IL-6 promotes the differentiation of a subset of naïve CD8+ T cells into IL-21–producing B helper CD8+ T cells.
Abstract
IL-6 is known to contribute to the differentiation of CD4+ T cells into different subsets of effector T helper cells. Less is known about the potential of IL-6 in regulating CD8+ T cell effector function. Here, we identify IL-6 as a master regulator of IL-21 in effector CD8+ T cells. IL-6 promotes the differentiation of a subset of naive CD8+ T cells that express IL-6R into a unique population of effector CD8+ T cells characterized by the production of high levels of IL-21 and low levels of IFN-γ. Similar to CD4+ T follicular helper (Tfh) cells, IL-21–producing CD8+ T cells generated in the presence of IL-6 directly provide help to B cells to induce isotype switching. CD8+ T cell–derived IL-21 contributes to the production of protective virus-specific IgG antibodies during influenza virus infection. Thus, this study reveals the presence of a new mechanism by which IL-6 regulates antibody production during viral infection, and a novel function of effector CD8+ T cells in the protection against viruses.
INTRODUCTION
IL-6 is a proinflammatory cytokine produced by multiple cell types in response to external stimuli, including trauma, stress, and infection (Kishimoto, 2005). IL-6 plays a crucial role in regulating CD4+ Th cell differentiation and effector functions (Dienz and Rincon, 2009). It enhances Th2 differentiation through an autofeedback by up-regulating IL-4 production (Diehl et al., 2002). IL-6 also inhibits IFN-γ production and Th1 differentiation through an independent mechanism (Diehl et al., 2000). In combination with TGF-β, IL-6 contributes to the differentiation of Th17 cells (Bettelli et al., 2006; Ivanov et al., 2006). Importantly, IL-6 by itself also induces IL-21 production in CD4+ T cells (Suto et al., 2008; Dienz et al., 2009; Diehl et al., 2012) and is required for the generation of T follicular helper (Tfh) cells (Nurieva et al., 2008). IL-6 indirectly promotes the production of antibodies by B cells by acting on CD4+ Tfh cells through the production of IL-21 (Dienz et al., 2009).
In contrast to CD4+ T cells, little is known about the potential effect of IL-6 on CD8+ T cells. Effector CD8+ T cells are high producers of IFN-γ and are also cytotoxic through the production of Granzyme and perforin, the two major functions by which these cells protect from virus infections (Russell and Ley, 2002). However, CD8+ Tc2 and Tc17 subsets have also been identified when placed in a complex cytokine environment (Croft et al., 1994; Hamada et al., 2009). No effect of IL-6 on Tc2 has been reported. Similar to CD4+ Th17 cells, IL-6 in combination with multiple other cytokines contributes to the generation of CD8+ Tc17 cells (Hamada et al., 2009). Tc17 cells play an important role in protecting against lethal influenza infection (Hamada et al., 2009). Indirect evidence through the use of class I–deficient mice suggested that CD8+ T cells may provide help for IgG production by B cells (Spriggs et al., 1992; Christianson et al., 1997). IL-4–producing CD8+ T cell clones have also been shown to promote B cell antibody production in vitro (Cronin et al., 1995). However, there is no direct evidence that CD8+ T cells promote antibody production.
Here, we show that IL-6 alone induces the differentiation of CD8+ T cells into IL-21–producing cells that provide B cell help to promote antibody production. Furthermore, IL-21 production by effector CD8+ T cells is required for an antibody response to influenza virus. Thus, through the IL-6–IL-21 axis, CD8+ T cells emerge as regulators of the antiviral antibody response.
RESULTS AND DISCUSSION
IL-6 induces the production of IL-21 in CD8+ T cells through Stat3
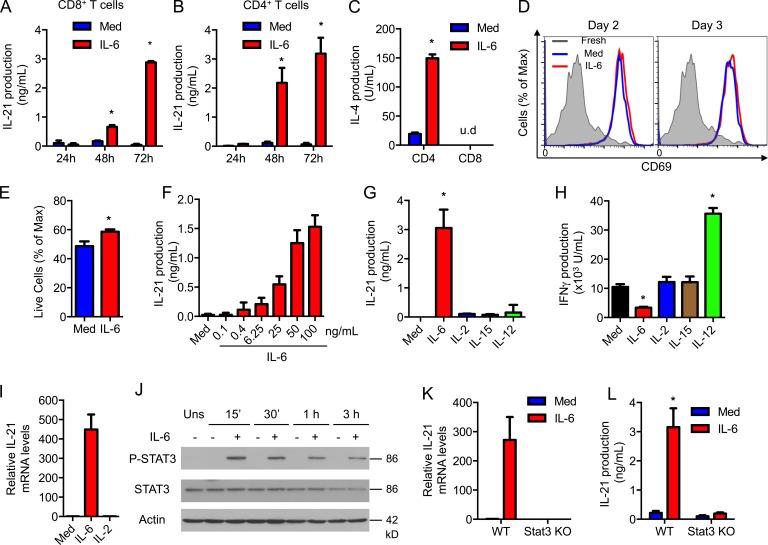
IL-6 is known to be major inducer of IL-21 in CD4+ T cells (Suto et al., 2008; Dienz et al., 2009; Diehl et al., 2012), but no previous studies have reported the effect of IL-6 on CD8+ T cells. To determine whether CD8+ T cells also produce IL-21 in response to IL-6, CD8+ T cells were activated with anti-CD3 and -CD28 antibodies in the presence or absence of IL-6 for different periods of time. High levels of IL-21 were produced only by CD8+ T cells activated in the presence of IL-6 (Fig. 1 A). The IL-21 levels induced by IL-6 in CD8+ T cells were comparable to those produced by CD4+ T cells (Fig. 1 B). We have shown that IL-6 can also promote the production of IL-4 during activation in CD4+ T cells (Diehl et al., 2002). However, IL-6 failed to induce IL-4 in CD8+ T cells (Fig. 1 C). In addition, IL-6 had no effect on the expression of activation markers, such as CD69 (Fig. 1 D), or cell proliferation (not depicted), and had only a marginal effect on cell survival (Fig. 1 E) of CD8+ T cells during activation. Together, these results indicate a selective effect of IL-6 on IL-21 production. The induction of IL-21 by IL-6 during activation of CD8+ T cells was dose dependent, and a low dose of IL-6 was sufficient to trigger IL-21 production in CD8+ T cells (Fig. 1 F), demonstrating the efficacy of IL-6 in inducing IL-21 production by CD8+ T cells.
Figure 1.
IL-6 induces the production of IL-21 in CD8+ T cells through Stat3. (A and B) CD8+ and CD4+ T cells were activated using anti-CD3/CD28 Abs in the absence or presence of IL-6. IL-21 production was determined by ELISA (n = 3). (C) IL-4 production after activation for 72 h (n = 3). (D) CD69 expression in fresh CD8+ T cells (shaded) or activated as in A in the absence (blue) or presence (red) of IL-6. (E) Percentage of live CD8+ T cells activated as in A for 2 d by flow cytometry (n = 3). (F) CD8+ T cells were activated as in A for 72 h with or without IL-6, as indicated. IL-21 production was determined (n = 3). (G and H) CD8+ T cells were activated in the absence (Med) or presence of cytokines. IL-21 (G) and IFN-γ (H) production were determined (n = 3). (I) Relative mRNA levels for IL-21 in CD8+ T cells activated with or without IL-6 or IL-2. (J) The levels of Tyr705-phosphorylated STAT3 (P-STAT3), total STAT3, or actin in CD8+ T cells were activated in the absence or presence of IL-6 for 0 min (Uns), 15 min, 30 min, 1 h, or 3 h. (K and L) Relative IL-21 mRNA levels (K) and IL-21 production (L) in WT or Stat3 T cell conditional KO (Stat3 KO) CD8+ T cells activated with or without IL-6 for 72 h (n = 3). Error bars represent the mean ± SD. *, P < 0.05, as determined by Student’s t test (E), one-way (F–H) or two-way ANOVA (A, B, C, and L). u.d., undetectable levels. Results are representative of two to three experiments.
