Delaunay et al. reveal the role of U34 tRNA-modifying enzymes in the regulation of specific mRNA translation to support cell invasion and metastasis.
Abstract
Quantitative and qualitative changes in mRNA translation occur in tumor cells and support cancer progression and metastasis. Posttranscriptional modifications of transfer RNAs (tRNAs) at the wobble uridine 34 (U34) base are highly conserved and contribute to translation fidelity. Here, we show that ELP3 and CTU1/2, partner enzymes in U34 mcm5s2-tRNA modification, are up-regulated in human breast cancers and sustain metastasis. Elp3 genetic ablation strongly impaired invasion and metastasis formation in the PyMT model of invasive breast cancer. Mechanistically, ELP3 and CTU1/2 support cellular invasion through the translation of the oncoprotein DEK. As a result, DEK promotes the IRES-dependent translation of the proinvasive transcription factor LEF1. Consistently, a DEK mutant, whose codon composition is independent of U34 mcm5s2-tRNA modification, escapes the ELP3- and CTU1-dependent regulation and restores the IRES-dependent LEF1 expression. Our results demonstrate that the key role of U34 tRNA modification is to support specific translation during breast cancer progression and highlight a functional link between tRNA modification– and IRES-dependent translation during tumor cell invasion and metastasis.
INTRODUCTION
Cancer metastases at distant organs require tumor cell adaption. Common adaptive responses include modifications in the tumor microenvironment, as well as changes in the proteome of the tumor itself to sustain cell invasion, survival, and establishment of a distal secondary tumor (Taddei et al., 2013; Oskarsson et al., 2014). Several signaling pathways are aberrantly activated in metastatic cancer cells. Among them, the WNT and TGFβ pathways are prominent in promoting metastasis (Massagué, 2008; Anastas and Moon, 2013). Indeed, the WNT–TCF pathway is essential for dissemination and relapse of metastatic lung adenocarcinoma (Nguyen et al., 2009) and in supporting breast cancer metastatic colonization (Malanchi, 2012; Jang et al., 2015). Downstream of WNT activation, the transcription factor LEF1 was further pointed out as a crucial effector to support formation of WNT-dependent metastases (Nguyen et al., 2009). LEF1 expression is also induced by other signals, including TGFβ and mediates breast tumor cells invasion (Nawshad and Hay, 2003; LaGamba et al., 2005; Nguyen et al., 2005; Medici et al., 2006). The expression of LEF1 protein is also regulated posttranscriptionally, through the presence of a regulatory internal ribosome entry site (IRES) sequence in its 5′ untranslated region (UTR; Jimenez et al., 2005). Indeed, the oncogenic LEF1 protein is predominantly produced by a cap-independent translational mechanism that requires the recruitment of several proteins to its IRES sequence, namely canonical translation initiation factors and IRES trans-acting factors (ITAFs; Jimenez et al., 2005; Tsai et al., 2011, 2014). However, the biological consequences of IRES-dependent regulation of LEF1 expression in the context of breast cancer remain poorly explored.
Translational control of protein expression is now recognized as a crucial component of human cancer development and progression (Silvera et al., 2010). mRNA translation requires the pairing between codons in the mRNA and the anticodons of transfer RNAs (tRNAs) within ribosomes. tRNAs are subjected to a variety of posttranscriptional modifications that affect their stability, affinity, and specificity (El Yacoubi et al., 2012). tRNA modifications at the wobble uridine 34 (U34) are highly conserved and contribute to translation fidelity by ensuring codon discrimination by tRNALys(UUU), tRNAGln(UUG), and tRNAGlu(UUC) target tRNAs (Karlsborn et al., 2015). The enzymatic cascade catalyzing the modifications at U34 (U34 tRNA anticodon modifying enzymes) has been characterized in yeast and also in higher eukaryotes (Karlsborn et al., 2015). The acetyltransferase complex Elongator, initially described as an RNA polymerase II–associated factor (Otero et al., 1999), is believed to act as the first enzyme in the U34 tRNA modification cascade (Karlsborn et al., 2015). ELP3, the catalytic subunit of the complex, catalyzes the formation of 5-carbamoylmethyluridine (cm5U; Chen et al., 2009; Lin et al., 2013; Selvadurai et al., 2014). The subsequent activities of the methyltransferase ALKBH8 (Alkylation repair homologue 8) and URM1 (ubiquitin-related modifier 1) pathway, which encloses CTU1/2 proteins (Thiouridylase proteins 1/2), are then required for addition of 5-methoxycarbonylmethyl (mcm5) and a 2-thio group (s2) to uridine, respectively (Kalhor and Clarke, 2003; Nakai et al., 2004, 2008; Björk et al., 2007; Dewez et al., 2008; Huang et al., 2008; Schlieker et al., 2008; Leidel et al., 2009; Noma et al., 2009; Songe-Møller et al., 2010). Ribosome profiling experiments with yeast strains lacking these U34 tRNA-modifying enzymes show ribosome accumulation at AAA and CAA codons, suggesting that tRNALys(UUU) and tRNAGln(UUG) are the targets with the highest biological impact (Zinshteyn and Gilbert, 2013; Nedialkova and Leidel, 2015), which is in line with data from tRNA overexpression (Esberg et al., 2006). Despite their biochemically predicted importance, the role of U34 tRNA modifications in terms of biological impact has been little studied. Recently, the loss of U34 modifications was found to trigger a failure in protein homeostasis in yeast and in nematodes (Nedialkova and Leidel, 2015). Importantly, the loss of Elp3 in cortical stem cells was shown to impair neurogenesis through induction of the unfolded protein response (UPR) resulting from decreased codon translation rates (Laguesse et al., 2015). In a cancer context, Elp3 expression was increased upon constitutive Wnt signaling in a mouse model of intestinal tumorigenesis, as well as in human colon adenocarcinoma (Ladang et al., 2015). Elp3 was critical for Wnt-driven intestinal tumor initiation, at least in part by promoting Sox9 protein translation in order to sustain a pool of Lgr5+/Dclk1+/Sox9+ intestinal cancer stem cells essential for tumor initiation (Ladang et al., 2015). Oncogenic pathways apart from constitutive Wnt signaling that rely on U34 tRNA modifications still need to be defined. Importantly, global tRNA levels are significantly elevated in tumors versus normal breast tissues (Pavon-Eternod et al., 2009), and some tRNAs were recently found to be up-regulatd in metastatic human breast cancer (Goodarzi et al., 2016).
Here, we show that the U34 tRNA-modifying enzymes ELP3 and CTU1/2 are up-regulated in human breast cancer, as well as in models of invasive breast cancer. Accordingly, expression of PyMT oncogene in mammary gland epithelial cells elevated Elp3 and Ctu1/2 expression, which correlated with enhanced abundance of thiolated tRNAs. Moreover, Elp3 is required for PyMT-induced breast cancer metastasis in vivo. In vitro, depletion of ELP3 or CTU1 reduces migratory activities of invasive breast cancer cells. Mechanistically, we demonstrate that the ITAF protein DEK, whose expression increases in invasive breast cancers (Privette Vinnedge et al., 2011; Liu et al., 2012), relies on U34 tRNA-modifying enzymes for its translation through its codon composition. As a result, DEK promotes LEF1 protein translation through its IRES sequence, a pathway that leads to the establishment of a LEF1 proinvasive transcriptomic signature in breast tumors. Finally, expression levels of ELP3, DEK, and LEF1 were positively correlated in breast cancer patient samples, and their increased expression was associated with increased risk of metastasis. Together, these data demonstrate that U34 tRNA modifications are required for breast cancer metastases, at least partly by linking the translation of DEK to IRES-dependent expression of LEF1.
RESULTS
U34 tRNA-modifying enzymes are up-regulated in breast cancers
We measured the expression of ELP3, CTU1, and CTU2 in normal breast tissue or in noninvasive (DCIS) and invasive (IDC) breast cancer patients using tissue microarray analysis. Their expression was enriched in cases of human breast cancer as compared with normal mammary gland tissue, with no preferential association to any specific subtype (Fig. 1, A–C; and not depicted). In normal mouse mammary gland, we detected a specific expression of Elp3, Ctu1, and Ctu2 in the luminal epithelial layer (Fig. 1 D). We aimed to explore the potential role of U34 tRNA modification in breast tumor development and metastasis by using the MMTV-PyMT luminal breast cancer mouse model, which spontaneously forms metastases in the lungs (Guy et al., 1992; Lin et al., 2003). We first assessed the expression of the U34 tRNA-modifying enzymes Elp3, Ctu1, and Ctu2 in PyMT primary tumors of 7-wk-old mice. Protein levels of these enzymes were increased in PyMT tumors compared with adjacent tissue, as shown by immunohistochemistry (IHC; Fig. 1 E). These observations were corroborated in vitro after exogenous expression of the oncoprotein PyMT in normal murine mammary gland (NMuMG) cells. Indeed, PyMT expression significantly increased Elp1, Elp3, Alkbh8, Ctu1, and Ctu2 protein levels (Fig. 1, F and G). Remarkably, changes in expression correlated with tRNA modification activity, as the thiolation (s2) of target tRNA tE(UUC) was increased in PyMT-expressing NMuMG cells (Fig. 1, H and I). These data indicate that expression of U34 tRNA-modifying enzymes is elevated in cases of human breast cancer and in PyMT breast tumors.
Figure 1.
U34 tRNA-modifying enzymes are up-regulated in human breast cancer. (A–C) Tissue microarray analysis of ELP3 (A), CTU2 (B), and CTU1 (C) protein expression in normal breast tissue (n = 23) and in samples of noninvasive or invasive human breast cancer (n = 35 and n = 7, respectively). Representative images are shown. The mean Elp3-specific signal has been quantified and plotted (mean values ± SD; Student’s t test; *, P < 0.05; ***, P < 0.001). (D) IHC staining for Elp3, Ctu1, and Ctu2 expression in normal mammary glands of 7-wk-old mice. (E) IHC staining for Elp3, Ctu1, and Ctu2 expression in mammary glands from 7-wk-old PyMT mice. (F) Western blot showing Elp3, Elp1, Ctu1/2, and Alkbh8 expression in control NMuMG cells or after stable expression the PyMT oncogene. pAkt is used as a control to prove that PyMT expression was functional. Total Akt and Hsp90 were used as loading controls. (G) RT-qPCR analysis of PyMT mRNA in control or PyMT-expressing NMUMG cells. Detection of the GAPDH mRNA was used as normalization. The expression of PyMT in control cells was set to 1, and values obtained from PyMT-NMUMG cells expressed relative to it (**, P ≤ 0.01, Student's t test). (H) Northern blot analysis assessing t:E(UUC) tRNA thiolation in control and PyMT-expressing NMuMG cells. (I) Quantification of t:E(UUC) tRNA thiolation, calculated as the ration of thiolated over nonthiolated t:E(UUC), in control and PyMT-expressing NMUMG cells. Values represent mean ± SD of three independent experiments (*, P ≤ 0.05, Student’s t test).
