Verdun and colleagues find that the uracil-DNA glycosylase UNG, which promotes DNA breaks in the immunoglobulin genes during class switch recombination and is required for AID-induced chromosomal translocations, protects telomeres from AID-induced DNA damage and subsequent dysfunction.
Abstract
Activation-induced deaminase (AID) initiates antibody gene diversification by creating G:U mismatches in the immunoglobulin loci. However, AID also deaminates nonimmunoglobulin genes, and failure to faithfully repair these off-target lesions can cause B cell lymphoma. In this study, we identify a mechanism by which processing of G:U produced by AID at the telomeres can eliminate B cells at risk of genomic instability. We show that telomeres are off-target substrates of AID and that B cell proliferation depends on protective repair by uracil-DNA glycosylase (UNG). In contrast, in the absence of UNG activity, deleterious processing by mismatch repair leads to telomere loss and defective cell proliferation. Indeed, we show that UNG deficiency reduces B cell clonal expansion in the germinal center in mice and blocks the proliferation of tumor B cells expressing AID. We propose that AID-induced damage at telomeres acts as a fail-safe mechanism to limit the tumor promoting activity of AID when it overwhelms uracil excision repair.
INTRODUCTION
The first exposure of mature naive B cells to cognate antigen within secondary lymphoid organs prompts the formation of germinal centers (GCs). Therein, antigen-stimulated B cells proliferate while modifying their Ig genes. The mechanisms of somatic hypermutation (SHM) and class switch recombination (CSR) increase the affinity for the antigen and endow the antibody with new biological properties, respectively. SHM introduces point mutations within the exon encoding the V region of each Ig gene. CSR is a deletional recombination event within the Ig heavy chain (Igh) locus of B cells that involves the replacement of the IgM-constant region (Cμ) with a downstream CH exon set (γ, α, or ε), determining the IgG, IgA, or IgE isotypes, respectively (Stavnezer et al., 2008).
Both SHM and CSR are initiated by enzyme activation-induced deaminase (AID), which is only expressed at high levels in GC B cells (Crouch et al., 2007). AID initiates CSR by deaminating cytosines into uracils over large repetitive cytosine-rich (C-rich) switch-region (S region) sequences located upstream of each CH region (Fig. 1 A). AID-generated uracils are recognized by either the uracil-DNA glycosylase (UNG) or the mismatch repair heterodimer MSH2/MSH6 (Rada et al., 2004). Subsequent processing of uracils in opposite DNA strands by DNA repair enzymes produces double-strand breaks, which are the substrates of the end-joining mechanisms that complete CSR by joining two separate S regions (Stavnezer et al., 2008).
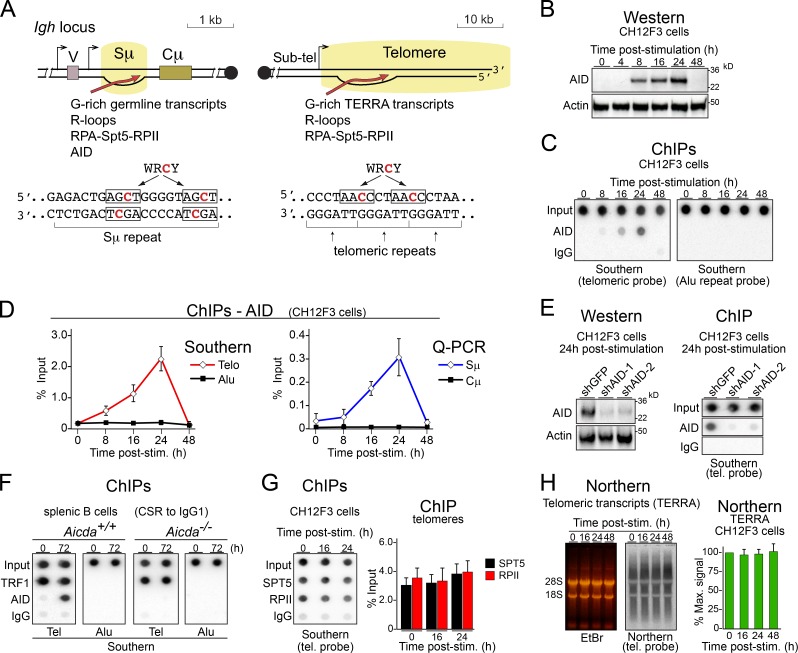
Figure 1.
AID interacts with telomeres in B cells during CSR. (A) Schematic depiction of similarities between telomeres and S regions and location of AID’s preferred target sequences (WRCY). Note that WRCY motifs are present in both S-region strands but exclusively in the C-rich strand in telomeres. Sub-tel, subtelomeric. RPA, replication protein A; V, variable region. (B) Western blot analysis of AID expression in CH12F3 cells after cytokine stimulation for CSR. (C) Representative dot blot analysis of ChIP assays using anti-AID and IgG control in stimulated CH12F3 B cells. Dot blots with 5% of the input or the immunoprecipitates were analyzed via Southern blot with telomeric or Alu repeat probes. (D) Quantification of AID accumulation at telomeres (Telo) and Alu repeats by dot blot, as in C, as well as Sμ and Cμ regions of the Igh locus (by quantitative PCR [Q-PCR]) in CH12F3 cells stimulated for CSR, from at least three independent experiments. post-stim., post-stimulation. Error bars represent SD. (E, left) Western blot analysis of AID expression in CH12F3 cells expressing the indicated shRNAs. (Right) Representative ChIPs in CH12F3 B cells with the indicated antibodies out of three independent experiments. Coimmunoprecipitated telomeric DNA was detected via Southern blot with a telomeric (tel.) probe in dot blots. (F) One representative of three independent ChIP assays, as in C but in splenic B cells purified from Aicda+/+ or Aicda−/− mice, and stimulated with LPS and IL-4 for 72 h. ChIP for the telomeric (Tel) protein TRF1 was included as a positive control. (G) ChIPs in CH12F3 B cells with the indicated antibodies. (Right) Quantification of the dot blot signals after hybridization with a telomeric probe. (H) Northern blot with a telomeric probe showing the level of telomeric transcripts in wild-type splenic B cells before and after stimulation for CSR. EtBr, ethidium bromide. (Right) Quantification of Northern signals. (G and H) Data show mean + SD values obtained at each time point from three independent experiments.
As a side effect of antibody gene diversification, AID produces off-target deaminations and DNA damage, which unless faithfully repaired can be oncogenic (Liu et al., 2008; Pasqualucci et al., 2008; Robbiani and Nussenzweig, 2013; Meng et al., 2014; Qian et al., 2014) or cytotoxic (Hasham et al., 2010; Zahn et al., 2014). UNG and MSH2/MSH6 modulate the mutagenic capacity of AID either by initiating error-free base excision repair (BER) and mismatch DNA repair (MMR), respectively, or by triggering mutagenic repair (Rada et al., 2004; Liu et al., 2008). The full extent of off-target AID activity and the repair mechanisms that control it are not yet known.
Telomeres, the natural ends of linear chromosomes, consist of kilobases of a hexanucleotide repeat (5′-TTAGGG-3′ in vertebrates) that protects the chromosome ends from being recognized as a DNA lesion (Arnoult and Karlseder, 2015). Telomeres that fail to hide their ends trigger a DNA damage response that leads to cell cycle arrest or cell death (d’Adda di Fagagna et al., 2003; Arnoult and Karlseder, 2015). Telomeres and S regions share many similarities: both are located downstream of an RNA polymerase II (RPII) promoter producing sterile transcripts (Schoeftner and Blasco, 2008; Storb, 2014) and have C-rich template DNA strands enriched in AID hotspot sequences (Fig. 1 A). Further, both regions form R-loops (RNA:DNA hybrid regions; Balk et al., 2013; Pfeiffer et al., 2013) and produce noncoding transcripts capable of forming G-quartets, which help recruiting AID to S regions (Zheng et al., 2015). Based on these similarities and the relevance of telomeres for genomic stability, we asked whether telomeres might be targeted by AID in activated B cells. We found this to be the case. We further uncovered a critical role of UNG in protecting the telomeres and the GC reaction. In the absence of UNG, a mismatch repair-mediated mechanism makes gaps in the C-rich strand of the telomeres deaminated by AID and leads to their sudden shortening, resulting in greatly reduced B cell proliferation. Indeed, we show that during an immune response, B cell clonal expansion and formation of the GC depend on the presence of UNG. Therefore, we propose that B cells use a novel mechanism for telomere homeostasis to control the impact of AID off-target activity. We finally show that this is an actionable mechanism to target tumor cells expressing AID.
RESULTS
AID at the telomeres in activated B cells
To test whether AID localizes to telomeres, we used chromatin immunoprecipitation (ChIP) on chromatin extracts of the CH12F3 B cell lymphoma line and mouse splenic B cells. CH12F3 cells showed increasing expression of AID starting from 8 h until 24 h after cytokine stimulation, as evaluated by Western blotting (Fig. 1 B). We found that AID associated to telomeres and Sμ after CSR stimulation with the same kinetics (Fig. 1, C and D). The association was specific: first, no AID signal was observed at Alu repeats or the Cμ (Fig. 1, C and D). Second, depleting AID in CH12F3 cells by shRNA (Fig. 1 E) or using AID-null splenic B cells (Fig. 1 F) eliminated the AID ChIP signal from the telomeres. Telomere occupancy by RPII and the transcription factor Spt5, which are necessary for recruiting AID to the DNA (Pavri et al., 2010; Storb, 2014), and steady-state levels of telomeric transcripts (Schoeftner and Blasco, 2008) did not change upon inducing CSR (Fig. 1, G and H), suggesting unaltered telomeric transcription. We conclude that AID interacts with and might deaminate telomeric DNA in B cells concomitantly with CSR.
