Tumor-associated macrophages are pivotal constructors of the tumoral ECM structure and molecular composition. In particular, they orchestrate the buildup of the tumorigenic collagenous ECM niche.
Abstract
Tumor-associated macrophages (TAMs) promote tumor development, invasion, and dissemination by various mechanisms. In this study, using an orthotopic colorectal cancer (CRC) model, we found that monocyte-derived TAMs advance tumor development by the remodeling of its extracellular matrix (ECM) composition and structure. Unbiased transcriptomic and proteomic analyses of (a) TAM-abundant and -deficient tumor tissues and (b) sorted tumor-associated and -resident colonic macrophage subpopulations defined a distinct TAM-induced ECM molecular signature composed of an ensemble of matricellular proteins and remodeling enzymes they provide to the tumor microenvironment. Remarkably, many of these ECM proteins are specifically increased in human CRC versus healthy colon. Specifically, we demonstrate that although differentiating into TAMs, monocytes up-regulate matrix-remodeling programs associated with the synthesis and assembly of collagenous ECM, specifically collagen types I, VI, and XIV. This finding was further established by advanced imaging showing that TAMs instruct the deposition, cross-linking, and linearization of collagen fibers during tumor development, especially at areas of tumor invasiveness. Finally, we show that cancer-associated fibroblasts are significantly outnumbered by TAMs in this model and that their expression of collagen XIV and I is reduced by TAM deficiency. Here, we outline a novel TAM protumoral function associated with building of the collagenous ECM niche.
INTRODUCTION
Tumor-associated macrophages (TAMs) are abundant in the tumor stroma at all stages of tumor progression. Clinical studies and experiments in mouse models clearly indicate that TAMs are typically polarized by the local tumor milieu to adopt a protumoral phenotype that promotes tumor cell invasion, motility, and intravasation (Biswas et al., 2013; Noy and Pollard, 2014). Macrophages also contribute to metastasis by priming the premetastatic site and enabling tumor cell extravasation, survival, and persistent growth (Qian et al., 2011). In particular, it has been established that TAMs orchestrate the so-called angiogenic switch by producing neoangiogenic molecules that increase vascular density (Lin and Pollard, 2007). Moreover, their release of inflammatory cytokines generates a chronic inflammatory environment permissive for tumor initiation and growth (Movahedi et al., 2010; Coussens et al., 2013; Noy and Pollard, 2014). It is, therefore not surprising that extensive TAM infiltration positively correlates with cancer metastasis and poor clinical prognosis in a variety of human cancers (Noy and Pollard, 2014).
The tumor cellular ecosystem is nourished by its extracellular matrix (ECM), comprising a three-dimensional (3D) supramolecular network of polysaccharides and proteins, including collagens, glycoproteins, and proteoglycans. The tumoral ECM actively promotes cancer by providing critical biomechanical and biochemical cues that drive tumor cell growth, survival, invasion, and metastasis and by regulating angiogenesis and immune function. It differs significantly from normal ECM, an outcome of aberrantly expressed or modified structural proteins and remodeling events orchestrated by specific proteolytic and protein cross-linking enzymes (Lu et al., 2012; Naba et al., 2012, 2016; Perryman and Erler, 2014; Pickup et al., 2014). Tumors are characterized by high levels of proteolytic degradation of physical barriers between cells that allow the invasion of malignant and endothelial cells and promote the activation and release of cryptic proteins, which directly stimulate tumor cell survival, proliferation, motility, and the neoangiogenic switch (Kessenbrock et al., 2010; Mason and Joyce, 2011). ECM deposition (desmoplasia) is also a hallmark of various solid tumors, and it ensues from altered deposition, cross-linking, and geometrical organization (e.g., linearization) of matrix proteins, especially of collagen fibers, the most abundant ECM scaffolding proteins in the tumor stroma (Provenzano et al., 2008; Levental et al., 2009; Lu et al., 2012; Pickup et al., 2014). Though it remains elusive, it has been suggested that dysregulated collagen deposition and metabolism resulting in increased fibrosis enhance tumor development and invasion (Levental et al., 2009; Lu et al., 2012; Pickup et al., 2014).
TAMs were suggested to participate in shaping the tumor stroma by producing proteolytic enzymes and matrix-associated proteins. Gene expression profiling of TAMs isolated from human ovarian carcinoma revealed their expression of various matrix proteolytic enzymes and matricellular proteins, suggesting their contribution to tumor growth and invasiveness (Liguori et al., 2011). Yet, the mode by which TAMs drive ECM remodeling and the resulting effect on tumor development remain largely unknown. Recently, there was a paradigm shift in the comprehension of macrophage ontogeny with the realization that most tissue-resident macrophages are established prenatally (Varol et al., 2015). In contrast, intestinal lamina propria macrophages (lpMFs) are mainly Ly6Chi monocyte derived during adulthood (Varol et al., 2007, 2009; Bogunovic et al., 2009; Zigmond et al., 2012; Bain et al., 2014). Similarly, TAMs depend on de novo Ly6Chi monocyte recruitment (Movahedi et al., 2010; Franklin et al., 2014; Shand et al., 2014), and the differentiation of both lpMFs and TAMs relies on CCR2 and CSF1 (Lin et al., 2001; Bogunovic et al., 2009; Varol et al., 2009; Qian et al., 2011; Zigmond et al., 2012; Franklin et al., 2014). Thus, given the shared ontogeny of TAMs and colonic lpMFs, colorectal cancer (CRC) serves as the perfect model for exploring the specific ECM signature acquired by monocytes upon their differentiation into TAMs versus tissue-resident macrophages.
Using an orthotopic CRC mouse model (Zigmond et al., 2011), we demonstrate that Ly6Chi monocytes, which massively infiltrate the tumors by virtue of their CCR2 expression, mature into TAMs and that their deficiency impairs tumor growth and ECM buildup. Using an integrated genomic and proteomic approach to define the ECM signature of colorectal TAMs together with advanced high-resolution optical imaging to visualize the tumoral ECM macromolecule network, we show that TAMs play a critical role in the deposition, cross-linking, and linearization of collagenous ECM, a feature that until now has been uniquely attributed to cancer-associated fibroblasts (CAFs; Kalluri and Zeisberg, 2006).
RESULTS
Characterization of TAM subsets in a mouse orthotopic model of CRC
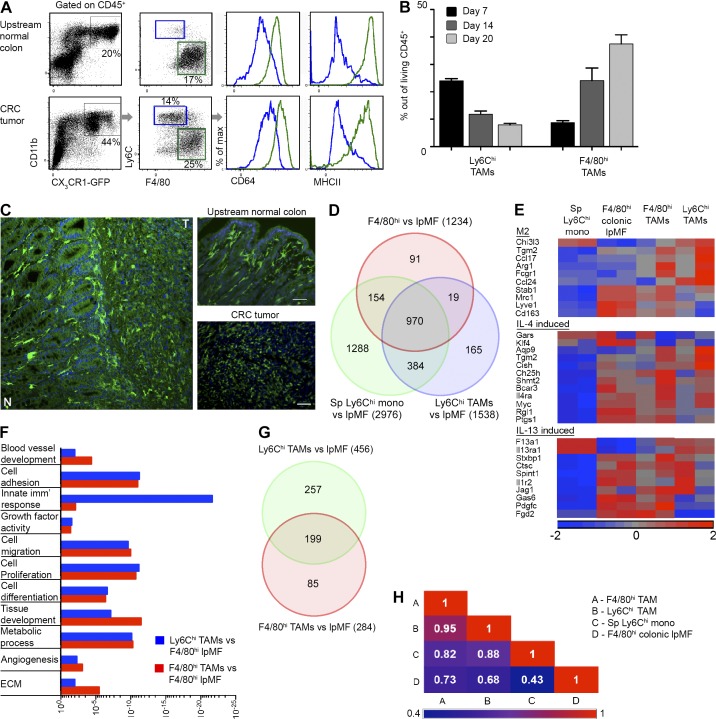
To define the role of TAMs in CRC, we used a mouse orthotopic model based on the endoscopic-guided colonic implantation of syngeneic CRC cells that is minimally invasive and highly reproducible (Zigmond et al., 2011). Given the significant expression of the fractalkine chemokine receptor CX3CR1 by monocyte-derived lpMFs (Varol et al., 2009; Zigmond et al., 2012) and TAMs in other models (Movahedi et al., 2010; Franklin et al., 2014), we characterized the TAM compartment in colorectal tumors implanted in Cx3cr1gfp/+ reporter mice (Jung et al., 2000). Flow cytometry analysis of upstream normal colonic lamina propria revealed a dominant population of resident lpMFs defined as CX3CR1-GFPhi cells that highly express CD11b, MHCII, and the macrophage lineage markers F4/80 and CD64 (FcγR1; Zigmond et al., 2012). Our assessment of colorectal tumors 2 wk after their implantation revealed the massive recruitment and accumulation of two CD11b+CX3CR1-GFP+ macrophage subsets: a Ly6ChiCD64loF4/80loMHCII− monocyte infiltrate and their Ly6CloF4/80hiMHCII+CD64hi mature TAM descendants (Fig. 1 A). Ly6Chi TAMs dominated the early tumor development phase (day 7), whereas F4/80hi TAMs were greater at the later phase (days 14–20) and constituted the major immune cell population (Fig. 1 B). Importantly, although colonic lpMFs and mature TAMs shared a similar expression pattern of macrophage-characteristic markers, immunofluorescent confocal imaging of the interface between the colorectal tumor and adjacent normal mucosa revealed clear morphological differences, with TAMs appearing smaller and round shaped, whereas lpMFs were larger and ramified (Fig. 1 C).
Figure 1.
Characterization of TAM subsets in an orthotopic model of CRC. (A) Flow cytometry analysis of living CD45+ leukocytes was performed at day 14 after tumor implantation in Cx3cr1gfp/+ mice. Representative images display the definition of colonic lpMFs within the upstream normal colon (top) and Ly6Chi and F4/80hi TAM subsets within the colorectal tumors (bottom). Percentages indicate the population fraction out of CD45+ cells. (B) Graphic summary showing the fraction of Ly6Chi and F4/80hi TAMs out of CD45+ living total tumor immune cells at days 7, 14, and 20 after tumor grafts. Data are presented as mean ± standard error of the mean. (C) Confocal fluorescence microscopy imaging of colorectal tumor margins was performed at day 14 after tumor implantation in Cx3cr1gfp/+ mice. Images show the interface between the tumor (T) and its surrounding normal tissue (N). Bars, 50 µm. (D) Venn diagram of monocytes (mono) and TAM subsets showing the distribution and number of differentially expressed genes in comparison with colonic resident lpMFs. Sp, splenic. (E) Heat map analysis showing the differential raw expression level of genes associated with M2 alternative macrophage activation phenotype and with IL-4– or Il-13–induced macrophage activation. (F) Graphic presentation of the significance (p-value) for the enrichment of selected GO categories out of GOEAST analyses performed for differentially expressed genes between Ly6Chi TAMs (blue) or F4/80hi TAMs (red) versus colonic lpMFs. Imm’, immune. (G) Venn diagram showing the distribution of functions found to be significantly enriched (P < 0.05) by the DAVID tool in the differentially expressed genes of F4/80hi TAMs and Ly6Chi TAMs versus colonic lpMFs. (H) Correlation matrix with Pearson correlation coefficient performed for all genes above background and above twofold change. Results are representative of one (B and C) or tens (A) of independent experiments with three to five mice in each experimental group. (D–H) Microarray data represent the average of two biological repeats, each extracted from a pool of mice (splenic monocytes, n = 5; TAMs and colonic lpMFs, n ≥ 10). GOEAST and DAVID analyses of differentially expressed genes (greater than or equal to twofold change; P < 0.05; ANOVA) use a hypergeometric test to assess the significantly enriched GO terms among a given gene list.
