptor expression. PROCR could thus potentially be targeted to therapeutically modulate the pathogenic phenotype of Th17 cells in autoimmune diseases. In addition to its expression on Th17 cells, our preliminary data suggests that Th1 cells, when chronically activated, also begin to express PROCR and therefore aPC may not only affect pathogenic Th17 cells but also encephalitogenic Th1 cells.
The latest generation of immunotherapies that target co-stimulatory pathways has shown remarkable promise for the treatment of pathologies caused by dysregulated immune responses. Anti–IL-17A and –IL-17R antibodies have been shown to have therapeutic efficacy in psoriasis, ankylosing spondylitis, and MS, but showed no benefit to or even exacerbated inflammatory bowel disease. The reason for this difference in efficacy in different autoimmune diseases has not directly been elucidated, but one could speculate that anti–IL-17 and –IL-17R antibodies may also affect nonpathogenic Th17 cells that decorate the lamina propria and promote gut barrier function. Based on its expression and regulatory effects on proinflammatory Th17 cells, PROCR could potentially serve as an interesting therapeutic target that could be used to modulate the phenotype of pathogenic Th17 without affecting beneficial effects of Th17 cells that help maintain mucosal barriers.
MATERIALS AND METHODS
Mice and induction of EAE
7–10-wk-old C57BL/6 mice were purchased from The Jackson Laboratory or bred in our animal facility at the Harvard Institutes of Medicine. 2D2 (Bettelli et al., 2003), Il23R−/− (Awasthi et al., 2009), and EPCRδ/δ mice (F. Castellino, University of Notre Dame, Notre Dame, IN; Castellino et al., 2002) have been described previously. EPCRδ/δ mice are >98.7% identical to the WT controls, with most of the differences found in the region of genetic modification, as determined by SNP analysis. Mice were induced for EAE by subcutaneous injection with 100 µl of an emulsion containing complete Freund’s adjuvant, 250 µg Mycobacterium tuberculosis extract H37-Ra (Difco), and 20 µg MOG35-55 peptide (QCB BioSource International). Mice received 200 ng pertussis toxin (List Biological Laboratories) i.p. on days 0 and 2. For passive induction of EAE, cells were differentiated and adoptively transferred as described (Jäger et al., 2009). Mice were housed in a specific pathogen–free animal facility at the Harvard Institutes of Medicine. All breeding and experiments were reviewed and approved by the Institutional Animal Care and Use Committee of Harvard Medical School.
In vitro T cell differentiation
Naive CD4+CD62L+ T cells were purified from mouse spleen or lymph node single-cell suspension using MACS magnetic beads (Miltenyi Biotec), followed by purification of naive T cells by flow cytometry. Cells were cultured in DMEM (Invitrogen) and supplemented as previously described (Lee et al., 2012). For Th cell differentiation, cells were activated with plate-bound antibody to CD3 (2 µg/ml; 145-2C11) and CD28 (2 µg/ml; PV-1) or anti-CD3 (2.5 µg/ml; 145-2C11) with irradiated splenocytes as APCs in the presence of the following cytokines: IL-12 (10 ng/ml) was added for Th1, IL-4 (10 ng/ml) for Th2, and TGF-β (2.5 ng/ml), IL-6 (25 ng/ml), IL-1β (20 ng/ml), and IL-23 (10 ng/ml) were added in multiple combinations for Th17 conditions. Where indicated, aPC (300 nM; a gift from J.H. Griffin, The Scripps Research Institute, La Jolla, CA) was added on day 0.
FACS and ELISA
For intracellular cytokine staining, cells were restimulated for 4 h with 50 ng/ml phorbol 12-myristate 13-acetate and 1 µM ionomycin (Sigma-Aldrich) in the presence of GolgiStop (BD). Cells were fixed with Cytofix/Cytoperm Fixation and Permeabilization Solution (BD) and stained for intracellular cytokines. Cells were analyzed on a FACSCalibur (BD). Antibodies to the following antigens were used: CD201 (EPCR; eBio1560; eBioscience), CD4 (RM4-5; BioLegend), IL-17A (TC11-18H10.1; BioLegend), IFN-γ (XMG1.2; BioLegend), CD62L (MEL-14; BioLegend), CD44 (IM7; BioLegend), IL-1R (JAMA-147; BioLegend), and GM-CSF (MP1-22E9; BioLegend). All flow cytometric data were analyzed using FlowJo Software (Tree Star). IL-17A concentration in the supernatant was determined by ELISA using standard protocols.