UNG protects B cells from AID-dependent telomere loss
Although AID-catalyzed deamination of telomeres could cause DNA damage, telomeres were normal in activated CH12F3 and splenic B cells, as judged by fluorescent in situ hybridization (FISH) of metaphase chromosomes (see paragraph below). This could mean that AID is recruited to telomeres but does not act on them or that the deaminations are repaired by error-free mechanisms.
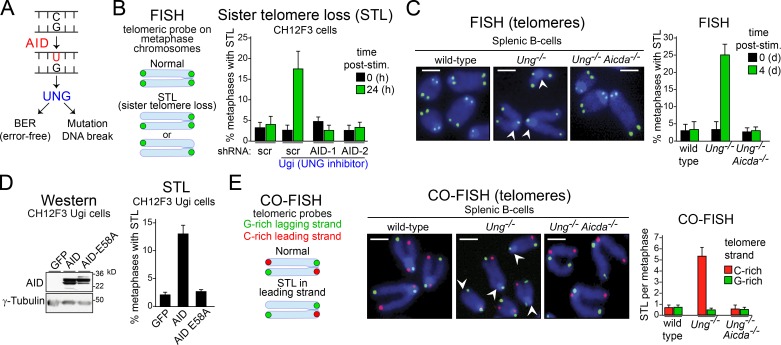
Although the repetitive nature of telomeres prevents their sequencing to detect point mutations, AID activity can be ascertained by interfering with cellular uracil-sensing factors. The major factor recognizing uracil at S regions is UNG, which can start either error-free BER or mutagenic processing (Fig. 2 A). To test the potential role of BER in repairing AID-induced damage at telomeres, we ablated UNG in stimulated B cells. Inhibiting UNG activity in CH12F3 cells through the expression of the specific inhibitor Ugi led to a fourfold increase in chromosomes with unequal telomere signals compared with control cells (Fig. 2 B). The majority of abnormal telomeres lacked a single chromatid, a phenotype known as sister telomere loss (STL, Fig. 2 B) that reflects a replication defect at the chromosome ends (Crabbe et al., 2004). Stimulated splenic B cells from Ung−/− mice also showed an eightfold increase in metaphases with STL-like phenotype over wild-type B cells (Fig. 2 C). Depleting AID by shRNAs in CH12F3 Ugi cells, as well as using mouse Ung−/− Aicda−/− splenic B cells, demonstrated that telomeric DNA loss in UNG-deficient B cells was AID dependent (Fig. 2, B and C). Finally, constitutive overexpression of AID in unstimulated CH12F3 Ugi cells was sufficient to increase the frequency of metaphases with STL-like phenotype, whereas the catalytic mutant AIDE58A did not cause that phenotype, despite being similarly expressed (Fig. 2 D). No increase in intrachromatid breaks was observed in CH12F3 Ugi or Ung−/− B cells (not depicted). No difference in single- or double-stranded telomeric repeats was observed by terminal restriction fragment analysis between activated Ung−/− and wild-type splenic B cells (not depicted), indicating that AID induces a sudden loss rather than an accelerated shortening of the telomeres. These results are consistent with the preference of AID to deaminate close to transcription initiation sites (Peters and Storb, 1996; Rada and Milstein, 2001; Ramiro et al., 2003; Taylor et al., 2014), which in telomeres is at the subtelomeric region (Fig. 1 A; Azzalin et al., 2007; Schoeftner and Blasco, 2008).
Figure 2.
AID induces telomere loss in UNG-deficient B cells. (A) Possible outcomes after AID-dependent DNA deaminations are processed by UNG in B cells. (B, left) Illustration of typical FISH staining with a telomere-specific probe in metaphase chromosomes from normal cells and cells with STL. (Right) Effect of UNG inhibition via Ugi expression on the proportion of metaphases with STL in different CH12F3 lines expressing scrambled (scr) control or two different shRNAs that deplete AID, before and after stimulating for CSR to IgA. Post-stim., post-simulation. (C, left) Representative pictures of FISH on metaphase chromosomes in wild-type, Ung−/−, and Ung−/− Aicda−/− mouse splenic B cells stimulated for CSR to IgG1. Telomeres were hybridized with an Alexa Fluor 488–[TTAGGG]4 probe (in green); total DNA was stained with DAPI (in blue). Arrowheads indicate missing telomere staining from single sister chromatids. Bars, 2 µm. (Right) Quantification of STL per metaphase after FISH analysis. Error bars represent mean + SD from at least three independent experiments. (D, left) Western analysis of wild-type AID or AIDE58A levels in CH12F3 Ugi cells. (Right) Quantification of metaphases with STL from CH12F3 Ugi cells expressing GFP, AID, or AIDE58A. (E, left) Illustration of CO-FISH staining. Leading-strand telomeres are shown in red, and lagging-strand telomeres are in green. (Middle) Representative pictures of CO-FISH in B cells at 4 d after stimulation with LPS and IL-4. Arrowheads indicate missing telomere staining from leading-strand telomeres. Bars, 2 µm. (Right) Quantification of STL per metaphase after CO-FISH analysis. (B, D, and E) Data show the mean + SD of three independent experiments, in which 50 metaphases per cell line were analyzed in each experiment.
Because STL is usually related to dysfunction in telomere replication and AID exclusively deaminates deoxycytosine, we used two-color chromosome orientation FISH (CO-FISH) to identify whether the loss of telomeric DNA reflected a defect in leading (C-rich) or lagging (G-rich) strand synthesis. Loss of signal in UNG-deficient B cells was restricted to the leading strand (Fig. 2 E), demonstrating that the AID-induced telomeric loss resulted from defects in replicating the C-rich telomeric strand.
Our data are consistent with a model where, in activated B cells, AID deaminates the telomeres, but these are efficiently protected by UNG from further DNA damage.
Mismatch repair mediates telomere loss in Ung-deficient B cells
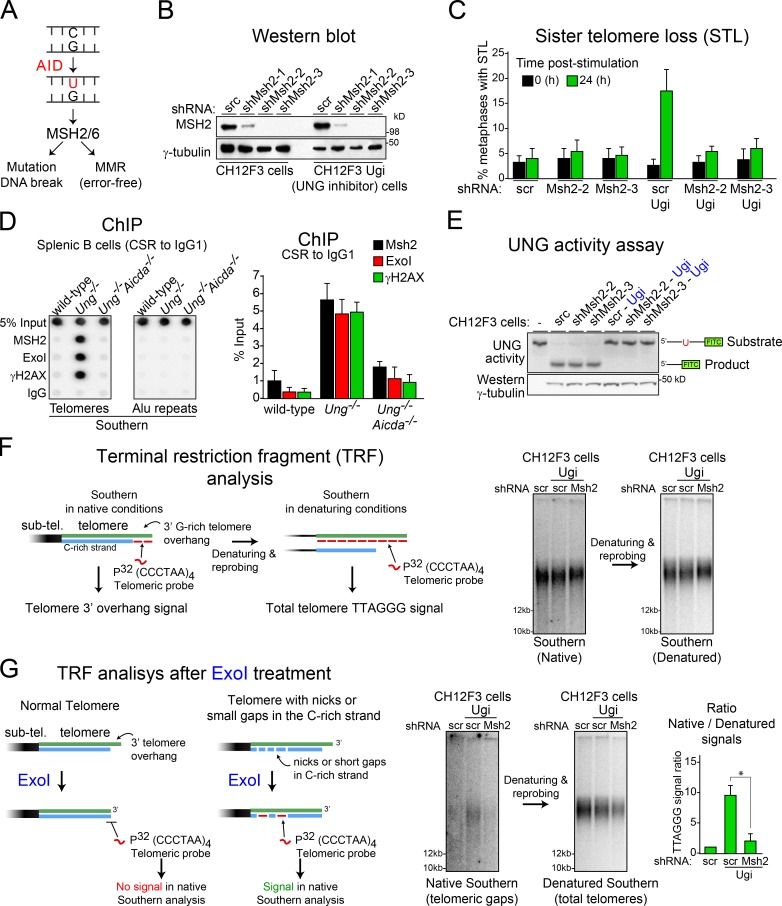
We then asked whether MSH2/MSH6, which can also detect AID-catalyzed uracil and initiate faithful or mutagenic DNA repair (Fig. 3 A; Rada et al., 2004; Liu et al., 2008), played any role at the telomeres of activated B cells. Contrary to its role in telomere maintenance observed in mouse embryonic fibroblasts (Campbell et al., 2006), depleting MSH2 did not affect telomere stability in stimulated CH12F3 cells. However, MSH2 knockdown prevented the increase in STL observed in CH12F3 Ugi cells (Fig. 3, B and C). Accordingly, ChIP assays demonstrated AID-dependent accumulation of the MMR factors MSH2 and exonuclease 1 at the telomeres only in stimulated Ung−/− primary B cells (Fig. 3 D) and stimulated CH12F3 Ugi cells (not depicted). UNG inhibition in CH12F3 Ugi cell lines was confirmed by activity assays (Fig. 3 E). These results indicate that UNG outcompetes MSH2/MSH6 in recognizing the uracils, which only accumulate and can be detected as mismatches in the absence of UNG activity. Terminal restriction fragment analysis showed that CH12F3 Ugi cells had a normal telomere G-rich 3′ overhang signal (Fig. 3 F). However, performing the same assay after treating the DNA with exonuclease to degrade this overhang revealed an increase in intratelomeric G-rich single-stranded DNA (ssDNA), indicative of ssDNA gaps, only in MSH2-depleted cells (Fig. 3 G). We conclude that, in the absence of UNG, MMR-dependent processing of AID lesions creates gaps in the telomeric C-rich strand, thereby mediating STL in replicating B cells.