We next sought to molecularly define the difference between the colorectal TAM subsets and colonic lpMFs. To that end, we performed a transcriptome microarray analysis of highly purified Ly6Chi and F4/80hi TAM subsets sorted from CRC tumors from Cx3cr1gfp/+ mice in comparison with colonic-resident lpMFs sorted from upstream normal mucosa to exclude TAM contamination (Fig. S1). These macrophage subsets were also compared with the shared circulating Ly6Chi monocyte precursors isolated from the splenic reservoir of the same mice. We identified 1,538 and 1,234 genes that were differentially expressed by Ly6Chi TAMs and F4/80hi TAMs, respectively, in comparison with colonic lpMFs (Fig. 1 D; greater than or equal to twofold and P < 0.05). Upon their differentiation toward Ly6Chi and F4/80hi CRC TAM subsets, the Ly6Chi monocyte precursors up-regulated genes previously defined as signature genes of alternatively activated M2 phenotype and of IL-4– or IL-13–induced macrophage activation (Fig. 1 E; Martinez et al., 2013; Murray et al., 2014; Xue et al., 2014). Some of these were also induced in colonic lpMFs, which further supports their previously defined antiinflammatory signature (Bain et al., 2013; Zigmond et al., 2014). GOEAST Gene Ontology (GO) analysis (Zheng and Wang, 2008) further provided function enrichment categorization of the differentially expressed genes between Ly6Chi TAMs and colonic lpMFs and between F4/80hi TAMs and colonic lpMFs (Fig. 1 F and Table S1). In both gene lists, there was a significant enrichment for ECM-associated genes. Additional function enrichment analysis was performed using the DAVID tool (Huang et al., 2009), and the resulting functions and pathways with enrichment scores of P < 0.05 were selected and visualized in Venn diagrams (Fig. 1 G); many were common to both gene lists and uncovered a significant enrichment for ECM genes (Table S2). In support of their tissue of origin, ingenuity disease and function pathway analysis of the differentially expressed genes between Ly6Chi TAMs or F4/80hi TAMs versus colonic lpMFs revealed highly significant enrichment for pathways associated with gastrointestinal tract cancer and tumors (Ly6Chi TAMs: p-value = 5.6E-12, activation z score = 2.68; F4/80hi TAMs: p-value = 1.1E-09, activation z score = 2.5) and with intestinal inflammation (Ly6Chi TAMs: p-value = 9.2E-11, activation z score = 2.38; F4/80hi TAMs: p-value = 8.3E-07, activation z score = 2.68; Table S3). Finally, a correlation matrix revealed clear gene expression similarity between TAM subsets with a Pearson correlation coefficient of 0.95. In contrast, there was gene expression variance between the resident lpMFs and Ly6Chi or F4/80hi CRC-TAM subsets, with a Pearson correlation coefficient of 0.68 for the former and 0.73 for the latter (Fig. 1 H). Overall, these results highlight that CRC-TAM subsets and colonic lpMFs are molecularly and functionally distinct.
TAM-deficient colorectal tumors display impaired tumor growth
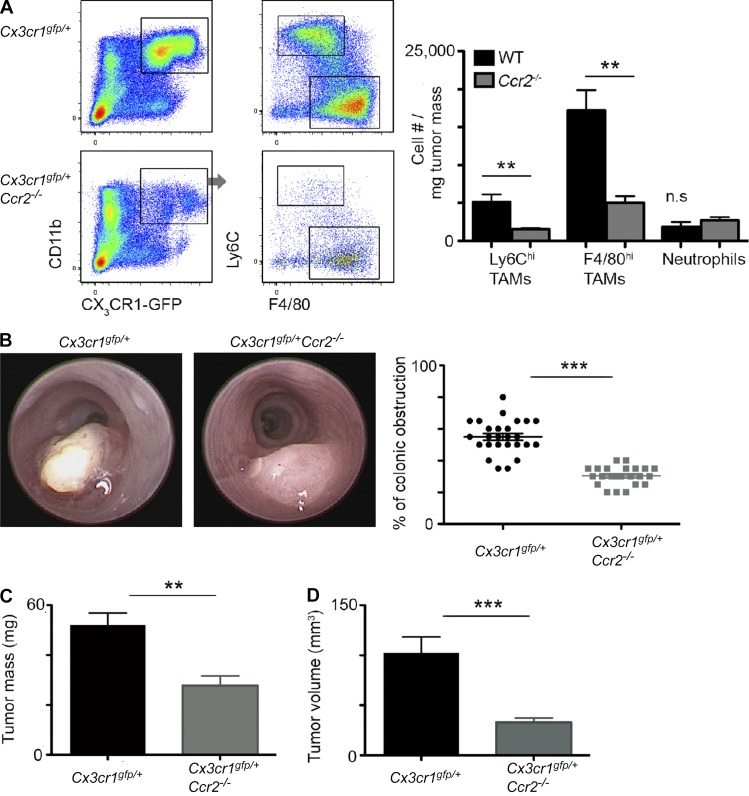
Monocyte recruitment to mammary tumors is CCR2 dependent (Franklin et al., 2014). Our comparative flow cytometry analysis of colorectal tumors implanted into Cx3cr1gfp/+ and Cx3cr1gfp/+Ccr2−/− mice revealed that CCR2 deficiency leads to a significant reduction in the amount of tumor-infiltrating Ly6Chi monocytes and their F4/80hi TAM descendants but has no effect on other tumor-infiltrating CCR2-negative myeloid cells, such as neutrophils (Fig. 2 A). Colonoscopy analysis at day 18 after colorectal tumor implantation revealed impaired tumor development in Ccr2−/− mice, manifested by a smaller degree of tumor obstruction of colonic lumen (Fig. 2 B) and significant reduction in tumor mass (Fig. 2 C) and volume (Fig. 2 D). These results uncover that TAMs play a direct critical protumoral role in CRC. They also point to Ccr2−/− mice as being a suitable model for studying the effects of TAM deficiency on tumoral ECM in the physiological colonic environment.
Figure 2.
Colorectal tumors established in Ccr2−/− mice display impaired TAM recruitment and tumor growth. (A) Flow cytometry analysis of living CD45+ leukocytes was performed at day 14 after tumor implantation in Cx3cr1gfp/+ and Cx3cr1gfp/+Ccr2−/− mice. The graph presents cell population number normalized per tumor mass of Ly6Chi and F4/80hi TAM subsets and of neutrophils in WT versus Ccr2−/− tumors. (B–D) Analysis of tumor growth was performed at day 18 after tumor implantation in WT and Ccr2−/− mice. Graphical summaries display colonic lumen obstruction as assessed by colonoscopy (B), tumor mass (C), and tumor volume (D). Results are representative of one (A) or four (B–D) independent experiments with 6 mice (A) or at least 15 mice (B–D) in each experimental group. Data were analyzed by unpaired, two-tailed Student’s t tests and are presented as mean ± standard error of the mean. **, P < 0.01; ***, P < 0.001.
TAM-deficient colorectal tumors display altered ECM composition
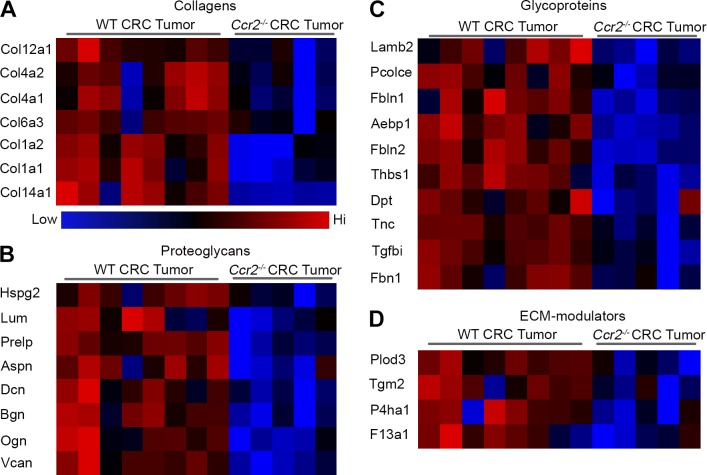
To characterize the influence of TAM deficiency on colorectal tumor ECM composition, we performed proteomic profiling of the tumors and categorized the ECM-core and -associated proteins based on the matrisome definition (Naba et al., 2012, 2016). Unbiased comparative liquid chromatography with mass spectrometry (MS; LC-MS/MS) analysis of whole protein contents from WT colorectal tumors and upstream normal colon tissue revealed 1,305 proteins that were differentially expressed (Student’s t test; positive false discovery rate [pFDR] < 0.05). Among them, 71 were ECM related, out of which 31 were also differentially expressed between WT and Ccr2−/− tumors, implying that TAMs are profoundly involved in promoting the altered ECM composition of colorectal tumors versus healthy colon (Fig. S2). In a direct comparison between WT and Ccr2−/− tumors, we found 46 significantly different ECM-related proteins out of the differentially expressed 348 proteins (enrichment factor = 2.72; pFDR = 1.37 × 10−12; Fig. 3, A–D). Many of these were reduced in the TAM-deficient tumors, including the core matrisome proteins collagen types Iα1, Iα2, IVα1, IVα2, VIα3, XIIα1, and XIVα1 (Fig. 3 A); the proteoglycans heparan sulfate proteoglycan 2 (Hspg2), lumican (Lum), prolargin (Prelp), asporin (Asp), decorin (Dcn), biglycan (Bgn), osteoglycin (Ogn), and versican (Vcan; Fig. 3 B); and the glycoproteins laminin β-2 (Lamb2), procollagen C–endopeptidase enhancer (Pcolce), fibulin 1 and 2 (Fbln1 and Fbln2), thrombospondin 1 (Thbs1), dermatopontin (Dpt), tenascin 1 (Tnc1), TGFβ-induced protein (Tgfbi), and fibrillin 1 (Fbn1; Fig. 3 C), as well as the ECM-associated modulators procollagen-lysine 2-oxoglutarate 5-dioxygenase 3 (Plod3), transglutaminase 2 (Tgm2), prolyl 4-hydroxylase, α polypeptide I (P4ha1), and the coagulation factor XIII subunit a1 (F13a1; Fig. 3 D). Thus, these results attribute a major direct or indirect role for TAMs in shaping ECM composition during colorectal tumor development.
Figure 3.