Breast cancer invasiveness and metastases rely on U34 tRNA-modifying enzymes
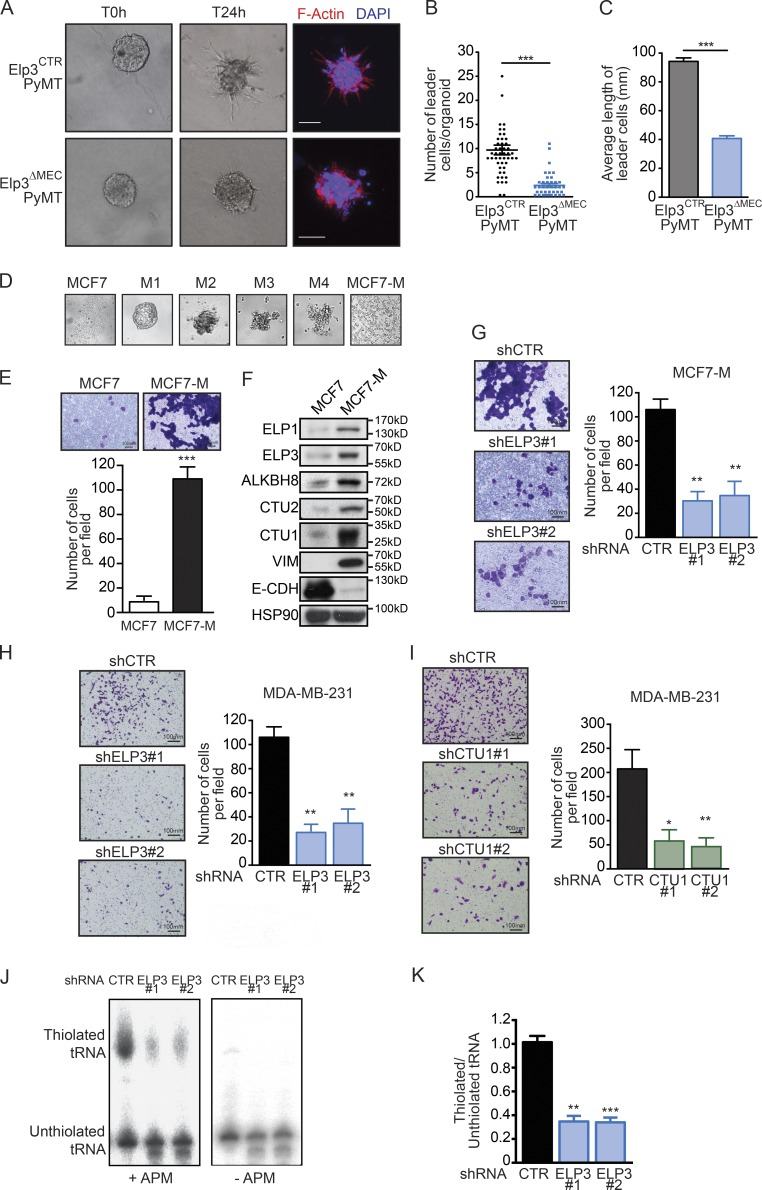
To further investigate the role of U34 tRNA modification in breast cancer development, we inactivated Elp3, the first enzyme of the cascade, in mammary epithelial cells (MEC) by crossing the Elp3loxP/loxP mice (referred to as Elp3CTR) with the MMTV-CRE mice (progeny referred to as Elp3ΔMEC; Fig. 2, A and B). The mammary gland development appeared normal in Elp3ΔMEC, with no apparent defect (Fig. 2, C and D). The Elp3CTR and Elp3ΔMEC mice were then crossed with transgenic MMTV-PyMT mice (Elp3CTRPyMT and Elp3ΔMECPyMT, respectively). Decreased expression of Elp3 in the mammary epithelium (Fig. 2 E) led to a significant delay in tumor appearance (average time for tumor appearance is 35 ± 2.64 d in Elp3CTRPyMT mice and 49 ± 2.78 d in Elp3ΔMECPyMT; Fig. 2 F). Also, tumor burden was significantly delayed in Elp3ΔMECPyMT mice compared with control Elp3CTRPyMT mice (Fig. 2, G and H). Interestingly, Elp3 deficiency did not affect the overall proliferative capacity of PyMT tumors (Fig. 2 I). PyMT-induced murine breast tumors produce pulmonary metastases in all cases after 100 d. IHC analyses revealed that Elp3 and Ctu1/Ctu2 proteins levels are strongly expressed in lung metastases of PyMT mice (Fig. 2 J). Strikingly, Elp3 deficiency in MECs dramatically impacted on metastasis formation. Indeed, whereas lung metastases were detected in 100% of 14-wk-old control PyMT mice, less than half of the Elp3ΔMECPyMT mice showed detectable lung metastases (Fig. 2 K). Moreover, the number and size of pulmonary metastases were dramatically decreased in the Elp3ΔMECPyMT mice to <15% of controls (Elp3CTRPyMT; Fig. 2, M and N). This defect was still very pronounced at 16 wk, when tumor burden (primary tumor) is similar to 14-wk-old control PyMT mice. Finally, the expression of the MEC-specific β-casein mRNA was used as a marker of breast cancer cell infiltration to the lung during tumor development. Whereas β-casein expression was detected from 9 wk of age onward in control mice (and 100% of 11-wk-old control mice were β-casein positive), it was only detected from week 14 in Elp3ΔMECPyMT mice (Fig. 2 O). Together, these data demonstrate that Elp3 is crucially required for metastasis formation in the PyMT model of breast cancer.
Figure 2.
Breast cancer metastasis relies on U34 tRNA modifications. (A) IHC staining for Elp3 expression in normal mammary glands from 7-wk-old Elp3CTR or Elp3ΔMEC mice. (B) RT-qPCR analysis of Elp3 mRNA levels in 7-wk Elp3CTR and Elp3ΔMEC mammary glands. Values represent mean ± SD of five different tumors performed in triplicate (n = 5; *, P ≤ 0.05, Student’s t test). (C) Representative whole mounted normal mammary glands from 8-wk Elp3CTR and Elp3ΔMEC mice. (D) Quantification of terminal end buds (TEB) from whole-mounted normal mammary glands from 8-wk Elp3CTR and Elp3ΔMEC mice (n = 5 and n = 7, respectively; ns, nonsignificant). (E) RT-qPCR analysis of Elp3 mRNA levels in 7-wk Elp3CTRPyMT and Elp3ΔMECPyMT tumors. Values represent mean ± SD of five different tumors performed in triplicate (**, P ≤ 0.01, Student’s t test). (F) Kaplan-Meier curve showing tumor appearance in Elp3CTRPyMT and Elp3ΔMECPyMT mice (n = 12 per condition; ***, P ≤ 0.001). (G) Quantification of total tumor burden in 14 wk Elp3CTRPyMT (n = 8) and Elp3ΔMECPyMT mice (n = 8) or in 16 wk Elp3ΔMECPyMT (n = 5). Tumor weight values are expressed as a percentage of the total mouse weight (*, P ≤ 0.05, Student’s t test). (H) Quantification of tumors surface in mammary gland of 5, 7, 9, and 11 wk Elp3CTRPyMT (n = 3 per time point) and Elp3ΔMECPyMT (n = 3 per time point) mice based on H&E staining (***, P ≤ 0.001, Student’s t test). (I) Quantification of Ki67 staining in tumors of 7 and 14 wk Elp3CTRPyMT and Elp3ΔMECPyMT mice. The percentage of Ki67-positive cells in tumors is expressed (mean values ± SD; five mice per condition). (J) Anti-Elp3, Ctu1, and Ctu2 IHC analyses in metastazed lungs from 14-wk-old Elp3CTRPyMT mice. A representative picture is shown. (K) Representative lungs and corresponding H&E staining of 14 wk Elp3CTRPyMT and Elp3ΔMECPyMT mice and of 16 wk Elp3ΔMECPyMT mice. (L) Quantification of the number of Elp3CTRPyMT and Elp3ΔMECPyMT mice with detectable lung metastases. (M and N) Quantification of the number of lung metastases (M) and surface metastases (N) in 14 wk Elp3CTRPyMT (n = 9) and Elp3ΔMECPyMT (n = 11) mice and in 16 wk Elp3ΔMECPyMT mice (n = 5; *, P ≤ 0.05; **, P ≤ 0.01, Student’s t test). (O) Quantification of the number of mice with β-casein–positive lung in 5, 7, 9, and 11 wk Elp3CTRPyMT (n = 3 per time point) and Elp3ΔMECPyMT (n = 3 per time point). β-casein mRNA was detected by RT-qPCR using the GAPDH mRNA for normalization purposes.
As a prominent mechanism supporting breast cancer metastasis, collective invasion has been recently assessed in three dimensional (3D) organoid assays using primary PyMT tumors and was shown to rely on cells exhibiting basal features that arise from luminal progenitors (Cheung et al., 2013; Ye et al., 2015). Elp3 inactivation strongly reduced metastasis of PyMT mammary tumors in mice, therefore tumors of similar size from 9-wk-old mice were dissociated into organoids and grown in 3D type I collagen gel to assess tumor cells invasion. Elp3 deficiency strongly impacted the number of leader tumor cells in organoids (Fig. 3, A and B). Moreover, the length of the very few tumor cell protrusions was dramatically smaller in Elp3ΔMECPyMT organoids (Fig. 3 C). These results indicate that Elp3 promotes tumor cells invasion in MMTV-PyMT mammary tumors in vitro.
Figure 3.
U34 tRNA modifications regulate breast cancer cell invasion. (A) Representative pictures of ex vivo 3D collagen I culture of organoids from similarly sized 9-wk-old Elp3CTRPyMT and Elp3ΔMECPyMT primary tumors. (B and C) Quantification of the number of leader cells per organoid (B) and the mean length of leader cells (C). Data are mean ± SD (n = 3 biological replicates per group; *, P ≤ 0.05; ***, P ≤ 0.001, Student’s t test). (D) MCF7 cells were cultured in nonadherent conditions for several weeks and passaged 4 times (M1 to M4). Resulting cells were then cultured in adherent conditions (MCF7-M). Representative pictures are shown. (E) Migration of control or ELP3-depleted MCF7-M cells toward a serum gradient was measured using Transwell assays. The results are expressed as number of migrating cells per field. Values represent mean ± SD of three independent experiments performed in triplicates (***, P ≤ 0.001). (F) Western blot showing ELP3, ELP1, CTU1/2, ALKBH8, Vimentin, and E-Cadherin expression in MCF7 and MCF7-M cells. HSP90 was detected for normalization purpose. (G) Migration of control and ELP3-depleted MCF7-M cells toward a serum gradient was measured using Transwell assays. The results are expressed as in E (**, P ≤ 0.01, Student’s t test). (H and I) Migration of control, ELP3- (H), or CTU1-depleted (I) MDA-MB-231 cells toward a serum gradient was measured using Transwell assays. The results are expressed as in E (*, P ≤ 0.05; **, P ≤ 0.01, Student’s t test). (J) Northern blot analysis assessing t:E(UUC) tRNA thiolation in control or ELP3-depleted MDA-MB-231 cells. (K) Quantification of t:E(UUC) tRNA thiolation, calculated as the ratio of thiolated over nonthiolated t:E(UUC), in control and ELP3-depleted cells. Values represent mean ± SD of three independent experiments (**, P ≤ 0.01; ***, P ≤ 0.001, Student’s t test).
To further support the idea that U34 tRNA-modifying enzymes are required for breast cancer cells invasion, we generated a highly invasive breast cancer cell line by growing luminal epithelial MCF7 cancer cells in low-adherent conditions for 4 wk (Guttilla et al., 2012; Fig. 3 D). The resulting adherent MCF7-M cells lost their epithelial features (i.e., expressed lower of E-cadherin), expressed mesenchymal-specific proteins (i.e., Vimentin), and were mostly CD44+/CD24− (Fig. 3 E and not depicted). Moreover, MCF7-M cells were more prone to cellular invasion than their MCF7 counterpart, as shown using transwell assays (Fig. 3 F). Remarkably, ELP3, ALKBH8, CTU1, and CTU2 protein levels were strongly increased in MCF7-M cells compared with their epithelial counterpart MCF7 cells (Fig. 3 E). Whereas Elp3 depletion in MCF7-M cells did not impact on the expression of EMT markers (unpublished data), it strongly reduced their invasive capacity, as well as their ability to grow as tumorspheres (Fig. 3 G and not depicted). These observations were corroborated using the invasive breast cancer MDA-MB-231 cell line. Indeed, Elp3 depletion in MDA-MB-231 cells did not interfere with the expression of EMT markers (unpublished data), but strongly decreased the motility of MDA-MB-231 cells, as well as colony formation in a tumorsphere formation assay (Fig. 3 H and not depicted). Strikingly, depletion of CTU1 led to similar phenotypes. Indeed, CTU1 deficiency did not impact on the expression of either epithelial or mesenchymal markers (unpublished data), but it strongly impaired motility and tumorsphere formation in MDA-MB-231 cells (Fig. 3 I and not depicted). Finally, Elp3 depletion in MDA-MB-231 cells significantly reduced the abundance of thiolated tRNAs, as evidenced by Northern blot analysis (Fig. 3, J and K).
