Significance
Psoriasis is a complex inflammatory disease with clear genetic contribution that affects roughly 2% of the population in Europe and North America. Inflammation of the skin, and in many cases the joints, leads to severe clinical symptoms, including disfiguration and disability. Immune cells and their inflammatory effector functions have been identified as critical factors for disease development; however, how genetic susceptibility contributes to disease remains largely unclear. Here we developed mouse models based on the gene TNIP1, whose loss-of-function in humans is linked to psoriasis. Based on these models, we provide evidence that nonimmune cells, specifically skin-resident keratinocytes, contribute causally to disease. This work shifts attention to keratinocytes as causal contributors and therapeutic targets in psoriasis.
Keywords: psoriasis, Tnip1, Abin1, keratinocytes, IL-17
Abstract
Psoriasis is a chronic inflammatory skin disease with a clear genetic contribution, characterized by keratinocyte proliferation and immune cell infiltration. Various closely interacting cell types, including innate immune cells, T cells, and keratinocytes, are known to contribute to inflammation. Innate immune cells most likely initiate the inflammatory process by secretion of IL-23. IL-23 mediates expansion of T helper 17 (Th17) cells, whose effector functions, including IL-17A, activate keratinocytes. Keratinocyte activation in turn results in cell proliferation and chemokine expression, the latter of which fuels the inflammatory process through further immune cell recruitment. One question that remains largely unanswered is how genetic susceptibility contributes to this process and, specifically, which cell type causes disease due to psoriasis-specific genetic alterations. Here we describe a mouse model based on the human psoriasis susceptibility locus TNIP1, also referred to as ABIN1, whose gene product is a negative regulator of various inflammatory signaling pathways, including the Toll-like receptor pathway in innate immune cells. We find that Tnip1-deficient mice recapitulate major features of psoriasis on pathological, genomic, and therapeutic levels. Different genetic approaches, including tissue-specific gene deletion and the use of various inflammatory triggers, reveal that Tnip1 controls not only immune cells, but also keratinocyte biology. Loss of Tnip1 in keratinocytes leads to deregulation of IL-17–induced gene expression and exaggerated chemokine production in vitro and overt psoriasis-like inflammation in vivo. Together, the data establish Tnip1 as a critical regulator of IL-17 biology and reveal a causal role of keratinocytes in the pathogenesis of psoriasis.
The etiology of psoriasis is complex and involves both genetic and environmental risk factors. The latter include physical stress and exogenous inflammatory triggers, which may lead to transient inflammation in healthy subjects; however, in genetically susceptible individuals, the same exogenous triggers lead to improper containment of inflammation and eventually psoriasis disease, characterized by skin infiltrations with various immune cell types and keratinocyte proliferation (1). Thus, genetic susceptibility provides the basis for inadequate interpretation and containment of inflammatory triggers.
Significant progress in the understanding of the pathogenesis and treatment of psoriasis has been made in the last several years (2). Detailed animal models and therapeutic studies in humans have revealed a key role of immune cells and the so-called “IL-23/IL-17 axis,” where activated myeloid cells, possibly on exposure to a less well-defined Toll-like receptor (TLR) agonist, produce IL-23, which activates specific T-cell subsets to produce IL-17 (3–5). Other major contributors to psoriasis are nonhematopoietic cells, specifically keratinocytes and fibroblasts, which produce various factors, including chemokines, particularly on IL-17 exposure. Chemokines, in turn, have various functions, including recruitment of immune cells into the skin, such as IL-23–producing myeloid cells and IL-17–producing T-cells, as well as neutrophilic granulocytes forming pathognomonic microabscesses (6–9). As such, two major entities—IL-23– and IL-17–producing immune cells and chemokine-producing nonhematopoietic cells—appear to be critical constituents of an amplifying feed forward loop that promotes disease (2, 10).
One major question is which of these processes are actually deregulated due to psoriasis-specific genetic alterations and which merely follow the physiological sequelae of inflammation biology. For example, it is currently unclear whether it is primarily immune cell biology that is deregulated (e.g., in form of exaggerated IL-23 and IL-17 production), or if keratinocyte biology is at the root of the problem (e.g., via increased production of chemokines). Although therapeutic approaches targeting key inflammatory effector mechanisms, such as IL-23 and IL-17, are producing important benefits in a large percentage of patients, it is likely that a better understanding of causative factors will be relevant to further improve therapeutic strategies, not least from the perspective of prevention (11–13).
An important advance in psoriasis research is the identification of various genetic psoriasis loci, which provide the basis for the aforementioned genetic susceptibility. Genes identified in these loci span an array of possible activities, including adaptive immune cell functions and cytokine regulation. Their precise functions and roles in various cell types are just beginning to emerge, however. In part, this limited understanding in disease causality is due to the just-starting implementation of respective mouse models that are based on human susceptibility factors (14, 15). One defined susceptibility locus is TNIP1 (TNFAIP3-interacting protein 1), which encodes a protein with established negative regulatory function in the TNFR and TLR pathways (16–19). We had previously identified TNIP1/ABIN1 (A20-binding inhibitor of NF-kappa-B activation 1) proteomically as part of the TLR signaling complex, and more detailed work based on macrophages derived from Tnip1−/− mice revealed a critical function of TNIP1/ABIN1 in the C/EBPβ pathway, controlling a small, selective number of TLR target genes (19). Genome-wide association studies (GWAS) revealed several psoriasis-specific single-nucleotide polymorphisms in the intergenic (noncoding) region upstream of TNIP1, and more detailed analysis of the skin of psoriasis patients with respective polymorphisms demonstrated significantly reduced Tnip1 expression, strongly suggesting loss of function of TNIP1 as a cause for disease susceptibility (16). As mentioned above, on the basis of such genetic predisposition, partially defined exogenous factors, such as physical stress or drug-mediated TLR7 activation, appear to instigate deregulated gene expression, resulting in exaggerated inflammation and overt disease flares. The hypothesis that reduced expression of TNIP1 provides a defined genetic susceptibility factor for psoriasis is supported by experiments based on deletion of Tnip1 in myeloid cells, resulting in increased production of TLR-induced cytokines, including IL-23, as well as increased skin inflammation on exposure to the TLR7 agonist imiquimod (IMQ) (19, 20).
Here we investigated mouse strains with germ line- or keratinocyte-specific deletion of Tnip1, resulting in loss of function of Tnip1 in all tissues or selectively in keratinocytes, respectively. Based on detailed pathological, immunological, transcriptional, and therapeutic analyses, we found that loss of Tnip1 function and exposure to proinflammatory triggers lead to an inflammatory skin disease with major characteristics of human psoriasis. Using these novel models, we investigated the contribution of different cell types and major inflammatory factors, including IL-17, in disease pathogenesis. We found that Tnip1 directly controls IL-17–mediated chemokine regulation in nonhematopoietic cells, which contribute causally to disease development.