TAM-deficient colorectal tumors display altered ECM composition. (A–D) LC-MS/MS analysis was performed on whole protein extracts from WT versus Ccr2−/− tumors. Color-coded heat maps show the differentially expressed ECM-related proteins categorized into collagens (A), proteoglycans (B), glycoproteins (C), and ECM modulators (D). Results are a summary of a single experiment with eight mice in the WT tumor group and five mice in the Ccr2−/− tumor group. Data were analyzed by unpaired, two-tailed Student’s t tests with pFDR = 0.05 and presented in z-score form.
Decellularized ECM fragments extracted from TAM-sufficient tumors, but not from TAM-deficient tumors or normal colon, are tumorigenic
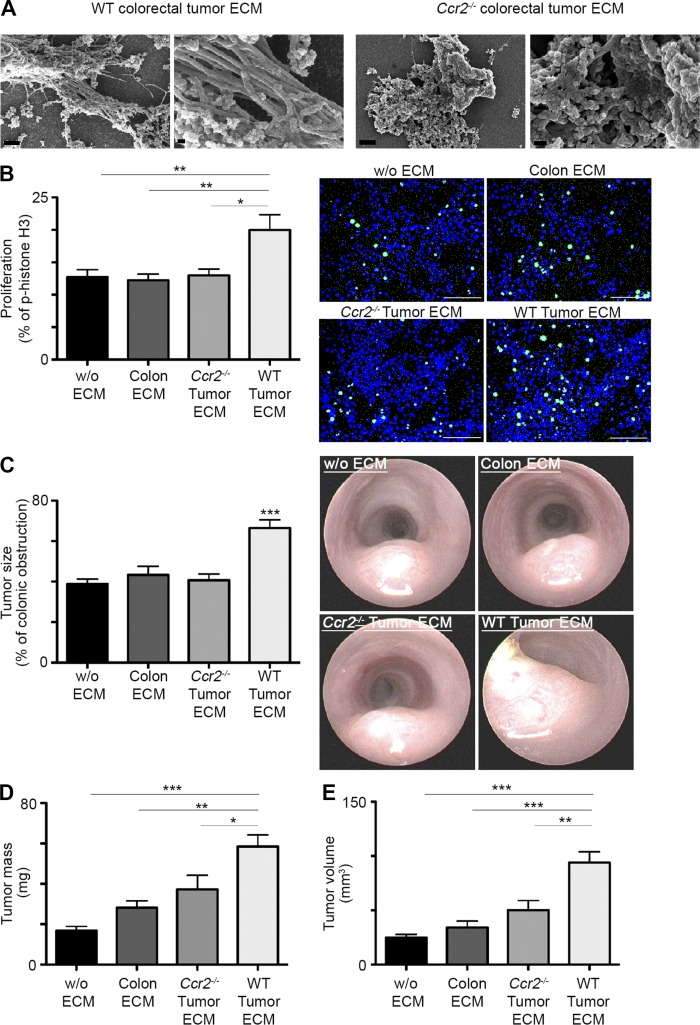
To study the effects of TAM-mediated direct or indirect remodeling of ECM composition on tumor growth, MC38 CRC cells were cultured with equal concentrations of decellularized 3D ECM fragments homogenously extracted from WT or Ccr2−/− tumors or upstream healthy colon. Scanning electron microscopy (SEM) analysis (Fig. 4 A) confirmed that the ECM fragments were indeed cell free, also preserving their unit structure. The effect on tumor cell proliferation was assessed 48 h later by immunofluorescence staining for phosphohistone 3 (p-histone H3+). Fluorescence microscopy analysis revealed an increased fraction of p-histone H3+ proliferating MC38 cells after their culture with ECM fragments from WT tumors but not from Ccr2−/− tumors or healthy colon tissue (Fig. 4 B). To further investigate this effect in the colonic native physiological environment, MC38 cells were orthotopically implanted together with decellularized 3D ECM fragments extracted from either WT or Ccr2−/− tumors or upstream healthy colon. Colonoscopy analysis 20 d later revealed a significant acceleration in tumor growth only in those mice implanted with WT tumor ECM fragments (Fig. 4 C). The observed increase in tumor growth was further validated by the measurement of tumor mass (Fig. 4 D) and volume (Fig. 4 E). These findings indicate that the TAM-governed overall effect on ECM composition is protumorigenic. Importantly, the tumorigenic effect can be derived from ECM-associated proteins, such as secreted growth factors trapped in the matrix scaffold, which were directly produced or released by TAMs or indirectly by TAM-mediated regulation of other cells in the tumor microenvironment.
Figure 4.
TAM-mediated remodeling of core and affiliated ECM protein composition is tumorigenic. (A) SEM imaging was performed on ECM fragments extracted from decellularized WT or Ccr2−/− tumors. Bars: (left and middle left) 1 µm; (middle right and right) 200 nm. (B) MC38 CRC cells were cultured without or with decellularized 3D ECM fragments extracted from normal colon, WT, or Ccr2−/− tumors, and their proliferation was assessed by staining for p-histone H3 (green) and DAPI (blue). The graph (left) and fluorescence microscopy images (right) show the fraction of actively proliferating MC38 cells. Bars, 100 µm. (C and D) Analysis of tumor growth was performed at day 20 after orthotopic implantation of MC38 CRC cells without or with decellularized 3D ECM fragments extracted from normal colon, WT, or Ccr2−/− tumors. Graphical summaries display colonic lumen obstruction as assessed by colonoscopy (C), tumor mass (D), and tumor volume (E). Results are representative of one (B) or two (A and C–E) independent experiments with 10 repeats per group (B) or at least 7 mice in each experimental group (C– E). Data were analyzed by unpaired, two-tailed Student’s t tests, comparing each time the WT tumor ECM with one of the other groups, and are presented as mean ± standard error of the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001. w/o, without.
TAMs provide a unique set of ECM proteins and modulators to the tumor microenvironment
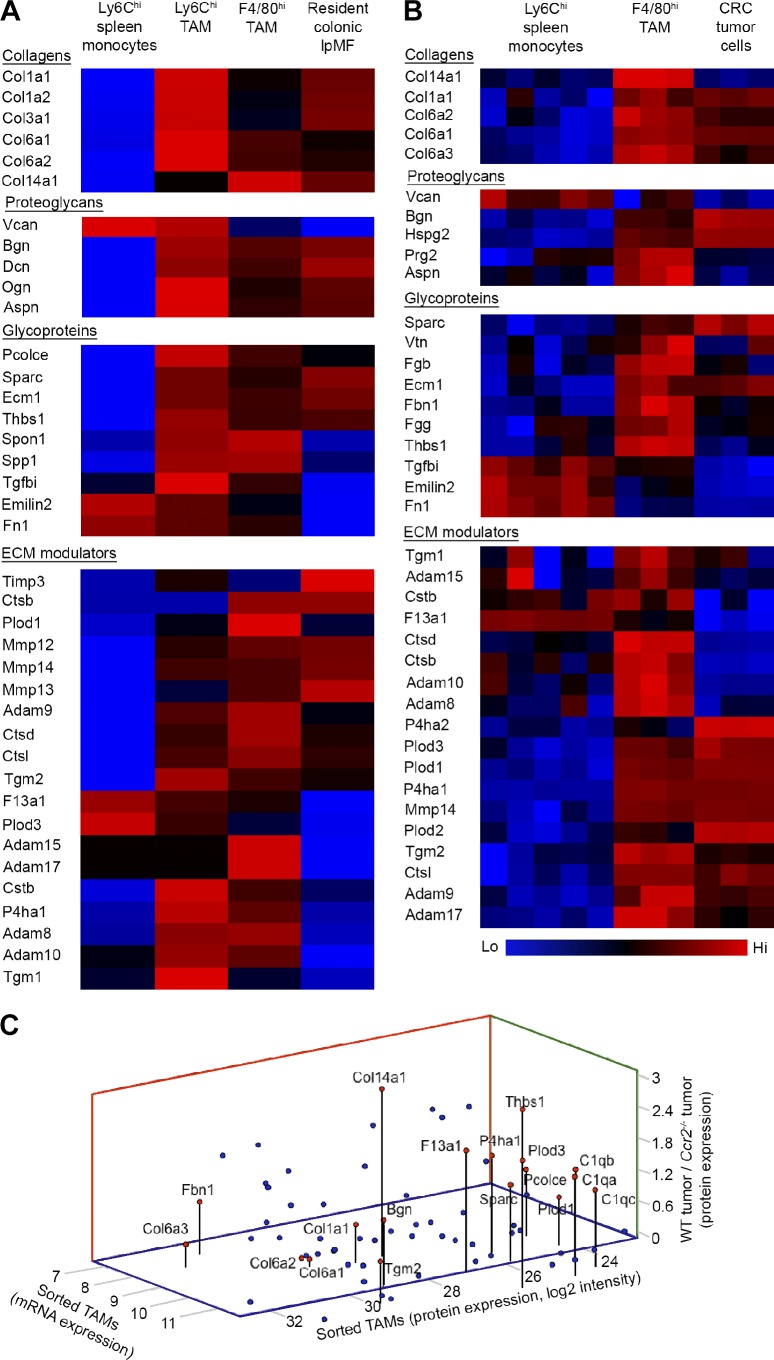
To define the direct contribution of TAMs to the ECM compositional changes observed in TAM-sufficient versus -deficient colorectal tumors (Fig. 3), we characterized the ECM-related molecular pathways that are uniquely activated in Ly6Chi monocytes within the colorectal tumor microenvironment. We first compared the gene expression signature of sorted Ly6Chi and F4/80hi TAM subsets with those of colonic lpMFs and the shared naive Ly6Chi monocyte precursors. Genes encoding for collagens types Iα1, Iα2, IIIα1, VIα1, VIα3, and XIVα1 were significantly up-regulated in Ly6Chi TAMs in comparison with their circulating monocyte precursors (Fig. 5 A). Remarkable was their expression of collagen XIVα1, which was further up-regulated upon their maturation into F4/80hi TAMs. Additional ECM-associated genes that were uniquely activated in TAMs included matrix enzymes involved with collagen synthesis and assembly such as PLOD1 and 3, P4HA1, and PCOLCE, as well as the ECM and cell surface protein modulators ADAMs (a disintegrin and metalloproteinase) 8, 9, 10, 15, and 17, cathepsins (Cts) B, D, and L, the protease inhibitor cystatin B (Cstb), and the matrix cross-linkers F13A1 and TGM-1 and -2. TAMs were also higher for the core ECM structural genes versican, asporin, osteoglycin, osteopontin (Spp1), THBS1, and spondin 1 (Spon1; Fig. 5 A). Given their shared monocytic ontogeny and colonic environment, various ECM-related genes were similarly expressed by both TAMs and colonic lpMFs (Fig. S3).
Figure 5.