ELP3 is required for the expression of LEF1 in PyMT tumors
To gain more information about the molecular mechanisms by which Elp3 promotes breast cancer invasion and metastasis, we profiled the transcriptome of similarly sized control and Elp3-deficient PyMT primary tumors to identify pathways specifically deregulated upon Elp3 deletion. Total RNAs were extracted from 7-wk-old Elp3ΔMECPyMT or Elp3CTRPyMT primary tumor epithelial cells and subjected to high throughput RNA sequencing (Fig. 4 A). RT-qPCR analysis further confirmed that a selection of identified target mRNAs was indeed down-regulated in Elp3-deficient PyMT tumor epithelial cells (Fig. 4 B). Interestingly, a gene set enrichment analysis (GSEA), using both the KEGG pathways and GO database, highlighted an Elp3-dependent pro-metastatic signature, supporting the fact that Elp3ΔMECPyMT tumors are less invasive than the Elp3CTRPyMT tumors (Fig. 4 C). We next looked for potential loss of oncogenic pathways in the GSEA analysis. We found that genes downstream of the PI3K–AKT and KRAS pathways (i.e., which are known to be hyperactivated after PyMT expression), as well as genes downstream of the WNT pathway, were significantly down-regulated in Elp3ΔMECPyMT samples (Fig. 4 D). Strikingly, these analyses highlighted a very significant and specific Lef1-dependent signature (normalized enrichment score = –1.88; P < 0.001) that was missing in Elp3-deficient PyMT tumor cells (Fig. 4, D and E). Given the described importance of Lef1 as mediator of chemotactic invasion during lung adenocarcinoma metastasis (Nguyen et al., 2009), as well as in breast cancer cells (Huang et al., 2012), we surmised that disrupted Lef1 expression or function may underlie the decreased invasiveness of tumor cells lacking Elp3. Remarkably, Lef1 protein but not mRNA levels were indeed strongly decreased in Elp3ΔMECPyMT tumors compared with Elp3CTRPyMT (Fig. 4, F and G). Consistently, mRNA levels of four Lef1 targets identified in our RNaseq experiments (Pik3r5, EphB2, PdgfrA, and Vcam) were strongly diminished in Elp3-deficient tumor cells (Fig. 4 G). Finally, in line with the previously described proinvasive role of LEF1 (Nguyen et al., 2009; Huang et al., 2012), its depletion indeed reduced the migration of MDA-MB-231 cells and also impaired their ability to grow as tumorspheres, similarly to ELP3 loss (Fig. 4, H and I; and not depicted). Together, these results demonstrate that loss of Elp3 impairs Lef1 expression at the protein level in PyMT breast tumor cells, and suggest that ELP3-dependent expression of LEF1 promotes breast cancer cell motility.
Figure 4.
Elp3-deficient PyMT tumors lack a Lef1-dependent gene signature. (A) Scatter plot of RNA-seq data obtained from in Elp3CTRPyMT and Elp3ΔMECPyMT tumor cells. (B) RT-qPCR analysis was used to assess the expression of the indicated gens in Elp3CTRPyMT and Elp3ΔMECPyMT tumor cells. Values represent mean ± SD of signals form three different mice, performed in triplicate (***, P ≤ 0.001, Student’s t test). (C) GSEA analyses of published metastasis- and invasion-related datasets for genes down-regulated in Elp3ΔMECPyMT tumors. NES, normalized enrichment score with adjusted p-value for each enrichment plots. (D) GSEA analysis using Oncogenic Pathway database. NES and adjusted p-values are added for each enrichment dataset. (E) Enrichment plot of the Lef1 dataset (with NES and p-value). Heatmap of the top 20 genes down-regulated in Elp3ΔMECPyMT from the Lef1 dataset. (F) Western blot analysis of Elp3, Lef1, and Ctu1 in similarly sized Elp3CTRPyMT and Elp3ΔMECPyMT tumors (14 wk). (G) RT-qPCR analysis of Lef1 mRNA and its target genes (Pik3r5, Ephb2, PdgfrA, and Vcam). Values represent mean ± SD of signals from five different 14 wk tumors, performed in triplicate (*, P ≤ 0.05, Student’s t test). (H) LEF1, ELP3, and CTU1 protein levels in control (CTR) or LEF1-depleted (LEF1#1 and LEF1#2) MDA-MB231 cells were assessed by Western blot analysis. (I) Migration of control (shCTR) or LEF1-depleted (shLEF1#1 and shLEF#2) MDA-MB-231 cells toward a serum gradient was measured using Transwell assays. The results are expressed as number of migrating cells per field. Values represent mean ± SD of three independent experiments performed in triplicates (*, P ≤ 0.05; **, P ≤ 0.01, Student’s t test).
ELP3 regulates LEF1 IRES-dependent translation to promote breast cancer cell migration
LEF1 protein levels correlated with those of ELP3 and CTU1/2 in two different cellular models of invasive breast cancer, further strengthening the link between LEF1 expression and U34 tRNA modification activity in transformed MECs. Indeed, prolonged TGFβ stimulation (for 13 d) of Py2T cells (extracted from PyMT tumors; TGFβ-long time-stimulated Py2T are referred as Py2T-LT), which has been shown to induce EMT-dependent cell motility (Waldmeier et al., 2012), increased Lef1, Elp3, Alkbh8, and Ctu1/Ctu2 mRNA and protein expression (Fig. 5 A). Similarly, this correlation was also observed in invasive MCF7-M cells as compared with their epithelial MCF7 counterpart cells (Fig. 5 B). Remarkably, ELP3 depletion in three cellular models of invasive breast cancer (MCF7-M, MDA-MB-231, and Py2T-LT) led to dramatically decreased LEF1 protein, but not mRNA levels (Fig. 5, C and D). U34 modification affects protein synthesis (Bauer et al., 2012; Fernández-Vázquez et al., 2013; Rezgui et al., 2013), suggesting that LEF1 translational elongation may rely on ELP3. To assess changes in translational control upon ELP3 deficiency, we measured polysomal distribution in control or ELP3-depleted MCF7-M cells. ELP3 deficiency resulted in a decrease in the number of polysomes (Fig. 5, E and F), suggesting that a selective translation reprogramming takes place. We next quantified the global translation rates by 35S-methionine incorporation measurements. Interestingly, we found that ELP3 deficiency in MCF7-M cells reduced methionine incorporation, confirming that its absence globally affects total protein production (Fig. 5 G). Recent studies in yeast and nematodes demonstrated that loss of U34 tRNA modification triggers proteotoxic stress, compromises the clearing of stress-induced protein aggregates (Nedialkova and Leidel, 2015), and leads to activation of the PERK-transduced UPR in the developing murine cortex (Laguesse et al., 2015). In contrast, the absence of ELP3 in breast cancer cells did not lead to any increase in pPERK-related UPR pathway, nor to differences in the expression of the canonical translation initiation proteins (i.e., eIF4A, eIF4E, and eIF4G; unpublished data). To further investigate the possibility that ELP3 promotes LEF1 protein synthesis, we examined the distribution of the LEF1 mRNA in the polysomal fractions of control or ELP3-depleted MCF7-M. Remarkably, the abundance of LEF1 transcripts in the polysomal fractions was severely reduced and increased in subpolysomal fractions (Fig. 5, H and I), suggesting that LEF1 mRNA translation efficiency is reduced upon ELP3 depletion in MCF7-M cells. As a control, the distribution of GAPDH (high translation), PABP (low translation), and SNAIL (intermediate translation) transcripts remained unchanged upon ELP3 deficiency in MCF7-M cells (unpublished data). Similarly, LEF1 mRNA distribution, but not that of GAPDH, PABP, and SNAIL mRNAs, was also modified toward reduced abundance in polysomal fractions in ELP3-depleted MDA-MB-231 cells (unpublished data).
Figure 5.
U34 tRNA modifications regulate LEF1 IRES translation to promote breast cancer cell migration. (A) Western blot analysis of indicated proteins in Py2T cells (extracted from PyMT tumors) and Py2T-LT (idem and long-time stimulated with TGFβ). HSP90 is used as loading control. (B) Western blot analysis of LEF1 and ELP3 in MCF7 and MCF7-M cells. HSP90 is used as loading control. (C and D) ELP3 and LEF-1 protein (C) or mRNA (D) levels in control (CTR) or ELP3-depleted (ELP3#1 and ELP3#2) MCF7-M, Py2T-LT, and MDA-MB-231 cells as indicated. Values represent mean ± SD of three experiments performed in triplicate (*, P ≤ 0.05; ***, P ≤ 0.001; ns, nonsignificant, Student’s t test). (E) Representative polysome profiles of control (shCTR) or ELP3-depleted (shELP3#1 and ELP3#2) MCF7-M cells. (F) As in E, but translation efficiency, calculated as the ratio of polysomal (P) compared with subpolysomal fractions (S), is shown in the graph. Data are mean ± SD (n = 3 per group; *, P ≤ 0.05, Student’s t test). (G) MCF7-M cells were pulsed for 30 min with 35S-labeled methionine/cysteine. Incorporation of 35S into protein was quantified by scintillation counting and normalized to total amount of proteins. Data are mean ± SD (n = 3 biological replicates per group; *, P ≤ 0.05, Student’s t test). (H) Distribution of LEF-1–specific mRNA in polysome fractions quantified by RT-qPCR. Data are mean ± SD of three independent experiments performed in triplicates. (I) Quantification of LEF-1–specific translation efficiency by calculation of the ratio of polysomal (P) compared with subpolysomal (S) fractions. Data are mean ± SD (n = 3 per group; *, P ≤ 0.05; ***, P ≤ 0.001, Student’s t test). (J) Western blot showing FLAG and ELP3 expression in control (shCTR) or ELP3-depleted (shELP3#1 and shELP3#2) MDA-MB-231 cells stably expressing empty vector (EV), IRES-LEF1 cDNA (IRES-LEF1-FLAG), or LEF1 cDNA (LEF1-FLAG). Numbers represent ratios between the respective FLAG and HSP90 signals. (K) Migration of control or ELP3-depleted MDA-MB-231 cells stably expressing IRES-LEF1 cDNA (IRES-LEF1-FLAG) or LEF1 cDNA (LEF1-FLAG) toward a serum gradient was measured using Transwell assays. Representative pictures are shown. Quantification of migrating cells is also shown in the graph (right). Results are expressed as number of migrating cells per field. Values represent mean ± SD of three representative experiments performed in triplicates (*, P ≤ 0.05; ***, P ≤ 0.001, Student’s t test).