Results
IMQ Triggers a Psoriasis-Like Disease in Tnip1−/− Mice.
IMQ, a synthetic TLR7 agonist, has been shown to trigger and exacerbate psoriasis flares in susceptible patients on topical application (21–24). In wildtype (WT) mice, repetitive skin application of IMQ at higher concentrations (here referred to as IMQhigh) was shown to induce skin inflammation that displays symptoms similar to psoriasis (25); however, more detailed, transcriptome-based cross-species comparison studies revealed that IMQhigh induced not only psoriasis-specific genes, but also a large number of unrelated genes (15). This observation is not entirely unexpected, because TLR activation results in extensive changes in gene expression, most likely exceeding psoriasis-specific deregulation in favor of a more general inflammatory immune response.
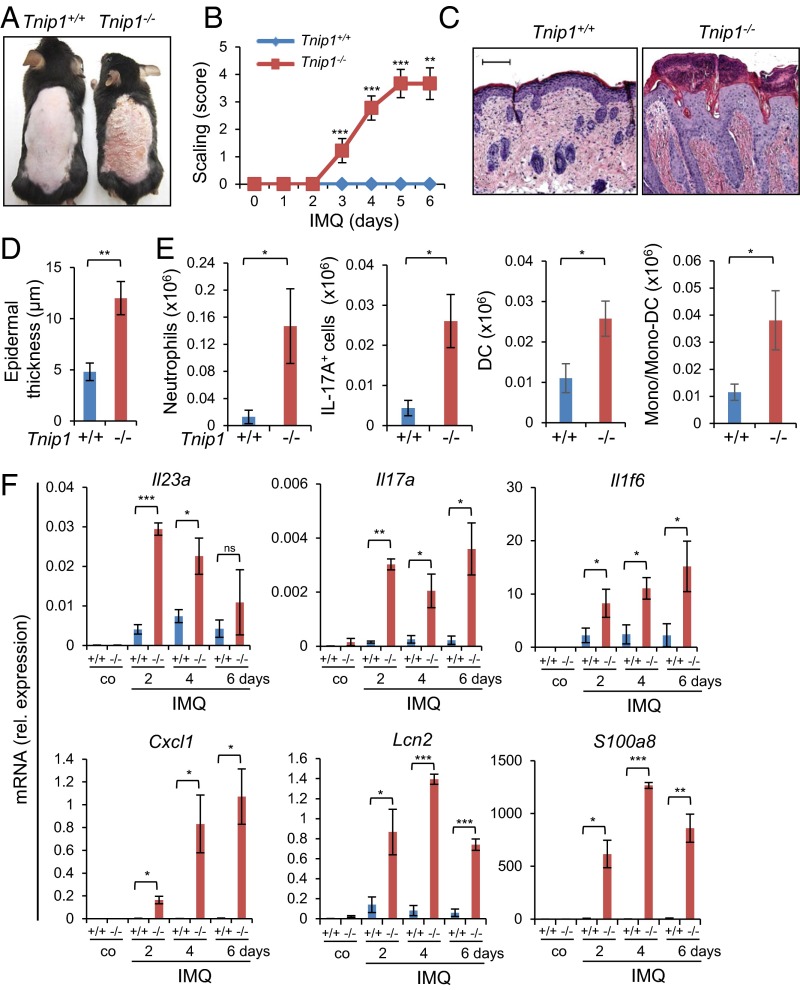
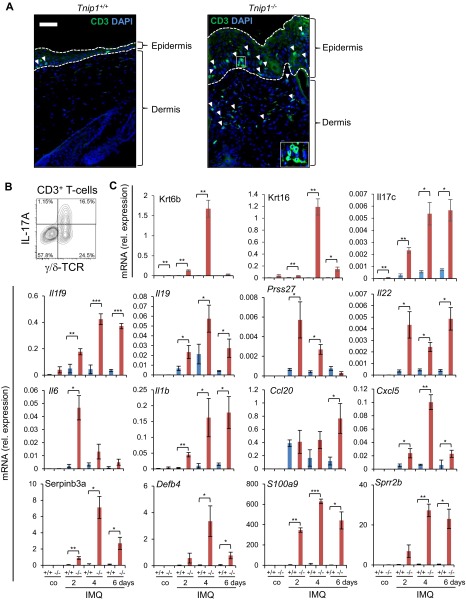
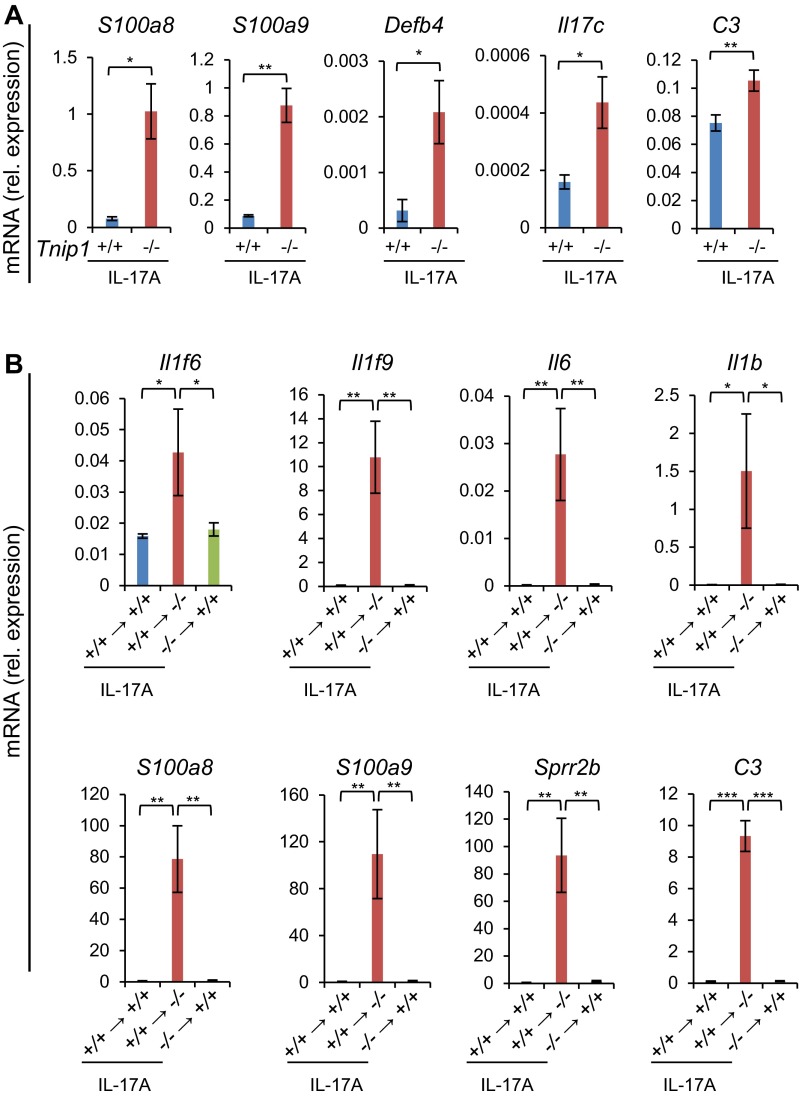
Given the clear genetic contribution to psoriasis and aforementioned observations in susceptible patients, we hypothesized that loss of Tnip1 function might contribute the psoriasis-specific genetic basis for increased sensitivity against IMQ. To test this hypothesis, we treated Tnip1−/− mice with a low concentration of IMQ that did not provoke overt disease symptoms in WT mice akin to healthy human subjects. Indeed, topical low-dose IMQ treatment of Tnip1−/− mice for 6 d resulted in progressive development of psoriasis-like skin symptoms, macroscopically apparent by increasing redness and scaling (Fig. 1 A and B). Microscopy revealed psoriasis-specific pathology characterized by epidermal thickening (acanthosis), elongated rete-like ridges, papillomatosis, retention of nuclei within corneocytes (parakeratosis), and infiltrations with different immune cell types, including neutrophils forming epidermal microabscesses (Fig. 1 C and D). CD3+ T cells were particularly accentuated in the epidermis but were also scattered throughout the dermis, similar to more recent reports (26–28) (Fig. S1A). Phenotyping and quantification of immune cell types by flow cytometry revealed significant increases in neutrophils, dendritic cells (DCs), monocytes and monocyte-derived DCs, as well as IL-17A+ T cells, mainly expressing γ/δ T-cell receptors (TCRs) (Fig. 1E and Fig. S1B). This T-cell subset dominates in mouse skin, but also has been identified in high numbers in human patients (26, 27). Neither IL-17+ T cells nor neutrophils were detected at increased numbers in the skin of untreated Tnip1−/− mice. mRNA analysis of skin samples by quantitative PCR (qPCR) over time revealed strong up-regulation of psoriasis-associated genes in Tnip1−/− mice already at 2 d after IMQ treatment, which was largely sustained until day 6 (Fig. 1F and Fig. S1C). Consistent with human disease, these included members of the IL-12, IL-17, IL-1, S100A, keratin, antimicrobial peptide (AMP), and chemokine families (Fig. 1F and Fig. S1C).