Quantitative RT-PCR
Total RNA was extracted using RNeasy reagents (QIAGEN) and reverse transcription was performed using iScript Reverse transcription (Bio-Rad Laboratories). Real-time PCR was done using validated TaqMan probe sets to mouse Procr (Mm00440992_m1), Il3 (Mm00439631_m1), Cxcl3 (Mm01701838_m1), Ccl4 (Mm00443111_m1), Tbx21 (Applied Biosystems, Mm00450960_m1), Icos (Mm00497600_m1), Csf2 (Mm01290062_m1), Il6st (Mm00439665_m1), Il22 (Mm00444241_m1), Il23r (Mm00519943_m1), Il1r1 (Mm00434237_m1), and GAPDH (Mm99999915_g1).
ChIP
Lysates were generated from WT Th17 cells differentiated for 4 d in the presence of TGF-β1 + IL-6 using the SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology) according to the manufacturer’s specifications. Immune precipitation was performed with 3 µg antibody (rabbit polyclonal anti-phospho STAT3 [#9134; Cell Signaling Technology], rat monoclonal anti-Rorγt [AFKJS-9; 14–6988-80; eBioscience], and goat polyclonal anti-IRF4 [sc-6059; Santa Cruz Biotechnology, Inc.] or the respective isotype controls from the same manufacturer). To quantify DNA, real-time qPCR was performed using the Fast SYBR Green Master Mix (Applied Biosystems) with the following primers: promoter region of Procr (promoter 1: 5′-CAGCACTCAGAAGGCAGGGTAAG-3′ and 5′-GCCAGGGCTACACAGAAAAACCC-3′), promoter region of Procr (promoter 2: 5′-GACTATGACCTGTCCGATGCTGAG-3′ and 5′-CGATGACGGCAGATCCAGGATTGG-3′), promoter and start codon region of Procr (promoter3+ORF1: 5′-AAGCCTGACTCCCTCTTCCTGCACGC-3′ and 5′-GCCTTGACCCAAACTCACCATCGG-3′), open reading frame of Procr (ORF2: 5′-GTGGAGGTTGTGATGGGAAGGAGG-3′ and 5′-GGCGCACCACAAGTCAATGTGTGC-3′), and open reading frame of Procr (ORF3: 5′-AACACTGCCAAAGCCCTTTCCTGGG-3′ and 5′-GGCCCTAGTTGCCTTTTCTGGCG-3′).
Procr promoter assay
The pGL4-Procr construct was generated by cloning a 1,040-bp fragment of the murine Procr promoter (-1040 – +1 bp from TSS) into the pGL4.10 vector (Promega) using KpnI/BglII sites, and verified by sequencing. HEK 293T cells were transiently cotransfected with the pGL4-Procr construct, an empty Renilla luciferase reporter vector, and the indicated transcription factor expression vectors using Fugene HD (Roche). Luciferase activity was analyzed 48 h after transfection using a dual luciferase assay kit (Promega).
Retroviral transduction
For retroviral production, HEK 293T cells were seeded at the density of 1.5 × 106 cells in 10-cm dishes 24 h before transduction. 4.5 µg pCMV5 containing the Procr mRNA along with 1 µg pcl and 2 µg gag/pol viral envelope constructs were transfected using Fugene HD (Promega). Virus containing supernatant was collected 48 h after transfection. For transduction, naive CD4+ T cells were activated with plate-bound anti-CD3 and anti-CD28 and spin-infected at 2,000 rpm for 45 min in the presence of polybrene (Sigma-Aldrich; final 8 ng/ml).