Transcriptomic and proteomic analyses of TAM’s ECM signature. (A) Color-coded Affymetrix gene array heat maps displaying the ECM-related gene expression in sorted Ly6Chi and F4/80hi TAMs (day-14 tumors) in comparison with their Ly6Chi monocyte precursors (splenic reservoir) and resident lpMFs sorted from upstream normal colon. Data were z scored and represent the average of two biological repeats; each was extracted from a pool of mice (splenic monocytes, n = 5; TAMs and colonic lpMFs, n ≥ 10). (B) Color-coded heat maps presenting ECM-related proteins found by LC-MS/MS analysis to be significantly and differentially expressed in sorted F4/80hi TAMs in comparison with their Ly6Chi monocyte precursors and colocalizing colorectal tumor cells. Ly6Chi monocytes: five biological repeats; each was extracted from a pool of three mice. F4/80hi TAMs and CRC cells: three biological repeats; each was extracted from a pool of five mice. Data were analyzed by unpaired, two-tailed Student’s t tests (pFDR = 0.05) and z scored. (C) Graphical summary showing ECM molecules that were mutually expressed (log2) at the protein level in sorted F4/80hi TAMs (x axis) and at the RNA level in sorted F4/80hi TAMs (y axis) and were higher in the TAM-sufficient (WT) versus TAM-deficient (Ccr2−/−) tumors (z axis). Each dot represents an ECM protein; red dots highlight proteins associated with fibrous ECM formation.
To define the ECM signature acquired by TAMs at the protein level, we performed a comparative LC-MS/MS analysis between sorted naive Ly6Chi monocytes, their F4/80hi TAM descendants, and CRC tumor cells (sorted from the same tumors). Overall, 5,261 proteins were identified in all sorted cells, out of which 108 were ECM related. Further analysis revealed 4,101 proteins that were differentially expressed between TAMs and either Ly6Chi monocytes or CRC tumors cells (Student’s t test; pFDR < 0.05), out of which 100 proteins were ECM related. In alignment with the gene expression profiling, TAMs up-regulated the protein expression of collagen types Iα1, VIα1, VIα2, VIα3, and XIVα1 and of proteins associated with collagen synthesis and assembly, such as PLOD1 and 3, P4HA1, PCOLCE, and secreted protein acidic and rich in cysteine (SPARC) compared with their monocyte precursors. Moreover, TAMs were higher for ECM modulators, such as ADAMs 8, 9, 10, 15, and 17; cathepsins B, D, and L; cystatin B; the ECM cross-linker enzymes F13A1 and TGM-1 and -2.; the proteoglycans asporin and proteoglycan 2 (Ptg2); and the glycoproteins ECM1, THBS1, vitronectin (Vtn), fibrinogen β and γ chains (Fgb and g), and fibulin 1 (Fig. 5 B). For a broader list of ECM-core and -associated proteins provided by TAMs into the tumor microenvironment, see Fig. S4. Of note, the ECM protein profile may be not complete because of technical limitations that stem from the processing that the cells were subjected to before the proteomic profiling.
We next integrated our transcriptomic and proteomic data of ECM molecules that were identified in TAMs at the mRNA and protein levels and exhibited a reduction at the protein level in Ccr2−/− versus WT tumors (Fig. 5 C). This analysis corroborated the expression of molecules associated with collagen synthesis, stability, assembly, and cross-linking. Among them were the α1 chains of collagen I and collagen XIV, the three α chains of collagen VI, the glycoprotein PCOLCE, the enzyme P4HA1, collagen cross-linkers PLOD1 and 3, the glycoprotein SPARC, and the proteoglycan biglycan. Notably, our integrative analysis also highlighted the TAM-enhanced expression of the ECM covalent cross-linker enzymes TGM2 and F13A1, the complement C1q complex, and THBS1. This detailed molecular profiling indicates that TAMs directly construct specific types of collagenous ECM.
Impaired construction of collagenous matrix in TAM-deficient colorectal tumors
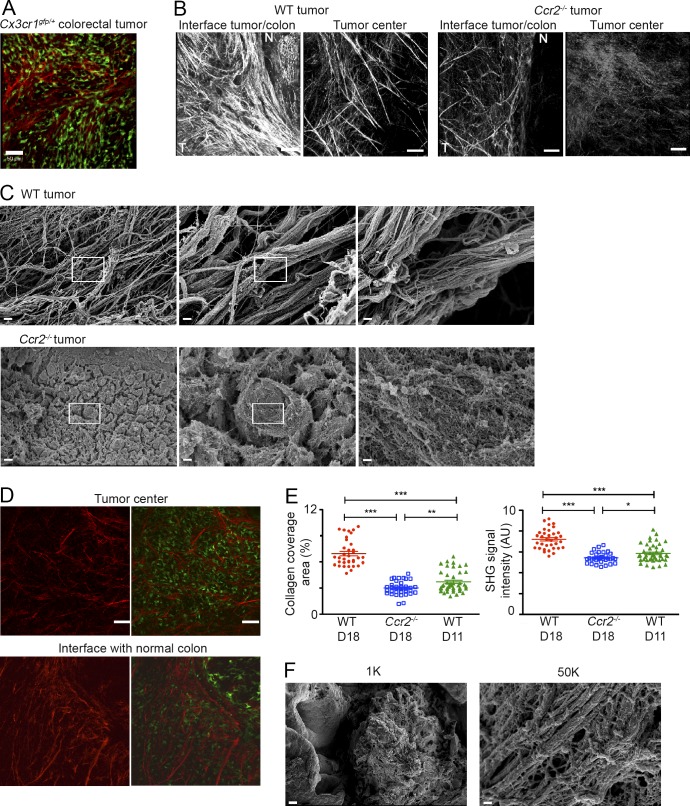
Solid-tumor ECMs are often associated with increased deposition, cross-linking, and linearization of collagen fibers, which is suggested to actively promote tumor growth and invasion (Levental et al., 2009; Lu et al., 2012; Pickup et al., 2014). Nonlinear microscopy techniques, such as second harmonic generation (SHG), provide powerful tools to image fibrillar collagen structures (especially collagen type I) in intact tissues and specifically tumors (Wyckoff et al., 2007; Provenzano et al., 2008). SHG signal imaging in unfixed tumors implanted in Cx3cr1gfp/+ mice revealed colocalization between CX3CR1-GFP+ TAMs and fully assembled collagen fibers at the tumor margins (Fig. 6 A). A comparison between WT and TAM-deficient tumors at day 18 after tumor implantation revealed profound differences in collagen density and assembly. Collagen fibers were more linearized, elongated, thicker, and abundant in WT tumors than in Ccr2−/− tumors (Fig. 6 B). This altered collagen structure appearance could be clearly detected at the tumor’s core but was even more pronounced at its collagen-rich borders. In these border regions, the collagen fibers, mostly consisting of type I, were oriented toward the normal tissue at areas of basement-membrane breakdown (Fig. 6 B), forming a distinct structural signature that has been reported to support cancer cell invasion (Provenzano et al., 2006). To obtain more detailed structural insights, next, we performed high-resolution SEM studies of decellularized 3D ECM scaffolds extracted from WT and Ccr2−/− tumors. Reinforcing our SHG results, the SEM analysis readily detected thick cross-linked and linearized collagen fiber assemblies in the WT ECM scaffolds but not in Ccr2−/− tumors (Fig. 6 C). Importantly, SHG imaging revealed that the buildup of collagenous matrix is already evident at an early developmental stage of WT tumors (day 11), when they have reached the same tumor size as day 18 Ccr2−/− TAM-deficient tumors (Fig. 6 D). Semiquantitative analysis of SHG signals depicted a significant reduction in collagen coverage area and intensity in day 18 Ccr2−/− tumors in comparison with both day 11 and day 18 WT tumors (Fig. 6 E). Notably, there was a profound increase in collagen signals between days 11 and 18 in WT tumors, suggesting a progressive buildup of collagenous matrix by TAMs or by other cells affected by their presence. Finally, SEM imaging of day 11 WT tumors provided clear evidence for collagen cross-linking and linearization already at an early developmental phase especially at areas of tumor invasiveness (Fig. 6 F), whereas these were not detected in day 18 Ccr2−/− tumors of the same size (Fig. 6 C).
Figure 6.
TAM-deficient colorectal tumors display aberrant deposition and organization of fibrillar collagen. (A–F) Colorectal tumors were excised and subjected to SHG and SEM imaging techniques at days 11 and 18 after their orthotopic implantation in Cx3cr1gfp/+ mice. (A) Two-photon SHG microscopy image focusing on the interface between the day 18 tumor and muscularis mucosa. SHG signal is represented as pseudocolor red (excitation: 900 nm; detection: 450 nm), and TAMs are GFP. Bar, 50 µm. (B) Representative two-photon SHG microscopy images of WT and Ccr2−/− colorectal tumor sections revealing collagen deposition and structures at the center of the tumor and at its margins. SHG signal is represented as pseudocolor white. N, normal tissue; T, tumor. Bars, 50 µm. (C) Representative SEM images of decellularized ECM scaffolds extracted from WT and Ccr2−/− colorectal tumors. Bars: (left) 10 µm; (middle) 2 µm; (right) 200 nm. (D) Representative two-photon SHG microscopy images of WT colorectal tumor sections extracted at earlier developmental stage (day 11), when tumors reach 30% of colonic obstruction as Ccr2−/− tumors at day 18. Bars, 50 µm. (E) Graphical summaries of semiquantitative SHG signal intensity (arbitrary units [AU]) and of collagen coverage area (percentage) in WT tumors extracted at days 11 and 18 and Ccr2−/− tumors extracted at day 18. Data were acquired from at least 33 images of at least 3 tumors, analyzed by unpaired, two-tailed Student’s t tests, and are presented as mean ± standard error of the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (F) Representative SEM images of decellularized ECM scaffolds extracted from day 11 WT tumors. Bars: (left) 10 µm; (right) 200 nm. Results are representative of three independent experiments with at least four mice in each experimental group (A–C) or of a single experiment with at least four mice in each group (D and F).
CAFs from TAM-deficient tumors exhibit reduced gene expression of collagen types I and XIV
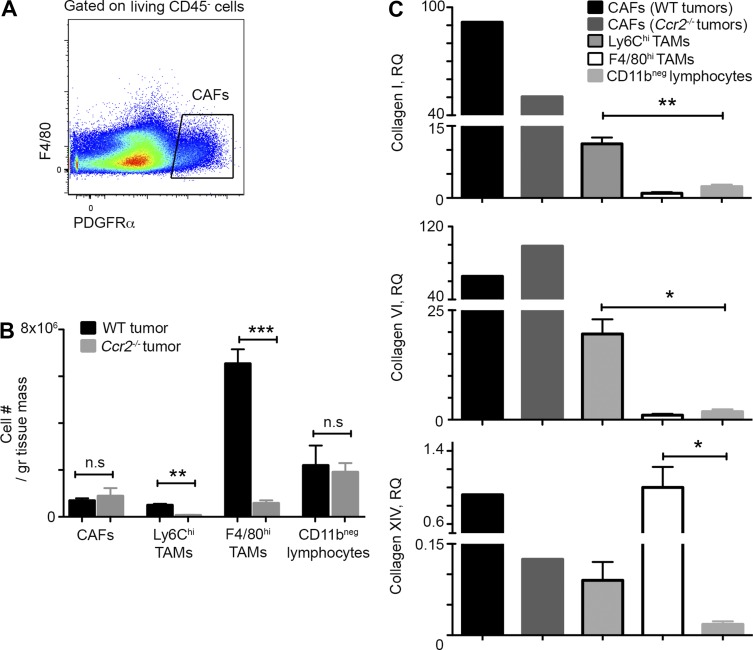
CAFs are considered to be the key producers of collagenous matrix in developing tumors (Kalluri and Zeisberg, 2006). PDGFRα is a surface marker for mouse and human CAFs (Erez et al., 2010; Sharon et al., 2013). Flow cytometry analysis revealed that PDGFRα+ CAFs are considerably outnumbered by TAMs in this CRC model. Moreover, TAM deficiency had no effect on CAFs numbers normalized for tumor mass, outlining that TAMs are not involved with CAF attraction or survival (Fig. 7, A and B). Because of the scarce number of CAFs, we pooled CRC tumors from 20 mice for their sorting and performed quantitative real-time PCR analysis for the gene expression of collagen types I, VI, and XIV, shown by us to be produced by TAMs (Fig. 5). The expression of collagens VI and I was greater in CAFs versus TAMs, but they similarly expressed collagen XIV. Notably, there was a marked reduction in the gene expression of collagens XIV and I concomitantly with elevation in collagen VI in CAFs sorted from Ccr2−/− tumors (Fig. 7 C). Of note, TAM deficiency did not affect the recruitment of CD45+CD11bneg lymphocytes to the tumors (Fig. 7 B), and their production of collagens I, VI, and XIV was significantly lower than TAMs (Fig. 7 C). Collectively, these findings suggest an additional mechanism by which TAMs can affect collagenous matrix remodeling through their regulation of collagen production by CAFs.