Fig. 1.
IMQ triggers a psoriasis-like disease in Tnip1−/− mice. Tnip1+/+ and Tnip1−/− mice were treated with IMQ for 6 d and analyzed by macroscopic appearance (A); skin scaling by macroscopic severity score (B); microscopy of H&E-stained sections of back skin (C); microscopic measurement of epidermal thickness of back skin (D); flow cytometry-based quantification of CD45+ CD11b+ Ly6G+ neutrophils, CD45+ CD3+ IL-17A+ T cells, MHC class II+ DCs, and CD11b+ monocytes/ monocyte-derived DCs from back skin (E); and qPCR analysis of mRNA levels in back skin (F). n = 5 mice per group. (Scale bars: 20 μm.) Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Co, control. Results from one representative experiment of two independent experiments are shown.
Fig. S1.
IMQ triggers a psoriasis-like disease in Tnip1−/− mice. (A) Representative images of back skin sections from Tnip1+/+ and Tnip1−/− mice that were treated with IMQ for 6 d, stained with antibodies to CD3 (green) and DAPI (blue), and analyzed by immunofluorescence microscopy (n = 5 mice per group. (Scale bars: 50 μm.) (B) Representative flow cytometry analysis of CD45+ CD3+ T cells isolated from the back skin of Tnip1−/− mice treated for 6 d with IMQ. Antibodies against γ/δ TCRs and intracellular IL-17A are indicated (n = 5 mice per group). (C) qPCR analysis of mRNA levels in the back skin of Tnip1+/+ (blue) and Tnip1−/− (red) mice during IMQ treatment (n = 5 mice per group). Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Co, control. Results from one representative experiment of two independent experiments are shown.
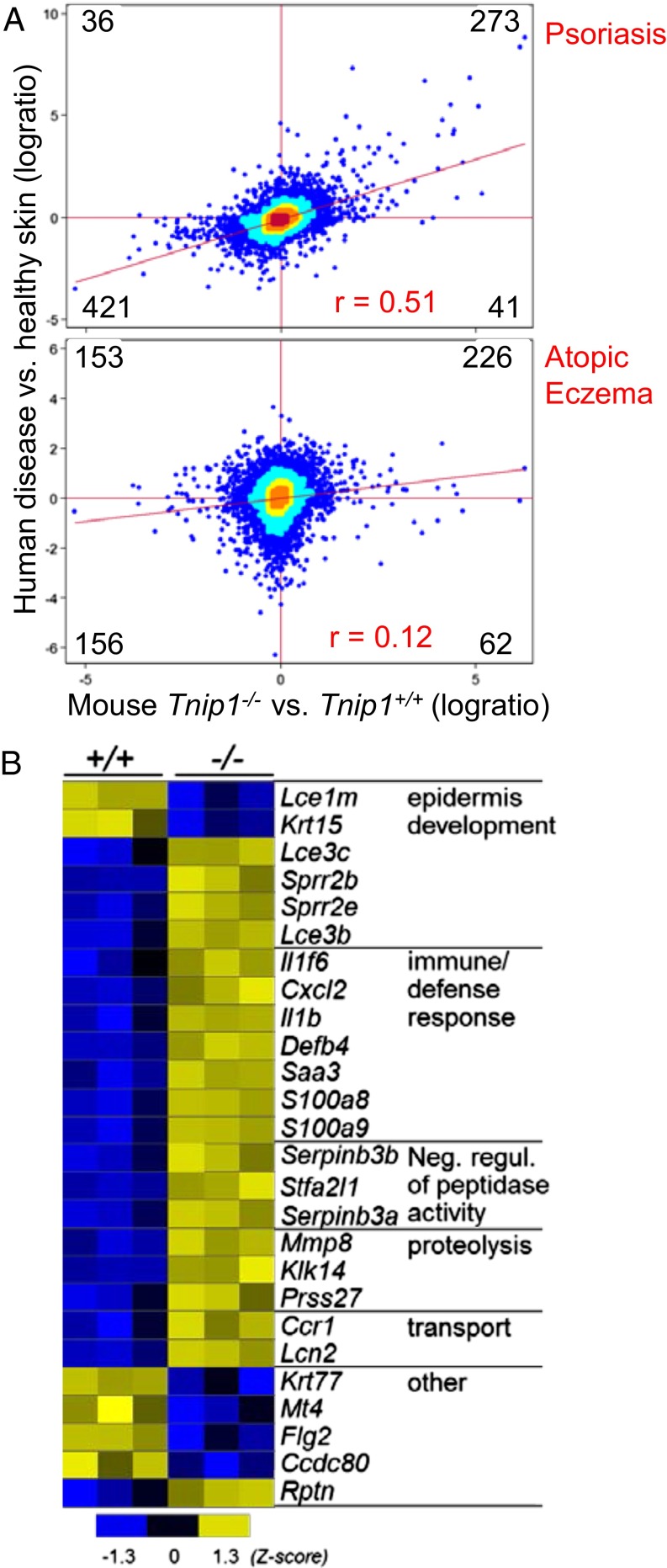
To further test the similarity with human disease, we analyzed the transcriptome data of IMQ-treated mice by RNA sequencing (RNAseq) and performed cross-species analyses with human transcriptome data derived from psoriasis patients and healthy subjects. Intriguingly, the vast majority of genes deregulated in Tnip1−/− mice correlated with their human counterparts, with respect to both up-regulated genes (273 genes) and down-regulated genes (421 genes) (Fig. 2A, Upper, and Table S1). Among the total number of genes deregulated in either the mouse model or human psoriasis patients, 90% displayed concordant regulation. Only a small set of genes (i.e., 36 and 41 genes) showed discordant regulation (Fig. 2A), supporting the interpretation that gene deregulation in Tnip1−/− mice reflects a psoriasis-specific pattern rather than being the consequence of an overall exaggerated inflammatory skin response. It is interesting to note that this correlation between mouse and human was not limited to genes expressed at high levels, but was rather evident for the entire set of genes, resulting in a tilting of the overall gene cloud toward a common