We next investigated the mechanisms underlying the regulation of LEF1 translation by U34 tRNA modification. We hypothesized that tRNA-dependent decoding impairment during LEF1 mRNA translation leads to defective LEF1 protein production in ELP3-depleted cells. Surprisingly, a construct expressing the LEF1 open reading frame (ORF) cDNA was found insensitive to ELP3 depletion in MDA-MB-231 cells (Fig. 5 J, lanes 7–9), implying that ELP3 may indirectly regulate LEF1 translation. Previous studies demonstrated that oncogenic LEF1 translation is regulated through its 5′ UTR IRES sequence (Jimenez et al., 2005; Tsai et al., 2014). We cloned the LEF1 5′UTR IRES-sequence upstream its ORF cDNA and expressed the LEF1 constructs containing (IRES-LEF1-FLAG) or not (LEF1-FLAG) its 5′ IRES sequence in control or ELP3-depleted MDA-MB-231 breast cancer cells. Remarkably, the presence of the IRES in the LEF1 5′ UTR was associated with a down-regulation of LEF1 protein expression in ELP3-depleted cells (Fig. 5 J, lanes 4–6). This experiment demonstrates that ELP3 indirectly controls LEF1 translation, through its 5′-IRES sequence. Finally, expression of the LEF1 construct lacking the 5′-IRES, which escaped ELP3-dependent regulation, could partially rescue the cell migration defects in ELP3-depleted MDA-MB-231 cells (Fig. 5 K). These data demonstrate that Elongator is critical to sustain LEF1 protein synthesis through its IRES to promote breast cancer cells motility.
The ITAF DEK is a direct target of U34 mcm5s2 tRNA modification
We next sought to investigate the missing mechanism linking U34 tRNA modification and IRES-LEF1 translation. The regulatory proteins that bind to the LEF1 IRES sequence, called ITAFs (IRES-trans-acting factors), have been identified in a proteomic analysis (Tsai et al., 2011). We analyzed the codon content of each of them and calculated the frequencies of the AAA, GAA, and CAA codons, which require the mcm5s2 U34 modification for decoding (Fig. 6 A). We hypothesized that some of the ITAFs that bind the LEF1 IRES sequence may be enriched in these codons and may rely on U34 tRNA modification for their translation. Among those, we found that the ITAF DEK was particularly enriched in U34 mcm5s2-sensitive codons, as compared with the expected frequencies calculated in the global human genome (i.e., 6.7%). Indeed, the mRNA sequence of DEK is composed at 19.9% of combined AAA/GAA/CAA codons (Fig. 6 A). Strikingly, ELP3 deficiency in MDA-MB-231 and MCF7-M cells correlated with decreased DEK protein but not mRNA levels, whereas no change in expression was observed for other IRES-LEF1 ITAFs (Fig. 6, B–E). DEK protein levels were similarly decreased after CTU1 depletion in MDA-MB-231 and MCF7-M cells, strengthening the requirement of U34 tRNA modification in supporting DEK protein synthesis (Fig. 6, F–I). We also assessed DEK expression in control and Elp3-deficient PyMT tumors and we observed a strong reduction in DEK protein levels, as judged by both Western blot and IHC experiments (Fig. 6, J–L). Finally, to formally validate this hypothesis, we generated a DEK cDNA sequence in which all AAA/GAA/CAA codons have been replaced by their cognate synonymous AAG/GAG/CAG codons (DEK-δAAA/GAA/CAA-FLAG), which do not require the U34 mcm5s2 tRNA modification for their translation (Fig. 6 M). Remarkably, although the expression of wild-type DEK (DEK-WT-FLAG) was decreased in ELP3- or CTU1-depleted MDA-MB-231 cells, the DEK-δAAA/GAA/CAA sequence escaped ELP3- and CTU1-dependent translation regulation (Fig. 6 N). Furthermore, LEF1 and DEK protein levels positively correlated in ELP3- or CTU1-depleted cells complemented with the DEK-δAAA/GAA/CAA construct (Fig. 6 N), suggesting that DEK expression regulates LEF1 protein levels. Together, these data demonstrate that U34 tRNA modification is required for DEK protein synthesis through its AAA/CAA/GAA codons. Moreover, these results suggest that reduced DEK expression impairs LEF1 protein translation and, consequently, breast cancer cells motility.
Figure 6.
The ITAF DEK is a direct target of U34 tRNA modifications. (A) Table of reported LEF1 ITAFs (Tsai et al., 2011). Proteins were ordered according to the combined frequency of AAA, GAA, and CAA codons in their ORF. (B) Western blot showing indicated proteins expression in control (shCTR) or ELP3-depleted (shELP3#1 and shELP3#2) MDA-MB-231 cells. HSP90 is used as loading control. (C) RT-qPCR assessing DEK mRNA levels in control or ELP3-depleted MDA-MB-231 cells. Values represent mean ± SD of three different experiments performed in triplicate. (D and E) Expression of LEF1 and DEK was assessed at protein levels by Western blot (D and F) and at mRNA levels by RT-qPCR (E and G) in control or ELP3-depleted MCF7-M cells. (F–I) DEK and LEF1 protein (F and H) and mRNA (G and I) expression were assessed in control or CTU-1–depleted MDA-MB-231 (F and G) and MCF7-M cells (H and I; ***, P < 0.001, Student’s t test). (J) Western blot showing DEK protein expression in extracts from Elp3CTRPyMT and Elp3ΔMECPyMT primary tumors of similar size. HSP90 is used as loading control. (K) IHC staining for Dek expression in tumorigenic mammary glands from 7-wk-old Elp3CTRPyMT or Elp3ΔMECPyMT mice. Images were generated from five different tumors/mouse, using three mice per condition. Representative images are shown. (L) The mean Dek-specific signal shown in M was quantified and plotted (mean values ± SD; Student’s t test; ***, P < 0.001). (M) Positions of AAA, CAA, and GAA codons were represented in the DEK ORF sequence. (N) DEK (FLAG), LEF1, ELP3, and CTU1 protein levels were assessed in control (shCTR), ELP3 (shELP3)-, or CTU1 (shCTU1)-depleted MDA-MB-231 stably expressing DEK-wild-type sequence (DEK-WT-FLAG) or a DEF mutant in which AAA, CAA, and GAA codons have been replaced by their synonymous codons (AGA, CAG, GAG, respectively; DEK-δAAA/GAA/CAA-FLAG).
The ITAF DEK is required for IRES-LEF1 protein synthesis
DEK is essential for breast cancer cell invasion and metastasis (Privette Vinnedge et al., 2011, 2014). Importantly, high levels of the prooncogene DEK in human breast tumors is associated with reduced metastasis-free survival (Fig. 7 A) and with high grades in human breast cancer (Fig. 7 B). DEK deficiency in MDA-MB-231 cells led to marked decrease in LEF1 protein but not mRNA levels (Fig. 7, C–E), which, together with previous evidences that DEK binds the LEF1 IRES sequence (Tsai et al., 2011), confirms a direct regulation of LEF1 by DEK in breast cancer. To further confirm the IRES-dependent regulation of LEF1 by ELP3 and DEK, we used a bicistronic construct with dual luciferase system in which the Renilla luciferase (RLuc) is used as the upstream cistron using cap-dependent translation, and firefly luciferase (FLuc) in the downstream position using cap-independent translation (Jimenez et al., 2005; Tsai et al., 2014). The full-length LEF1 5′UTR sequence (1.178 kb) was inserted upstream the Fluc ORF (Fig. 7 F). Remarkably, the Fluc activity was significantly decreased upon ELP3 or DEK depletion in MDA-MB-231 cells compared with control cells, and no change was observed in Rluc activity (Fig. 7, G and H). To exclude cryptic promoter or alternative splicing occurrence, resulting in change of the bicistronic mRNA expression that could interfere with luciferase activity, we amplified PCR products from different regions of the bicistronic sequence and showed that their abundance did not vary between control and ELP3- or DEK-depleted cells (Fig. 7, I and J). Also, using a promoter less bicistronic construct, we confirmed that no alternative transcript could be produced, leading to a strong (almost complete) reduction of the Fluc activity, and production of transcripts (Fig. 7, K–M). We finally found that DEK depletion strongly affects cell migration in MDA-MB-231 cells (Fig. 7 N), further strengthening our model that the ELP3-dependent regulation of DEK, which is required for LEF1 protein synthesis promotes breast cancer cells motility.
Figure 7.
The ITAF DEK is required for IRES-LEF1 protein synthesis. (A) Kaplan-Meier curves showing distant metastasis-free survival according to DEK mRNA expression using GOBO (Gene expression-based Outcome for Breast cancer Online; Ringnér et al., 2011). Black, low expression; gray, medium expression; red, high expression (n represents the number of patients included in the analysis). (B) Box plot graph showing DEK mRNA expression in breast cancers of different grades according to GOBO. (C) Western blot showing expression of indicated proteins in MDA-MB-231 cells transfected with esiRNAs control or targeting DEK. (D) Same as C, but detecting LEF1 and DEK mRNA levels by RT-qPCR. (E) Western blot showing expression of indicated proteins in MDA-MB-231 cells stably expressing shRNA control or specific to DEK (DEK#1 and DEK#2). (F–H) Luciferase reporter assay using a bicistronic Renilla luciferase-IRES-LEF1-firefly luciferase construct (i.e., pRSTF-LEF1) in control (CTR) and ELP3- (G) or DEK-depleted (H) MDA-MB-231 cells (ELP3#1, ELP3#2 and DEK#1, and DEK#2). Values represent mean ± SD of three independent experiments performed in triplicate. (I and J) RT-qPCR showing the ratio of mRNA levels of the firefly amplicon (Fire1), selected close to the AUG initiation codon, on two renilla amplicons (Ren1 and Ren2) in control (CTR) and ELP3- (G) or DEK-depleted (H) MDA-MB-231 cells (ELP3#1, ELP3#2 and DEK#1, and DEK#2). Values represent mean ± SD of three independent experiments performed in triplicate. (K and L) Luciferase reporter assay using pRTSF-LEF1 construct or the corresponding promoterless construct pRSTF-LEF1ΔSV40 in MDA-MB-231 cells. Data are expressed as the relative light unit of firefly, normalized to β-gal levels, as a control of transfection efficiency. Values represent mean ± SD of three different experiments performed in triplicate. (M) RT-qPCR showing mRNA levels of the firefly mRNA normalized to GAPDH levels in MDA-MB-231 cells, transfected with pRSTF-LEF1 or pRSTF-LEF1ΔSV40. Values represent mean ± SD of three different experiments performed in triplicate. (N) Migration of control (shCTR) or DEK-depleted (DEK#1 and DEK#2) MDA-MB-231 cells toward a serum gradient was measured using Transwell assays. The results are expressed as number of migrating cells per field. Values represent mean ± SD of three independent experiments performed in triplicates.