Figure 7.
TAM deficiency reduces collagen XIV and I gene expression in CAFs. (A) Flow cytometry analysis was performed in day 14 tumors, and CAFs were identified as living CD45−CD11b−F4/80−PDGFRα+ cells (Erez et al., 2010; Sharon et al., 2013). (B) Graphic summary of flow cytometry results presenting cell population number normalized for tumor mass comparing WT with Ccr2−/− tumors. n ≥ 3 tumors in each group. gr, gram. (C) Quantitative real-time PCR analysis showing the relative gene expression of collagens I, VI, and XIV in comparison with F4/80hi TAMs. CAFs were sorted out of pool of 20 WT or Ccr2−/− tumors. For the other populations, data were extracted from three biological repeats; each was extracted from pool of six to seven mice. RQ, relative quantification. Data were analyzed by unpaired, two-tailed Student’s t tests and are presented as mean ± standard error of the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
TAM-defined ECM proteins are increased in human CRC
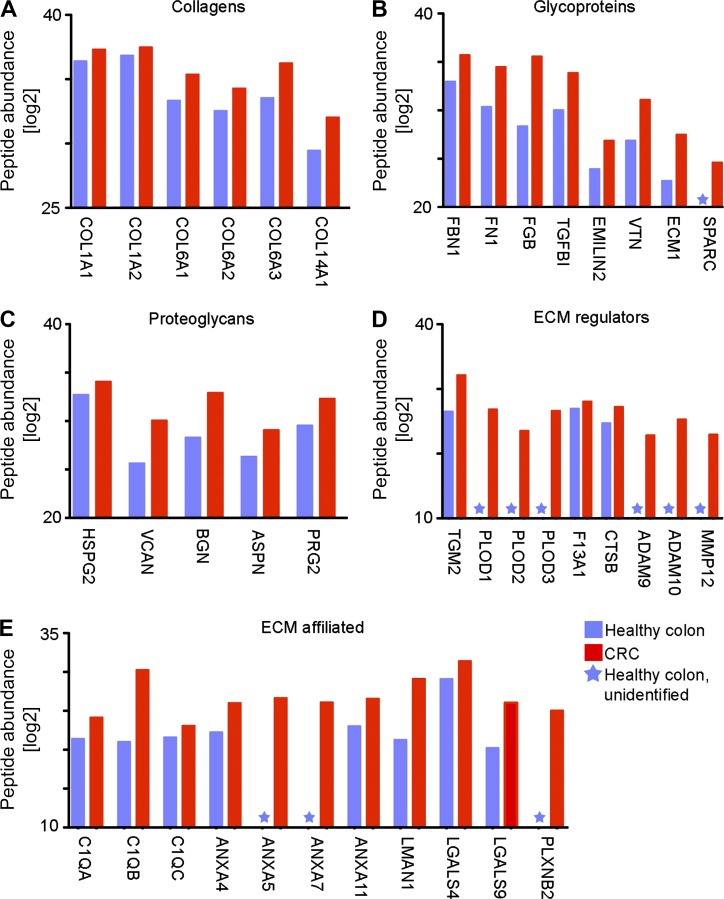
The ECM protein signature of human CRC has been recently defined (Naba et al., 2014). Data mining out of this comprehensive database revealed that many of the ECM proteins, shown by us to be up-regulated after monocyte differentiation into TAMs (Fig. 5), are also increased in the transition from healthy colon to CRC; some are even unidentified in the healthy colon (Fig. 8). Specifically with respect to the collagenous matrix, there was an increase in the abundance of collagen types I, VI, and XIV. Moreover, ECM proteins involved with collagen stability and assembly, which are undetected in the healthy colon, were increased in CRC including the glycoprotein SPARC and the collagen cross-linkers PLOD1, 2, and 3. These results highlight the clinical relevance of the TAM-defined ECM signature.
Figure 8.
TAM-defined ECM proteins are increased in human CRC. (A–E) Graphic representation of protein expression levels of collagens (A), glycoproteins (B), proteoglycans (C), ECM regulators (D), and ECM-affiliated proteins (E). Data are presented as protein abundance (log2 modified) in normal human colon (blue) versus CRC (red) and correspond to the sum of specific peptide abundance across independent samples (three independent samples for each tissue type). Unidentified proteins are marked with a star. These results were analyzed out of a published database (Naba et al., 2014).
DISCUSSION
The view of the ECM as a supporting scaffold upon which tissues are organized has been dramatically extended over the last decades, particularly in cancer, with studies showing that the ECM provides critical biochemical and biomechanical cues that modulate virtually every acquired behavioral hallmark of the tumor cells and associated stromal cells (Pickup et al., 2014). These cues originate from aberrantly expressed or modified structural proteins and remodeling events orchestrated by specific matrix enzymes and are essential for tumor development and dissemination. TAMs, too, play a pivotal protumoral role in primary tumors and during metastasis (Biswas et al., 2013; Noy and Pollard, 2014). They are believed to vigorously participate in protumoral remodeling of the ECM by providing remodeling proteases and matricellular proteins (Liguori et al., 2011). Here, we shed new light on their function as constructors of tumoral ECM structure and molecular composition. By combining unbiased genomics with proteomic approaches, we were able to define the distinct TAM-induced ECM signature, composed from a repertoire of matrix cross-linking and proteolytic enzymes and matricellular proteins introduced by these cells into the tumor microenvironment. In particular, we show that TAMs promote collagen fibrillogenesis by directly contributing to matrix deposition, cross-linking, and linearization of fibrillar collagens, a function that has been uniquely attributed to CAFs (Kalluri and Zeisberg, 2006). Moreover, we show reduced expression of collagens XIV and I in CAFs extracted from TAM-deficient tumors, implying that beyond their direct role, TAMs regulate collagen-remodeling activity by CAFs.
Macrophages are known to display remarkable plasticity, which allows them to efficiently adjust to rapidly changing environmental signals (Varol et al., 2015). TAMs and colonic lpMFs share the same Ly6Chi monocyte precursor (Varol et al., 2009, 2015; Movahedi et al., 2010; Franklin et al., 2014; Shand et al., 2014). In agreement with a previous study in a mammary tumor model (Franklin et al., 2014), we show that Ly6Chi monocytes massively infiltrate CRC tumors in a CCR2-dependent manner and give rise to mature F4/80hi TAMs. Both Ly6Chi and F4/80hi TAM subsets are morphologically distinct from colonic lpMFs and exhibit a distinct gene expression profile. Our results show that circulating monocytes up-regulate the expression of signature genes previously attributed to IL-4– and IL-13–induced M2 activation. Given the antiinflammatory signature of colonic lpMFs (Bain et al., 2013; Zigmond et al., 2014), it was not surprising that many of these genes are also induced during the differentiation of the same monocyte precursors into lpMFs. GOEAST and DAVID functional enrichment analyses of genes that were differentially expressed between the TAM subsets and colonic lpMFs revealed a significant enrichment for ECM genes implying distinct ECM remodeling abilities. Ingenuity analysis uncovered a significant enrichment for genes associated with CRC development and intestinal inflammation in the colorectal TAMs in comparison with macrophages sorted from normal adjacent colon. These changes highlight a tissue specialization program that Ly6Chi monocytes undergo upon their differentiation toward TAMs versus lpMFs. Tumor development in Ccr2−/− mice was severely impaired, thus substantiating a pivotal protumoral role for TAMs in orthotopic colorectal tumors. Importantly, the impaired development of TAM-deficient tumors may be the result of TAM-governed cell-intrinsic mechanisms and also via their regulation of other cells in the tumor microenvironment.
Malignant tissue is typically stiffer than its healthy counterpart. This altered biomechanical property is primarily caused by elevated deposition, cross-linking, and collagen linearization events, especially of collagen I fibers, and it has been linked to increased tumor growth, invasiveness, and metastasis (Provenzano et al., 2008; Levental et al., 2009; Lu et al., 2012; Pickup et al., 2014). Intravital imaging studies in mammary tumors revealed an abundance of TAMs at the tumor’s collagen-rich border (Wyckoff et al., 2007). However, the synthesis and deposition of stromal collagen has been primarily attributed to CAFs (Kalluri and Zeisberg, 2006). Here, we unravel a complement and yet critical role for TAMs in phenotypic ECM buildup. Specifically, we show that TAMs accumulate at the invasive margins of colorectal tumors and significantly contribute to the deposition and geometrical organization of collagenous ECM. With the aid of advanced SHG imaging, we were able to visualize a notable increase in collagen density within WT colorectal tumors, especially at their interface with normal colonic tissue where the basement membrane is breached. SEM imaging revealed that the collagen fibers were straightened and aligned in heavily cross-linked bundles. In contrast, TAM-deficient Ccr2−/− tumors lack this collagen signature, displaying lower collagen density and shorter, thinner, and randomly ordered fibers. These structural collagen arrangements are already evident at an earlier developmental stage of WT tumors, especially at the invasive front area, suggesting that TAM-mediated remodeling of the collagenous matrix is integrated within the tumor developmental program.