axis and a Pearson correlation coefficient of 0.51 (Fig. 2A). Taken together, the foregoing data demonstrate that loss of Tnip1 function leads to genetic susceptibility against IMQ, akin to human disease. As expected, Gene Ontology (GO) analysis highlighted gene sets known to be affected in psoriasis, such as epidermal development, immune/defense response, proteolysis, and response to wounding (Fig. 2B). A complete list of genes and GO categories is provided in Table S2.
Fig. 2.
Correlation of genome-wide RNA expression between Tnip1−/− mice and patients with psoriasis. (A, Upper) RNAseq-based cross-species comparison of skin gene expression between IMQ-treated (for 2 d) Tnip1−/− vs. Tnip1+/+ mice (x axis; n = 3 mice per group) and skin gene expression between patients with psoriasis vs. healthy subjects (y axis; n = 92 psoriasis patients; n = 82 heathy subjects). (A, Lower) RNAseq-based cross-species comparison of skin gene expression between the mouse model in the upper panel and patients with atopic eczema vs. healthy subjects (y axis; n = 15 atopic eczema patients; n = 15 heathy subjects. The number of genes in respective quadrants (log ratio > 0.5) and Pearson correlation indices (r) are indicated. The genes are listed in Table S1. The color display is based on bin counts (bc; Materials and Methods): blue, bc <10; cyan, bc 10–49; yellow, bc 50–99; orange, bc 100–250; red, bc ≥250. (B) Heat map of selected genes and GO categories based on RNAseq from skin of Tnip1+/+ and Tnip1−/− mice that were treated for 2 d with IMQ (P < 0.05, log ratio > 2). A complete list of genes is provided in Table S2.
To further establish how specifically the gene expression profile of Tnip1−/− mice matched psoriasis, we performed a cross-species comparison with another human inflammatory skin disease, atopic eczema. Hardly any correlation was observed between Tnip1−/− mice and this human disease (Fig. 2A, Lower). A lack of correlation was obvious for highly deregulated genes, which were distributed almost equally among the four quadrants; it was also apparent for less strongly deregulated genes, which followed a more symmetrical distribution throughout the four quadrants. Lack of correlation between the Tnip1−/− mouse model and human atopic eczema translated to a low Pearson correlation coefficient of 0.12 (Fig. 2A, Lower). These observations further support the interpretation that the skin inflammation observed in IMQ-treated Tnip1−/− mice corresponds to human psoriasis, rather than reflecting a nonspecific skin pathology following a common inflammatory insult. In summary, Tnip1−/− mice are susceptible to the development of a psoriasis-like disease (triggered by IMQ) with major characteristics of human disease on both pathological and gene regulatory levels.
Lymphocytes and IL-17 Are Critically Involved in Psoriasis-Like Disease in Tnip1−/− Mice.