Although IL-2 and -15 are critical regulators of CD8+ T cell proliferation, survival, and differentiation (Waldmann, 2006), neither IL-2 nor -15 induced IL-21 production in CD8+ T cells (Fig. 1 G). Similarly, IL-12 did not trigger the production of IL-21 (Fig. 1 G), but it enhanced the production of IFN-γ (Fig. 1 H). In contrast to IL-21, IL-6 reduced IFN-γ production (Fig. 1 H). None of the combinations of IL-2, IL-15, and IL-12 induced IL-21 production in CD8+ T cells (unpublished data). Thus, induction of IL-21 production in CD8+ T cells during activation is relatively selective to IL-6.
To examine whether IL-21 production in CD8+ T cells by IL-6 was caused by the induction of IL-21 gene expression, Il21 mRNA levels were measured by real-time RT-PCR. The levels of IL-21 mRNA were markedly higher in CD8+ T cells activated in the presence of IL-6 (Fig. 1 I). Stat3 is a transcription factor required for Il21 gene expression in CD4+ T cells (Nurieva et al., 2007; Ray et al., 2014). Analysis of Stat3 phosphorylation by Western blot showed a clear activation of Stat3 by IL-6 in CD8+ T cells (Fig. 1 J). We examined Il21 expression in CD8+ T cells from WT and T cell conditional Stat3 knockout (Stat3 KO) mice and found no Il21 mRNA in Stat3 KO CD8+ T cells activated in the presence of IL-6 (Fig. 1 K). Accordingly, IL-6 failed to induce IL-21 production in Stat3-deficient CD8+ T cells (Fig. 1 L). Thus, IL-6 is a selective and powerful inducer of IL-21 in CD8+ T cells via a Stat3-dependent mechanism.
IL-6 differentiates CD8+ T cells into IL-21–producing effector cells
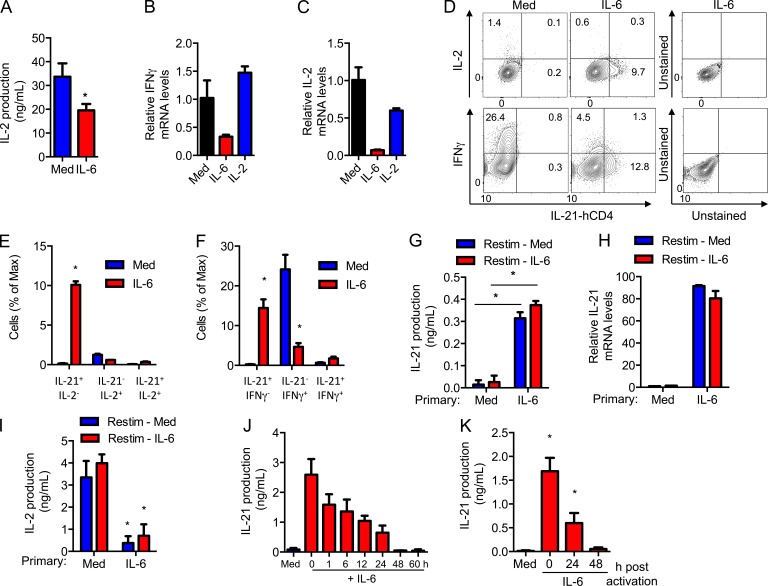
The aforementioned data show that high levels of IL-21, but low levels of IFN-γ, are produced by CD8+ T cells activated with IL-6. The levels of IL-2 produced by CD8+ T cells activated with IL-6 were also lower (Fig. 2 A). IL-6 also reduced the levels of Ifng (Fig. 2 B) and Il2 mRNA (Fig. 2 C), indicating that this effect is not caused by increased consumption. Thus, although IL-6 induces IL-21, it also has a negative effect on the expression of IFN-γ and IL-2 in CD8+ T cells.
Figure 2.
IL-6 reprograms effector CD8+ T cells into IL-21–producing effector cells. (A) IL-2 production from CD8+ T cells activated using anti-CD3/CD28 Abs in the absence or presence of IL-6 for 72 h (n = 3). (B and C) Relative mRNA levels for IFN-γ (B) and IL-2 (C) in CD8+ T cells activated with or without IL-6 or IL-2. (D) Intracellular staining for IFN-γ and IL-2, and surface staining for hCD4 (surrogate for IL-21) on activated CD8+ IL-21-hCD4 reporter T cells. Numbers indicate the percentage of cells in each quadrant. (E and F) Percentage of different cytokine-producing populations as determined in D (n = 3). (G–I) CD8+ T cells were activated in the absence (primary, Med) or presence (primary, IL-6) of IL-6 for 2 d, harvested, and restimulated using anti-CD3 Ab in the absence (Restim, Med) or presence (Restim, IL-6) of IL-6. After 24 h, IL-21 production (G), the relative IL-21 mRNA levels (H), and IL-2 production (I) were determined (n = 3). (J and K) CD8+ T cells were activated with or without IL-6 that was added at the indicated time after activation. After 72 h of activation (J) and 72 h after the addition of IL-6 (K), IL-21 production was determined (n = 3). Error bars represent the mean ± SD. *, P < 0.05, as determined by Student’s t test (A), one-way ANOVA (B, C, J, and K), or two-way ANOVA (E–I). Results are representative of two to three experiments.
These results suggested that IL-6 could promote the differentiation of CD8+ T cells into effector CD8+ T cells characterized by high IL-21, low IFN-γ, and low IL-2 production. We therefore performed IL-2 and IFN-γ intracellular staining in CD8+ T cells from IL-21-reporter mice expressing a truncated human CD4 (hCD4) in the Il21 locus (unpublished data). No hCD4 was detected in CD8+ T cells activated in the absence of IL-6 (Fig. 2 D). However, a subset of hCD4 (IL-21)-expressing CD8+ T cells was present when cells were activated with IL-6 (Fig. 2 D). The percentage of IFN-γ– and IL-2–producing cells were markedly reduced in CD8+ T cells activated with IL-6 (Fig. 2 D). Importantly, most IL-21–producing cells did not make IL-2 (Fig. 2 E) or IFN-γ (Fig. 2 F). Thus, IL-6 promotes the generation of an effector CD8+ T cell subpopulation that produces high levels of IL-21, but minimal levels of IFN-γ and IL-2.
We then investigated whether this IL-21–producing subset of effector CD8+ T cells represented a relatively committed subset. CD8+ T cells were activated for 2 d with anti-CD3/CD28 antibodies (Abs) in the presence or absence of IL-6 (primary activation), extensively washed, and an equal number of cells was then restimulated with anti-CD3 Ab only or anti-CD3 Ab plus IL-6. IL-21 levels were determined in the supernatant 24 h later. CD8+ T cells where primary activation was done in the absence of IL-6 did not produce IL-21 after restimulation even when IL-6 was added during the restimulation phase (Fig. 2 G). In contrast, high levels of IL-21 were induced after restimulation with anti-CD3 Abs alone if effector CD8+ T cells were initially activated in the presence of IL-6 (Fig. 2 G). Similar results were obtained when Il21 mRNA levels were assayed (Fig. 2 H), indicating that the absence of IL-21 was not caused by increased consumption. Moreover, CD8+ T cells that received IL-6 during the primary activation still produced low levels of IL-2 after restimulation (Fig. 2 I). These data show that the presence of IL-6 during the primary activation promotes the differentiation of CD8+ T cells into a unique effector CD8+ T cell subset that remains capable of producing IL-21 upon activation.