In correlation with these remarkable visualized changes in tumor collagen remodeling induced by the presence of TAMs, our integrated genomic and proteomic findings provide direct evidence that monocytes intrinsically up-regulate matrix-remodeling programs associated with the synthesis of collagen types I, VI, and XIV as they differentiate into TAMs. Collagen type I is synthesized and processed using a platform of matrix enzymes, including P4HA1, 2, and 3 and PLOD 1, 2, and 3 (Gilkes et al., 2014). Our results show that TAMs express the mRNA and protein of P4HA1, PLOD1, and PLOD3 and that Ccr2−/− tumors are significantly lower for collagen I, P4HA1, and PLOD3 proteins. Moreover, we found that TAMs express PCOLCE, which enhances the activity of procollagen C–proteinase, involved in collagen maturation. MS analysis also revealed that Ly6Chi monocytes significantly up-regulate the three α chains of collagen VI (α1, 2, and 3) upon their polarization into TAMs. Most interesting, the collagen VIα3 chain was significantly reduced in Ccr2−/− tumors, suggesting that TAMs are a major source of this collagen type. Indeed, the production of this collagen subtype has been documented before in the human monocyte cell line (Schnoor et al., 2008). The cleavage product of collagen VIα3, called endotrophin, promotes mammary tumor growth (Park and Scherer, 2012). We found that TAMs also express collagen XIVα1 and that it is significantly reduced in TAM-deficient tumors. This collagen belongs to a family of fibril-associated collagens that do not form fibrils. Rather, collagen XIVα1 interacts predominantly with collagen I to promote fiber assembly (Gerecke et al., 2003), thus possibly affecting tumors’ matrix density. In addition, TAMs expressed the proteoglycan biglycan, a key regulator of lateral assembly of collagen fibers that has been shown to specifically interact with collagen I (Schönherr et al., 1995). Previous studies demonstrated the importance of TAM-derived SPARC in the deposition and assembly of collagenous ECM (Sangaletti et al., 2008). We show that SPARC expression is indeed induced in tumor-infiltrating Ly6Chi monocytes and their mature TAM descendants. Importantly, the ability of macrophages to acquire a fibroblast-like phenotype involved with their direct synthesizing of collagen is further supported by studies in renal fibrosis (Nikolic-Paterson et al., 2014; Wang et al., 2016).
Accumulating evidence in various inflammatory settings suggest that infiltrating monocytes become integral effector cells within their host tissue (Varol et al., 2015). Our results are in alignment with this up-to-date paradigm, as we molecularly prove a distinct gene expression profile, and specifically ECM signature, among the circulating Ly6Chi monocyte precursors, their tumor infiltrating effector Ly6Chi TAMs, and their mature F4/80hi TAM descendants. With respect to the collagenous matrix, we show that Ly6Chi TAMs are higher for genes involved with collagen deposition and remodeling such as collagens I, III, and VI, PCOLCE, SPARC, PLOD3, and P4HA1, whereas F4/80hi TAMs are higher for collagen XIV and PLOD1. Moreover, collagenous matrix remodeling is already evident at an early tumor development phase when the Ly6Chi TAMs still dominate the tumor macrophage compartment. Therefore, our results support the idea that Ly6Chi TAMs are directly involved in this process.
TAMs promote tumor development by various mechanisms (Biswas et al., 2013; Noy and Pollard, 2014), and hence, their deficiency may induce changes in ECM composition and structure that are not directly associated with their repertoire of ECM structural proteins and remodeling enzymes. Specifically, TAMs may indirectly contribute to the production and organization of collagenous matrix through their regulation of CAFs. Our results show that in this orthotopic CRC model, CAFs are outnumbered by TAMs. Although their representation in the tumor is not affected by TAM deficiency, their production of collagen types XIV and I is reduced. Although the mechanisms remain elusive, these results suggest an additional indirect mechanism by which TAMs contribute to the remodeling of collagenous matrix during tumor development. Of note, because of the scarce number of CAFs, we could only appreciate their collagen production at the gene expression level, whereas in the future, it will be important to delineate TAM-governed regulation of CAFs-derived collagen at the level of protein deposition, assembly, and stability. Interestingly, although TAMs are lower than CAFs in the expression of collagen types VI and I, they express similar levels of collagen type XIV, highlighting them as a major source of this collagen given their significant favorable representation. Recently it has been shown in a skin injury model that macrophages in response to IL-4 polarization induce the activity of the collagen cross-linker enzyme lysyl hydroxylase 2 (PLOD2) in adjacent fibroblasts (Knipper et al., 2015). We show that TAMs up-regulate IL-4 polarization signature genes. Therefore, it remains elusive whether TAMs can induce PLOD2-governed collagen–cross-linking activity in CAFs during tumor development. Moreover, we show that TAMs themselves express PLOD1 and PLOD3 collagen cross-linkers, suggesting a task division between them and CAFs. The levels of tumor-infiltrating CD11bneg lymphocytes were not disturbed by TAM deficiency, and their production of these collagen subtypes was considerably lower than TAMs. Collectively, these results further highlight TAMs as pivotal players in collagen remodeling.
Our integrative analysis also defined the repertoire of matrix-remodeling enzymes brought by TAMs into the tumor microenvironment. We show that TAMs express uniquely high levels of TGM2, a marker for an alternatively activated phenotype in both human and mouse macrophages (Martinez et al., 2013). Increased expression of TGM2 in various cancer-cell types has been linked to increased drug resistance, cell survival, invasiveness, metastasis, and poor patient survival (Mehta et al., 2010). Specifically, prior study has demonstrated that TGM2 induces cross-linking between fibronectin and collagen I (Collighan and Griffin, 2009). Matrix metalloproteinases (MMPs) represent the most prominent family of proteinases associated with tumorigenesis (Kessenbrock et al., 2010). We found that TAMs express the genes encoding for various MMPs, though at the protein level they express mainly MMP14, an efficient collagenase associated with tumor angiogenesis, invasion, and progression. Closely related to the MMPs are the ADAM family of proteinases, most of which are anchored to the membrane and function in the pericellular space with implications for many aspects of tumorigenesis (Murphy, 2008). We discovered that TAMs were uniquely high for several ADAMs including 8, 9, 10, 15, and 17. Another family of lysosomal matrix proteases is the cysteine cathepsins, shown to function in proteolytic pathways that increase neoplastic progression (Mohamed and Sloane, 2006). Our findings indicate that TAMs express a decent repertoire of cathepsins; some are already expressed at the monocyte precursor level (A, C, E, H, O, S, and Z) and others are up-regulated within the tumor (B, D, and L).
Using the matrisome ECM protein expression database (Naba et al., 2014), we show that many of the ECM signature proteins of TAMs, including proteins involved with collagen production and assembly, are also increased in human CRC versus normal colon. Moreover, a progressive increase in collagen linearization and cross-linking and tissue stiffness is evident in the transition from healthy colon to colorectal carcinoma in humans (Nebuloni et al., 2016). In this study, Nebuloni et al. (2016) have suggested that the increased stiffness and cross-linking of the ECM is predisposing an environment suitable for CRC invasion. Therefore, these results highlight a clinical relevance for the TAM-induced ECM remodeling reported here in the mouse model. Altogether, we provide a set of evidence to support a novel protumoral mechanism of TAMs that is associated with the production and remodeling of collagenous ECM.
MATERIALS AND METHODS
Mice
C57BL/6 WT mice were purchased from Envigo. The Cx3cr1gfp/+Ccr2−/−, Cx3cr1gfp/+, and Ccr2−/− mice were bred at the Weizmann Institute of Science (WIS) and Tel-Aviv Sourasky Medical Center (TASMC) animal facilities. All mice were maintained under specific pathogen-free conditions. All experiments and procedures were approved by the WIS and TASMC animal care and use committees, protocol numbers 06351012-2 and 06150913-2 and 4-1-11 and 7-3-13, respectively.
Orthotopic mouse CRC model
In this study, we used a previously established endoscopy-guided orthotopic CRC model in mice (Zigmond et al., 2011). In brief, mice were anesthetized and injected with 5 × 104 syngeneic CRC MC38 cells suspended in 50 µl of sterile saline into the colonic lamina propria. Injection was performed using custom-made needles (Injecta) and guided by a high-resolution mouse video endoscopic system (SPEIS; Karl Storz).
Flow cytometry and sorting
Isolation of colonic lpMFs was performed as previously reported (Zigmond et al., 2012). In brief, normal colons of Cx3cr1gfp/+ mice located upstream of the tumor were carefully separated from the surrounding fat and lymphatic vessels and flushed of their luminal fluids with cold PBS−/−. Then, 0.5-cm colon pieces were cut and immersed into HBSS (without Ca2+ and Mg2+) containing 5% FBS, 2 mM EDTA, and 1 mM dithiothreitol (Sigma-Aldrich) and placed for 40 min at 250 rpm shaking at 37°C. Colon pieces were then digested in PBS+/+ containing 5% FBS, 1 mg/ml collagenase VIII (Sigma-Aldrich), and 0.1 mg/ml DNase I (Roche) for 40 min at 250 rpm shaking at 37°C. TAM subsets and MC38 CRC cells were isolated from CRC tumors established in Cx3cr1gfp/+ mice. The tumors were carefully separated from the surrounding normal colonic and fat tissues, cut into 1–4 mm3 pieces, and digested in PBS+/+ containing 5% FBS, 1 mg/ml collagenase VIII (Sigma-Aldrich), and 0.1 mg/ml DNase I (Roche) for 40 min at 250 rpm shaking at 37°C. For the sorting of tumor cells, we implanted MC38 cells genetically manipulated by lentiviral transduction to express RFP (Zigmond et al., 2011) and sorted them based on their expression of this reporter gene. Splenic monocytes were obtained from spleens of Cx3cr1gfp/+ mice, mashed through a 100-µm cell strainer, and lysed for erythrocytes using an ACK buffer (0.15 M NH4Cl, 0.1 M KHCO3, and 1 mM EDTA in PBS). Antibodies used to characterize colonic lpMFs, CRC TAMs, and neutrophils included: CD45 (30-F11), Ly6C (HK1.4), CD11b (M1/70), CD64 (X54-5/7.1), IAb (AF6-120.1), and Ly6G (1A8; all from BioLegend) and F4/80 (CI: A3-1; AbD Serotec). For splenic monocytes, we also used CD115 antibody (AFS98; BioLegend). Cells were analyzed with an LSRFortessa or FACSCanto II flow cytometer (BD) and sorted with a FACSAria flow cytometer (BD). Flow cytometry analysis was done with FlowJo software (Tree Star).
Confocal fluorescence microscopy
CRC tumors were extracted and fixed in 2% paraformaldehyde overnight at 4°C before impregnating in 30% sucrose (in PBS−/−) for 48 h. Sequentially, the tumors were frozen in an optimal cutting temperature buffer (Tissue-Tek) in isopentane cooled with liquid nitrogen and then cut with a cryostat to 12-µm thick sections. Slides were observed with a confocal laser-scanning microscope (LSM700; ZEISS), and image acquisition was conducted with ZEN imaging software.
SHG imaging
Tumors from WT or Ccr2−/− mice were harvested and frozen in an optimal cutting temperature buffer (Tissue-Tek). Sections of 150 µm were imaged using a two-photon microscope (equipped with a broadband Mai Tai-HP-femtosecond single box tunable Ti-sapphire oscillator with automated broadband wavelength tuning 100–1,020 nm for two-photon excitation; LSM 510 META NLO; ZEISS). Two-photon excitation was performed at a wavelength of 900 nm and detected using a META detector; for collagen second harmonic imaging, detection wavelength was set to 450 nm, whereas Cx3cr1gfp/+ macrophages were detected at 500–550 nm wavelengths. Images were acquired from the tumor’s margins and central area using a Plan Apochromat 20×/0.8 objective and processed using LSM Image browser software (ZEISS). Calculations of collagen coverage area and SHG signal intensity were done by first transforming each image, using only the SHG channel into grayscale (8 bit) using Photoshop (Adobe). Images were then binarized using the global (histogram derived) threshold tool in ImageJ (National Institutes of Health). Subsequently, the Measure tool in ImageJ was applied to calculate the area fraction on collagen signal and its intensity in the images.