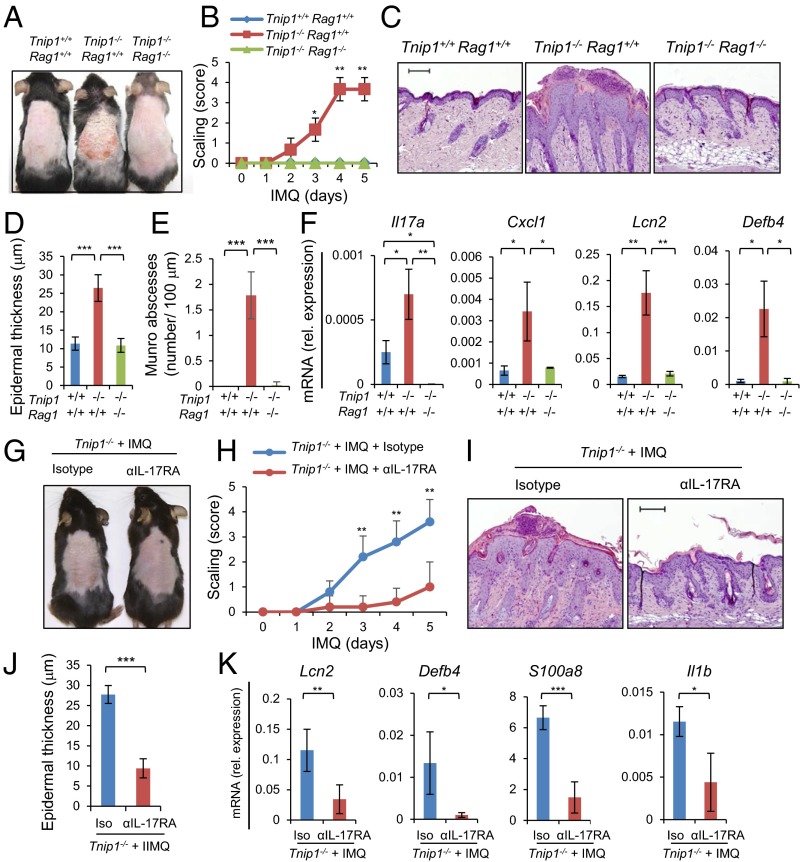
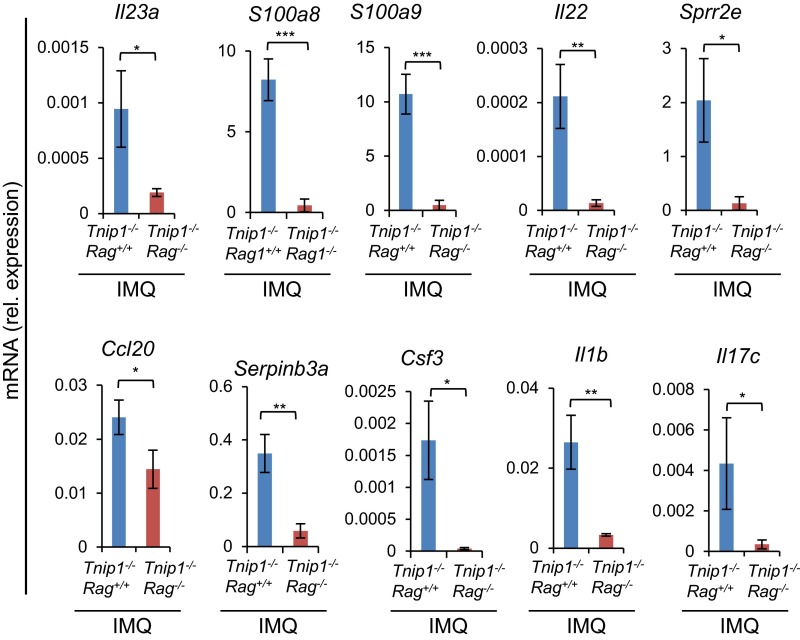
As mentioned earlier, T lymphocytes have been identified as important contributors to psoriasis in human disease (29). To evaluate the contribution of lymphocytes in general, we crossed Tnip1−/− mice to Rag1-deficient mice, which lack mature B and T cells (30). After IMQ treatment, all disease parameters measured, including redness, scaling, histological parameters (i.e., elongated rete-like ridges, papillomatosis, parakeratosis, inflammation, epidermal thickening, formation of microabscesses) and gene deregulation were significantly ameliorated in Rag1-deficient Tnip1−/− mice, consistent with human disease (Fig. 3 A–F and Fig. S2).
Fig. 3.
Lymphocytes and IL-17 control psoriasis-like disease in Tnip1−/− mice. (A–F) Tnip1+/+Rag1+/+, Tnip1−/− Rag1+/+, and Tnip1−/− Rag1−/− mice (n = 5 mice per group) were treated with IMQ for 5 d and analyzed by macroscopic appearance (A), skin scaling by macroscopic severity score (B), microscopy of H&E-stained sections of back skin (C), microscopic measurement of epidermal thickness of back skin (D), microscopic quantification of microabscesses (E), and qPCR analysis of mRNA levels in back skin (F). (G–K) Tnip1−/− mice (n = 5 mice per group) were treated with IMQ for 5 d in the presence of IL-17RA–neutralizing antibodies or isotype control antibodies and analyzed by macroscopic appearance (G), skin scaling by macroscopic severity score (H), microscopy of H&E-stained sections of back skin (I), microscopic measurement of epidermal thickness of back skin (J), and qPCR analysis of mRNA levels in back skin (K). (Scale bars: 20 μm.) Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Results from one representative experiment of two independent experiments are shown.
Fig. S2.
Lymphocytes control psoriasis-like disease in Tnip1−/− mice. qPCR analysis of mRNA levels in the back skin of Tnip1−/− Rag1+/+ and Tnip1−/− Rag1−/− mice after 5 d of IMQ treatment (n = 5 mice per group). Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Results from one representative experiment of two independent experiments are shown.
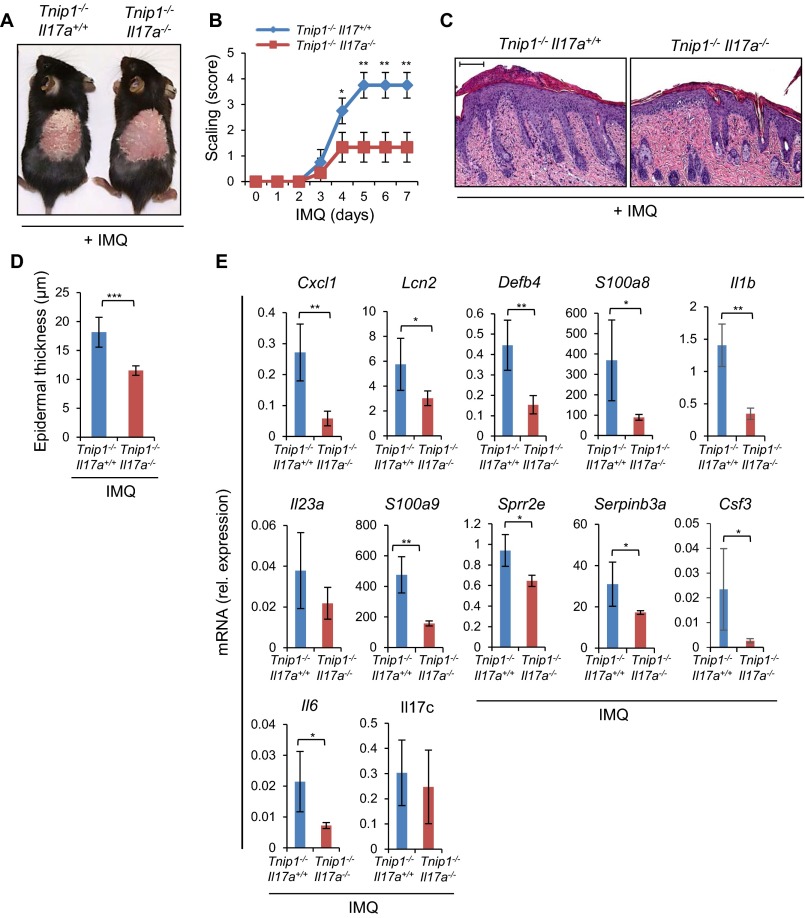
To directly test the role of IL-17A, we crossed Tnip1−/− mice to Il17a−/− mice, followed by an IMQ challenge. Consistent with an important role of IL-17A in skin inflammation, disease symptoms, including gene deregulation, were reduced, although not completely prevented, in the Tnip1−/− Il17a−/− mice (Fig. S3). It is interesting to note that deregulation of Il17c, a known contributor to psoriasis that is produced primarily by keratinocytes, was also prevented in Tnip1−/− Rag1−/− mice, but not significantly reduced in Il17a−/− mice (Figs. S2 and S3E). As such, IL-17C activation appears to follow lymphocyte-derived but IL-17A–independent signals, which may explain the partial protection from psoriasis by selective deletion of Il17a (31).