To better dissect the period of time during activation when CD8+ T cells remain responsive to IL-6 and produce IL-21, CD8+ T cells were activated with anti-CD3/CD28 Abs, and IL-6 was administered at different periods of time after stimulation. IL-21 levels were measured 3 d after the stimulation. The maximum production of IL-21 was achieved when IL-6 was added at the initiation of stimulation (Fig. 2 J). Delayed addition of IL-6 resulted in progressively decreased production of IL-21 by CD8+ T cells (Fig. 2 J). Addition of IL-6 48 h after activation did not induce IL-21 production when examined a day later (Fig. 2 J) or even when cells were incubated for 3 additional days after addition of IL-6 (Fig. 2 K). Thus, the differentiation of CD8+ T cells into IL-21–producing effector cells requires IL-6 signaling early during differentiation.
IL-6R defines a CD8+ T cell subset capable of producing IL-21
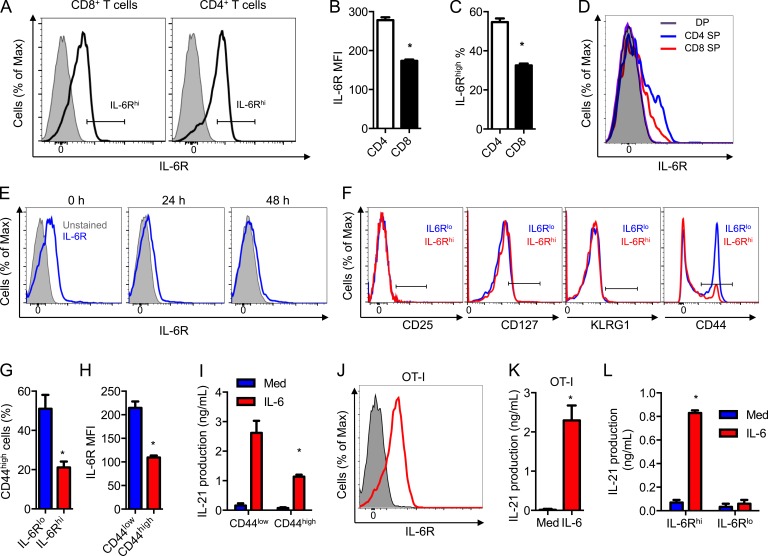
IL-6 induces signals by binding to its membrane IL-6R, which then associates with gp130, a common signal transducer that activates the Jak/Stat3 pathway (Heinrich et al., 2003). To determine the fraction of CD8+ T cells that could produce IL-21 by responding to IL-6, we analyzed the cell surface levels of IL-6R by flow cytometry. Only a subset of CD8+ T cells had detectable levels of IL-6R (IL-6Rhigh; Fig. 3 A). The levels of surface expression of IL-6R in CD8+ T cells were lower than in CD4+ T cells, both in intensity (Fig. 3 B) and frequency of expressing cells (Fig. 3 C). Analysis of IL-6R expression during thymocyte development showed that single-positive CD4+ thymocytes already expressed higher levels of IL-6R than single-positive CD8+ thymocytes (Fig. 3 D), whereas no IL-6R expression could be found in double-positive thymocytes (Fig. 3 D). Expression of IL-6R on the cell surface has been shown to be down-regulated in CD4+ T cells during activation (Rincón et al., 1997). Analysis of IL-6R during activation of CD8+ T cells also showed a down-regulation (Fig. 3 E). Down-regulation of IL-6R with activation correlates with the failure of IL-6 to induce Stat3 phosphorylation (unpublished data) and IL-21 production (Fig. 2, J and K) in CD8+ T cells when administrated later during activation.
Figure 3.
IL-6R defines a CD8+ T cell subset capable of producing IL-21. (A) IL-6R expression in fresh CD4+ and CD8+ T cells. IL-6R KO T cells were used as negative controls (shaded). The gate defines the IL-6Rhigh (IL-6Rhi) population. (B and C) IL-6R MFI (B) and percentage of IL-6Rhigh (C) as in A (n = 3). (D) IL-6R expression on CD4/CD8 double positive (DP) and single CD4- or CD8-positive (CD4 SP or CD8 SP). Unstained DP thymocytes were used as negative control (shaded). (E) IL-6R expression on fresh CD8+ T cells (0 h) or after activation for the indicated periods of time. The unstained cells were used as negative controls (shaded). (F) Expression of CD25, KLRG1, CD127, and CD44 on IL-6Rhigh (IL-6Rhi) and IL-6Rlow (IL-6Rlo) CD8+ T cells. The gate based on an unstained negative control is shown. (G) Percentage of CD44high cells as gated in F (n = 3). (H) IL-6R MFI on CD44high and CD44low CD8+ T cells (n = 3). (I) FACS sorted CD44low and CD44high CD8+ T cells were activated in the absence or presence of IL-6. IL-21 production (72 h) was determined (n = 3). (J) Expression of IL-6R on OT-I CD8+ T cells (shaded histogram shows unstained cells). (K) IL-21 production from OT-I CD8+ T cells activated in the absence or presence of IL-6 for 72 h (n = 3). (L) FACS sorted IL-6Rhi and IL-6Rlo CD8+ T cells were activated in the absence or presence of IL-6. IL-21 production after 72 h was determined (n = 3). Error bars represent mean ± SD. *, P < 0.05, as determined by Student’s t test (B, C, G, H, and K) and two-way ANOVA (I and L). Results are representative of two to three experiments.
We performed a phenotypic analysis of the CD8+ T cell subset expressing detectable IL-6R (IL-6Rhi) before activation. No CD25 (IL-2Rα) expression could be detected in either the IL-6Rhi or IL-6Rlo subsets (Fig. 3 F). Only minimal KLRG1 (short-lived effector cell marker) and low levels of CD127 (IL-7R) were detected, but there was no difference between the two subsets (Fig. 3 F). However, the levels of CD44 were different between IL-6Rhi and IL-6Rlo CD8+ T cell subsets (Fig. 3 F). The IL-6Rhi CD8+ T cell subset contained a lower frequency of cells expressing high levels of CD44 (CD44high; Fig. 3 G). Similarly, analysis of IL-6R expression in gated CD44low (naive) and CD44high (memory/activated) showed higher expression of IL-6R in naive CD8+ T cells (Fig. 3 H). Accordingly, analysis of IL-21 production in FACS-sorted CD44high and CD44low CD8+ T cells showed higher levels of IL-21 in CD44low CD8+ T cells (Fig. 3 I). To further confirm the production of IL-21 in bona fide naive CD8+ T cells, we used CD8+ T cells from OT-I mice that express a TCR recognizing OVA. Similar to WT CD8+ T cells, IL-6R was expressed on most OT-I CD8+ T cells (Fig. 3 J). Whereas no IL-21 was detected in the absence of IL-6, OT-I CD8+ T cells activated with IL-6 produced high levels of IL-21 (Fig. 3 K). Thus, naive CD8+ T cells express IL-6R and can produce high levels of IL-21 during activation when IL-6 is present.
To determine whether the cell surface expression of IL-6R could be used as a marker to identify the subset of CD8+ T cells that can differentiate into IL-21–producing cells, IL-6Rhi and IL-6Rlo CD8+ T cells were FACS sorted and activated with or without IL-6. IL-6Rhi CD8+ T cells produced high levels of IL-21 in response to IL-6 (Fig. 3 L), whereas IL-21 was almost undetectable in IL-6Rlo CD8+ T cells (Fig. 3 L). Thus, the expression of IL-6R defines the subset of CD8+ T cells capable of producing IL-21.