Figure 3.
Mismatch repair factors mediate AID-induced STL in Ung-deficient B cells. (A) Possible outcomes of MSH2/MSH6-initiated repair of AID-induced DNA deaminations in B cells. (B) Western blot analysis of MSH2 in CH12F3 cells expressing the indicated shRNAs. scr, scrambled. (C) Quantification of the proportion of STL per 50 metaphases in each of the different CH12F3 lines expressing or not expressing Ugi and scrambled control or two different shRNAs that deplete MSH2, before and after stimulation of CSR to IgA. Error bars represent mean + SD from at least three independent experiments. (D, left) Representative ChIP performed with the indicated antibodies in wild-type, Ung−/−, and Ung−/− Aicda−/− splenic B cells stimulated for CSR to IgG1 and analyzed by dot blotting using telomeric or Alu probes. (Right) Plot of the mean + SD dot blot signals for the telomeric probe from three independent experiments. (E) UNG activity assay using a fluorescein-labeled oligonucleotide containing a single dU, incubated with cell extracts (10 µg protein) from the indicated CH12F3 lines used in C. Substrate and product, indicated on the left, were resolved on 15% TBE-urea polyacrylamide gels. Western blot of γ-tubulin level was used as a loading control. (F) Terminal restriction fragment analysis of TTAGGG repeats in stimulated CH12F3 and CH12F3-Ugi cells expressing the indicated shRNAs via Southern blotting in native or denatured conditions. sub-tel., subtelomeric. (G, left) Diagram showing the expected outcomes after treatment of genomic DNA with exonuclease I before the TRF analyses of TTAGGG repeats. The 3′ to 5′ single-strand exonuclease activity of ExoI will remove the telomeric 3′ G-rich overhang. Therefore, the signal for single-stranded TTAGGG repeats will be lost in a TRF analysis in native conditions. However, in telomeres with short gaps or nicks in the C-rich strand, ExoI activity will expose G-rich single-stranded gaps that can be detected in a TRF analysis in native conditions. (Right) Representative Southern blots of TRF after ExoI treatment in native and denatured conditions and quantification of telomeric ssDNA/dsDNA ratio in ExoI-treated genomic DNA. Error bars represent mean + SD from at least three independent experiments. *, P < 0.003 (Student’s t test).
Short telomeres in Ung-deficient B cells trigger a DNA damage response
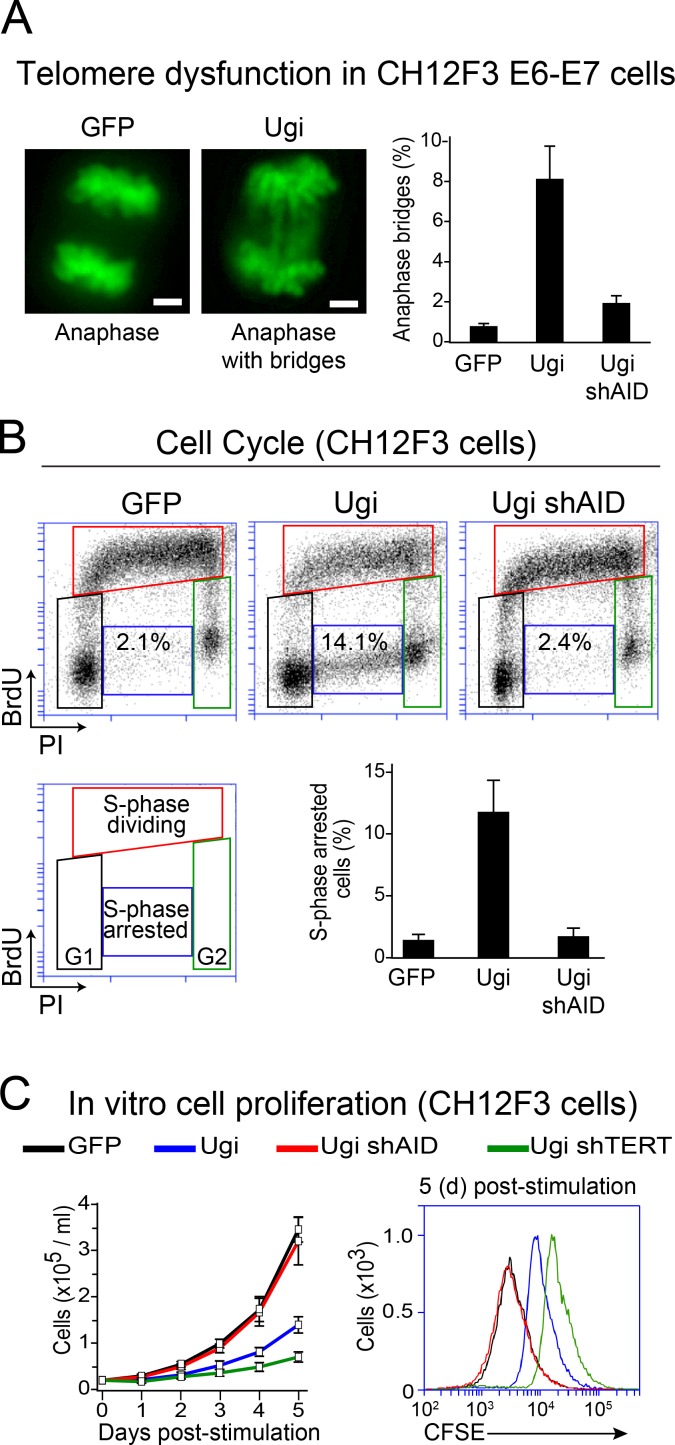
Excessive loss of telomeric DNA induces a DNA damage response at the chromosome ends (d’Adda di Fagagna et al., 2003; Arnoult and Karlseder, 2015). Indeed, we detected AID-dependent accumulation of phospho-ser139-H2AX (a marker of DNA damage often found at dysfunctional telomeres; d’Adda di Fagagna et al., 2003) at telomeres in stimulated Ung−/− B cells (Fig. 3 D). By suppressing the p53- and p16INK4a/pRb-dependent pathways via the expression of papillomavirus proteins E6 and E7 to prevent B cell death, we found that CH12F3 Ugi cells significantly accumulated anaphase bridges compared with CH12F3 Ugi expressing an shRNA against AID (shAID) and control GFP cells (Fig. 4 A). Thus, AID expression causes telomere dysfunction in UNG-deficient B cells.
Figure 4.
Compromised proliferation of Ung-deficient B cells expressing AID. (A, left) Representative microscopy pictures of anaphases from CH12F3 cells expressing the HPV16 E6 and E7 oncoproteins and GFP control, Ugi, or Ugi shAID. Total DNA was stained with DAPI (shown in green). Bars, 2 µm. (Right) Mean + SD proportion of anaphases showing chromosome bridges in CH12F3 GFP and CH12F3 Ugi cells from at least three independent experiments. 50 anaphases were analyzed for each cell line per experiment. (B) Cell cycle profile analysis by BrdU incorporation and propidium iodide (PI) staining in the indicated CH12F3 cells 24 h after stimulating CSR. (C) Cell number (left) and CFSE staining (right) were used to evaluate cellular proliferation of dCH12F3 and CH12F3 Ugi cells expressing the indicated shRNAs after stimulation for CSR. Error bars represent mean + SD from at least three independent experiments. shTERT, shRNA against the telomerase reverse transcriptase (TERT).
A DNA damage response caused by telomere dysfunction usually causes cell division defects (d’Adda di Fagagna et al., 2003; Arnoult and Karlseder, 2015). Accordingly, cell cycle profiling revealed that stimulated UNG-deficient B cells had an approximately sixfold increase in cells arrested in synthesis phase (S phase), which was AID dependent (Fig. 4 B). These results predicted that UNG-deficient B cells expressing AID should have reduced proliferation capacity. Indeed, CH12F3 Ugi cells stimulated for CSR proliferated less than the control, as evaluated by total cell number or CFSE dilution assay, which was prevented by AID knockdown (Fig. 4 C). In addition, consistent with the role of telomerase in protecting against cell proliferation defects caused by excessive telomere shortening (Verdun and Karlseder, 2007), knockdown of the catalytic subunit of telomerase in CH12F3 Ugi cells further decreased their proliferation (Fig. 4 C). We conclude that in the absence of Ung, AID induces telomere dysfunction and a DNA damage response that compromises B cell proliferation.