SEM of ECM scaffolds
Tumors from WT or Ccr2−/− mice were decellularized (20 mM EDTA and 2% triton in double-distilled water [DDW]) for 24 h in gentle shake, followed by multiple washes in DDW, to obtain a cell-free ECM scaffold. These scaffolds were then fixed using 4% paraformaldehyde overnight at 4°C, stained with 4% sodium silicotungstate, pH7, for 45 min, and dehydrated through ascending concentrations of ethanol from 30–100%. Samples were subsequently dried in a critical point dryer and gold sputtered for imaging by SEM (Ultra 55 Feg; ZEISS).
LC-MS/MS analysis
Tissue slices from WT or Ccr2−/− tumors or normal upstream colons were immersed in 100 mM ammonium bicarbonate (Sigma-Aldrich) and coarsely homogenized by repeated cycles of boiling, freezing, and sonication. An equivalent volume of ammonium bicarbonate with 8 M urea was then added to reach a final concentration of 4 M, followed by vigorous shaking overnight at room temperature. Subsequently, 10 mM dithiothreitol (Sigma-Aldrich) was added and incubated at 30°C with shaking for 30 min, followed by the addition of 25 mM iodoacetamide (Sigma-Aldrich) and shaking for 30 min at 30°C. Urea was then diluted to a concentration of 2 M followed by addition of LysC (1:100 weight/weight; Wako Pure Chemical Industries) and shaking at 37°C for 2 h. Sequencing-grade modified trypsin (Promega) was sequentially added at a 1:50 ratio (weight/weight) overnight and, on the following morning, added again at a 1:100 ratio for 4 h. Peptide mixtures were purified on C18 stageTips. Eluted peptides were analyzed on a high-performance liquid chromatography system (EASY-nLC-1000; Thermo Fisher Scientific) coupled to a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific). MS data were analyzed with MaxQuant software and the Andromeda search engine (Cox et al., 2011), with a 1% pFDR threshold on the peptide and protein levels. Bioinformatics analysis was performed with the Perseus program. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD002094.
Calculation of the enrichment factor for ECM matrisome proteins in the proteomic profiling of WT and Ccr2−/− tumors was performed as follows: 2,367 proteins were identified in both WT and Ccr2−/− tumors, among which 115 were ECM related (4.85%). When comparing between WT and Ccr2−/− tumors, 348 proteins were significantly different in their abundance (Student’s t test; pFDR < 0.05), out of which 46 were ECM related (13.21%). Therefore, the enrichment factor for ECM proteins within significantly differently expressed proteins between WT and Ccr2−/− tumors is: 13.21/4.85 = 2.72.
Microarray analysis
Total RNA was isolated from sorted cells using a miRNeasy Mini kit (QIAGEN). RNA purity was determined using Nanodrop (ND-100; Peqlab) and Bioanalyzer 2100 (Agilent Technologies). The cDNA was prepared, labeled, and hybridized to a GeneChip mouse gene (1.0 ST; Affymetrix) according to the manufacturer’s protocol. Scanning of hybridized chips was performed with a GeneChip plus scanner (300 7G; Affymetrix). Data analysis and heat map hierarchical clustering were generated using Partek Genomics Suite software with Pearson’s dissimilarity correlation, average linkage methods, and present genes that were differentially expressed with at least a twofold change with P < 0.05 (ANOVA test). Functional enrichment analyses were performed using DAVID (Huang et al., 2009), GOEAST (Zheng and Wang, 2008), and Ingenuity (QIAGEN) tools. All microarray data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus public database under accession no. GSE67953.
ECM fragmentation
Tumors from WT or Ccr2−/− mice and normal colons were decellularized (20 mM EDTA and 2% triton in DDW) for 24 h in gentle shake, followed by multiple washes in DDW, to obtain a cell-free ECM scaffold. Decellularized 3D ECM scaffolds were weighted, cultured with 0.1 mg/ml DNase I (Roche) in PBS+/+ at 37°C for 1 h, washed, and transferred into PBS with 10× penicillin-streptomycin for overnight incubation. Subsequently, tissues were washed four times with PBS for 5 min each and rinsed in PBS for an estimated concentration of 100 mg tissue/ml to normalize tissue concentration during homogenization. Then, they were homogenized in a soft tissue–homogenizing tube containing 1.4-mm ceramic beads (Precellys; KT03961-1-003.2; Bertin Corp.) using a Bead Ruptor homogenizer (Omni International Inc.) for six cycles of 45 s at 5.56 rpm, until the solution appeared homogeneous. Then, concentration was evaluated again using Nanodrop (to normalize ECM concentrations between samples), and a fragment from each tissue was plated for 3 d to ensure ECM fragments do not contain bacterial contamination. ECM was maintained at −20°C for no more than 2 wk before co-culture with cells or orthotopically coinjected with cells.
Tumor cell proliferation assay
104 MC38 CRC cells were cultured for 48 h with 0.1 mg of decellularized 3D ECM fragments from WT tumors, Ccr2−/− tumors, or normal colon or without ECM fragments. Cells were subsequently fixed and stained according to a standard manufacturer protocol with a primary antibody against phosphohistone H3 (sc-8656-R; Santa Cruz Biotechnology, Inc.) and then with a fluorescently labeled secondary antibody (ab150073; Abcam) and DAPI. Cells were viewed under a fluorescent microscope (eclipse 90i; Nikon), and pictures were taken with a digital camera (1310; DVC). The fraction of phosphohistone H3+ cells out of total DAPI+ cells was calculated in randomly chosen fields using ImageJ software.
Orthotopic co-implantation of tumor cells and ECM fragments
MC38 cells alone or a mixture of 5 × 104 MC38 cells and 0.8 mg ECM fragments (WT, Ccr2−/− tumor, or normal colon) were orthotopically injected into the colonic lamina propria of WT recipients in comparison with MC38 cells only.
Quantitative real-time PCR
Ly6chi and F4/80hi TAMs, CD45+CD11b− lymphocytes, and CD45−PDGFRα+ were sorted from pools of WT tumors (three pools of six tumors each for TAM subsets and lymphocytes and a pool of 18 tumors for CAFs). CAFs were also sorted from the pool of 18 tumors implanted into Ccr2−/− mice. RNA from the sorted cells was isolated with an RNeasy Micro kit (QIAGEN) and reverse transcribed with a High Capacity cDNA Reversed transcription kit (Applied Biosystems). PCRs were performed with the SYBER green PCR Master Mix (Applied Biosystems). Quantification was done with Step One software (V2.2). The genes of interest, collagen I, collagen VI, and collagen XIV were compared with ribosomal protein, large PO (RPLPO) housekeeping gene. Primer sequences (forward and reverse, respectively) were: RPLPO, 5′-TCCAGCAGGTGTTTGACAAC-3′ and 5′-CCATCTGCAGACACACACT-3′; collagen I, 5′-GAGAGCATGACCGATGGATT-3′ and 5′-CCTTCTTGAGGTTGCCAGTC-3′; collagen VI, 5′-GATGAGGGTGAAGTGGGAGA-3′ and 5′-CACTCACAGCAGGAGCACA-3′; and collagen XIV, 5′-CTTTTGAAGGACCCGACATC-3′ and 5′-TGCCTTCTGACCAACTTCCT-3′.
Online supplemental material
Fig. S1 shows the sorting strategy of Ly6Chi monocytes, resident colonic lpMFs, and CRC TAMs. Fig. S2 shows that CRC tumors acquire a unique ECM protein signature in comparison with healthy upstream colon that is altered in the absence of TAMs. Fig. S3 shows that TAMs express a unique signature of ECM-related genes. Fig. S4 shows proteomic profiling of sorted F4/80hi TAMs in comparison with their Ly6Chi monocyte precursors and colocalizing CRC tumor cells. Table S1 is included as an Excel file and shows a GOEAST function enrichment analyses of the differentially expressed genes between Ly6Chi TAMs and colonic lpMFs and between F4/80hi TAMs and colonic lpMFs. Table S2 is included as an Excel file and shows a DAVID function enrichment analyses of the differentially expressed genes between Ly6Chi TAMs and colonic lpMFs and between F4/80hi TAMs and colonic lpMFs. Table S3 is included as an Excel file shows an ingenuity pathway analysis of the differentially expressed genes between F4/80hi TAMs and colonic lpMFs.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by a collaborative Weizmann Institute of Science–Tel Aviv Sourasky Medical Center grant in biomedical research (to C. Varol and I. Sagi) and by the Israel Science Foundation (grant no. 1226/13), The Thompson Family Foundation, and the Geraldo Rosenkranz Fund (to I. Sagi). I. Sagi is the incumbent of the Pontecorvo professorial chair.
The authors declare no competing financial interests.
Author contributions: The corresponding senior authors confirm that all authors have agreed to be so listed and approved the manuscript submission. R. Afik, E. Zigmond, I. Sagi, and C. Varol designed, performed, and analyzed all experiments and wrote the manuscript. M. Vugman and R. Afik performed the sorting of macrophage subsets and the extraction of mRNA and protein. M. Vugman, M. Klepfish, and E. Shimshoni performed all CAF experiments and ECM imaging of early stage tumors. M. Pasmanik-Chor analyzed the gene expression microarray data. A. Shenoy and T. Geiger greatly contributed to the design and analysis of MS experiments. E. Bassat performed the staining of p-histone H3+ MC38 cells. Z. Halpern contributed to the establishment of the orthotopic CRC model.