IL-21–producing CD8+ T cells promote IgG isotype switching in B cells in vitro
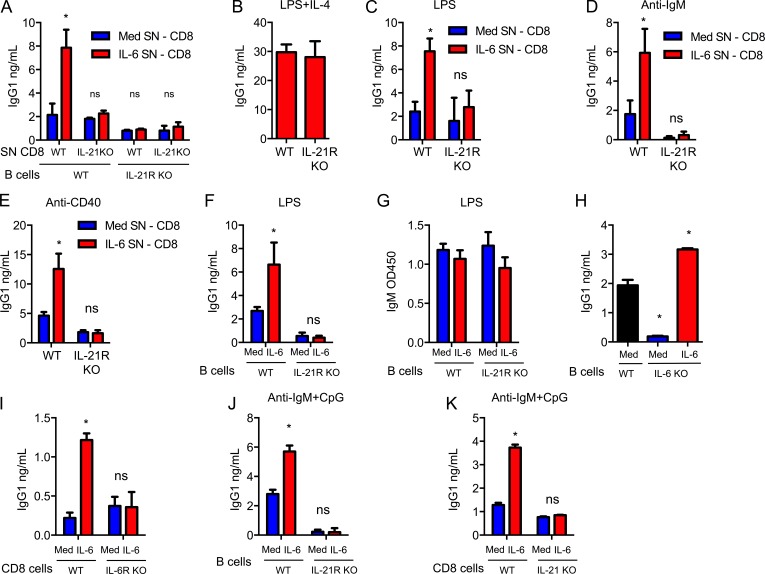
IL-21 has an essential role in the B cell antibody responses (Ozaki et al., 2002), as it induces isotype switching to IgG isotypes, germinal center (GC) formation, and plasma cell differentiation (Ettinger et al., 2005; Linterman et al., 2010). IL-21 is produced primarily by CD4+ Tfh cells (Linterman et al., 2010). We investigated whether CD8+ T cells could also acquire a B cell helper function through IL-21. WT and IL-21R KO B cells were activated with LPS and an anti-CD40 Ab in the absence or presence of supernatant from WT or IL-21 KO CD8+ T cells previously activated with or without IL-6. Culture supernatants from WT CD8+ T cells activated with IL-6 significantly increased the production of IgG1 in WT B cells (Fig. 4 A). In contrast, supernatants from IL-21 KO CD8+ T cells had no effect on IgG1 production (Fig. 4 A). Supernatants from WT CD8+ T cells activated with IL-6 had no effect on IgG1 production by IL-21R KO B cells (Fig. 4 A), although these B cells still could produce IgG1 in response to IL-4 (Fig. 4 B). Similar results were found when B cells were activated with LPS alone (Fig. 4 C), an anti-IgM Ab alone (Fig. 4 D), or an anti-CD40 Ab alone (Fig. 4 E). Thus, IL-21 produced by CD8+ T cells in vitro is sufficient to promote IgG1 production by B cells when activated through different pathways.
Figure 4.
IL-6 reprograms CD8+ T cells to become B cell helpers through their production of IL-21 in vitro. (A) Culture supernatants (SN) were collected from WT and IL-21 KO CD8+ T cells activated in absence (Med SN, CD8) or presence (IL-6 SN, CD8) of IL-6 for 3 d. IgG1 production by WT and IL-21R KO B cells activated for 6 d with LPS and anti-CD40 Abs using these SN was determined (n = 3). (B) WT or IL-21R KO B cells were activated using LPS+IL-4 for 6 d in SN from WT CD8+ T cells activated for 3 d in the presence of IL-6. IgG1 production was determined (n = 3). (C–E) WT or IL-21R KO B cells were activated using LPS (C), anti-IgM Ab (D), or anti-CD40 Ab (E) in the presence of SN obtained from WT CD8+ T cells activated as in A. After 6 d, IgG1 production was determined (n = 3). (F and G) In co-culture system, WT or IL-21R KO B cells were activated with LPS while WT CD8+ T cells were activated using anti-CD3/CD28 Abs with or without IL-6. After 6 d, the levels of IgG1 (F) and IgM (G) in the co-cultures were determined (n = 3). (H) IgG1 levels in 6 d co-cultures of WT or IL-6 KO B cells with WT CD8+ T cells activated as in F (n = 3). (I) IgG1 levels in 6-d co-cultures of IL-6 KO B cells with WT or IL-6R KO CD8+ T cells activated as in F (n = 3). (J) Co-cultures of B cells and CD8+ T cells were established as described in F, but a combination of an anti-IgM Ab and CpG was used to activate B cells. IgG1 levels in the co-cultures were determined (n = 3). (K) IgG1 levels in 6 d co-cultures of WT B cells with WT or IL-21 KO CD8+ T cells activated as in J (n = 3). Error bars represent the mean ± SD. *, P < 0.05, as determined by Student’s t test (B), one-way ANOVA (H), or two-way ANOVA test (A, C, D, E–G, and I–K). Results are representative of two to three experiments.
We then established co-cultures of CD8+ T cells with B cells where CD8+ T cells were activated with anti-CD3/CD28 Abs with or without IL-6, and B cells with LPS. WT B cells, but not IL-21R KO B cells, co-cultured with CD8+ T cells in the presence of IL-6 produced higher levels of IgG1 than those co-cultured in the absence of IL-6 (Fig. 4 F), indicating that the effect of IL-6 on IgG1 production was dependent on IL-21 acting on B cells. IL-21 has no effect on the production of IgM (Ozaki et al., 2002). Accordingly, IgM levels were comparable between WT and IL-21R KO B cells co-cultured with CD8+ T cells, either with or without IL-6 (Fig. 4 G), showing that IL-21R KO and WT B cells are equally activated. In the absence of IL-6, the baseline IgG1 produced in co-cultures of IL-21R KO B cells was lower than in co-cultures with WT B cells (Fig. 4 F). Because TLR ligands can trigger IL-6 production in B cells (Barr et al., 2007), this could be a result of endogenous B cell–derived IL-6 inducing IL-21 in CD8+ T cells. To address this question, we established co-cultures using WT and IL-6 KO B cells with WT CD8+ T cells. The levels of IgG1 were practically undetectable in co-cultures containing IL-6 KO B cells (Fig. 4 H). Addition of exogenous IL-6 to those co-cultures was able to restore the effect of CD8+ T cells in promoting antibody production by B cells (Fig. 4 H). Moreover, addition of IL-6 to co-cultures with IL-6R KO CD8+ T cells failed to promote the production of IgG1 (Fig. 4 I). In addition to TLR4 (LPS receptor), CD8+ T cells also enhanced IgG isotype switching in B cells activated with an anti-IgM Ab and the TLR9 ligand unmethylated CpG when IL-6 is present (Fig. 4 J), and this help was dependent on their ability to produce IL-21 (Fig. 4 K). Together, these results demonstrate that CD8+ T cells activated in the presence of IL-6 promote IgG isotype switching of B cells in vitro through their ability to produce IL-21.