ELP3, DEK, and LEF1 protein levels correlate in human breast cancer patients
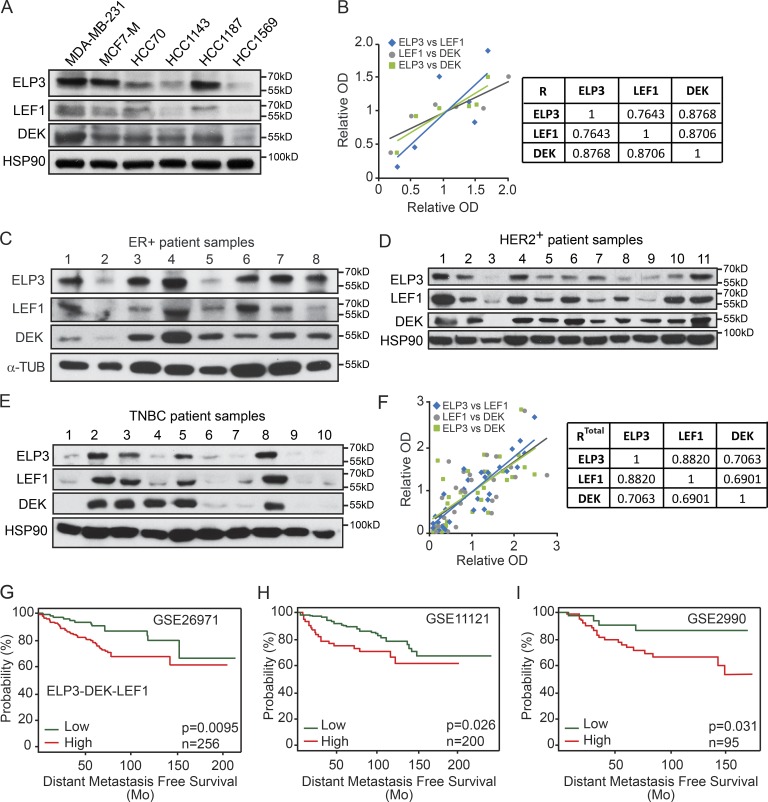
Having established that ELP3 promotes the expression of the DEK–LEF1 axis to support breast cancer cell invasion and metastasis, we further assessed the expression of these proteins in extracts from various breast cancer cell lines. Interestingly, we found that ELP3, DEK, and LEF1 protein levels correlated in breast cell lines, whereas no correlation was found with the transcription factor TCF3 (Fig. 8, A and B; and not depicted). We then tested if such correlation could be highlighted in samples from human breast cancer patients. Samples were collected from different subtypes of human breast cancer (i.e., ER+, HER2+, and Triple-negative breast cancers), and protein levels of ELP3, DEK, and LEF1 were quantified. Remarkably, the expressions of ELP3, DEK, and LEF1 were significantly correlated in human breast cancer patients (Fig. 8, C–E). Interestingly, this correlation was observed independently of the breast cancer subtype (Fig. 8 F), suggesting that the integrity of ELP3 is critical for the expression of the DEK–LEF1 axis as a general molecular pathway relevant in any human breast cancer. Finally, we searched in silico for breast cancer-related gene expression datasets with links to outcome. The GSE26971 (Filipits et al., 2011), GSE11121 (Schmidt et al., 2008), and GSE2990 (Sotiriou et al., 2006) datasets containing 256, 200, and 95 cases, respectively, showed that elevated concomitant expression of ELP3-DEK-LEF1 was associated with reduced metastasis-free survival (Fig. 8, G–I). Together, these data validate our molecular model and indicate that U34 tRNA modification supports the translational elongation of DEK and the subsequent IRES-dependent LEF1 translational initiation during metastasis.
Figure 8.
Expression of ELP3, LEF1, and DEK correlates in human breast cancer patients. (A) Western blot analysis of ELP3, LEF1, and DEK protein levels in the indicated human breast cancer cell lines. HSP90 is detected for normalization purpose. (B) Expression values (relative OD) of all human breast cancer samples were plotted and paired correlations were calculated. The table summarizes the correlation values (R). (C–E) ELP3, LEF1, and DEK protein levels were assessed in ER+ (C), HER2+ (D), and Triple-negative breast cancer (TNBC; E) human breast cancer samples. α-Tubulin and HSP90 were used as loading controls. (F) Expression values (relative OD) of all human breast cancer samples were plotted and paired correlations were calculated. The table summarizes the correlation values (R). (G–I) Kaplan-Meier curves for distant metastasis-free survival to all sites according to concomitant ELP3, DEK, and LEF1 mRNA expression, using GEO accession nos. GSE26971 (G), GSE11121 (H), GSE2990 (I) datasets of breast cancer patients. Patients were stratified into low and high expression based on autoselect best cutoff. Green, low expression; red, high expression. P-values were calculated with log-rank (Mantel–Cox) test.
DISCUSSION
The mechanisms supporting tumor cellular adaptation impact on metastasis formation and patient outcome (Valastyan and Weinberg, 2011; Klein, 2013). Here, we show that U34-modifying enzymes Elp3 and Ctu1/2, which catalyze the mcm5s2-U34 tRNA modification, are up-regulated in human breast cancer samples, as well as in models of invasive breast cancer cells, and support breast cancer metastasis. The genetic ablation of Elp3 in PyMT-induced metastatic breast cancer dramatically reduces metastases formation in mice and affects leader cells invasion in ex vivo culture of mammary tumors. Mechanistically, we demonstrate that the ITAF protein DEK, enriched in AAA, GAA, and CAA codons, requires ELP3 and CTU1 for its translational elongation, which in turn promotes IRES-dependent translational initiation of LEF1 to support motility (see model in Fig. 9). Finally, the expression of ELP3, DEK, and LEF1 correlates in breast cancer patient samples and is associated with high risk of developing metastasis. Therefore, our results highlight the importance of the U34 tRNA-modifying enzymes in establishing a proinvasive signature in breast cancers by linking the tRNA-dependent translation of an ITAF to the IRES-dependent translation of a key oncogenic protein.
Figure 9.
Model for the implication of the U34 tRNA modifications in breast cancer metastasis. The U34 tRNA-modifying enzymes ELP3 and CTU1/2 are induced in models of invasive breast cancer and are critical for the translation of DEK, which in turn regulates the IRES-dependent translation of the oncogenic LEF-1 mRNA to promote breast cancer cells motility and metastasis. This mechanism is essential for the establishment of a LEF-1–dependent prometastatic genes signature in breast tumors.
We found that expressing the oncoprotein PyMT in mammary epithelial cells led to up-regulation of ELP3, ALKBH8, and CTU1/2, which correlated with enhanced tRNA thiolation. PyMT activates both RAS and PI3K pathways (Raptis et al., 1991; Webster et al., 1998). How the activation of the oncogenic RAS–PI3K pathways leads to increased expression of U34 tRNA-modifying enzymes deserves further investigation. Interestingly, global tRNA levels are significantly increased in tumors compared with normal tissues in the breast (Pavon-Eternod et al., 2009; Goodarzi et al., 2016). Also, many oncogenic pathways, including RAS–PI3K–dependent cascades converge on RNA Polymerase III, responsible for tRNA production (Grewal, 2015). As substrates of the U34 tRNA modification enzymes, increased tRNA levels could directly or indirectly induce U34 modification enzymes levels and/or activation. Understanding the relationship between U34 modification enzymes and their substrates in cancers is an area that still needs to be studied.
In a model of intestinal cancer, we previously showed that Elp3 is induced upon constitutive Wnt signaling and sustains a pool of Lgr5+/Dclk1+/Sox9+ cancer stem cells to promote tumor initiation (Ladang et al., 2015). In breast cancer, recent studies showed that Wnt signaling is concentrated in the breast cancer stem cells population and is required for metastasis, but dispensable for primary tumor growth (Malanchi, 2012; Cai et al., 2013; Jang et al., 2015). Elp3 deficiency in the PyMT-induced breast cancer model slightly impacted on tumor appearance (2 wk delay) but led to a strong reduction of lung metastases. Moreover, we found that LEF1 expression, a mediator of Wnt-induced cellular invasion and metastasis (Nguyen et al., 2009; Cai et al., 2013), relies on ELP3 and CTU1. These data suggest that Wnt signaling involves Elp3 and U34 tRNA modification to promote breast cancer metastasis.
LEF1 mRNA translation depends on U34 tRNA modification for its synthesis in PyMT tumors, as well as in various breast cancer cell models. Downstream of the Wnt–TCF pathway, Lef1 promotes lung adenocarcinoma metastasis (Nguyen et al., 2009), mediates chemotactic invasion (Huang et al., 2012), and is associated with colon cancer progression and metastasis (Wang et al., 2013). Our data support these conclusions, as LEF1 depletion in breast cancer cells strongly decreases their chemotactic potential in a manner similar to ELP3 deficiency. Unexpectedly, we found that U34 modification enzymes regulate LEF1 protein expression through an IRES-dependent mechanism. Strikingly, the oncogenic LEF1 mRNA was shown to mainly use a cap-independent mechanism for translation and to be produced via IRES-directed translation (Jimenez et al., 2005; Tsai et al., 2014). Interestingly, recent studies described that IRES-mediated translation of Laminin B1 is enhanced during malignant EMT (Petz et al., 2012) and that VEGF-C IRES-dependent translation was higher in metastatic tumor cells than in the primary tumors (Morfoisse et al., 2014), suggesting that IRES-dependent translation could support cancer progression and metastasis. Our data link tRNA modification to IRES-dependent translation initiation. Surprisingly, the expression of other proteins whose translation has been described to require a specific IRES sequence (i.e., c-MYC, c-IAP, VEGF-α, CDK1, SRC, and EGFR) was also tested and was not found to be ELP3 or DEK dependent (unpublished data). Further studies are required to explore whether U34 modification enzymes promote a more general translation initiation of IRES-containing transcripts.
We define the ITAF protein DEK as a direct substrate of the mcm5s2 modified tRNAs. Our results show that ELP3 and CTU1 are critical for DEK protein expression in PyMT tumors as well as in cellular models of invasive breast cancer. DEK is an oncogenic protein whose expression is elevated in several solid tumors (Kondoh et al., 1999; Khodadoust et al., 2009; Shibata et al., 2010), including breast cancer where DEK expression is correlated with clinical features of the disease (Wise-Draper et al., 2009; Liu et al., 2012). A recent report demonstrated that DEK supports breast cancer metastasis in vivo through promotion of β-catenin signaling (Privette Vinnedge et al., 2014). Functionally, DEK is predominately found in the nucleus, but a pool of DEK also binds to cytoplasmic mRNAs (McGarvey et al., 2000; Kappes et al., 2001; Le Hir et al., 2001; Soares et al., 2006). More particularly, DEK was identified as a protein bound to the LEF1-IRES sequence in a proteomic analysis (Tsai et al., 2011). We establish a functional link between the ITAF DEK and the regulation of LEF1 IRES-dependent translation in breast cancer models as DEK depletion impairs LEF1 protein production, without affecting LEF1 mRNA levels. Remarkably, the expression of ELP3, DEK, and LEF1 was significantly correlated in samples of breast cancers and is associated with a higher risk of developing metastasis. Together, these results demonstrate that Elp3, through U34 tRNA modification, is critical for the translation of DEK mRNA to promote IRES-dependent translation of LEF1 and to support breast cancer invasion and metastasis.
The activity of the Elongator complex is required for cell migration of multiple primary and transformed cells (Close et al., 2006, 2012; Johansen et al., 2008; Creppe et al., 2009; Lee et al., 2009). Our data show that invasive breast cancer cells that underwent EMT show elevated levels of U34 modification enzymes. Surprisingly, EMT maintenance does not rely on them, as the expression of mesenchymal markers (i.e., Vimentin, Slug, Zeb1, and Twist) and the percentage of CD44+/CD24− cells were not changed upon ELP3 or CTU1 deficiency (unpublished data). Importantly, depletion of ELP3 or CTU1 in invasive breast cancer cells drastically reduces their migratory and tumorsphere formation capacities, two typical features of mesenchymal-like cells. In PyMT tumors, the number of K14+ cells, which have undergone basal differentiation and support tumor invasion (Cheung et al., 2013; Ye et al., 2015), is not significantly reduced upon Elp3 deficiency (unpublished data). These data suggest that U34 modification enzymes are crucial for the motility of cells that have undergone EMT, whereas they are dispensable for the maintenance of the EMT program.