Compromised GC B cell expansion in Ung−/− mice
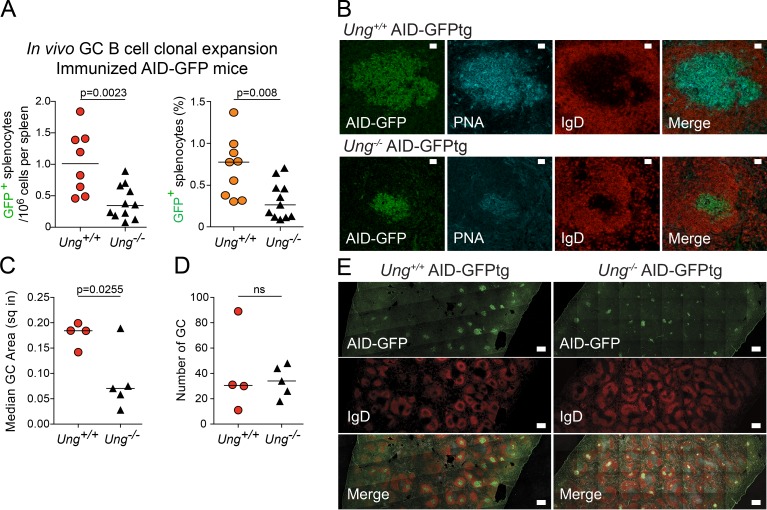
B cells must expand clonally while expressing AID during the GC reaction (Victora and Nussenzweig, 2012). To evaluate whether AID expression affected the clonal proliferation of UNG-deficient GC B cells in vivo, we enumerated AID+ GC B cells after acute antigenic challenge using an AID-GFP transgenic (AID-GFPtg) reporter (Crouch et al., 2007; Zahn et al., 2013). Despite wild-type and Ung−/− mice having similar quiescent splenic B cell populations (Zahn et al., 2013), 8 d after immunization, the spleen of Ung−/− AID-GFPtg mice showed ∼50% less AID+ B cells than AID-GFPtg controls (Fig. 5 A). Splenic AID-GFP cells are largely GC B cells (Crouch et al., 2007) and were confirmed by the colocalization of AID-GFP with peanut agglutininhigh (PNAhigh) IgD− cells inside B cell follicles (Fig. 5 B). This reduction in AID+ GC B cells was explained by an approximately threefold smaller average GC size in Ung−/− versus Ung+/+ AID-GFPtg mice, rather than any difference in the number of GCs (Fig. 5, C–E). Thus, UNG deficiency impairs the proliferation of B cells expressing AID, which surely contributes to the severe defect in antibody responses of Ung-null mice and humans (Imai et al., 2003; Zahn et al., 2013). We conclude that UNG plays a critical role in the GC reaction by protecting AID-expressing B cells during clonal expansion.
Figure 5.
Compromised clonal expansion of Ung−/− GC B cells in vivo. (A) Absolute number (left) and proportion (right) of AID-GFP+ cells in the spleen of AID-GFPtg and Ung−/− AID-GFPtg mice 8 d after immunization, as calculated from flow cytometry analyses of three independent experiments. (B) Representative confocal images of GCs in the spleen of AID-GFPtg and Ung−/− AID-GFPtg mice stained for IgD and PNA. Bars, 20 µm. (C) Quantification of GC size. Each symbol represents the median area in square inches (sq in) of all GCs observed in splenic sections from four wild-type and five Ung−/− AID-GFPtg mice coming from two of the experiments represented in A. Data are derived from three independent experiments. (D) The number of GCs per splenic section were counted in the same mice as in C. (A, C, and D) The p-values from unpaired two-tailed Student’s t tests are shown, and horizontal bars indicate median values. (E) Representative images used to measure AID-GFP+ GC number and area. Partial composite sections of immunized spleens made of multiple overlapping images obtained by spinning disk confocal microscopy are shown for each genotype. Bars, 0.25 mm.
UNG deficiency leads to proliferation defects in AID+ B cell lymphoma cells
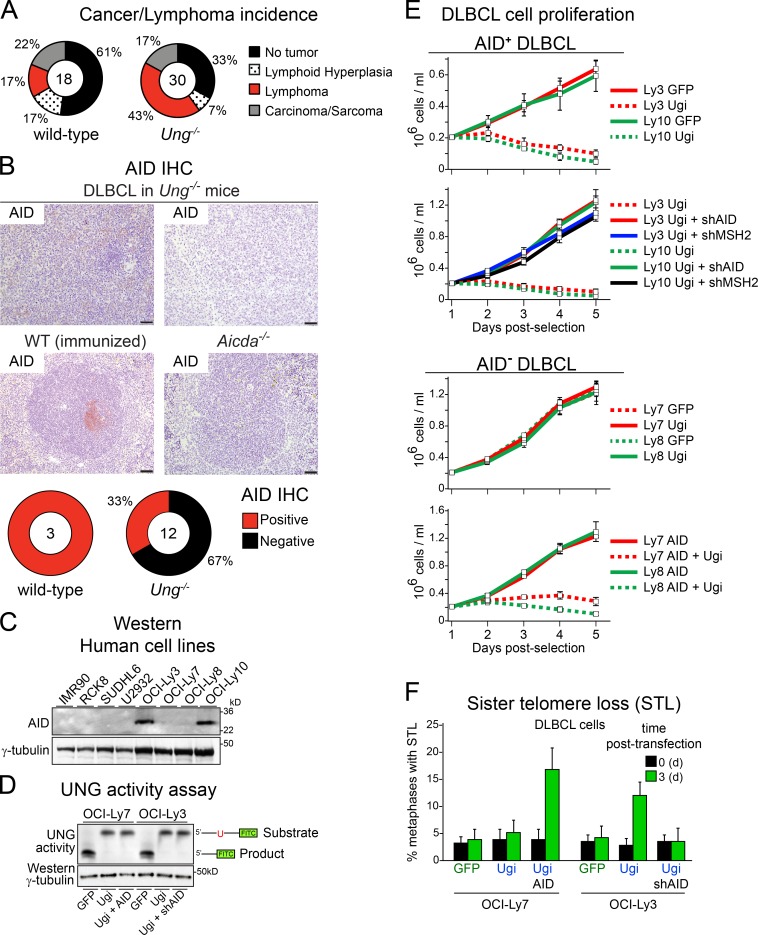
Our results suggested that the proliferation of lymphoma cells expressing high levels of AID might depend on UNG. Old Ung−/− mice are prone to develop B cell lymphomas (Nilsen et al., 2003; Andersen et al., 2005). Although it has been speculated that AID might be etiological in those tumors, the AID status of the actual lymphomas has not been studied. We therefore analyzed lymphomas that developed spontaneously in wild-type and Ung−/− mice. Similar to previous studies (Nilsen et al., 2003; Andersen et al., 2005), 13 out of 30 (43%) Ung−/− mice but only 3 out of 18 (17%) control mice developed lymphoma (Fig. 6 A), which in most cases had a histopathology consistent with mature B cell lymphoma (not depicted). However, the majority (67%) of Ung−/− lymphomas were mostly negative for AID by immunohistochemistry (IHC; Fig. 6 B). Although these data do not exclude the possibility that AID plays a role in the origin of these lymphomas, the low or negative expression of AID in Ung−/− B cell lymphomas is consistent with the notion that high AID expression is not well tolerated by UNG-deficient B cells during clonal expansion.
Figure 6.
The proliferation of malignant B cells expressing AID depends on UNG. (A) Incidence of spontaneous lymphoma, lymphoid hyperplasia, or any other tumor in wild-type and Ung−/− mice. The total number of mice analyzed is shown at the center of each pie. (B, top) Representative IHC pictures of AID staining in B cell lymphomas found in Ung−/− mice. Positive and negative staining controls are shown using spleen from immunized wild-type and Aicda−/− mice, respectively. (Bottom) AID status of the lymphoma cells, as judged from IHC, for each of the mice diagnosed with B cell lymphoma. Bars, 50 µm. (C) Western blot analysis of AID levels in human DLBCL cell lines and control fibroblasts (IMR90). (D) UNG activity assay in cell extracts (10 µg protein) of the indicated DLBCL cell lines using a fluorescein-labeled oligonucleotide with a single dU residue as the substrate. The reaction products resolved by TBE-urea PAGE are indicated on the left. A Western blot of γ-tubulin was used as the loading control. (E) The proliferation of the indicated human DLBCL cell lines expressing GFP control, AID, or Ugi and the indicated shRNAs was measured by growth curves. (F) Quantification of metaphases with STL in the same cell lines analyzed in E. (E and F) Data are mean + SD from three independent experiments.
In contrast to GC B cells in which AID is acutely induced, human non–Hodgkin’s lymphoma (NHL) cells such as diffuse large B cell lymphoma (DLBCL) can steadily express AID to various levels (Lossos et al., 2004; Pasqualucci et al., 2004). We therefore stratified human DLBCL cell lines on the basis of their AID protein levels into high (DLBCL AID+)- or low (DLBCL AID−)-expressing lines (Fig. 6 C) and established derivatives expressing Ugi for each one. DLBCL AID+ cells showed a lower rate of cell proliferation when compared with DLBCL AID− cells (Fig. 6 E). Expressing Ugi only impaired the growth of DLBCL AID+ cells (Fig. 6 E). The poor growth and sensitivity to Ugi of DLBCL AID+ cells was both AID- and MMR-dependent, as knockdown of AID or MSH2 allowed their proliferation at levels comparable with DLBCL AID− cells (Fig. 6 E). Furthermore, DLBCL AID− cells became sensitive to UNG inhibition after transfection with AID (Fig. 6 E). Importantly, we observed a significant increase in the number of metaphases with STL only when DLBCL cells expressed AID and Ugi but not with Ugi alone (Fig. 6 F), despite having a similar level of UNG inhibition (Fig. 6 D). We conclude that human DLBCL cells expressing high levels of AID and active MMR depend on UNG activity for maintenance of telomeres and their proliferation capacity.
DISCUSSION
We identify telomeres as novel off targets of AID in normal and cancer B cells. We also describe an ensuing mechanism that involves error-free and DNA-damaging actions of UNG and MMR, respectively, at the telomeres (Fig. 7). UNG normally protects B cells expressing AID from telomere dysfunction and proliferation defects, thereby permitting a normal GC reaction and antibody response. The fundamentally protective role of UNG at telomeres contrasts with the primary role it plays in generating DNA breaks for CSR and chromosomal translocations (Rada et al., 2004; Ramiro et al., 2006), uncovering an example of locus-specific repair that has critical relevance for cell viability.