IL-21–producing CD8+ T cells promote IgG isotype switching in B cells in vivo during influenza infection
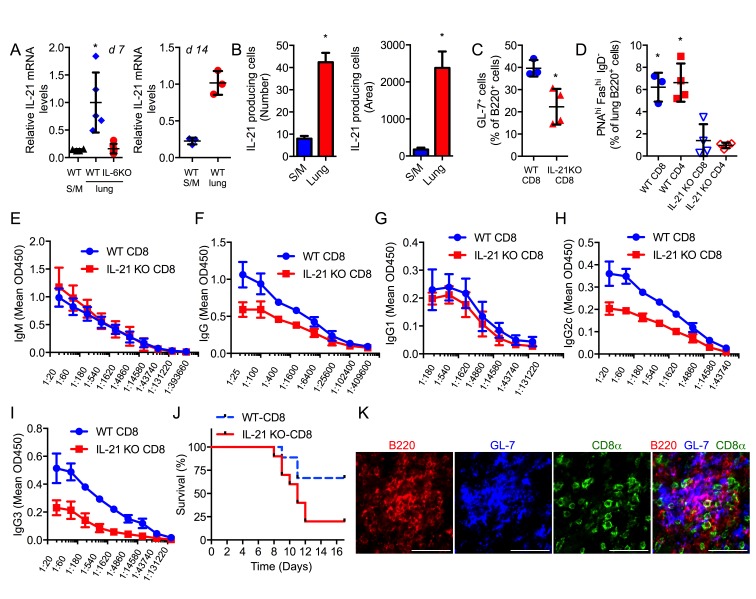
To determine whether CD8+ T cells can act as helper for B cells in vivo during virus infection, we first investigated whether CD8+ T cells produce IL-21 in vivo. WT mice and IL-6 KO mice were i.n. infected with a sublethal dose of the PR8 influenza A virus. Il21 gene expression was nearly undetectable in CD8+ T cells isolated from the spleen and mediastinal lymph node (MLN) 7 d after infection (Fig. 5 A). In contrast, lung CD8+ T cells from infected WT mice expressed high levels of IL-21 (Fig. 5 A). Il21 expression was almost undetectable in CD8+ T cells from lungs of infected IL-6 KO mice indicating that IL-21 expression is dependent on IL-6 (Fig. 5 A). Similar results were found 14 d after infection (Fig. 5 A). In addition, ELISPOT assay further confirmed higher levels of IL-21 being produced by CD8+ T cells from the lungs than from spleen/MLN (Fig. 5 B). Thus, similar to the in vitro studies, during influenza infection in vivo, CD8+ T cells recruited to the lungs produce IL-21, but only if IL-6 is present.
Figure 5.
IL-21–producing CD8+ T cells promote isotype switching in B cells in vivo during influenza virus infection. (A) WT or IL-6 KO mice were infected with a sublethal dose of H1N1 PR8 virus. 7 d (d 7) or 14 d (d 14) after infection, relative IL-21 mRNA levels in CD8+ T cells from lung or spleen/MLN (S/M) were determined. For 7 d, WT and IL-6 KO lungs, n = 5 and WT S/M, n = 3; for 14 d, WT lungs and S/M, n = 3. (B) CD8+ T cells from lungs or S/M of mice infected with influenza virus, as in A, for 14 d were assayed for the number of IL-21–producing cells (number of spots) and the total area of spots (area) per well by ELISPOT assay (n = 3). (C) IL-21 KO mice as host mice received WT CD8+ or IL-21 KO CD8+ T cells and were infected with a sublethal dose of H1N1 PR8 virus. After 14 d, lung tissue was immunostained and imaged. Percentage of GL-7+ cells relative to the number of B220+ cells in lungs of these mice was determined. (D) Percentage of GC B cells (PNAhi FAShi IgD−) within the B220 population in lungs from influenza virus infected (day 14) IL-21 KO mice that had received WT CD8+, WT CD4+, IL-21 KO CD8+, or IL-21 KO CD4+ T cells (n = 4). (E–I) Sera from IL-21 KO recipients as described in C were analyzed for influenza-specific IgM (E; P = 0.5703), IgG (F; P = 0.0012), IgG1 (G; P = 0.2278), IgG2c (H; P < 0.0001), and IgG3 (I; P = 0.0002) levels by ELISA. (J) Sera from IL-21 KO recipients as described in C were i.p. transferred to WT mice 1 d after infection with a lethal dose of influenza virus. (WT-CD8, blue, n = 9; IL-21 KO-CD8, red, n = 10). Kaplan-Meier survival curve is shown. P = 0.05. (K) Confocal microscopy images of lungs from influenza virus–infected WT mice (day 12) showing the staining for B220, GL-7, and CD8 individually or as a merged image in lymphoid structures. Bars, 50 µm. Error bars represent the mean ± SD. *, P < 0.05, as determined by Student’s t test. (B and C) One-way ANOVA (A and D), two-way ANOVA (E–I), or log-rank test (J). Results are representative of two to three experiments.
IgG antibody response against influenza virus is strongly attenuated in IL-21 KO mice, whereas IgM antibody response is not affected (Dienz et al., 2009; Rasheed et al., 2013). To investigate whether IL-21–producing CD8+ T cells could restore antibody response, we performed adoptive transfer of WT or IL-21 KO CD8+ T cells into IL-21 KO mice. Recipient mice were then infected with influenza virus. 2 wk later, activated germinal center (GC) B cells (Fig. S1, A and B) from the spleen and lung were analyzed. There was no difference in the frequency of activated GC B cells in the spleen (unpublished data), but there was an increased frequency of activated GC B cells in lungs of IL-21 KO mice reconstituted with WT CD8+ T cells (Fig. 5 C). To further demonstrate the contribution of IL-21 derived from CD8+ T cells compared with CD4+ T cells to the development of lung GC B cells, similar adoptive transfer experiments were performed with WT or IL-21 KO CD4+ and CD8+ T cells. Analysis of GC B cells in the lungs of infected mice by flow cytometry (Fig. S1 C) showed that WT CD8+ T cells were as efficient in promoting GC B cells in the lung as WT CD4+ T cells (Fig. 5 D). Together, these data show the relevant role that CD8+ T cells have on GC B cells in the lung through IL-21 production during influenza virus infection.
We also measured the levels of PR8 virus-specific antibodies in serum using ELISA. As expected, IgM response to influenza virus was comparable between the two groups of IL-21 KO recipient mice (Fig. 5 E). In contrast, higher levels of PR8-specific total IgG were found in IL-21 KO mice that received WT CD8+ T cells (Fig. 5 F). Analysis of the different virus-specific IgG isotypes showed no statistically significant difference in IgG1 titers (Fig. 5 G). However, the levels of virus-specific IgG2c (Fig. 5 H) and IgG3 (Fig. 5 I) were markedly decreased in mice that received IL-21 KO CD8+ T cells. Importantly, serum transfer experiments into WT mice showed that the sera from IL-21 KO mice that had received WT CD8+ T cells conferred more protection against a lethal dose of influenza virus than the sera from IL-21 KO mice that had received IL-21 KO CD8+ T cells (Fig. 5 J). Thus, CD8+ T cells contribute to the shaping of antibody response against influenza virus and antibody-mediated protection in virus infection through their ability to produce IL-21.
The presence of inducible bronchus associated lymphoid tissue has been reported as a secondary lymphoid tissue in the lungs of influenza virus infected mice (Moyron-Quiroz et al., 2004). Interestingly, immunostaining analysis in influenza virus infected lungs also showed the presence of CD8+ T cells in close contact with B cells and GC B cells (GL-7 cells) in these lymphoid structures (Fig. 5 K), further supporting that CD8+ T cells can provide help to B cells in the lung.
In summary, our studies here reveal that CD8+ T cells can produce high levels of IL-21 when IL-6 is present during their activation both in vitro and in vivo. More importantly, in addition to their known cytotoxic effector functions, here we demonstrate a novel function of CD8+ T cells as helpers of B cells in antibody production.