The molecular functions of Elongator have been the subject of extensive debate. Interestingly, a recent study biochemically established that Elp3 is a tRNA acetyltransferase that catalyzes the addition of 5′-carboxymethyluridine (cm5U) on tRNA molecules, through its KAT (lysine acetyltransferase) and radical SAM (S-adenosylmethionine) domains (Selvadurai et al., 2014). We show here that ELP3, as well as its partner enzymes in U34 tRNA modification CTU1/2, are up-regulated in breast cancers as well as upon induction of invasiveness in breast cancer cells. Moreover, our data identify DEK as a translational target of the mcm5s2 tRNA modification. Indeed, the DEK transcript sequence is highly enriched in the target AAA, GAA, and CAA codons. Strikingly, the replacement of the mcm5s2-tRNA-sensitive codons in the DEK sequence by their cognate, U34 modification–insensitive, synonymous codon is sufficient to restore DEK protein expression in ELP3-deficient cells. This formally demonstrates that Elongator regulates DEK protein levels through U34 mcm5s2 tRNA modification. This is further strengthened by the fact that depletion of CTU1 in invasive breast cancer cells perfectly phenocopies Elp3 deficiency; it also affects DEK protein but not mRNA expression levels and strongly reduces breast cancer cells motility, as well as their ability to grow as tumorspheres. Taken together, these results show that the tRNA modification function of Elongator is critical to promote breast cancer cell invasion and metastasis. It is likely that the lack of U34 tRNA modification affects the expression of other proteins than DEK, which could be essential for invasion and metastasis. A better understanding of the criteria that define a U34 tRNA modification mRNA substrate, as well as high-throughput proteomic analysis, are essential to provide a general view of their contribution in cancer.
Little is known about the role of U34 modification in cancer. Recently, we demonstrated that Elp3 is induced upon Wnt signaling activation and promotes Wnt-driven tumor initiation in the intestine (Ladang et al., 2015). We now highlight the specific regulation of U34 modifications in invasive breast cancer cells and demonstrate their crucial role in promoting breast cancer invasion and progression to metastasis. Further studies are needed to define the oncogenic pathways that rely on U34 modification activity to support cancer development and progression. This still poorly studied enzymatic cascade may reveal very promising features relevant for the development of future anticancer therapies.
MATERIALS AND METHODS
Plasmids, constructs, and antibodies
The LEF1-WT-FLAG, IRES-LEF1-FLAG, DEK-WT-FLAG, and DEK-δAAA/GAA/CAAA-FLAG sequences were cloned into LeGO-iG2 vector (GeneScript). The pRSTF-LEF1 bicistronic construct and pRSTF-LEF1ΔSV40 was a generous gift of M. Waterman (University of California, Irvine, CA). pBABE-Hygro control and pBABE-PyMT plasmids were obtained from Addgene (Pylayeva et al., 2009). Antibodies used for Western blotting were as follows: monoclonal anti-ELP3, anti-LEF1, anti-HUR and polyclonal anti-pAKT, and anti-AKT (Cell Signaling Technology); monoclonal anti-hnRNPA1 and polyclonal anti-ELP1, anti-HSP90, and anti-TCF3 (Santa Cruz Biotechnology, Inc.); monoclonal anti-CTU2 and polyclonal anti-ALKBH8, and anti-CTU1 (Abcam); monoclonal anti-Vimentin (Dako); monoclonal anti–E-cadherin and anti–N-cadherin (BD); polyclonal anti-FLAG and anti-hnRNPC (Sigma-Aldrich); and polyclonal anti-SFPQ/PSF, anti-NONO, anti-hnRNPK, and anti-YB1 (Bethyl).
Cell culture, transfections, and infection
Cells were typically grown in Dulbecco’s modified Eagle’s Medium (Gibco) containing 10% of fetal bovine serum and 1% glutamine, under humidified 5% CO2 atmosphere. MCF7-M cells were produced by nonadherent (Mammocult medium) culture of MCF7 cells for 4 wk, with weekly passages. They were then cultured in DMEM 10% FBS in adherent conditions. Cells of maximum eight passages were used to make sure the mesenchymal phenotype is kept intact. MDA-MB-231, MCF7, and NMuMG cells were obtained from the ATCC. The Py2T cell line was a generous gift from G. Christofori (University of Basel, Basel, Switzerland) and was previously described (Waldmeier et al., 2012). Py2T cells were stimulated for 15 d with TGF-β at 2 ng/ml (R&D Systems) to generate Py2T-LT cells.
Cells were transfected with esiRNA control or targeting DEK mRNA, using Mirus and Optimem (Thermo Fisher Scientific), according to the protocol specified by the manufacturer. Medium was changed after 18 h and cells were lysed after 48 h.
The shRNAs control or targeting ELP3, CTU1, or LEF1 were purchased from Sigma-Aldrich. Lentiviral infections in MCF7-M, Py2TLT, or MDA-MB-231 cells were performed as previously described (Creppe et al., 2009).
pBABE-WT or pBABE-PyMT were transfected in phoenix-ECO cells (ATCC) using Mirus and Opti-MEM (Thermo Fisher Scientific). After 48 h, the supernatants were used to transduce NMuMG cells.
Generation of Elp3ΔMECPyMT mouse
Elp3loxP/loxP mouse was generated as previously described (Ladang et al., 2015). The MMTV-CRE mouse strain was a gift from T. Hochepied (UGent-VIB Research, Ghent University, Ghent, Belgium). The constitutive KO allele was obtained after CRE-mediated recombination. Mice were backcrossed to the FVB/N genetic background for eight generations. The MMTV-PyMT mouse (FVB/N-Tg[MMTV-PyVT]634Mul/J) was purchased from The Jackson Laboratory. To delete Elp3 in the mammary epithelium, the Elp3loxP/loxP was crossed with the MMTVCRE/+strain (Elp3ΔMEC). All experiments were approved by the local Ethical Committee (University of Liège, Liège, Belgium).
RNA isolation and RT-qPCR
Total RNA was extracted from PyMT breast tumors using TRIzol (Life Technologies) and from indicated cell lines using RNeasy mini kit (QIAGEN) according to the manufacturer’s protocol and treated with DNase I (Roche). For cDNA synthesis, 1 µg of RNA was reverse transcribed using the RevertHaid H Minus First Strand kit (Thermo Fisher Scientific). Real-Time Quantitative PCR (RT-qPCR) was performed with LightCycler 480 (Roche), using 1 µl of cDNA newly synthetized and Sybrgreen (Sybr Premix Taq, ROX Plus; Takara Bio Inc.), in a final mix of 20 µl. Each quantification of mRNA was done using an endogenous control gene for normalization (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]). Data are plotted as mean values ± SD of three independent experiments performed in triplicates. All primer sequences used are available upon request.
Northern blot analysis
0.5 µg total RNA was resolved on 8% acrylamide gels containing 0.5X TBE, 7 M urea, and 50 µg/ml ([N-acryloylamino]phenyl)mercuric chloride (APM). Northern blot analysis was performed essentially as previously described (Leidel et al., 2009), with the following probe for tE(UUC): 5′-TTCCCATACCGGGAGTCGAACCCG-3′.
Migration assay
Cells were grown for 24 h in 1% FBS DMEM medium. Cells were then trypsinized, counted, resuspended in 1% FBS DMEM medium and placed in a Transwell insert (Costar; 104 MDA-MB-231 cells and 2 × 104 MCF7-M cells). The well was filled with 10% FBS DMEM medium. After 24 h, migrating cells were fixed and colored with Methanol 80% crystal violet for 10 min. Cells on the upper side of the filters were removed with cotton-tipped swabs and the filters were washed with PBS. Cells on the underside of the filters were viewed and quantified; five representative pictures per insert were taken. Each experimental condition was performed in triplicate. Data are plotted as mean values ± SD of three independent experiments.
Tumorsphere assay
Cells were resuspended in MammoCult medium human kit (STEMCELL Technologies), supplemented with Heparin (4 µg/ml; STEMCELL Technologies) and Hydrocortisone (0.5 µg/ml). Cells were then counted and cells (104 cells for MDA-MB-231, 2 × 104 cells for MCF7-M) were placed in an Ultra-low adherent culture dish (STEMCELL Technologies) with the same medium. After 7 d, number of tumorspheres was counted and representative pictures were taken for each condition. Each experimental condition was performed in triplicate. Data are plotted as mean value ± SD of three independent experiments.
Immunohistochemistry
For IHC analyses, samples were fixed overnight in 4% formalin solution and embedded in paraffin. Briefly, 3-µm-thick sections of FFPE tumors were deparaffinized and antigen retrieval was performed by boiling the sections in citrate buffer at pH 6.0 or EDTA at pH 9.0 for 20 min. Primary antibodies used were as follows: monoclonal anti-Elp3 (Cell Signaling Technology), polyclonal anti-Dek (Bethyl Laboratories), and monoclonal anti-Ctu1 and anti-Ctu2 (Abcam). Corresponding secondary antibody detection kits for reduced background on murine tissue were used (Histofine Simple Stain Mouse MAX PO; Medac) and stained on an automated stainer (LabVision Autostainer 480S; Thermo Fisher Scientific). All slides were scanned with a Panoramic 250 slide scanner (3DHISTECH Ltd.).
For quantification of IHC signals, total H-DAB–positive areas of each slide were calculated using the ImageJ software. Specifically, H-DAB positivity was quantified from the transformed images by ImageJ IHC Toolbox. Measurements were calculated in pixels. Data are expressed as percentage of H-DAB–positive pixels on total corresponding tissue structure pixels per slide.
Ex vivo organoid culture assay
Tumors of similar size were isolated from 9-wk-old mammary gland of control or Elp3-deficient PyMT mice. Tumors were chopped and placed for 15 h at 37°C, shacking at 125 rpm, in DMEM-F12 10% Gentle Collagenase (STEMCELL Technologies) for chemical digestion. Digested samples were then shacked and centrifuged at 1,000 rpm. The pellets were resuspended with 1:4 of mixture of HBSS (STEMCELL Technologies) supplemented with 2% of FBS and ammonium chloride (STEMCELL Technologies) and centrifuged. The pellets were then placed in DMEM-F12 FBS 5% and filtered through a 70-µm filter. The remaining solution was then centrifuged at 1,000 rpm for 5 min and the pellet was used for 3D collagen I culture. Organoids were cultured ex vivo in 3D collagen I culture kit (EMD Millipore) for 24 h. Pictures of organoids were taken using a Leica DMIL microscope. Quantification of leader cell was done on three different mice for each experimental condition (Elp3CTRPyMT and Elp3ΔMECPyMT). Mean lengths of leader cells were measured using the ImageJ software, on the same pictures. Representative organoids were stained with DAPI and Rhodamine-phaloidine (R415; Life Technologies) by immunofluorescence using the SP5 confocal microscope Leica.
RNA sequencing
Tumor organoids were extracted from PyMT tumors of similar size, in three different mice for each experimental condition and use for the generation of unicellular suspension. The organoid pellets were then trypsinized in 2 ml Trypsin-EDTA 0.25% for 5 min, centrifuged, and resuspended in prewarmed Dispase (5 mg/ml) and 0.2 ml of DNase I (1 mg/ml; STEMCELL Technologies). The samples were then centrifuged and resuspended in DMEM-F12 FBS 10% to form unicellular suspension. The EasySep Mouse Mammary Epithelial Cell Enrichment Kit (STEMCELL Technologies) was used to select epithelial cells. Total RNAs from mammary epithelial tumor cells were extracted using TRIzol (Life Technologies), according to the manufacturer’s protocol. RNA integrity was verified on the Bioanalyser 2100 with RNA 6000 Nano chips, and RIN scores were >9 for all samples. The Illumina Truseq RNA Sample Preparation kit V2 was used to prepare libraries from 500 ng total RNAs. PolyA RNAs were purified with polyT-coated magnetic beads, chemically fragmented, and used as template for cDNA synthesis using random hexamers. cDNA ends were subsequently end-blunted, adenylated at 3′OH extremities, and ligated to indexed adaptors. Finally, the adapters ligated library fragments were enriched by PCR following Illumina’s protocol and purified. Libraries were validated on the Bioanalyser DNA 1000 chip and quantified by qPCR using the KAPA library quantification kit. Sequencing was performed on HiSeq2000 in paired-end 2 × 100 base protocol. For data analysis, Fastq files were not trimmed for adaptor sequences. The reads were aligned with Tophat 2.0.9 to the mouse genome mm10. Cufflinks 2.1.1 suite was used to normalize data and generate FPKM values and CuffDiff was used to identify significantly differentially expressed genes. Generated lists were used to run GSEA was used in order to match various gene sets curated from the literature. Data is available from GEO under accession no. GSE85810.