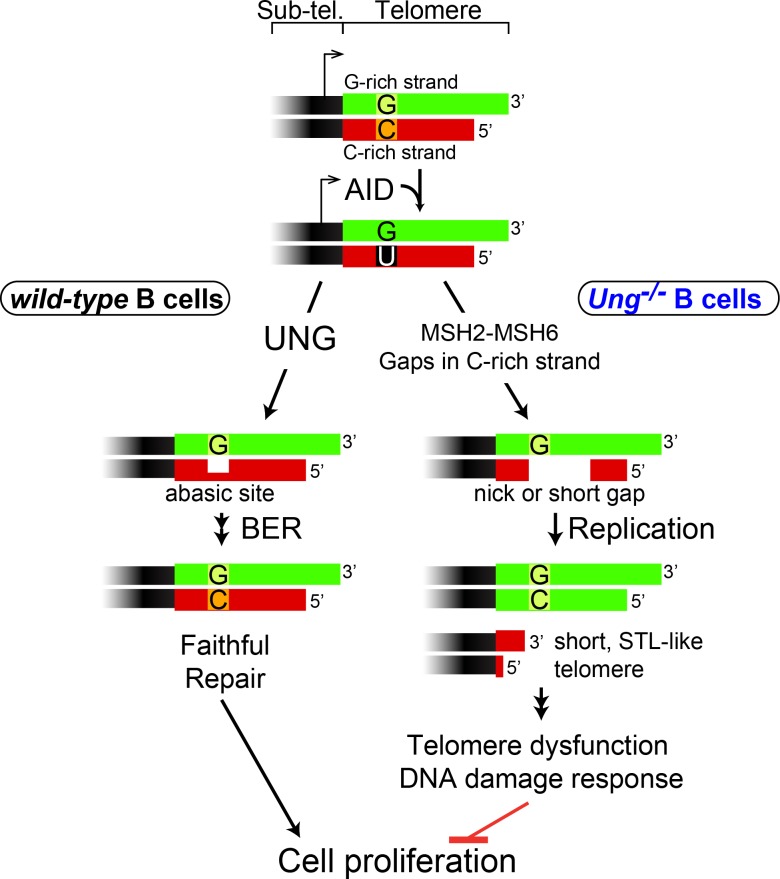
Figure 7.
Proposed molecular mechanism of the alternative processing of AID-induced telomeric damage in B cells. AID is induced in activated B cells and stochastically targets some telomeres, likely through association with the RPII at the subtelomeric (sub-tel.) promoter. UNG initiates error-free BER of the C-rich telomeres deaminated by AID, thereby preventing any telomeric damage and protecting cell proliferation. In UNG-deficient B cells, the uracils made by AID at telomeres are recognized instead as dG:dU mismatches by MMR and processed into a nick or short gap. This nick or short gap could stall leading strand synthesis and produce a very short telomere in one sister chromatid after replication, hampering cell proliferation potential.
The proposed molecular model for telomere maintenance after AID activation (Fig. 7) raises new interesting questions. First, why are uracils at the telomeres not detected by MMR in UNG-sufficient cells? One possibility is that UNG efficiently outcompetes MMR. Another one is that this is determined by the cell cycle stage when deamination and/or repair occurs. For instance, late-replicating DNA can accumulate more mutations because of reduced MMR activity in the late S phase (Supek and Lehner, 2015). Differential MMR activity could underlie the preferential access of UNG to late-replicating, deaminated telomeres (Arnoult et al., 2010). Additionally, AID could facilitate UNG recruitment (Ranjit et al., 2011; Zahn et al., 2014). Second, could other BER glycosylases act as backup for UNG at the telomeres? SMUG1, which can faithfully repair AID-catalyzed uracils (Di Noia et al., 2006), could partly explain the moderate impact of UNG deficiency on the size of GCs in immunized mice. However, in human cells, SMUG1 might not be enough to act as a backup to UNG, as its activity constitutes only 1% of uracil excision activity (Doseth et al., 2012). In support of a dominant role of UNG for processing uracil at the telomeres, it was recently shown that, in mouse hematopoietic cells, UNG is important for long-term maintenance of telomeres by protecting them from uracylation (Vallabhaneni et al., 2015). Yet, in that case, uracils originate from 2′-deoxyuridine 5′-triphosphate (dUTP) misincorporation during replication, which do not produce mismatches. SMUG1 can function upstream of MMR during CSR (Dingler et al., 2014). Thus, SMUG1 could actually induce the nicks needed for MMR-dependent deletion of deaminated telomeres. Finally, the reason why MMR fails to faithfully repair AID-damaged telomeres is unclear. Recently described noncanonical MMR pathways, which can produce genomic instability and create DNA breaks when processing uracils, are attractive possibilities (Peña-Diaz et al., 2012; Bregenhorn et al., 2016).
Our results are likely to explain one or more B cell phenotypes that lack mechanistic observation. UNG deficiency causes immunodeficiency in mice and humans, which has so far been exclusively attributed to reduced isotype switching (Imai et al., 2003; Zahn et al., 2013). The key role of UNG in CSR surely contributes to the hyper-IgM phenotype in the UNG-deficient background. We now uncover a mechanism by which UNG protects the GC reaction. Previously, we had shown that Ung−/− mice have normal B cell populations in the spleen and form GCs with a normal overall architecture (Zahn et al., 2013) but had not quantitatively analyzed GCs in these mice. We now find that although the number of splenic GCs in Ung−/− mice are similar to wild type, those GCs are smaller. The reduced number of AID+ cells in Ung−/− GCs is most likely explained by the telomere dysfunction we describe in activated Ung−/− B cells. This mechanism by which AID can eliminate B cells may contribute to constraining the size of GCs (Robbiani et al., 2009; Zaheen et al., 2009). It might also underpin the mechanism by which AID contributes to self-tolerance (Kelsoe, 2014). Although the handful of UNG-deficient patients available do not display defects in tolerance (Cantaert et al., 2015), Ung−/− mice do show autoantibodies (Zahn et al., 2013). Species-specific differences or clinical bias in the detection of UNG patients may explain this result. Our data also suggest an explanation for the much more profound immunodeficiency seen in UNG-deficient humans relative to mice (Imai et al., 2003; Zahn et al., 2013). Indeed, telomeres in humans are about five-times shorter than in inbred mice (Kipling and Cooke, 1990) and may be more susceptible to AID-induced STL. Finally, our results also provide another plausible explanation to the observation that GC B cells express high levels of telomerase compared with other B cell subsets (Norrback et al., 1996; Hu et al., 1997). Indeed, we show that telomerase helps to moderate telomere dysfunction in UNG-deficient B cells (Fig. 4 C), which probably contributes to moderating the effect of UNG deficiency on GC size.
The off-target mutagenic activity of AID and its ability to initiate chromosomal translocations can be oncogenic (Pasqualucci et al., 2008; Robbiani and Nussenzweig, 2013; Meng et al., 2014; Qian et al., 2014). We demonstrate that AID off-target activity can also induce telomere loss and a DNA damage response at the chromosomes ends. Because short or dysfunctional telomeres are tumor suppressors (Deng et al., 2008), the loss of telomeres induced by AID in UNG-deficient B cells or after UNG inhibition can act as a de facto tumor suppressor mechanism. The lack of AID expression in the B cell lymphomas that grow in UNG-deficient mice does not prove but is consistent with this view. We speculate that AID-mediated telomere loss may act as a fail-safe mechanism to eliminate GC B cells that have too much AID activity and/or are unable to repair their off-target activity and would thereby be at higher oncogenic risk. Thus, our results predict that UNG deficiency could in fact protect from GC-derived B cell lymphoma. Indeed, a recent study found that UNG deficiency protected Iμ-HABcl6 mice from DLBCL (Gu et al., 2016), which was previously shown to be AID dependent in that mouse model (Pasqualucci et al., 2008). In contrast, Iμ-HABcl6 Ung−/− Msh2−/− and Iμσ-HABcl6 Msh2−/− mice developed DLBCL faster than Iμ-HABcl6 mice (Gu et al., 2016). Our results provide the molecular explanation to those observations by indicating that UNG can contribute to lymphomagenesis by protecting the telomeres from AID- and MMR-induced dysfunction in B cell lymphoma cells. This mechanism can be harnessed to kill AID+ cancer cells. We show that cancerous human B cells expressing AID require UNG for proliferation. Many NHL, including a large proportion of DLBCL, the most common aggressive subtype of NHL, express AID (Lossos et al., 2004; Pasqualucci et al., 2004). DLBCL cells rarely lose and sometimes overexpress UNG, whereas MMR factors are frequently mutated in DLBCL (Couronné et al., 2013; de Miranda et al., 2013). Both these characteristics would help transformed cells cope with AID-induced telomere damage. Measuring UNG and MMR integrity would thus be useful for the stratification of B cell lymphomas and other cancers expressing AID. In conclusion, our data delineates a new mechanism protecting the GC reaction from the antibody diversification machinery and provide a rationale for targeting UNG as a means to delay the growth of AID+ cancers.
MATERIALS AND METHODS
Mice, mouse cohorts, and immunization
C57BL6/J mice were from The Jackson Laboratory. AID-GFPtg mice (Crouch et al., 2007), a gift from R. Casellas (National Cancer Institute, Bethesda, MD), Aicda−/− mice, a gift from T. Honjo (Kyoto University, Kyoto, Japan), and Ung−/− mice (Nilsen et al., 2000), a gift from H. Krokan (Norwegian University of Science and Technology, Trondheim, Norway), were all in C57BL6/J background. Aicda−/− Ung−/− mice were bred at the IRCM (Institut de Recherches Cliniques de Montréal) animal facility. Experimental cohorts for lymphoma follow up were observed daily for spontaneous signs of malaise or visible tumors and sacrificed when reaching one of the predefined endpoints or at 30 mo old. Spleens, enlarged lymph nodes, and/or any other tumors were harvested at necropsy and prepared for histology or analyzed by flow cytometry. Where indicated, mice were immunized with 50 µg NP18-CGG (LGC Biosearch Technologies) in Imject Alum adjuvant (Thermo Fisher Scientific) intraperitoneally and analyzed by flow cytometry 8 d later. All experiments were approved by the animal protection committee at the IRCM, according to the guidelines of the Canadian Council on Animal Care.