Although B cells are most commonly activated by antigen in the presence of CD40 ligand provided by activated CD4+ T cells, neither CD4+ T cells or CD40/CD40L are essential for Ab production and virus clearance during influenza virus infection (Lee et al., 2005). B cells can also be activated directly by different TLR agonists independently of CD40 (Genestier et al., 2007; Heer et al., 2007; Simchoni and Cunningham-Rundles, 2015). However, although TLRs can synergize with BCR for activation of B cells, TLR signaling is not highly efficient in inducing Ig class switching without additional T cell–derived cytokines. Here, we reveal that CD8+ T cells can promote Ig class switching and production of antibodies in B cells activated by TLR signals through the production of IL-21 when IL-6 is present.
Interestingly, our studies here in influenza virus infection also reveal that CD8+ T cells in the lung, but not CD8+ T cells from spleen and lymph nodes, produce IL-21. This raises the possibility that CD8+ T cells act as B cell helpers primarily in the nonlymphoid tissues where IL-6 production is high, although CD4+ Tfh cells are the predominant B cell helpers in spleen or lymph node follicles. Thus, lung B cells stimulated through specific TLRs may receive help from IL-21–producing CD8+ T cells activated in the lung through virus recognition on lung epithelial cells. IL-21–producing CD8+ T cells present in tissues with chronic inflammation may also provide help to the inflammatory B cells in these tissues (e.g., kidney in lupus). Overall, our studies provide new insight for the role of CD8+ T cells in the immune response and they can have a major clinical impact in the vaccine development and cancer immunology fields.
MATERIALS AND METHODS
Mice and influenza virus infection
Null IL-6 and IL-21 KO mice were previously described (Poli et al., 1994; Dienz et al., 2009). IL-21 hCD4 reporter mice were generated by C. Weaver and D. Randolph (University of Alabama at Birmingham, Birmingham, AL). T cell specific–Stat3 conditional knockout (Stat3 KO) mice were generated by crossing the homozygous floxed Stat3 mice (Stat3loxp/loxp; Takeda et al., 1998) with T cell–specific Lck-Cre transgenic [B6.Cg-Tg(Lck-cre)1Cwi N9] mice (Lee et al., 2001). OT-I TCR transgenic mice were previously described (Hogquist et al., 1994). All the mice were fully backcrossed into the C57BL/6J background. WT C57BL/6J mice were purchased from Jackson ImmunoResearch Laboratories.
For adoptive transfer experiments, 12 × 106 WT or IL-21 KO CD8+ T cells were adoptively transferred to IL-21 KO recipient mice. For influenza A virus infection, mice were infected i.n. with sublethal doses (3 × 103 EIU) of Puerto Rico A/PR/8/34 H1N1 (PR8) influenza A virus as previously described (Dienz et al., 2012). Mice were euthanized 7 or 14 d after infection for real-time RT-PCR analysis and 13 d after infection for PR8-specific antibody analysis. For analysis of serum-mediated protection, 13 d after sublethal influenza virus infection of IL-21 KO mice that had received WT or IL-21 KO CD8+ T cells, sera were harvested. Sera were i.p. administered (30 µl) into WT mice 1 d after infection with a lethal dose (104 EIU) of Puerto Rico A/PR/8/34 H1N1 (PR8) influenza A virus. Weight loss and survival were followed up for 17 d after infection. All mice were housed under sterile conditions at the Association for Assessment and Accreditation of Laboratory Animal Care–approved animal care facility at the University of Vermont. All procedures performed on the mice were approved by the University of Vermont Institutional Animal Care and Use Committee.
Cell purification and activation in vitro
In most experiments, CD8+ T cells were isolated from spleen and lymph nodes by negative selection, as previously described (Diehl et al., 2002). CD8+ T cells isolated by negative selection are typically >85% in purity and have <2% CD4+ T cell contaminants. CD8+ T cells from Stat3 conditional KO mice and IL-21 reporter mice and their corresponding WT controls, as well as CD8+ T cells from WT and IL-21 KO mice for adoptive transfer experiments, were purified by positive selection using anti-CD8 magnetic microbeads (Miltenyi Biotec). Positive selection resulted in >92% CD8+ T cell purity, with <0.5% CD4+ T cell contamination. CD8+ T cells from the lung were isolated using the lung dissociation kit (Miltenyi Biotec) as recommended by the manufacturer, followed by positive selection (Miltenyi Biotec). B cells from WT or IL-21R KO mice were purified by positive selection (Miltenyi Biotec; typically >92% in purity). IL-6Rhi and IL-6Rlo CD8+ T cells, and CD44high and CD44low CD8+ T cells, were isolated by immunostaining and cell sorting (FACSAria; BD). IL-6R KO cells stained with anti–IL-6R Abs were used as the negative control for IL-6Rhi and IL-6Rlo CD8+ T cell gating.
CD8+ T cells were activated with plate-bound anti-CD3 (145-2C11; 5 µg/ml) and soluble anti-CD28 (1 µg/ml; BioXCell) Abs in the presence or absence of IL-6 (50 ng/ml; Miltenyi Biotec), except where indicated. Human recombinant IL-2 (50 U/ml; BioLegend), mouse recombinant IL-12 (5 ng/ml; Amgen), and IL-15 (20 ng/ml; Amgen) were used. B cells were activated as listed below: first, LPS (1 µg/ml; Sigma-Aldrich); second, anti-IgM F(ab’)2 (10 µg/ml; Jackson ImmunoResearch Laboratories) alone; third, anti-CD40 Ab (1 µg/ml; BD) alone; fourth, LPS (1 µg/ml) and IL-4 (100 U/ml); fifth anti-CD40 Ab (1 µg/ml) with LPS (1 µg/ml); and sixth, anti-IgM F(ab’)2 (10 µg/ml) with CpG (1 µg/ml; Sigma-Aldrich).
Flow cytometry analysis
For IL-6R staining experiments, splenocytes were stained with anti-CD4 Ab (BioLegend), anti-CD8 Ab (Miltenyi), anti-CD44 Ab (BioLegend), anti–IL-6Rα Ab (BD), anti-KLRG1 Ab (eBioscience), anti-CD25 Ab (BioLegend), and anti-CD127 Ab (BD). For GC B cell staining, lung cell homogenates were generated using the lung dissociation kit (Miltenyi Biotec), as recommended by the manufacturer. Lung cells or spleen cells were stained with anti-CD8 Ab (Miltenyi Biotec), anti-Fas Ab (BD), anti-B220 Ab (BioLegend), anti-CD24 Ab (BioLegend), anti-IgD Ab (BD), and biotin-peanut agglutinin (Vector Laboratories), followed by secondary staining using Streptavidin staining Abs (BD). The immunostaining was always performed in the presence of Fc block (BD). For intracellular cytokine staining experiments, CD8+ T cells from IL-21 hCD4 reporter mice were activated for 3 d with or without IL-6. Cells were harvested, stained with anti-CD8 Ab (Miltenyi Biotec), anti-hCD4 Ab (BioLegend), and live-dead staining (Invitrogen). Cells were then fixed, permeabilized using saponin buffer (Sigma-Aldrich) as previously described (Yang et al., 1998), and stained with anti–IFN-γ Ab (BioLegend) or anti–IL-2 Ab (BioLegend). Expression of CD69 on activated cells was determined by immunostaining with an anti-CD69 Ab (BioLegend) and flow cytometry analysis. CD8+ T cell survival was determined by LIVE/Dead Viability kit, as recommended by the manufacturer (Molecular Probes) and flow cytometry analysis. CD8+ T cell proliferation was determined by CFSE staining (Molecular Probes) and flow cytometry analysis. All flow samples were run on FACS-LSR II (BD) and FlowJo Version 10.1 (FlowJo).