Fig. S3.
IL-17A controls psoriasis-like disease in Tnip1−/− mice. (A) Representative images of the back skin of Tnip1−/− Il17a+/+ and Tnip1−/− Il17a−/− mice treated with IMQ for 7 d (n = 5 mice per group). (B) Analysis of skin scaling by macroscopic severity score during IMQ treatment (n = 5 mice per group). (C) Representative images of H&E-stained sections of the back skin of Tnip1−/− Il17a+/+ and Tnip1−/− Il17a−/− mice treated with IMQ for 7 day (n = 5 mice per group) (Scale bars: 20 μm.) (D) Epidermal thickness of the back skin of Tnip1−/− Il17a+/+ and Tnip1−/− Il17a−/− mice treated with IMQ for 7 d (n = 5 mice per group). (E) qPCR analysis of mRNA levels in the back skin of Tnip1−/− Il17a+/+ and Tnip1−/− Il17a−/− mice after IMQ treatment for 7 d (n = 5 mice per group). Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Results from one representative experiment of two independent experiments are shown.
To investigate this possibility, we treated Tnip1−/− mice with IMQ in the presence of neutralizing antibodies against the IL-17 receptor IL-17RA, which transduces signaling by both IL-17A and IL-17C (31). IL-17RA blockade indeed almost completely prevented IMQ-induced psoriasis in Tnip1−/− mice (Fig. 3 G–K), close to the effectiveness seen in Rag1-deficient mice and more effectively than in Il17a−/− mice (Fig. 3 and Fig. S3). These data are consistent with the interpretation that different IL-17 family members, most likely IL-17A and IL-17C, contribute to the psoriasis-like skin inflammation in Tnip1−/− mice. Taken together, these results demonstrate a critical role for immune cells, specifically lymphocytes, and IL-17 for disease development in Tnip1−/− mice, and are consistent with a pathogenically important function of T cells similar to human psoriasis.
Tnip1 Controls IL-23– and IL-17A–Mediated Skin Disease.
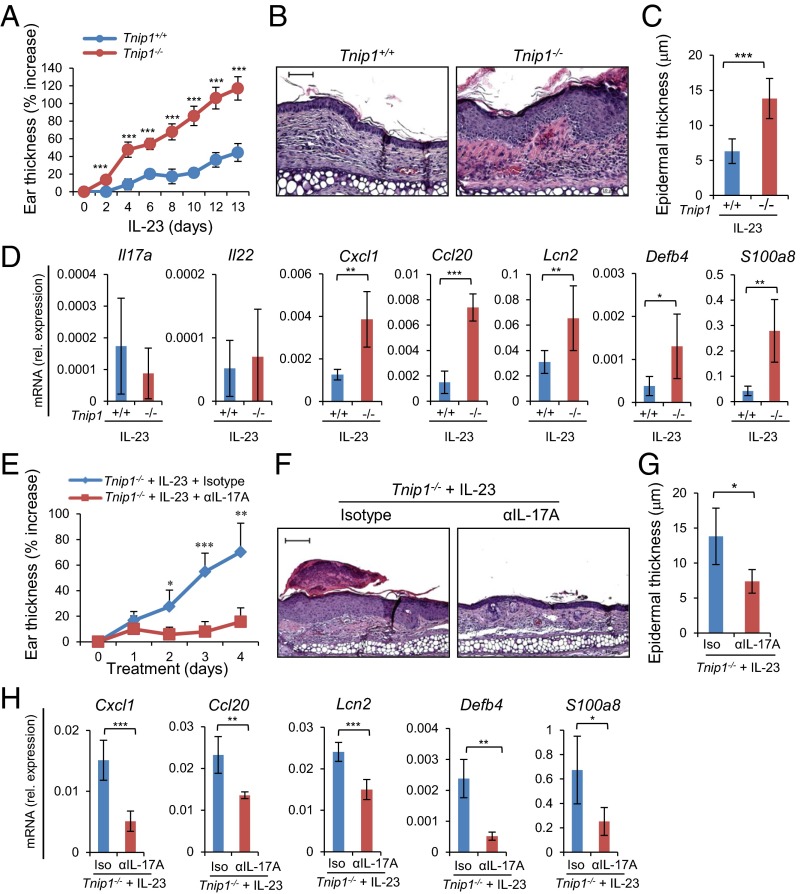
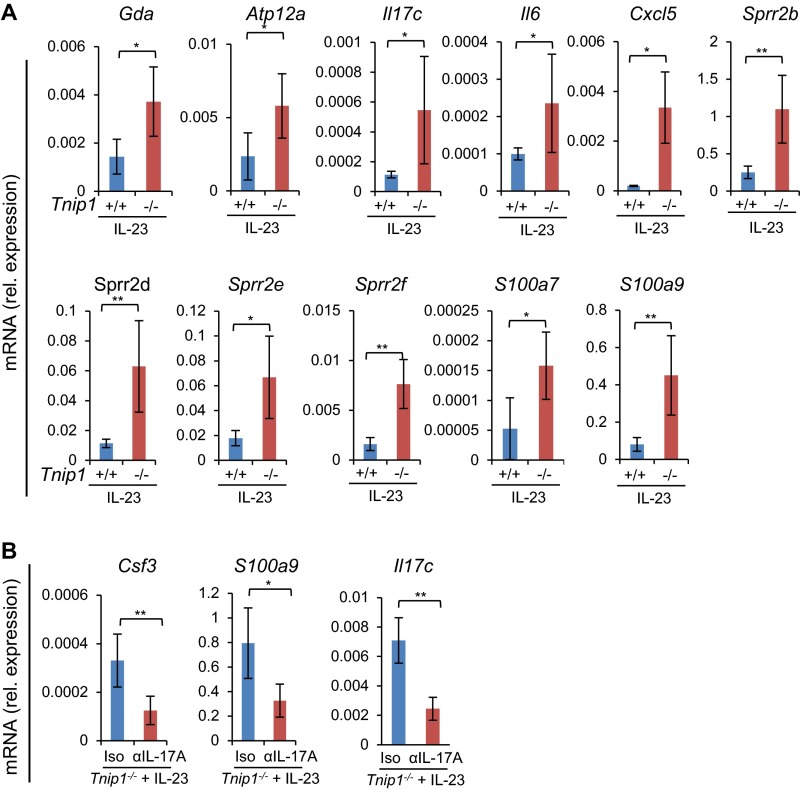
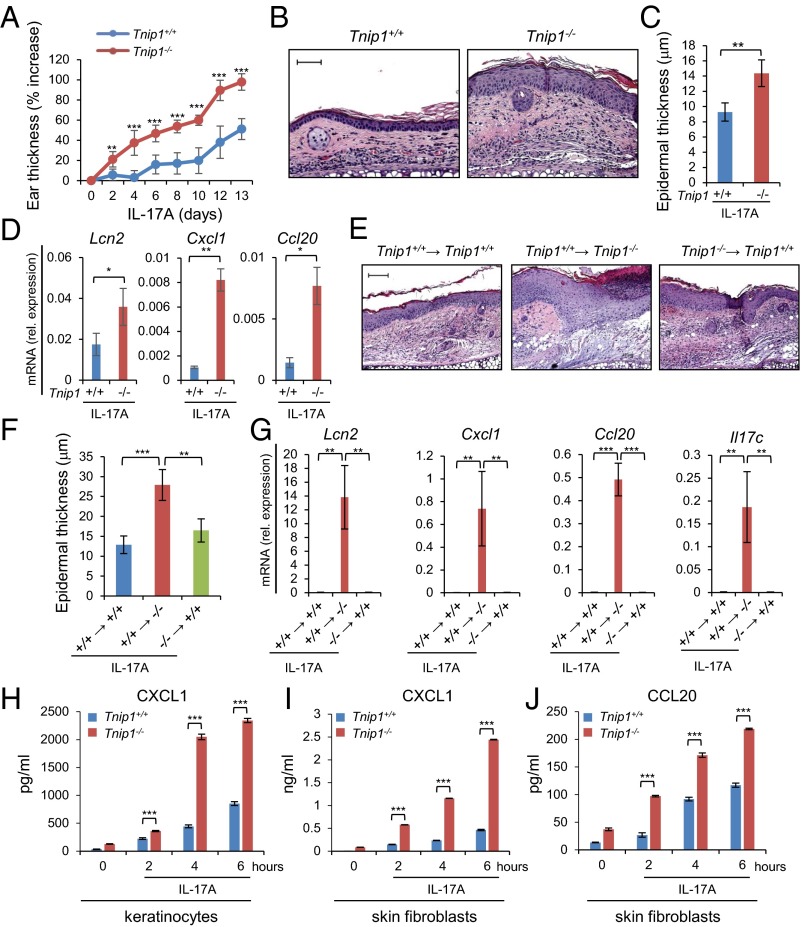
Given the established function of IL-23 for IL-17A production in psoriasis and in IMQ-induced skin inflammation in mice, we hypothesized that IL-23 also may result in increased inflammation in Tnip1−/− mice, similar to IMQ (25, 32–35). We tested this hypothesis by injecting IL-23 directly into the ear skin of WT and Tnip1−/− mice, followed by analysis of psoriasis-related