Antibodies sources and Western blotting
Western blotting for the different proteins was performed as previously described (Verdun et al., 2005). The following antibodies were used in this study: actin and γ-tubulin (Sigma-Aldrich), AID (Active Motif), EXO1 (Novus Biologicals), γH2AX (EMD Millipore), MSH2 and SPT5 (Santa Cruz Biotechnology, Inc.), and TRF1, a gift from J. Karlseder (Salk Institute, La Jolla, CA). The antibodies used for flow cytometry are indicated in the Flow cytometry and cell cycle analysis section, and the antibodies used for IHC are indicated in the Histological analysis section.
Cell lines and CSR analysis
DLBCL cell lines, a gift from I. Lossos (University of Miami, Miami, Florida), were cultured in IMDM medium supplemented with 20% of human plasma. CH12F3-2 cells were cultured in RPMI medium supplemented with 10% FBS, 0.05% 2-mercaptoethanol, and 5% NCTC 109 (Sigma-Aldrich). CH12F3-2 cells stably expressing Ugi were obtained by retroviral delivery using a pMIG vector as previously described (Cortizas et al., 2013). Class switching induction to IgA in CH12F3-2 cells was performed as previously described (Cortizas et al., 2013). CSR to IgA in CH12F3-2 cells was determined with a flow cytometer (Accuri C6; BD). Mouse B cells were purified from freshly isolated splenocytes using anti-CD43 magnetic beads as previously described (Zahn et al., 2014). CSR was induced using 5 µg/ml LPS (Sigma-Aldrich) and 20 ng/ml IL-4 (PeproTech). Cells were harvested for ChIP or metaphase spreads and FISH at 24 h after stimulation.
ChIP assays
ChIPs to evaluate interaction with the telomeric chromatin or the Ig locus were performed as described previously (Cortizas et al., 2013). In brief, cells were cross-linked with 1% formaldehyde for 20 min at room temperature, and the reaction was stopped by the addition of glycine to 125 mM final concentration. Cells were washed twice with cold PBS, harvested, and kept at −80°C overnight (ON). Samples were resuspended in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% [vol/vol] Igepal CA-630, 0.5% [wt/vol] sodium deoxycholate, 0.1% [wt/vol] SDS, 50 mM Tris-HCl, pH 8, 5 mM EDTA, pH 8, and protease and phosphatase inhibitors) and sonicated to generate DNA fragments <500 bp using a Bioruptor Next Gen sonication device (Diagenode).
To increase the sensitivity of the ChIP assays and evaluate the presence of AID at the telomeres, we performed the immunoprecipitations with fractions enriched in cross-linked chromatin. For this goal, B cells fixed with 1% formaldehyde as described in the previous paragraph were resuspended in RIPA buffer and sonicated for 90 s using the Bioruptor Next Gen sonication device, and the lysates were then spun down briefly (2,000 g for 5 min at 4°C) to remove debris. Then, sucrose was added to the extract (5% final concentration) and layered onto 20% sucrose in RIPA buffer for ultracentrifugation at 40,000 rpm for 1.5 h using a rotor (TLA-100; Beckman Coulter). The chromatin-enriched pellets were then sonicated with the Bioruptor Next Gen sonication device to generate DNA fragments <500 bp and used for the immunoprecipitation step. Samples were then clarified by centrifugation at 20,000 rpm and 4°C, and their protein content was measured using a DC protein assay (Bio-Rad Laboratories).
For immunoprecipitation, 0.5 mg (2 mg/ml) of protein extract or 0.2 mg (1 mg/ml) of chromatin-enriched extract was precleared for 2 h with 30 µl of 50% G protein–Sepharose slurry before addition of the indicated antibodies. Between 2 and 5 µg of each antibody was added to the samples and incubated ON at 4°C. Immunocomplexes were eluted from agarose A/G plus (Santa Cruz Biotechnology, Inc.) for 10 min at 65°C with 100 µl of elution buffer (1% [wt/vol] SDS), and cross-linking was reversed by adjusting to 200 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol and incubating ON at 65°C in the presence of 5 µg proteinase K. DNA was purified using a QIAquick PCR purification kit (QIAGEN), and DNA resuspended in 60 µl of 10 mM Tris-HCl, pH 8, was used as a template in real-time PCR reactions to evaluate coimmunoprecipitation of Igh DNA or Southern blotting with a telomeric probe or ALU repeat probe as described previously (Verdun et al., 2005). IgG and input DNA values were used to subtract/normalize the values from ChIP samples. All primer sequences used for the ChIP analyses are available upon request.
FISH
For metaphase analysis, cells were incubated with 50 ng/ml colcemide in cell culture media for 3 h, harvested by trypsinization, incubated for 10 min at room temperature in 75 mM KCl, and fixed in freshly prepared methanol/glacial acetic acid (3:1 vol/vol). Cells were stored at 4°C and, when needed, dropped onto wet slides and air dried. For FISH analysis of the metaphases, the cells were pretreated with 0.05% wt/vol pepsin in 10 mM HCl for 10 min at 37°C. After washes with 1× PBS, cells were fixed with 1% formaldehyde in 1× PBS for 10 min at room temperature, washed again with 1× PBS, and dehydrated with an ethanol series (70-90-100%; 2 min each at room temperature), and air dried ON. Then, cells were denatured with hybridization solution (70% deionized formamide, 2.5% 50× Denhardt solution, 10 mM Tris, pH 7.5, and 1.5 mM MgCl2) containing Alexa Fluor 488–conjugated PNA probe (Alexa Fluor 488–OO-[CCCTAA]3) for 2 min at 80°C on a heat block. After 4-h incubation at room temperature in the dark, the samples were washed twice with wash solution (70% deionized formamide and 10 mM Tris-HCl, pH 7.2) at room temperature and then twice with PBS. For DAPI staining of DNA, slides with metaphase spreads were incubated 10 min in 0.5 µg/ml DAPI (Sigma-Aldrich) in PBS, washed with PBS for 2 min, and mounted in SlowFade Gold antifade mounting reagent (Thermo Fisher Scientific). Finally, the samples were analyzed as described in Results. Where indicated, metaphases were spread as previously described (Verdun et al., 2005).
Anaphase cells were visualized by DAPI staining of cells fixed with 2% paraformaldehyde in 1× PBS, attached to slides pretreated with polylysine, and analyzed as described previously (Crabbe et al., 2004). A total of 50 anaphases were analyzed in each cell line per experiment.
CO-FISH
CO-FISH was performed as previously described (Crabbe et al., 2004), with the variation that cells were incubated with BrdU and BrdC simultaneously for 16 h, and hybridization was performed with CY5-OO-(TTAGGG)3 and Alexa Fluor 488–OO-(CCCTAA)3 probes (Panagene). In brief, BrdU and BrdC were incorporated into chromosomes throughout one S phase, metaphases were spread on slides, the BrdU-substituted DNA strands were degraded with exonuclease III, and the remaining strands were hybridized with fluorescence-labeled DNA probes of different colors, specific either for the G-rich telomere strand ([TTAGGG]n, polymerized by lagging strand synthesis) or the C-rich telomere strand ([CCCTAA]n, polymerized by leading strand synthesis). Hybridization with PNA probes and DAPI staining were performed as described in the FISH section. The resulting chromosomes show dual staining and allow distinction between leading and lagging strands. Chromosomes were visualized using a microscope (DMI6000B; Leica Biosystems). A total of 50 metaphases were analyzed in each cell line per experiment.
Flow cytometry and cell cycle analysis
Mononuclear cells from mouse spleens or other organs were extracted using a cell strainer and stained with anti-B220–APC and anti-CD3–PE (BD). Propidium iodide was used to gate out dead cells. Results were acquired using an LSR I flow cytometer (BD) and analyzed in FlowJo software (Tree Star).
For BrdU incorporation analysis, CH12F3 cells (2 × 105/ml) were incubated for 30 min in culture medium containing 10 µM BrdU. Then, cells were harvested, washed twice with PBS, and fixed in cold 70% ethanol ON at 4°C. After removal of ethanol, DNA was denatured with 2 N HCl/0.5% Triton X-100 for 30 min at room temperature, neutralized with two washes of 0.1 M sodium tetraborate, pH 9, and resuspended in 70% ethanol. Cells were recovered by centrifugation, washed once with PBS, resuspended in 100 µl of blocking buffer (0.5% Tween 20 and 1% BSA in PBS) containing 10 µl mouse anti-BrdU antibody (BD), and incubated at room temperature for 30 min. After a wash with PBS, cells were incubated for 15 min at room temperature with goat anti–mouse Alexa Fluor 647 antibody diluted in blocking buffer. Finally, cells were washed with PBS once, resuspended in PBS containing 5 µg/ml propidium iodide, and analyzed using an Accuri flow cytometer.