RNA isolation and real-time RT-PCR
Total RNA was isolated using the micro RNeasy kit (QIAGEN), as recommended by manufacture. cDNA synthesis was performed as previously described (Yang et al., 2015). cDNA was used for real-time RT-PCR assay using specific probe/primer sets for mouse Il21, Il2, Ifng, and β-microglobulin (Assays-on-Demand; Applied Biosystems). We followed the comparative CT method (2−ΔΔCT) as described by the manufacturer (Applied Biosystems; Livak and Schmittgen, 2001). In brief, Il21, Il2, and Ifng mRNA levels relative to the expression of β-microglobulin as housekeeping gene were obtained (ΔCT). ΔΔCT values were then obtained by subtracting the ΔCt value from a given reference sample (Figs. 1, I and K; and 2, B, C, and H) as calibrator to the rest of the samples. For Fig. 5 A, the mean of the ΔCT value for the five individual mice in the WT/Lung CD8+ T cell group was used as calibrator. The final relative expression data are provided as 2−ΔΔCT, defined as RQ value (relative quantitation).
ELISPOT assay
The detection of IL-21–producing CD8+ T cells by ELISPOT was performed as previously described (Dienz et al., 2009). In brief, purified CD8+ T cells (5 × 104 cells/well) were plated in an anti–IL-21 Ab–coated ELISPOT plate and incubated (9 h) in the presence of PMA (20 ng/ml) and ionomycin (1 μg/ml). Plates were developed as previously described (Dienz et al., 2009). The number of spots (number of IL-21–producing cells) per well, as well as the total area of spots (overall IL-21 production) per well, were measured.
Western blot analysis
Western blot analyses were performed as previously described (Yang et al., 2015), using the following antibodies: anti-actin (Santa Cruz Biotechnology, Inc.), anti-pSTAT3 Tyr705 (Cell Signaling Technology), and anti-STAT3 (Cell Signaling Technology), anti–rabbit HRP (Jackson ImmunoResearch Laboratories), and anti–goat HRP (Santa Cruz Biotechnology, Inc.).
Lung imaging
Lungs frozen in Tissue-Tek O.C.T. compound (Sakura Finetek) were sectioned using a CM1850 cryostat (Leica). 30-µm frozen sections were blocked for 30 min with Background Buster (Innovex Biosciences) at room temperature, stained 6 h at 4°C in a humidified chamber with anti-CD8α (BD), anti-GL-7 (BioLegend), and anti-B220 (BioLegend) in PBS with 2% goat serum. Alternatively (for lymphoid structures in the lung), whole mount imaging was performed on 250 µm scalpel cut lung sections that were then fixed with 2% PFA (1 h at 4°C), blocked, and stained with the antibody cocktail mentioned above and anti-CD4 Ab (BD). Immunofluorescence confocal microscopy was performed with the Zeiss 780 laser scanning microscope (Carl Zeiss; air objective 20× Plan-Apochromat with NA [numerical aperture] 0.5 or water objective 10x Plan-Apochromat with NA 0.30) using multichannel frame scans. Image processing was performed using Imaris 8.1 software. Images were analyzed using LSM5 image browser and cells in three to four images from each mouse were counted using ImageJ (National Institutes of Health) cell counter plugin.
ELISA
IL-21, IL-2, IL-4, and IFN-γ levels in cell culture supernatants were determined by ELISA, as previously described (Diehl et al., 2000; Yang et al., 2015). IgG1 levels in cell culture supernatants were determined by ELISA, as previously described (Dienz et al., 2009). Levels of Influenza A PR8-specific IgG, IgM, IgG1, IgG2c, and IgG3 were determined using UV-inactivated influenza PR8 as antigen, as previously described (Dienz et al., 2009). Antibody levels in serially diluted serum samples were analyzed using HRP-conjugated Abs specific for mouse IgM, IgG, IgG1, IgG2c, and IgG3 (SouthernBiotech).
Online supplemental material
Fig. S1 shows the gating strategy for GC B cell staining in lungs and spleen and confocal image of GC B cell staining in lungs.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Hunter and J. Hoffman for help with real-time PCR analysis (DNA Analysis Facility, University of Vermont, Burlington, VT), R. del Rio-Guerra for help with flow cytometry analysis (Flow Cytometry Facility, University of Vermont, Burlington, VT), Brian Silverstrim for technical help, and Roberta Pelanda and Raul Torres for helpful discussion (Department of Immunology, University of Colorado, Denver, CO).
This work was supported by National Institutes of Health grants R56AI094027 (to M. Rincon) and P20GM103496 (to M. Rincon and R. Yang). R. Yang was supported through the American Association of Immunologists Careers in Immunology Fellowship Program.
The authors declare no competing financial interests.
Author contributions: R. Yang, A.R. Masters, K.A. Fortner, D.P. Champagne, N. Yanguas-Casás, and D.J. Silberger performed experiments; R. Yang, and M. Rincon interpreted data; R. Yang, L. Haynes, C.T. Weaver, and M. Rincon designed experiments; and R. Yang, L. Haynes, C.T. Weaver, and M. Rincon wrote the paper.
Footnotes
Abbreviations used:
- GC
- germinal center
- KLRG1
- killer cell lectin like receptor G1
- Tfh
- T follicular helper
References
- Barr T.A., Brown S., Ryan G., Zhao J., and Gray D.. 2007. TLR-mediated stimulation of APC: Distinct cytokine responses of B cells and dendritic cells. Eur. J. Immunol. 37:3040–3053. 10.1002/eji.200636483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., and Kuchroo V.K.. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238. 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Christianson G.J., Brooks W., Vekasi S., Manolfi E.A., Niles J., Roopenian S.L., Roths J.B., Rothlein R., and Roopenian D.C.. 1997. Beta 2-microglobulin-deficient mice are protected from hypergammaglobulinemia and have defective antibody responses because of increased IgG catabolism. J. Immunol. 159:4781–4792. [PubMed] [Google Scholar]
- Croft M., Carter L., Swain S.L., and Dutton R.W.. 1994. Generation of polarized antigen-specific CD8 effector populations: reciprocal action of interleukin (IL)-4 and IL-12 in promoting type 2 versus type 1 cytokine profiles. J. Exp. Med. 180:1715–1728. 10.1084/jem.180.5.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin D.C. II, Stack R., and Fitch F.W.. 1995. IL-4-producing CD8+ T cell clones can provide B cell help. J. Immunol. 154:3118–3127. [PubMed] [Google Scholar]
- Diehl S., Anguita J., Hoffmeyer A., Zapton T., Ihle J.N., Fikrig E., and Rincón M.. 2000. Inhibition of Th1 differentiation by IL-6 is mediated by SOCS1. Immunity. 13:805–815. 10.1016/S1074-7613(00)00078-9 [DOI] [PubMed] [Google Scholar]