symptoms. Indeed, the Tnip1−/− mice developed a significantly stronger psoriasis-like disease than the WT animals, as determined by ear thickness and histopathology, including inflammation and epidermal thickness (Fig. 4 A–C). Interestingly, particularly keratinocyte-derived genes, such as Lcn2, Il17c, S100a- and Sprr family members, and chemokines (Cxcl1 and Ccl20) were up-regulated in the skin of Tnip1−/− mice, and genes expressed primarily in T cells, such as Il-17a and IL-22, were expressed at a similar level (Fig. 4D and Fig. S4A). These data indicate that the activity of IL-23, acting directly on T cells to trigger IL-17A and IL-22 production, was comparable in WT and Tnip1−/− cells. In contrast, biological effects mediated by IL-17A, specifically keratinocyte activation, appeared to be deregulated in Tnip1−/− mice.
Fig. 4.
Tnip1 controls IL-23–mediated skin disease. (A–D) The ears of Tnip1+/+ and Tnip1−/− mice (n = 7 mice per group) were injected with IL-23 for 14 d and analyzed by measurement of ear thickness (A), microscopy of H&E-stained sections of ear skin on day 14 (B), microscopic measurement of epidermal thickness of ear skin on day 14 (C), and qPCR analysis of mRNA levels in ear skin on day 14 (D). (E–H) The ears of Tnip1−/− mice (n = 5 mice per group) were injected with IL-23 for 4 d in the presence of IL-17A–neutralizing antibodies or isotype control antibodies and analyzed by measurement of ear thickness (E), microscopy of H&E-stained sections of ear skin on day 4 (F), microscopic measurement of epidermal thickness of ear skin on day 4 (G), and qPCR analysis of mRNA levels in ear skin on day 4 (H). (Scale bars: 20 μm.) Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Results from one representative experiment of two independent experiments are shown.
Fig. S4.
Tnip1 controls IL-23-mediated skin disease. (A) qPCR analysis of mRNA levels in the ears of Tnip1+/+ and Tnip1−/− mice at day 14 during IL-23 injections (n = 7 mice per group). (B) qPCR analysis of mRNA levels in the ears of Tnip1−/− mice at day four during IL-23 injections in the presence of IL-17A–neutralizing antibodies or isotype control antibodies (n = 5 mice per group). Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Results from one representative experiment of two independent experiments are shown.