Histological analysis
Tissues were fixed with 4% formaldehyde ON at room temperature and embedded in paraffin. Sections were stained with hematoxylin and eosin. IHC was performed on 5-µm sections of paraffin-embedded tissues, deparaffinized, rehydrated, and subjected to antigen retrieval. After blocking endogenous peroxidase activity with 3% hydrogen peroxide, sections were incubated ON at 4°C with rat anti–mouse CD45R (1:50; BD) or AID (1:100; eBioscience) followed by goat bio–anti–rat IgG or goat bio–anti–rabbit IgG (1:200; Vector Laboratories) or directly with 20 µg/ml biotinylated PNA (Vector Laboratories) for 60 min at room temperature and developed using avidin, bio-HRP, and HRP substrate included in the ImmPACT NovaRED HRP kit (Vector Laboratories). Formalin-fixed and paraffin-embedded tissues were analyzed by a pathologist after hematoxylin and eosin and IHC staining. Lymphoma was defined by the presence of nodular and/or diffuse lymphoid aggregates composed by a relatively monomorphic population of intermediate to large lymphoid cells with cytological atypia and increased number of mitoses. Features of cytological atypia included a combination of the following: increased size of the lymphoid cells, presence of irregular nuclear contours, and one or more enlarged nucleoli. Nodular lymphoid hyperplasia was defined by the presence of expanded lymphoid follicles, with or without GCs that were composed by lymphoid cells predominantly small in size and without cytological atypia. For visualization of AID-GFP in the spleen of AID-GFPtg mice, organs were embedded in CryoMatrix (Thermo Fisher Scientific) and snap frozen before sectioning. Sections were fixed in 3.7% (wt/vol) paraformaldehyde for 10 min, permeabilized with 0.5% Triton X-100 for 10 min, washed, and incubated for 15 min with DAPI before mounting in Aqua Mount (Thermo Fisher Scientific). Some cryosections were stained with anti-IgD–PE (1/200; BioLegend) and/or biotinylated PNA (1:200; Vector Laboratories) + streptavidin-APC (1:100; BD). Images of GCs were taken with a confocal microscope (LSM700; ZEISS) fitted with 555-nm, 488-nm, and 639-nm lasers. Images of whole tissue sections were composed from multiple fields imaged using a spinning disk confocal microscope (LSM700) with a Cell Observer SD and a CSU-X1 Yokogawa head (ZEISS). The GFP+ areas inside B cell follicles were scored in ImageJ (National Institutes of Health).
shRNA knockdowns and Ugi expression
We generated stable shRNA-mediated knockdowns in CH12F3 cells following protocols from the RNAi Consortium. Transfected cells were selected with puromycin at 1 µg/ml final concentration. For double knockdowns, CH12F3 cells were first transfected with a pLKO.1 neomycin version of the shRNA (shAID or shGFP), selected and then transduced with the second shRNA, and selected again with puromycin before analysis. The shRNAs used in this study are available upon request. CH12F3 Ugi cells transduced with pMIG-Ugi-IRES (internal ribosomal entry site)–GFP were generated as described previously (Cortizas et al., 2013), human lymphoma Ugi cells were generated by retroviral delivery using a pLPC-PURO system (pLPC-Ugi-PURO), and transduced cells were selected with 1 µg/ml of puromycin. The Ugi protein from bacteriophage PBS2 and its ability to inhibit eukaryotic UNG have been described previously (Di Noia and Neuberger, 2002).
Ung activity assay
The UNG activity assay was performed as previously described (Di Noia and Neuberger, 2002) with minor modifications. In brief, exponentially growing cells were washed in 1× PBS buffer, resuspended in HED buffer (25 mM Hepes, 5 mM EDTA, 1 mM dithiothreitol, and 10% glycerol, pH 7.8) with complete protease inhibitors (Roche), and lysed by sonication (five pulses of 30 s) in a Bioruptor (Diagenode). After centrifugation at 14,000 rcf for 20 min at 4°C, the supernatant was frozen in aliquots into liquid nitrogen and stored at 80°C. UNG assays were performed in HED buffer by mixing 10 µg of cell extract with 1 pmol of fluorescein-labeled oligonucleotide substrate in a final volume of 10 µl for 3 h at 37°C. The double-stranded oligonucleotide with a single deoxyuridine/deoxyguanosine mismatch was made by annealing 5′-ATTATTATTATTCCGUGGATTTATTTATTTATTTATTTATTT-fluorescein to the complementary oligonucleotide, 5′-AAATAAATAAATAAATAAATAAATCCGCGGAATAATAATAAT-3′. The reaction was terminated by the addition of 10 µl of formamide loading dye, and the products were resolved on 15% Tris/borate/EDTA (TBE)-urea polyacrylamide gels. The apyrimidinic endonuclease activity present in the extracts was sufficient to cleave all the abasic substrate generated by UNG during the reaction. The fluorescein signal of the reaction products was visualized using a Typhoon PhosphorImager (GE Healthcare).
ACKNOWLEDGMENTS
We are indebted to Dr. H. Krokan, T. Honjo, and R. Casellas for mouse strains, Dr. I. Lossos for DLBCL cell lines and advice in mouse tumor analysis, M. Cawthorn for technical assistance with mice, D. Fillon and S. Methot for help with confocal microscopy and image analysis, S. Terouz for histological preparations, the University of Miami Sylvester Cancer Center Oncogenomics facility for DNA sequencing assistance, Dr. D. Muñoz for advice on AID IHC, and Dr. J. Karlseder, Dr. N. Francis, and Dr. C. Rada for comments on the manuscript.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (R56-AI106894-01A1), the Miami Center for AIDS Research (1P30AI073961 to R.E. Verdun), and the Canadian Institutes of Health Research (MOP130535 to J.M. Di Noia). J.M. Di Noia holds a Canada Research Chair tier 2.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AID
- activation-induced deaminase
- AID-GFPtg
- AID-GFP transgenic
- BER
- base excision repair
- ChIP
- chromatin immunoprecipitation
- CO-FISH
- chromosome orientation FISH
- CSR
- class switch recombination
- DLBCL
- diffuse large B cell lymphoma
- FISH
- fluorescent in situ hybridization
- GC
- germinal center
- IHC
- immunohistochemistry
- MMR
- mismatch DNA repair
- NHL
- non–Hodgkin’s lymphoma
- ON
- overnight
- PNA
- peanut agglutinin
- RIPA
- radioimmunoprecipitation assay
- S phase
- synthesis phase
- S region
- switch region
- SHM
- somatic hypermutation
- ssDNA
- single-stranded DNA
- STL
- sister telomere loss
- TBE
- Tris/borate/EDTA
- UNG
- uracil-DNA glycosylase
References
- Andersen S., Ericsson M., Dai H.Y., Peña-Diaz J., Slupphaug G., Nilsen H., Aarset H., and Krokan H.E.. 2005. Monoclonal B-cell hyperplasia and leukocyte imbalance precede development of B-cell malignancies in uracil-DNA glycosylase deficient mice. DNA Repair (Amst.). 4:1432–1441. 10.1016/j.dnarep.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Arnoult N., and Karlseder J.. 2015. Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 22:859–866. 10.1038/nsmb.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N., Schluth-Bolard C., Letessier A., Drascovic I., Bouarich-Bourimi R., Campisi J., Kim S.H., Boussouar A., Ottaviani A., Magdinier F., et al. 2010. Replication timing of human telomeres is chromosome arm–specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 6:e1000920 10.1371/journal.pgen.1000920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., and Lingner J.. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 318:798–801. 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- Balk B., Maicher A., Dees M., Klermund J., Luke-Glaser S., Bender K., and Luke B.. 2013. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 20:1199–1205. 10.1038/nsmb.2662 [DOI] [PubMed] [Google Scholar]
- Bregenhorn S., Kallenberger L., Artola-Borán M., Peña-Diaz J., and Jiricny J.. 2016. Non-canonical uracil processing in DNA gives rise to double-strand breaks and deletions: relevance to class switch recombination. Nucleic Acids Res. 44:2691–2705. 10.1093/nar/gkv1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.R., Wang Y., Andrew S.E., and Liu Y.. 2006. Msh2 deficiency leads to chromosomal abnormalities, centrosome amplification, and telomere capping defect. Oncogene. 25:2531–2536. 10.1038/sj.onc.1209277 [DOI] [PubMed] [Google Scholar]
- Cantaert T., Schickel J.N., Bannock J.M., Ng Y.S., Massad C., Oe T., Wu R., Lavoie A., Walter J.E., Notarangelo L.D., et al. 2015. Activation-induced cytidine deaminase expression in human B cell precursors is essential for central B cell tolerance. Immunity. 43:884–895. 10.1016/j.immuni.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortizas E.M., Zahn A., Hajjar M.E., Patenaude A.M., Di Noia J.M., and Verdun R.E.. 2013. Alternative end-joining and classical nonhomologous end-joining pathways repair different types of double-strand breaks during class-switch recombination. J. Immunol. 191:5751–5763. 10.4049/jimmunol.1301300 [DOI] [PubMed] [Google Scholar]
- Couronné L., Ruminy P., Waultier-Rascalou A., Rainville V., Cornic M., Picquenot J.M., Figeac M., Bastard C., Tilly H., and Jardin F.. 2013. Mutation mismatch repair gene deletions in diffuse large B-cell lymphoma. Leuk. Lymphoma. 54:1079–1086. 10.3109/10428194.2012.739687 [DOI] [PubMed] [Google Scholar]
- Crabbe L., Verdun R.E., Haggblom C.I., and Karlseder J.. 2004. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 306:1951–1953. 10.1126/science.1103619 [DOI] [PubMed] [Google Scholar]