Footnotes
Abbreviations used:
- 3D
- three-dimensional
- ADAM
- a disintegrin and metalloproteinase
- CAF
- cancer-associated fibroblast
- CRC
- colorectal cancer
- DDW
- double-distilled water
- ECM
- extracellular matrix
- LC-MS/MS
- liquid chromatography with MS
- lpMF
- lamina propria macrophage
- MMP
- matrix metalloproteinase
- MS
- mass spectrometry
- PCOLCE
- procollagen C–endopeptidase enhancer
- pFDR
- positive false discovery rate
- PLOD
- procollagen-lysine 2-oxoglutarate 5-dioxygenase
- SEM
- scanning electron microscopy
- SHG
- second harmonic generation
- SPARC
- secreted protein acidic and rich in cysteine
- TAM
- tumor-associated macrophages
- TGM
- transglutaminase
References
- Bain C.C., Scott C.L., Uronen-Hansson H., Gudjonsson S., Jansson O., Grip O., Guilliams M., Malissen B., Agace W.W., and Mowat A.M.. 2013. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 6:498–510. 10.1038/mi.2012.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain C.C., Bravo-Blas A., Scott C.L., Gomez Perdiguero E., Geissmann F., Henri S., Malissen B., Osborne L.C., Artis D., and Mowat A.M.. 2014. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15:929–937. 10.1038/ni.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S.K., Allavena P., and Mantovani A.. 2013. Tumor-associated macrophages: functional diversity, clinical significance, and open questions. Semin. Immunopathol. 35:585–600. 10.1007/s00281-013-0367-7 [DOI] [PubMed] [Google Scholar]
- Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M., et al. . 2009. Origin of the lamina propria dendritic cell network. Immunity. 31:513–525. 10.1016/j.immuni.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collighan R.J., and Griffin M.. 2009. Transglutaminase 2 cross-linking of matrix proteins: biological significance and medical applications. Amino Acids. 36:659–670. 10.1007/s00726-008-0190-y [DOI] [PubMed] [Google Scholar]
- Coussens L.M., Zitvogel L., and Palucka A.K.. 2013. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 339:286–291. 10.1126/science.1232227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Erez N., Truitt M., Olson P., Arron S.T., and Hanahan D.. 2010. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-κB-dependent manner. Cancer Cell. 17:135–147. 10.1016/j.ccr.2009.12.041 [DOI] [PubMed] [Google Scholar]
- Franklin R.A., Liao W., Sarkar A., Kim M.V., Bivona M.R., Liu K., Pamer E.G., and Li M.O.. 2014. The cellular and molecular origin of tumor-associated macrophages. Science. 344:921–925. 10.1126/science.1252510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecke D.R., Meng X., Liu B., and Birk D.E.. 2003. Complete primary structure and genomic organization of the mouse Col14a1 gene. Matrix Biol. 22:209–216. 10.1016/S0945-053X(03)00021-0 [DOI] [PubMed] [Google Scholar]
- Gilkes D.M., Semenza G.L., and Wirtz D.. 2014. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat. Rev. Cancer. 14:430–439. 10.1038/nrc3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., and Littman D.R.. 2000. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114. 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R., and Zeisberg M.. 2006. Fibroblasts in cancer. Nat. Rev. Cancer. 6:392–401. 10.1038/nrc1877 [DOI] [PubMed] [Google Scholar]
- Kessenbrock K., Plaks V., and Werb Z.. 2010. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 141:52–67. 10.1016/j.cell.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipper J.A., Willenborg S., Brinckmann J., Bloch W., Maaß T., Wagener R., Krieg T., Sutherland T., Munitz A., Rothenberg M.E., et al. . 2015. Interleukin-4 receptor α signaling in myeloid cells controls collagen fibril assembly in skin repair. Immunity. 43:803–816. 10.1016/j.immuni.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F., Csiszar K., Giaccia A., Weninger W., et al. . 2009. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 139:891–906. 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori M., Solinas G., Germano G., Mantovani A., and Allavena P.. 2011. Tumor-associated macrophages as incessant builders and destroyers of the cancer stroma. Cancers (Basel). 3:3740–3761. 10.3390/cancers3043740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E.Y., and Pollard J.W.. 2007. Tumor-associated macrophages press the angiogenic switch in breast cancer. Cancer Res. 67:5064–5066. 10.1158/0008-5472.CAN-07-0912 [DOI] [PubMed] [Google Scholar]
- Lin E.Y., Nguyen A.V., Russell R.G., and Pollard J.W.. 2001. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 193:727–740. 10.1084/jem.193.6.727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Weaver V.M., and Werb Z.. 2012. The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 196:395–406. 10.1083/jcb.201102147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F.O., Helming L., Milde R., Varin A., Melgert B.N., Draijer C., Thomas B., Fabbri M., Crawshaw A., Ho L.P., et al. . 2013. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. 121:e57–e69. 10.1182/blood-2012-06-436212 [DOI] [PubMed] [Google Scholar]
- Mason S.D., and Joyce J.A.. 2011. Proteolytic networks in cancer. Trends Cell Biol. 21:228–237. 10.1016/j.tcb.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta K., Kumar A., and Kim H.I.. 2010. Transglutaminase 2: a multi-tasking protein in the complex circuitry of inflammation and cancer. Biochem. Pharmacol. 80:1921–1929. 10.1016/j.bcp.2010.06.029 [DOI] [PubMed] [Google Scholar]
- Mohamed M.M., and Sloane B.F.. 2006. Cysteine cathepsins: multifunctional enzymes in cancer. Nat. Rev. Cancer. 6:764–775. 10.1038/nrc1949 [DOI] [PubMed] [Google Scholar]
- Movahedi K., Laoui D., Gysemans C., Baeten M., Stangé G., Van den Bossche J., Mack M., Pipeleers D., In’t Veld P., De Baetselier P., and Van Ginderachter J.A.. 2010. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 70:5728–5739. 10.1158/0008-5472.CAN-09-4672 [DOI] [PubMed] [Google Scholar]
- Murphy G. 2008. The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer. 8:932–941. 10.1038/nrc2459 [DOI] [PubMed] [Google Scholar]
- Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., et al. . 2014. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 41:14–20. 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A., Clauser K.R., Hoersch S., Liu H., Carr S.A., and Hynes R.O.. 2012. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteomics. 11:M111.014647 10.1074/mcp.M111.014647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A., Clauser K.R., Whittaker C.A., Carr S.A., Tanabe K.K., and Hynes R.O.. 2014. Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer. 14:518 10.1186/1471-2407-14-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A., Clauser K.R., Ding H., Whittaker C.A., Carr S.A., and Hynes R.O.. 2016. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 49:10–24. 10.1016/j.matbio.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebuloni M., Albarello L., Andolfo A., Magagnotti C., Genovese L., Locatelli I., Tonon G., Longhi E., Zerbi P., Allevi R., et al. . 2016. Insight on colorectal carcinoma infiltration by studying perilesional extracellular matrix. Sci. Rep. 6:22522 10.1038/srep22522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic-Paterson D.J., Wang S., and Lan H.Y.. 2014. Macrophages promote renal fibrosis through direct and indirect mechanisms. Kidney Int Suppl (2011). 4:34–38. 10.1038/kisup.2014.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy R., and Pollard J.W.. 2014. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 41:49–61. 10.1016/j.immuni.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., and Scherer P.E.. 2012. Adipocyte-derived endotrophin promotes malignant tumor progression. J. Clin. Invest. 122:4243–4256. 10.1172/JCI63930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perryman L., and Erler J.T.. 2014. Lysyl oxidase in cancer research. Future Oncol. 10:1709–1717. 10.2217/fon.14.39 [DOI] [PubMed] [Google Scholar]
- Pickup M.W., Mouw J.K., and Weaver V.M.. 2014. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 15:1243–1253. 10.15252/embr.201439246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano P.P., Eliceiri K.W., Campbell J.M., Inman D.R., White J.G., and Keely P.J.. 2006. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4:38 10.1186/1741-7015-4-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano P.P., Inman D.R., Eliceiri K.W., Knittel J.G., Yan L., Rueden C.T., White J.G., and Keely P.J.. 2008. Collagen density promotes mammary tumor initiation and progression. BMC Med. 6:11 10.1186/1741-7015-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B.Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L.R., Kaiser E.A., Snyder L.A., and Pollard J.W.. 2011. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 475:222–225. 10.1038/nature10138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangaletti S., Di Carlo E., Gariboldi S., Miotti S., Cappetti B., Parenza M., Rumio C., Brekken R.A., Chiodoni C., and Colombo M.P.. 2008. Macrophage-derived SPARC bridges tumor cell-extracellular matrix interactions toward metastasis. Cancer Res. 68:9050–9059. 10.1158/0008-5472.CAN-08-1327 [DOI] [PubMed] [Google Scholar]
- Schnoor M., Cullen P., Lorkowski J., Stolle K., Robenek H., Troyer D., Rauterberg J., and Lorkowski S.. 2008. Production of type VI collagen by human macrophages: a new dimension in macrophage functional heterogeneity. J. Immunol. 180:5707–5719. 10.4049/jimmunol.180.8.5707 [DOI] [PubMed] [Google Scholar]
- Schönherr E., Witsch-Prehm P., Harrach B., Robenek H., Rauterberg J., and Kresse H.. 1995. Interaction of biglycan with type I collagen. J. Biol. Chem. 270:2776–2783. 10.1074/jbc.270.6.2776 [DOI] [PubMed] [Google Scholar]
- Shand F.H., Ueha S., Otsuji M., Koid S.S., Shichino S., Tsukui T., Kosugi-Kanaya M., Abe J., Tomura M., Ziogas J., and Matsushima K.. 2014. Tracking of intertissue migration reveals the origins of tumor-infiltrating monocytes. Proc. Natl. Acad. Sci. USA. 111:7771–7776. 10.1073/pnas.1402914111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon Y., Alon L., Glanz S., Servais C., and Erez N.. 2013. Isolation of normal and cancer-associated fibroblasts from fresh tissues by fluorescence activated cell sorting (FACS). J. Vis. Exp. e4425 10.3791/4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C., Landsman L., Fogg D.K., Greenshtein L., Gildor B., Margalit R., Kalchenko V., Geissmann F., and Jung S.. 2007. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J. Exp. Med. 204:171–180. 10.1084/jem.20061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C., Vallon-Eberhard A., Elinav E., Aychek T., Shapira Y., Luche H., Fehling H.J., Hardt W.D., Shakhar G., and Jung S.. 2009. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 31:502–512. 10.1016/j.immuni.2009.06.025 [DOI] [PubMed] [Google Scholar]
- Varol C., Mildner A., and Jung S.. 2015. Macrophages: development and tissue specialization. Annu. Rev. Immunol. 33:643–675. 10.1146/annurev-immunol-032414-112220 [DOI] [PubMed] [Google Scholar]
- Wang S., Meng X.M., Ng Y.Y., Ma F.Y., Zhou S., Zhang Y., Yang C., Huang X.R., Xiao J., Wang Y.Y., et al. . 2016. TGF-β/Smad3 signalling regulates the transition of bone marrow-derived macrophages into myofibroblasts during tissue fibrosis. Oncotarget. 7:8809–8822. 10.18632/oncotarget.6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff J.B., Wang Y., Lin E.Y., Li J.F., Goswami S., Stanley E.R., Segall J.E., Pollard J.W., and Condeelis J.. 2007. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 67:2649–2656. 10.1158/0008-5472.CAN-06-1823 [DOI] [PubMed] [Google Scholar]
- Xue J., Schmidt S.V., Sander J., Draffehn A., Krebs W., Quester I., De Nardo D., Gohel T.D., Emde M., Schmidleithner L., et al. . 2014. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 40:274–288. 10.1016/j.immuni.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., and Wang X.J.. 2008. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 36:W358–W363. 10.1093/nar/gkn276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E., Halpern Z., Elinav E., Brazowski E., Jung S., and Varol C.. 2011. Utilization of murine colonoscopy for orthotopic implantation of colorectal cancer. PLoS One. 6:e28858 10.1371/journal.pone.0028858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E., Varol C., Farache J., Elmaliah E., Satpathy A.T., Friedlander G., Mack M., Shpigel N., Boneca I.G., Murphy K.M., et al. . 2012. Ly6Chi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 37:1076–1090. 10.1016/j.immuni.2012.08.026 [DOI] [PubMed] [Google Scholar]
- Zigmond E., Bernshtein B., Friedlander G., Walker C.R., Yona S., Kim K.W., Brenner O., Krauthgamer R., Varol C., Müller W., and Jung S.. 2014. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 40:720–733. 10.1016/j.immuni.2014.03.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.