- Diehl S., Chow C.W., Weiss L., Palmetshofer A., Twardzik T., Rounds L., Serfling E., Davis R.J., Anguita J., and Rincón M.. 2002. Induction of NFATc2 expression by interleukin 6 promotes T helper type 2 differentiation. J. Exp. Med. 196:39–49. 10.1084/jem.20020026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl S.A., Schmidlin H., Nagasawa M., Blom B., and Spits H.. 2012. IL-6 triggers IL-21 production by human CD4+ T cells to drive STAT3-dependent plasma cell differentiation in B cells. Immunol. Cell Biol. 90:802–811. 10.1038/icb.2012.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienz O., and Rincon M.. 2009. The effects of IL-6 on CD4 T cell responses. Clin. Immunol. 130:27–33. 10.1016/j.clim.2008.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienz O., Eaton S.M., Bond J.P., Neveu W., Moquin D., Noubade R., Briso E.M., Charland C., Leonard W.J., Ciliberto G., et al. 2009. The induction of antibody production by IL-6 is indirectly mediated by IL-21 produced by CD4+ T cells. J. Exp. Med. 206:69–78. 10.1084/jem.20081571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienz O., Rud J.G., Eaton S.M., Lanthier P.A., Burg E., Drew A., Bunn J., Suratt B.T., Haynes L., and Rincon M.. 2012. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol. 5:258–266. 10.1038/mi.2012.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger R., Sims G.P., Fairhurst A.M., Robbins R., da Silva Y.S., Spolski R., Leonard W.J., and Lipsky P.E.. 2005. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J. Immunol. 175:7867–7879. 10.4049/jimmunol.175.12.7867 [DOI] [PubMed] [Google Scholar]
- Genestier L., Taillardet M., Mondiere P., Gheit H., Bella C., and Defrance T.. 2007. TLR agonists selectively promote terminal plasma cell differentiation of B cell subsets specialized in thymus-independent responses. J. Immunol. 178:7779–7786. 10.4049/jimmunol.178.12.7779 [DOI] [PubMed] [Google Scholar]
- Hamada H., Garcia-Hernandez M.L., Reome J.B., Misra S.K., Strutt T.M., McKinstry K.K., Cooper A.M., Swain S.L., and Dutton R.W.. 2009. Tc17, a unique subset of CD8 T cells that can protect against lethal influenza challenge. J. Immunol. 182:3469–3481. 10.4049/jimmunol.0801814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heer A.K., Shamshiev A., Donda A., Uematsu S., Akira S., Kopf M., and Marsland B.J.. 2007. TLR signaling fine-tunes anti-influenza B cell responses without regulating effector T cell responses. J. Immunol. 178:2182–2191. 10.4049/jimmunol.178.4.2182 [DOI] [PubMed] [Google Scholar]
- Heinrich P.C., Behrmann I., Haan S., Hermanns H.M., Müller-Newen G., and Schaper F.. 2003. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 374:1–20. 10.1042/bj20030407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist K.A., Jameson S.C., Heath W.R., Howard J.L., Bevan M.J., and Carbone F.R.. 1994. T cell receptor antagonist peptides induce positive selection. Cell. 76:17–27. 10.1016/0092-8674(94)90169-4 [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., and Littman D.R.. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Kishimoto T. 2005. Interleukin-6: from basic science to medicine—40 years in immunology. Annu. Rev. Immunol. 23:1–21. 10.1146/annurev.immunol.23.021704.115806 [DOI] [PubMed] [Google Scholar]
- Lee B.O., Rangel-Moreno J., Moyron-Quiroz J.E., Hartson L., Makris M., Sprague F., Lund F.E., and Randall T.D.. 2005. CD4 T cell-independent antibody response promotes resolution of primary influenza infection and helps to prevent reinfection. J. Immunol. 175:5827–5838. 10.4049/jimmunol.175.9.5827 [DOI] [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Linterman M.A., Beaton L., Yu D., Ramiscal R.R., Srivastava M., Hogan J.J., Verma N.K., Smyth M.J., Rigby R.J., and Vinuesa C.G.. 2010. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J. Exp. Med. 207:353–363. 10.1084/jem.20091738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Moyron-Quiroz J.E., Rangel-Moreno J., Kusser K., Hartson L., Sprague F., Goodrich S., Woodland D.L., Lund F.E., and Randall T.D.. 2004. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat. Med. 10:927–934. 10.1038/nm1091 [DOI] [PubMed] [Google Scholar]
- Nurieva R., Yang X.O., Martinez G., Zhang Y., Panopoulos A.D., Ma L., Schluns K., Tian Q., Watowich S.S., Jetten A.M., and Dong C.. 2007. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 448:480–483. 10.1038/nature05969 [DOI] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Hwang D., Yang X.O., Kang H.S., Ma L., Wang Y.H., Watowich S.S., Jetten A.M., Tian Q., and Dong C.. 2008. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 29:138–149. 10.1016/j.immuni.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K., Spolski R., Feng C.G., Qi C.F., Cheng J., Sher A., Morse H.C. III, Liu C., Schwartzberg P.L., and Leonard W.J.. 2002. A critical role for IL-21 in regulating immunoglobulin production. Science. 298:1630–1634. 10.1126/science.1077002 [DOI] [PubMed] [Google Scholar]
- Poli V., Balena R., Fattori E., Markatos A., Yamamoto M., Tanaka H., Ciliberto G., Rodan G.A., and Costantini F.. 1994. Interleukin-6 deficient mice are protected from bone loss caused by estrogen depletion. EMBO J. 13:1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed M.A., Latner D.R., Aubert R.D., Gourley T., Spolski R., Davis C.W., Langley W.A., Ha S.J., Ye L., Sarkar S., et al. 2013. Interleukin-21 is a critical cytokine for the generation of virus-specific long-lived plasma cells. J. Virol. 87:7737–7746. 10.1128/JVI.00063-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray J.P., Marshall H.D., Laidlaw B.J., Staron M.M., Kaech S.M., and Craft J.. 2014. Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity. 40:367–377. 10.1016/j.immuni.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón M., Anguita J., Nakamura T., Fikrig E., and Flavell R.A.. 1997. Interleukin (IL)-6 directs the differentiation of IL-4–producing CD4+ T cells. J. Exp. Med. 185:461–469. 10.1084/jem.185.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J.H., and Ley T.J.. 2002. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 20:323–370. 10.1146/annurev.immunol.20.100201.131730 [DOI] [PubMed] [Google Scholar]
- Simchoni N., and Cunningham-Rundles C.. 2015. TLR7- and TLR9-responsive human B cells share phenotypic and genetic characteristics. J. Immunol. 194:3035–3044. 10.4049/jimmunol.1402690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs M.K., Koller B.H., Sato T., Morrissey P.J., Fanslow W.C., Smithies O., Voice R.F., Widmer M.B., and Maliszewski C.R.. 1992. Beta 2-microglobulin-, CD8+ T-cell-deficient mice survive inoculation with high doses of vaccinia virus and exhibit altered IgG responses. Proc. Natl. Acad. Sci. USA. 89:6070–6074. 10.1073/pnas.89.13.6070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto A., Kashiwakuma D., Kagami S., Hirose K., Watanabe N., Yokote K., Saito Y., Nakayama T., Grusby M.J., Iwamoto I., and Nakajima H.. 2008. Development and characterization of IL-21–producing CD4+ T cells. J. Exp. Med. 205:1369–1379. 10.1084/jem.20072057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Kaisho T., Yoshida N., Takeda J., Kishimoto T., and Akira S.. 1998. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol. 161:4652–4660. [PubMed] [Google Scholar]
- Waldmann T.A. 2006. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat. Rev. Immunol. 6:595–601. 10.1038/nri1901 [DOI] [PubMed] [Google Scholar]
- Yang D.D., Conze D., Whitmarsh A.J., Barrett T., Davis R.J., Rincón M., and Flavell R.A.. 1998. Differentiation of CD4+ T cells to Th1 cells requires MAP kinase JNK2. Immunity. 9:575–585. 10.1016/S1074-7613(00)80640-8 [DOI] [PubMed] [Google Scholar]
- Yang R., Lirussi D., Thornton T.M., Jelley-Gibbs D.M., Diehl S.A., Case L.K., Madesh M., Taatjes D.J., Teuscher C., Haynes L., and Rincón M.. 2015. Mitochondrial Ca²+ and membrane potential, an alternative pathway for interleukin 6 to regulate CD4 cell effector function. eLife. 4:4 10.7554/eLife.06376 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.