- Crouch E.E., Li Z., Takizawa M., Fichtner-Feigl S., Gourzi P., Montaño C., Feigenbaum L., Wilson P., Janz S., Papavasiliou F.N., and Casellas R.. 2007. Regulation of AID expression in the immune response. J. Exp. Med. 204:1145–1156. 10.1084/jem.20061952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., and Jackson S.P.. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 426:194–198. 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- de Miranda N.F., Peng R., Georgiou K., Wu C., Falk Sörqvist E., Berglund M., Chen L., Gao Z., Lagerstedt K., Lisboa S., et al. 2013. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J. Exp. Med. 210:1729–1742. 10.1084/jem.20122842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Chan S.S., and Chang S.. 2008. Telomere dysfunction and tumour suppression: the senescence connection. Nat. Rev. Cancer. 8:450–458. 10.1038/nrc2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingler F.A., Kemmerich K., Neuberger M.S., and Rada C.. 2014. Uracil excision by endogenous SMUG1 glycosylase promotes efficient Ig class switching and impacts on A:T substitutions during somatic mutation. Eur. J. Immunol. 44:1925–1935. 10.1002/eji.201444482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J., and Neuberger M.S.. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48. 10.1038/nature00981 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Rada C., and Neuberger M.S.. 2006. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J. 25:585–595. 10.1038/sj.emboj.7600939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doseth B., Ekre C., Slupphaug G., Krokan H.E., and Kavli B.. 2012. Strikingly different properties of uracil-DNA glycosylases UNG2 and SMUG1 may explain divergent roles in processing of genomic uracil. DNA Repair (Amst.). 11:587–593. 10.1016/j.dnarep.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Gu X., Booth C.J., Liu Z., and Strout M.P.. 2016. AID-associated DNA repair pathways regulate malignant transformation in a murine model of BCL6-driven diffuse large B-cell lymphoma. Blood. 127:102–112. 10.1182/blood-2015-02-628164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasham M.G., Donghia N.M., Coffey E., Maynard J., Snow K.J., Ames J., Wilpan R.Y., He Y., King B.L., and Mills K.D.. 2010. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat. Immunol. 11:820–826. 10.1038/ni.1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.T., Lee S.C., Marin E., Ryan D.H., and Insel R.A.. 1997. Telomerase is up-regulated in human germinal center B cells in vivo and can be re-expressed in memory B cells activated in vitro. J. Immunol. 159:1068–1071. [PubMed] [Google Scholar]
- Imai K., Slupphaug G., Lee W.I., Revy P., Nonoyama S., Catalan N., Yel L., Forveille M., Kavli B., Krokan H.E., et al. 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 4:1023–1028. 10.1038/ni974 [DOI] [PubMed] [Google Scholar]
- Kelsoe G. 2014. Curiouser and curiouser: the role(s) of AID expression in self-tolerance. Eur. J. Immunol. 44:2876–2879. 10.1002/eji.201445102 [DOI] [PubMed] [Google Scholar]
- Kipling D., and Cooke H.J.. 1990. Hypervariable ultra-long telomeres in mice. Nature. 347:400–402. 10.1038/347400a0 [DOI] [PubMed] [Google Scholar]
- Liu M., Duke J.L., Richter D.J., Vinuesa C.G., Goodnow C.C., Kleinstein S.H., and Schatz D.G.. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845. 10.1038/nature06547 [DOI] [PubMed] [Google Scholar]
- Lossos I.S., Levy R., and Alizadeh A.A.. 2004. AID is expressed in germinal center B-cell-like and activated B-cell-like diffuse large-cell lymphomas and is not correlated with intraclonal heterogeneity. Leukemia. 18:1775–1779. 10.1038/sj.leu.2403488 [DOI] [PubMed] [Google Scholar]
- Meng F.L., Du Z., Federation A., Hu J., Wang Q., Kieffer-Kwon K.R., Meyers R.M., Amor C., Wasserman C.R., Neuberg D., et al. 2014. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 159:1538–1548. 10.1016/j.cell.2014.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen H., Steinsbekk K.S., Otterlei M., Slupphaug G., Aas P.A., and Krokan H.E.. 2000. Analysis of uracil-DNA glycosylases from the murine Ung gene reveals differential expression in tissues and in embryonic development and a subcellular sorting pattern that differs from the human homologues. Nucleic Acids Res. 28:2277–2285. 10.1093/nar/28.12.2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen H., Stamp G., Andersen S., Hrivnak G., Krokan H.E., Lindahl T., and Barnes D.E.. 2003. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 22:5381–5386. 10.1038/sj.onc.1206860 [DOI] [PubMed] [Google Scholar]
- Norrback K.F., Dahlenborg K., Carlsson R., and Roos G.. 1996. Telomerase activation in normal B lymphocytes and non-Hodgkin’s lymphomas. Blood. 88:222–229. [PubMed] [Google Scholar]
- Pasqualucci L., Guglielmino R., Houldsworth J., Mohr J., Aoufouchi S., Polakiewicz R., Chaganti R.S., and Dalla-Favera R.. 2004. Expression of the AID protein in normal and neoplastic B cells. Blood. 104:3318–3325. 10.1182/blood-2004-04-1558 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Bhagat G., Jankovic M., Compagno M., Smith P., Muramatsu M., Honjo T., Morse H.C. III, Nussenzweig M.C., and Dalla-Favera R.. 2008. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 40:108–112. 10.1038/ng.2007.35 [DOI] [PubMed] [Google Scholar]
- Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V., et al. 2010. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 143:122–133. 10.1016/j.cell.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Diaz J., Bregenhorn S., Ghodgaonkar M., Follonier C., Artola-Borán M., Castor D., Lopes M., Sartori A.A., and Jiricny J.. 2012. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell. 47:669–680. 10.1016/j.molcel.2012.07.006 [DOI] [PubMed] [Google Scholar]
- Peters A., and Storb U.. 1996. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 4:57–65. 10.1016/S1074-7613(00)80298-8 [DOI] [PubMed] [Google Scholar]
- Pfeiffer V., Crittin J., Grolimund L., and Lingner J.. 2013. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 32:2861–2871. 10.1038/emboj.2013.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Wang Q., Dose M., Pruett N., Kieffer-Kwon K.R., Resch W., Liang G., Tang Z., Mathé E., Benner C., et al. 2014. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 159:1524–1537. 10.1016/j.cell.2014.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C., and Milstein C.. 2001. The intrinsic hypermutability of antibody heavy and light chain genes decays exponentially. EMBO J. 20:4570–4576. 10.1093/emboj/20.16.4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C., Di Noia J.M., and Neuberger M.S.. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171. 10.1016/j.molcel.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Stavropoulos P., Jankovic M., and Nussenzweig M.C.. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456. 10.1038/ni920 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Callen E., Difilippantonio S., Chen H.T., McBride K.M., Eisenreich T.R., Chen J., Dickins R.A., Lowe S.W., et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109. 10.1038/nature04495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjit S., Khair L., Linehan E.K., Ucher A.J., Chakrabarti M., Schrader C.E., and Stavnezer J.. 2011. AID binds cooperatively with UNG and Msh2-Msh6 to Ig switch regions dependent upon the AID C terminus. J. Immunol. 187:2464–2475. 10.4049/jimmunol.1101406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., and Nussenzweig M.C.. 2013. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu. Rev. Pathol. 8:79–103. 10.1146/annurev-pathol-020712-164004 [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Bunting S., Feldhahn N., Bothmer A., Camps J., Deroubaix S., McBride K.M., Klein I.A., Stone G., Eisenreich T.R., et al. 2009. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell. 36:631–641. 10.1016/j.molcel.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S., and Blasco M.A.. 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10:228–236. 10.1038/ncb1685 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E., and Schrader C.E.. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292. 10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U. 2014. Why does somatic hypermutation by AID require transcription of its target genes? Adv. Immunol. 122:253–277. 10.1016/B978-0-12-800267-4.00007-9 [DOI] [PubMed] [Google Scholar]
- Supek F., and Lehner B.. 2015. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 521:81–84. 10.1038/nature14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B.J., Wu Y.L., and Rada C.. 2014. Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes. eLife. 3:e03553 10.7554/eLife.03553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni H., Zhou F., Maul R.W., Sarkar J., Yin J., Lei M., Harrington L., Gearhart P.J., and Liu Y.. 2015. Defective repair of uracil causes telomere defects in mouse hematopoietic cells. J. Biol. Chem. 290:5502–5511. 10.1074/jbc.M114.607101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun R.E., and Karlseder J.. 2007. Replication and protection of telomeres. Nature. 447:924–931. 10.1038/nature05976 [DOI] [PubMed] [Google Scholar]
- Verdun R.E., Crabbe L., Haggblom C., and Karlseder J.. 2005. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell. 20:551–561. 10.1016/j.molcel.2005.09.024 [DOI] [PubMed] [Google Scholar]
- Victora G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Zaheen A., Boulianne B., Parsa J.Y., Ramachandran S., Gommerman J.L., and Martin A.. 2009. AID constrains germinal center size by rendering B cells susceptible to apoptosis. Blood. 114:547–554. 10.1182/blood-2009-03-211763 [DOI] [PubMed] [Google Scholar]
- Zahn A., Daugan M., Safavi S., Godin D., Cheong C., Lamarre A., and Di Noia J.M.. 2013. Separation of function between isotype switching and affinity maturation in vivo during acute immune responses and circulating autoantibodies in UNG-deficient mice. J. Immunol. 190:5949–5960. 10.4049/jimmunol.1202711 [DOI] [PubMed] [Google Scholar]
- Zahn A., Eranki A.K., Patenaude A.M., Methot S.P., Fifield H., Cortizas E.M., Foster P., Imai K., Durandy A., Larijani M., et al. 2014. Activation induced deaminase C-terminal domain links DNA breaks to end protection and repair during class switch recombination. Proc. Natl. Acad. Sci. USA. 111:E988–E997. 10.1073/pnas.1320486111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Vuong B.Q., Vaidyanathan B., Lin J.Y., Huang F.T., and Chaudhuri J.. 2015. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell. 161:762–773. 10.1016/j.cell.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]