Single-cell RNA-Seq analysis
The experimental protocol for the single-cell RNA-seq data acquisition, as well as alignment, normalization, and filtering of the data are provided in the original study (Gaublomme et al., 2015). In total, 755 Th17 cells, either harvested in vivo or differentiated in vitro, were profiled transcriptionally based on mRNA SMART-Seq libraries derived from single cells using microfluidic chips (Fluidigm C1). At least two independent biological replicates for each condition were analyzed. RNA-seq reads were aligned to the National Center for Biotechnology Information Build 37 (UCSC mm9) of the mouse genome using TopHat (Trapnell et al., 2009). The resulting alignments were processed by Cufflinks to evaluate the abundance (using FPKM) of transcripts from RefSeq (Pruitt et al., 2007). We used log transform and quantile normalization to further normalize the expression values (fragments per kilobase of exon per million; FPKM) within each batch of samples (i.e., all single cells in a given run). To account for low (or zero) expression values, we added a value of 1 before log transform. We filtered the set of analyzed cells by a set of quality metrics (such as sequencing depth), and added an additional normalization step specifically controlling for these quantitative confounding factors, as well as batch effects. Our analysis is based on ∼7,000 appreciably expressed genes (FPKM > 10 in at least 20% of cells in each sample) for in vitro experiments and ∼4,000 for in vivo ones. We also developed a strategy to account for expressed transcripts that are not detected (false negatives) due to the limitations of single-cell RNA-seq (Deng et al., 2014; Shalek et al., 2014). Our analysis (e.g., computing signature scores) down-weighted the contribution of less reliably measured transcripts. To compute the pathogenic signature score (Fig. 4 A), we first standardized the expression matrix for each gene, then, for each cell, we define the score as the weighted mean of the normalized values of the propathogenic genes in the signature minus the nonpathogenic genes.
ACKNOWLEDGMENTS
We would like to thank F. Castellino for the kind provision of the EPCRδ/δ mice, J. Griffin for reagents and advice on PROCR, D. Kozoriz for cell sorting, Jessica Kilpatrick for genotyping of the EPCRδ/δ mice, and members of the Kuchroo Group for discussions.
This work was supported by the National Institutes of Health (grants RO1 NS30843, AI093671, AI056299, and AI073748 to V.K. Kuchroo; and K01 DK090105 to S. Xiao); the Arthritis National Research Foundation (to N. Joller); the Swiss National Science Foundation (grant PP00P3_150663 to N. Joller); the European Research Council (grant no. 677200 to N. Joller); and the Alliance for Lupus Research (to S. Xiao).
During the course of the research, V.K. Kuchroo had an ownership interest in Tempero Pharmaceuticals; his ownership ended due to a buy-out in October 2014. V.K. Kuchroo’s interests were reviewed and are managed by the Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict-of-interest policies. Y. Kishi and T. Kondo are employees of Mitsubishi Tanabe Pharma Corporation and were supported by its Scholarship Program. The authors declare no additional competing financial interests.
Author contributions: Y. Kishi and T. Kondo designed and performed experiments, analyzed the data, and revised the manuscript. J. Gaublomme, N. Yosef, A. Regev, and V.K. Kuchroo conceived and performed the original single-cell analysis. N. Yosef and A. Regev devised the computational analyses. S. Xiao performed the ChIP-PCR and luciferase assays. C. Wu and C. Wang contributed to experiments. N. Joller and V.K. Kuchroo conceived and designed the study, analyzed the data, wrote the manuscript, and directed the research.