Polysome profiling
5 × 106 cells were lysed in ice-cold 150 mM NaCl, 5 mM MgCl2, 15 mM Tris, pH 7.5, and 0.1 mg/ml cycloheximide, containing Superase-In 25 U/ml (Life Technologies), and supplemented with 1% Triton X-100. Lysates were then layered into 10 ml 10–50% (wt/vol) sucrose gradient containing the same buffer without Triton X-100. Gradients were centrifuged at 38,000 rpm, 4°C for 2 h using SW41Ti rotor (Beckman Coulter) and fractionated through a live UV detector (Teledyne Isco), with detection at 254 nm. Comparison between peaks was done based on the area under the curve. RNA from different fractions was precipitated using guanidine hydrochloride 7.7 ml and ethanol. After centrifugation, total RNAs were resuspended in distillated H2O and transferred in 3 M sodium acetate, pH 5.2, and Ethanol. After centrifugation and EtOH 75% wash, the pellets were air-dried and resuspended in nuclease-free water.
Global protein synthesis determination assay
Cells were treated with 16.5 µCi/ml 35S-methionine radiolabeled (NEG072; Perkin-Elmer) for 30 min in a prewarmed DMEM medium 10% dialyzed FBS and lysed. Precipitated proteins were concentrated on a filter paper using trichloroacetic acid 25%, and washed with EtOH 70% and with acetone. Result was obtained by scintillation of triplicates in each experimental condition. Total protein determination in each condition (Thermo Fisher Scientific) was done for standardization between conditions. Data are plotted as mean values of three independent experiments ± SD.
Luciferase reporter assay
2 × 105 MDA-MB-231 cells were plated in 6-wells plate. Cells were transfected with the bicistronic construct Renilla-firefly (0.5 µg), containing SV40 promoter (pRSTF-LEF1) or not (pRSTF-LEF1ΔSV40), using Mirus. Firefly and Renilla luciferase activities were measured 24 h after transfection using Dual-Luc Reporter kit (Promega). Firefly signals were normalized using Renilla and β-galactosidase. Data are plotted as mean values of three independent experiments ± SD, performed in triplicates.
Human samples
Samples from breast cancer patients were retrieved from Biothèque of the University of Liege (Belgium). The study was conducted according to the regulation of the Ethics Committee and the specimens were anonymized and analyzed in a blinded manner.
The Kaplan-Meier survival curves were plotted with publicly deposited gene expression data (EGA and TCGA), using Kaplan-Meier plotter and GOBO, which integrates statistical analysis. The percentiles of the patients between the upper and lower quartiles were autoselected based on the computed best performing thresholds as cutoffs. 221094_s_at, 200934_at, and 221557_s_at probe sets were used in all analyses for ELP3, DEK, and LEF1, respectively.
Acknowledgments
We thank Marian Waterman (University of California, Irvine, CA, USA) for the gift of the pRSTF-LEF1 and pRSTF-LEF1ΔSV40 constructs and Gerhard Christofori (University of Basel, Basel, Switzerland) for Py2T cells. We thank Michel Georges and Benoit Charloteaux (GIGA-Genetics, University of Liege, Liege, Belgium) with their help with processing the RNA-seq data. We are grateful to the GIGA imaging, genomics and viral vector facilities (GIGA, University of Liege), and to the Biothèque of the University of Liege for their assistance.
This study was supported by an Incentive Grant for Scientific Research (MIS F:4532.13) from the Belgian National Funds for Scientific Research, grants from the Concerted Research Action Program (Bio-Acet and tRAME) and Special Research Funds (FSR) at the University of Liege, the Belgian Foundation against Cancer, as well as by the Walloon Excellence in Life Sciences and Biotechnology (WELBIO) and the Max Planck Society. We are also grateful to the “Fonds Leon Fredericq” and the “Centre Anticancéreux” of the CHU Liege for their financial support. S. Delaunay, A. Chariot, C. Desmet, L. Nguyen, and P. Close are Research Fellow, Research Director and Research Associates at the FNRS, respectively.
The authors declare no competing financial interests.
Author contributions: conceptualization and methodology: S. Delaunay, A. Chariot, and P. Close; software and formal analysis: S. Delaunay, and Z. Zhou; Investigation; S. Delaunay, F. Rapino, K. Shostak, M. Termathe, I. Klevernic, L. Tharun, A. Florin, H. Desmecht, and L. Heukamp; resources: C.J. Desmet, L. Nguyen, S.A. Leidel, A. Willis, and R. Büttner; writing: S. Delaunay, A. Chariot, and P. Close; supervision and funding acquisition: A. Chariot and P. Close.
Footnotes
Abbreviations used:
- 3D
- three dimensional
- GSEA
- gene set enrichment analysis
- mcm5s2
- 5-methoxycarbonylmethyl-2-thio-
- IHC
- immunohistochemistry
- IRES
- internal ribosome entry site
- ITAF
- IRES-trans-acting factor
- tRNA
- transfer RNA
- U34
- uridine 34
- UTR
- untranslated region
- ORF
- open reading frame
References
- Anastas J.N., and Moon R.T.. 2013. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer. 13:11–26. 10.1038/nrc3419 [DOI] [PubMed] [Google Scholar]
- Bauer F., Matsuyama A., Candiracci J., Dieu M., Scheliga J., Wolf D.A., Yoshida M., and Hermand D.. 2012. Translational control of cell division by Elongator. Cell Reports. 1:424–433. 10.1016/j.celrep.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk G.R., Huang B., Persson O.P., and Byström A.S.. 2007. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 13:1245–1255. 10.1261/rna.558707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Guan H., Fang L., Yang Y., Zhu X., Yuan J., Wu J., and Li M.. 2013. MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J. Clin. Invest. 123:566–579. 10.1172/JCI65871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Tuck S., and Byström A.S.. 2009. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 5:e1000561 10.1371/journal.pgen.1000561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Gabrielson E., Werb Z., and Ewald A.J.. 2013. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 155:1639–1651. 10.1016/j.cell.2013.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close P., Hawkes N., Cornez I., Creppe C., Lambert C.A., Rogister B., Siebenlist U., Merville M.P., Slaugenhaupt S.A., Bours V., et al. . 2006. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell. 22:521–531. 10.1016/j.molcel.2006.04.017 [DOI] [PubMed] [Google Scholar]
- Close P., Gillard M., Ladang A., Jiang Z., Papuga J., Hawkes N., Nguyen L., Chapelle J.P., Bouillenne F., Svejstrup J., et al. . 2012. DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator. J. Biol. Chem. 287:32535–32545. 10.1074/jbc.M112.402727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creppe C., Malinouskaya L., Volvert M.L., Gillard M., Close P., Malaise O., Laguesse S., Cornez I., Rahmouni S., Ormenese S., et al. . 2009. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 136:551–564. 10.1016/j.cell.2008.11.043 [DOI] [PubMed] [Google Scholar]
- Dewez M., Bauer F., Dieu M., Raes M., Vandenhaute J., and Hermand D.. 2008. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc. Natl. Acad. Sci. USA. 105:5459–5464. 10.1073/pnas.0709404105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B., Bailly M., and de Crécy-Lagard V.. 2012. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 46:69–95. 10.1146/annurev-genet-110711-155641 [DOI] [PubMed] [Google Scholar]
- Esberg A., Huang B., Johansson M.J.O., and Byström A.S.. 2006. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 24:139–148. 10.1016/j.molcel.2006.07.031 [DOI] [PubMed] [Google Scholar]
- Fernández-Vázquez J., Vargas-Pérez I., Sansó M., Buhne K., Carmona M., Paulo E., Hermand D., Rodríguez-Gabriel M., Ayté J., Leidel S., and Hidalgo E.. 2013. Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 9:e1003647 10.1371/journal.pgen.1003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipits M., Rudas M., Jakesz R., Dubsky P., Fitzal F., Singer C.F., Dietze O., Greil R., Jelen A., Sevelda P., et al. EP Investigators . 2011. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 17:6012–6020. 10.1158/1078-0432.CCR-11-0926 [DOI] [PubMed] [Google Scholar]
- Goodarzi H., Nguyen H.C.B., Zhang S., Dill B.D., Molina H., and Tavazoie S.F.. 2016. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell. 165:1416–1427. 10.1016/j.cell.2016.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.S. 2015. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim. Biophys. Acta. 1849:898–907. 10.1016/j.bbagrm.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Guttilla I.K., Phoenix K.N., Hong X., Tirnauer J.S., Claffey K.P., and White B.A.. 2012. Prolonged mammosphere culture of MCF-7 cells induces an EMT and repression of the estrogen receptor by microRNAs. Breast Cancer Res. Treat. 132:75–85. 10.1007/s10549-011-1534-y [DOI] [PubMed] [Google Scholar]
- Guy C.T., Cardiff R.D., and Muller W.J.. 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12:954–961. 10.1128/MCB.12.3.954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Lu J., and Byström A.S.. 2008. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 14:2183–2194. 10.1261/rna.1184108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.-I., Chen Y.-L., Chang C.-N., Yuan R.-H., and Jeng Y.-M.. 2012. Hepatocyte growth factor activates Wnt pathway by transcriptional activation of LEF1 to facilitate tumor invasion. Carcinogenesis. 33:1142–1148. 10.1093/carcin/bgs131 [DOI] [PubMed] [Google Scholar]
- Jang G.-B., Kim J.-Y., Cho S.-D., Park K.-S., Jung J.-Y., Lee H.-Y., Hong I.-S., and Nam J.-S.. 2015. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci. Rep. 5:12465 10.1038/srep12465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J., Jang G.M., Semler B.L., and Waterman M.L.. 2005. An internal ribosome entry site mediates translation of lymphoid enhancer factor-1. RNA. 11:1385–1399. 10.1261/rna.7226105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen L.D., Naumanen T., Knudsen A., Westerlund N., Gromova I., Junttila M., Nielsen C., Bøttzauw T., Tolkovsky A., Westermarck J., et al. . 2008. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J. Cell Sci. 121:854–864. 10.1242/jcs.013722 [DOI] [PubMed] [Google Scholar]
- Kalhor H.R., and Clarke S.. 2003. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 23:9283–9292. 10.1128/MCB.23.24.9283-9292.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes F., Burger K., Baack M., Fackelmayer F.O., and Gruss C.. 2001. Subcellular localization of the human proto-oncogene protein DEK. J. Biol. Chem. 276:26317–26323. 10.1074/jbc.M100162200 [DOI] [PubMed] [Google Scholar]
- Karlsborn T., Tükenmez H., Mahmud A.K.M.F., Xu F., Xu H., and Byström A.S.. 2015. Elongator, a conserved complex required for wobble uridine modifications in Eukaryotes. RNA Biol. 11:1519–1528. 10.4161/15476286.2014.992276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodadoust M.S., Verhaegen M., Kappes F., Riveiro-Falkenbach E., Cigudosa J.C., Kim D.S.L., Chinnaiyan A.M., Markovitz D.M., and Soengas M.S.. 2009. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 69:6405–6413. 10.1158/0008-5472.CAN-09-1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C.A. 2013. Selection and adaptation during metastatic cancer progression. Nature. 501:365–372. 10.1038/nature12628 [DOI] [PubMed] [Google Scholar]
- Kondoh N., Wakatsuki T., Ryo A., Hada A., Aihara T., Horiuchi S., Goseki N., Matsubara O., Takenaka K., Shichita M., et al. . 1999. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 59:4990–4996. [PubMed] [Google Scholar]