Footnotes
Abbreviations used:
- aPC
- activated protein C
- ChIP
- chromatin immunoprecipitation
- EAE
- experimental autoimmune encephalomyelitis
- EPCR
- endothelial PROCR
- MS
- multiple sclerosis
- PROCR
- protein C receptor
References
- Awasthi A., Riol-Blanco L., Jäger A., Korn T., Pot C., Galileos G., Bettelli E., Kuchroo V.K., and Oukka M.. 2009. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J. Immunol. 182:5904–5908. 10.4049/jimmunol.0900732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauquet A.T., Jin H., Paterson A.M., Mitsdoerffer M., Ho I.C., Sharpe A.H., and Kuchroo V.K.. 2009. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10:167–175. 10.1038/ni.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Pagany M., Weiner H.L., Linington C., Sobel R.A., and Kuchroo V.K.. 2003. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J. Exp. Med. 197:1073–1081. 10.1084/jem.20021603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Oukka M., and Kuchroo V.K.. 2007. T(H)-17 cells in the circle of immunity and autoimmunity. Nat. Immunol. 8:345–350. 10.1038/ni0407-345 [DOI] [PubMed] [Google Scholar]
- Cao C., Gao Y., Li Y., Antalis T.M., Castellino F.J., and Zhang L.. 2010. The efficacy of activated protein C in murine endotoxemia is dependent on integrin CD11b. J. Clin. Invest. 120:1971–1980. 10.1172/JCI40380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino F.J., Liang Z., Volkir S.P., Haalboom E., Martin J.A., Sandoval-Cooper M.J., and Rosen E.D.. 2002. Mice with a severe deficiency of the endothelial protein C receptor gene develop, survive, and reproduce normally, and do not present with enhanced arterial thrombosis after challenge. Thromb. Haemost. 88:462–472. [PubMed] [Google Scholar]
- Chung Y., Chang S.H., Martinez G.J., Yang X.O., Nurieva R., Kang H.S., Ma L., Watowich S.S., Jetten A.M., Tian Q., and Dong C.. 2009. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 30:576–587. 10.1016/j.immuni.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M., et al. . 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cua D.J., Sherlock J., Chen Y., Murphy C.A., Joyce B., Seymour B., Lucian L., To W., Kwan S., Churakova T., et al. . 2003. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 421:744–748. 10.1038/nature01355 [DOI] [PubMed] [Google Scholar]
- Della Valle P., Pavani G., and D’Angelo A.. 2012. The protein C pathway and sepsis. Thromb. Res. 129:296–300. 10.1016/j.thromres.2011.11.013 [DOI] [PubMed] [Google Scholar]
- Deng Q., Ramsköld D., Reinius B., and Sandberg R.. 2014. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 343:193–196. 10.1126/science.1245316 [DOI] [PubMed] [Google Scholar]
- Esmon C.T. 2012. Protein C anticoagulant system—anti-inflammatory effects. Semin. Immunopathol. 34:127–132. 10.1007/s00281-011-0284-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L.M., Sage P.T., and Sharpe A.H.. 2010. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 236:219–242. 10.1111/j.1600-065X.2010.00923.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaublomme J.T., Yosef N., Lee Y., Gertner R.S., Yang L.V., Wu C., Pandolfi P.P., Mak T., Satija R., Shalek A.K., et al. . 2015. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. 163:1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Laurence A., Yang X.P., Hirahara K., and O’Shea J.J.. 2011. T helper 17 cell heterogeneity and pathogenicity in autoimmune disease. Trends Immunol. 32:395–401. 10.1016/j.it.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson E.M., O’Donnell J.S., and Preston R.J.. 2012. The endothelial cell protein C receptor: cell surface conductor of cytoprotective coagulation factor signaling. Cell. Mol. Life Sci. 69:717–726. 10.1007/s00018-011-0825-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.H., Hwang S.I., Roy D.B., Lundgren D.H., Price J.V., Ousman S.S., Fernald G.H., Gerlitz B., Robinson W.H., Baranzini S.E., et al. . 2008. Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature. 451:1076–1081. 10.1038/nature06559 [DOI] [PubMed] [Google Scholar]
- Hirahara K., Ghoreschi K., Yang X.P., Takahashi H., Laurence A., Vahedi G., Sciumè G., Hall A.O., Dupont C.D., Francisco L.M., et al. . 2012. Interleukin-27 priming of T cells controls IL-17 production in trans via induction of the ligand PD-L1. Immunity. 36:1017–1030. 10.1016/j.immuni.2012.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger A., Dardalhon V., Sobel R.A., Bettelli E., and Kuchroo V.K.. 2009. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J. Immunol. 183:7169–7177. 10.4049/jimmunol.0901906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschen E., Hernandez I., Zogg M., Jia S., Hessner M.J., Fernandez J.A., Griffin J.H., Huettner C.S., Castellino F.J., and Weiler H.. 2010. Activated protein C targets CD8+ dendritic cells to reduce the mortality of endotoxemia in mice. J. Clin. Invest. 120:3167–3178. 10.1172/JCI42629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M., and Kuchroo V.K.. 2009. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27:485–517. 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- Kwon H., Thierry-Mieg D., Thierry-Mieg J., Kim H.P., Oh J., Tunyaplin C., Carotta S., Donovan C.E., Goldman M.L., Tailor P., et al. . 2009. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 31:941–952. 10.1016/j.immuni.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Awasthi A., Yosef N., Quintana F.J., Xiao S., Peters A., Wu C., Kleinewietfeld M., Kunder S., Hafler D.A., et al. . 2012. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13:991–999. 10.1038/ni.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleer J.P., and Kolls J.K.. 2011. Mechanisms controlling Th17 cytokine expression and host defense. J. Leukoc. Biol. 90:263–270. 10.1189/jlb.0211099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy M.J., Bak-Jensen K.S., Chen Y., Tato C.M., Blumenschein W., McClanahan T., and Cua D.J.. 2007. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat. Immunol. 8:1390–1397. 10.1038/ni1539 [DOI] [PubMed] [Google Scholar]
- Montes R., Puy C., Molina E., and Hermida J.. 2012. Is EPCR a multi-ligand receptor? Pros and cons. Thromb. Haemost. 107:815–826. 10.1160/TH11-11-0766 [DOI] [PubMed] [Google Scholar]
- Peters A., Pitcher L.A., Sullivan J.M., Mitsdoerffer M., Acton S.E., Franz B., Wucherpfennig K., Turley S., Carroll M.C., Sobel R.A., et al. . 2011. Th17 cells induce ectopic lymphoid follicles in central nervous system tissue inflammation. Immunity. 35:986–996. 10.1016/j.immuni.2011.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., and Maglott D.R.. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35(Database issue, Database):D61–D65. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek A.K., Satija R., Shuga J., Trombetta J.J., Gennert D., Lu D., Chen P., Gertner R.S., Gaublomme J.T., Yosef N., et al. . 2014. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 510:363–369. 10.1038/nature13437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton C., Brereton C., Keogh B., Mills K.H., and Lavelle E.C.. 2006. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J. Exp. Med. 203:1685–1691. 10.1084/jem.20060285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., and Salzberg S.L.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Yosef N., Yang J., Wang Y., Zhou L., Zhu C., Wu C., Baloglu E., Schmidt D., Ramesh R., et al. . 2014. Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 40:477–489. 10.1016/j.immuni.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.O., Nurieva R., Martinez G.J., Kang H.S., Chung Y., Pappu B.P., Shah B., Chang S.H., Schluns K.S., Watowich S.S., et al. . 2008. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 29:44–56. 10.1016/j.immuni.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H., Yang L., Qiao G., Li Z., Zhang L., Yin F., Xie D., and Zhang J.. 2010. Cutting edge: CTLA-4–B7 interaction suppresses Th17 cell differentiation. J. Immunol. 185:1375–1378. 10.4049/jimmunol.0903369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N., Shalek A.K., Gaublomme J.T., Jin H., Lee Y., Awasthi A., Wu C., Karwacz K., Xiao S., Jorgolli M., et al. . 2013. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 496:461–468. 10.1038/nature11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski C.E., Mele F., Aschenbrenner D., Jarrossay D., Ronchi F., Gattorno M., Monticelli S., Lanzavecchia A., and Sallusto F.. 2012. Pathogen-induced human TH17 cells produce IFN-γ or IL-10 and are regulated by IL-1β. Nature. 484:514–518. 10.1038/nature10957 [DOI] [PubMed] [Google Scholar]