- Ladang A., Rapino F., Heukamp L.C., Tharun L., Shostak K., Hermand D., Delaunay S., Klevernic I., Jiang Z., Jacques N., et al. . 2015. Elp3 drives Wnt-dependent tumor initiation and regeneration in the intestine. J. Exp. Med. 212:2057–2075. 10.1084/jem.20142288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGamba D., Nawshad A., and Hay E.D.. 2005. Microarray analysis of gene expression during epithelial-mesenchymal transformation. Dev. Dyn. 234:132–142. 10.1002/dvdy.20489 [DOI] [PubMed] [Google Scholar]
- Laguesse S., Creppe C., Nedialkova D.D., Prévot P.-P., Borgs L., Huysseune S., Franco B., Duysens G., Krusy N., Lee G., et al. . 2015. A Dynamic Unfolded Protein Response Contributes to the Control of Cortical Neurogenesis. Dev. Cell. 35:553–567. 10.1016/j.devcel.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Lee G., Papapetrou E.P., Kim H., Chambers S.M., Tomishima M.J., Fasano C.A., Ganat Y.M., Menon J., Shimizu F., Viale A., et al. . 2009. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 461:402–406. 10.1038/nature08320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield D., Izaurralde E., and Moore M.J.. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20:4987–4997. 10.1093/emboj/20.17.4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Pedrioli P.G.A., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., and Peter M.. 2009. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 458:228–232. 10.1038/nature07643 [DOI] [PubMed] [Google Scholar]
- Lin E.Y., Jones J.G., Li P., Zhu L., Whitney K.D., Muller W.J., and Pollard J.W.. 2003. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163:2113–2126. 10.1016/S0002-9440(10)63568-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.J., Shen L., Jang C.W., Falnes P.O., and Zhang Y.. 2013. Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet. 9:e1003516 10.1371/journal.pgen.1003516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Wang X., Sun F., Kong J., Li Z., and Lin Z.. 2012. DEK overexpression is correlated with the clinical features of breast cancer. Pathol. Int. 62:176–181. 10.1111/j.1440-1827.2011.02775.x [DOI] [PubMed] [Google Scholar]
- Malanchi I. 2012. Abstract SY28-02: Interactions between cancer stem cells and their niche govern metastatic colonization. Cancer Res. 72(8 Supplement):1–12. 10.1158/1538-7445.AM2012-SY28-02 [DOI] [PubMed] [Google Scholar]
- Massagué J. 2008. TGFbeta in Cancer. Cell. 134:215–230. 10.1016/j.cell.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T., Rosonina E., McCracken S., Li Q., Arnaout R., Mientjes E., Nickerson J.A., Awrey D., Greenblatt J., Grosveld G., and Blencowe B.J.. 2000. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J. Cell Biol. 150:309–320. 10.1083/jcb.150.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici D., Hay E.D., and Goodenough D.A.. 2006. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol. Biol. Cell. 17:1871–1879. 10.1091/mbc.E05-08-0767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfoisse F., Kuchnio A., Frainay C., Gomez-Brouchet A., Delisle M.B., Marzi S., Helfer A.C., Hantelys F., Pujol F., Guillermet-Guibert J., et al. . 2014. Hypoxia induces VEGF-C expression in metastatic tumor cells via a HIF-1α-independent translation-mediated mechanism. Cell Reports. 6:155–167. 10.1016/j.celrep.2013.12.011 [DOI] [PubMed] [Google Scholar]
- Nakai Y., Umeda N., Suzuki T., Nakai M., Hayashi H., Watanabe K., and Kagamiyama H.. 2004. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J. Biol. Chem. 279:12363–12368. 10.1074/jbc.M312448200 [DOI] [PubMed] [Google Scholar]
- Nakai Y., Nakai M., and Hayashi H.. 2008. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J. Biol. Chem. 283:27469–27476. 10.1074/jbc.M804043200 [DOI] [PubMed] [Google Scholar]
- Nawshad A., and Hay E.D.. 2003. TGFbeta3 signaling activates transcription of the LEF1 gene to induce epithelial mesenchymal transformation during mouse palate development. J. Cell Biol. 163:1291–1301. 10.1083/jcb.200306024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova D.D., and Leidel S.A.. 2015. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell. 161:1606–1618. 10.1016/j.cell.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A., Rosner A., Milovanovic T., Hope C., Planutis K., Saha B., Chaiwun B., Lin F., Imam S.A., Marsh J.L., and Holcombe R.F.. 2005. Wnt pathway component LEF1 mediates tumor cell invasion and is expressed in human and murine breast cancers lacking ErbB2 (her-2/neu) overexpression. Int. J. Oncol. 27:949–956. [PubMed] [Google Scholar]
- Nguyen D.X., Chiang A.C., Zhang X.H.F., Kim J.Y., Kris M.G., Ladanyi M., Gerald W.L., and Massagué J.. 2009. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 138:51–62. 10.1016/j.cell.2009.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A., Sakaguchi Y., and Suzuki T.. 2009. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 37:1335–1352. 10.1093/nar/gkn1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T., Batlle E., and Massagué J.. 2014. Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 14:306–321. 10.1016/j.stem.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G., Fellows J., Li Y., de Bizemont T., Dirac A.M., Gustafsson C.M., Erdjument-Bromage H., Tempst P., and Svejstrup J.Q.. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 3:109–118. 10.1016/S1097-2765(00)80179-3 [DOI] [PubMed] [Google Scholar]
- Pavon-Eternod M., Gomes S., Geslain R., Dai Q., Rosner M.R., and Pan T.. 2009. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 37:7268–7280. 10.1093/nar/gkp787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petz M., Them N., Huber H., Beug H., and Mikulits W.. 2012. La enhances IRES-mediated translation of laminin B1 during malignant epithelial to mesenchymal transition. Nucleic Acids Res. 40:290–302. 10.1093/nar/gkr717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privette Vinnedge L.M., McClaine R., Wagh P.K., Wikenheiser-Brokamp K.A., Waltz S.E., and Wells S.I.. 2011. The human DEK oncogene stimulates β-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene. 30:2741–2752. 10.1038/onc.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privette Vinnedge L.M., Benight N.M., and Wagh P.K.. 2014. The DEK oncogene promotes cellular proliferation through paracrine Wnt signaling in Ron receptor-positive breast cancers. Oncogene. 34:2325–2536. 10.1038/onc.2014.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva Y., Gillen K.M., Gerald W., Beggs H.E., Reichardt L.F., and Giancotti F.G.. 2009. Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J. Clin. Invest. 119:252–266. 10.1172/JCI37160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raptis L., Marcellus R., Corbley M.J., Krook A., Whitfield J., Anderson S.K., and Haliotis T.. 1991. Cellular ras gene activity is required for full neoplastic transformation by polyomavirus. J. Virol. 65:5203–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezgui V.A.N., Tyagi K., Ranjan N., Konevega A.L., Mittelstaet J., Rodnina M.V., Peter M., and Pedrioli P.G.. 2013. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl. Acad. Sci. USA. 110:12289–12294. 10.1073/pnas.1300781110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringnér M., Fredlund E., Häkkinen J., Borg Å., and Staaf J.. 2011. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 6:e17911 10.1371/journal.pone.0017911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C.D., Van der Veen A.G., Damon J.R., Spooner E., and Ploegh H.L.. 2008. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc. Natl. Acad. Sci. USA. 105:18255–18260. 10.1073/pnas.0808756105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Böhm D., von Törne C., Steiner E., Puhl A., Pilch H., Lehr H.A., Hengstler J.G., Kölbl H., and Gehrmann M.. 2008. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 68:5405–5413. 10.1158/0008-5472.CAN-07-5206 [DOI] [PubMed] [Google Scholar]
- Selvadurai K., Wang P., Seimetz J., and Huang R.H.. 2014. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem. Biol. 10:810–812. 10.1038/nchembio.1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Kokubu A., Miyamoto M., Hosoda F., Gotoh M., Tsuta K., Asamura H., Matsuno Y., Kondo T., Imoto I., et al. . 2010. DEK oncoprotein regulates transcriptional modifiers and sustains tumor initiation activity in high-grade neuroendocrine carcinoma of the lung. Oncogene. 29:4671–4681. 10.1038/onc.2010.217 [DOI] [PubMed] [Google Scholar]
- Silvera D., Formenti S.C., and Schneider R.J.. 2010. Translational control in cancer. Nat. Rev. Cancer. 10:254–266. 10.1038/nrc2824 [DOI] [PubMed] [Google Scholar]
- Soares L.M.M., Zanier K., Mackereth C., Sattler M., and Valcárcel J.. 2006. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 312:1961–1965. 10.1126/science.1128659 [DOI] [PubMed] [Google Scholar]
- Songe-Møller L., van den Born E., Leihne V., Vågbø C.B., Kristoffersen T., Krokan H.E., Kirpekar F., Falnes P.Ø., and Klungland A.. 2010. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 30:1814–1827. 10.1128/MCB.01602-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., Nordgren H., Farmer P., Praz V., Haibe-Kains B., et al. . 2006. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 98:262–272. 10.1093/jnci/djj052 [DOI] [PubMed] [Google Scholar]
- Taddei M.L., Giannoni E., Comito G., and Chiarugi P.. 2013. Microenvironment and tumor cell plasticity: an easy way out. Cancer Lett. 341:80–96. 10.1016/j.canlet.2013.01.042 [DOI] [PubMed] [Google Scholar]
- Tsai B.P., Wang X., Huang L., and Waterman M.L.. 2011. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol. Cell. Proteomics. 10:007385 10.1074/mcp.M110.007385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B.P., Jimenez J., Lim S., Fitzgerald K.D., Zhang M., Chuah C.T.H., Axelrod H., Wilson L., Ong S.T., Semler B.L., and Waterman M.L.. 2014. A novel Bcr-Abl-mTOR-eIF4A axis regulates IRES-mediated translation of LEF-1. Open Biol. 4:140180 10.1098/rsob.140180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S., and Weinberg R.A.. 2011. Tumor metastasis: molecular insights and evolving paradigms. Cell. 147:275–292. 10.1016/j.cell.2011.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmeier L., Meyer-Schaller N., Diepenbruck M., and Christofori G.. 2012. Py2T murine breast cancer cells, a versatile model of TGFβ-induced EMT in vitro and in vivo. PLoS One. 7:e48651 10.1371/journal.pone.0048651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.J., Yao Y., Jiang L.L., Hu T.H., Ma J.Q., Liao Z.J., Yao J.T., Li D.F., Wang S.H., and Nan K.J.. 2013. Knockdown of lymphoid enhancer factor 1 inhibits colon cancer progression in vitro and in vivo. PLoS One. 8:e76596 10.1371/journal.pone.0076596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M.A., Hutchinson J.N., Rauh M.J., Muthuswamy S.K., Anton M., Tortorice C.G., Cardiff R.D., Graham F.L., Hassell J.A., and Muller W.J.. 1998. Requirement for both Shc and phosphatidylinositol 3′ kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis. Mol. Cell. Biol. 18:2344–2359. 10.1128/MCB.18.4.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise-Draper T.M., Mintz-Cole R.A., Morris T.A., Simpson D.S., Wikenheiser-Brokamp K.A., Currier M.A., Cripe T.P., Grosveld G.C., and Wells S.I.. 2009. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 69:1792–1799. 10.1158/0008-5472.CAN-08-2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Tam W.L., Shibue T., Kaygusuz Y., Reinhardt F., Ng Eaton E., and Weinberg R.A.. 2015. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 525:256–260. 10.1038/nature14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B., and Gilbert W.V.. 2013. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 9:e1003675 10.1371/journal.pgen.1003675 [DOI] [PMC free article] [PubMed] [Google Scholar]