To confirm that IL-23 mediates its inflammatory effects via IL-17A, we challenged mice by IL-23 injection in the presence of neutralizing antibodies against IL-17A. Histopathology, including measurement of epithelial thickness and neutrophil recruitment, and measurement of ear thickness and gene expression collectively confirmed the critical role of IL-17A in IL-23–induced disease in Tnip1−/− mice (Fig. 4 E–H and Fig. S4B). Given the significant deregulation of keratinocyte-derived genes on IL-23 injection and its dependence on IL-17A, we tested whether IL-17A–induced biology itself might be affected by loss of Tnip1 function. Indeed, direct injection of IL-17A into the ear skin led to a significantly increased appearance of disease symptoms, including ear thickness, epidermal thickness, inflammation, and activation of IL-17A target genes, such as Cxcl1, Ccl20, and Lcn2 (8) (Fig. 5 A–D and Fig. S5A). In summary, data based on IL-23 and IL-17A injection and neutralizing antibodies against IL-17A confirm that IL-23 acts via IL-17A; however, the data also suggest that IL-17A–mediated biology itself, most likely acting on skin-resident nonhematopoietic cells, may be controlled by Tnip1.
Fig. 5.
Tnip1 activity in nonhematopoietic cells is required to protect from IL17A-mediated skin disease. (A–D) The ears of Tnip1+/+ and Tnip1−/− mice (n = 7 mice per group) were injected with IL-17A for 14 d and analyzed by measurement of ear thickness (A), microscopy of H&E-stained sections of ear skin on day 14 (B), microscopic measurement of epidermal thickness of ear skin on day 14 (C), and qPCR analysis of mRNA levels in ear skin on day 14 (D). (E–G) Tnip1+/+ and Tnip1−/− mice (n = 5 mice per group) were lethally irradiated and then reconstituted with BM from Tnip1+/+ and Tnip1−/− mice, as indicated. Reconstituted mice were challenged by intradermal injection with IL-17A for 4 d, followed by microscopic analysis of H&E-stained sections of ear skin (E), measurement of epidermal thickness (F), and qPCR analysis of mRNA from ear skin (G). (H–J) Keratinocytes (H) and dermal fibroblasts (I and J) were isolated from Tnip1+/+ and Tnip1−/− mice and stimulated in vitro with IL-17A for the indicated time periods, followed by measurement of released chemokines in the supernatant. n = 3 biological replicates per group and time point. (Scale bars: 20 µm.) Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Results from one representative experiment of two independent experiments are shown.
Fig. S5.
Tnip1 activity in nonhematopoietic cells is required to protect from IL17A-mediated skin disease. (A) qPCR analysis of mRNA levels in the ear skin of Tnip1+/+ and Tnip1−/− mice at day 14 after IL-17A injection (n = 7 mice per group). (B) qPCR analysis of mRNA from ear skin of BM chimeric mice described in Fig. 5 E–G, which were treated by intradermal IL-17A injection for 4 d (n = 5 mice per group). Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Results from one representative experiment of two independent experiments are shown.
Tnip1 Activity in Nonhematopoietic Cells Is Required for Protection from IL-17A–Mediated Skin Disease.
To test the hypothesis that Tnip1 expression in nonhematopoietic cells contributes to IL-17–driven skin disease, we generated bone marrow (BM) chimeric mice by transfer of WT or Tnip1−/− BM cells into Tnip1+/+ or Tnip1−/− recipient mice, followed by IL-17A injection into the skin of the ear. IL-17A injection into Tnip1−/− recipient mice reconstituted with WT BM induced the strongest disease symptoms, as determined by histopathology and epidermal thickness (Fig. 5 E and F). Disease was much less apparent in WT mice reconstituted with Tnip1−/− BM and in WT mice reconstituted with WT BM (Fig. 5 E and F). Increased pathology was mirrored by strongly increased gene deregulation in Tnip1−/− recipient mice (Fig. 5G and Fig. S5B). Genes deregulated in Tnip1−/− recipient mice included Il17c, whose autoregulatory function along with IL-36γ is likely to amplify inflammation (Fig. 5G) (31, 36). As such, these data strongly suggest that IL-17A mediates skin pathology via nonhematopoietic cells lacking Tnip1 function.
To directly investigate whether Tnip1 controls IL-17A–induced gene regulation in nonhematopoietic cells, we isolated primary keratinocytes and fibroblasts from WT and Tnip1−/− mice and assayed CXCL1 and CCL20 release on IL-17A stimulation. IL-17A induced measurable protein levels of CXCL1 on both cell types and CCL20 on fibroblasts in a time-dependent manner. As expected from the in vivo experiments, Tnip1−/− cells indeed displayed strongly up-regulated chemokine expression, which was apparent throughout the time period investigated (Fig. 5 H–J). As such, these data and the foregoing BM chimeric experiments demonstrate that Tnip1 controls IL-17A–mediated gene regulation in keratinocytes and fibroblasts, and highlight the contribution of gene deregulation in skin-resident nonhematopoietic cells to psoriatic skin inflammation.
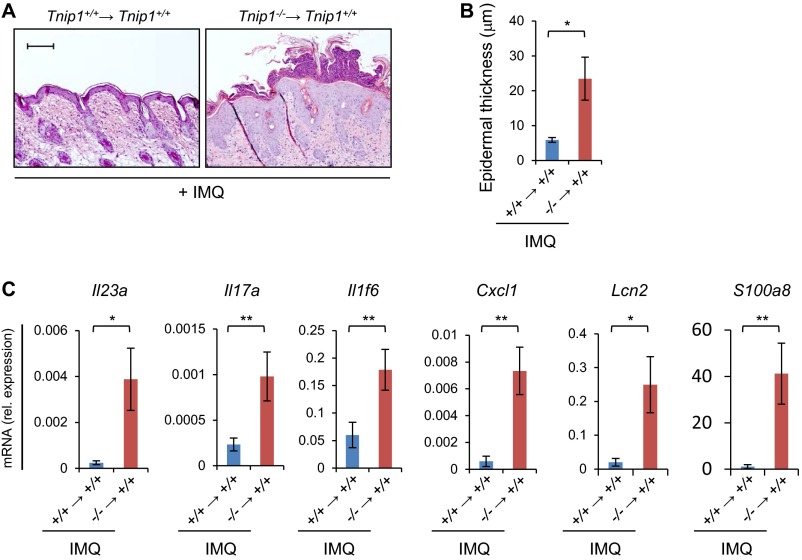
As mentioned earlier, CD11c-driven deletion of Tnip1, which deletes Tnip1 expression most likely in myeloid cells, has been shown to mediate susceptibility to skin inflammation on IMQ treatment (20). Thus, we also tested the effect of IMQ in BM chimeric mice. Tnip1+/+ mice reconstituted with BM from Tnip1−/− mice and treated with IMQ showed increased inflammation compared with mice reconstituted with WT BM (Fig. S6). These data confirm results obtained by Callahan et al. (20) and suggest that Tnip1 acts in immune cells, most likely myeloid cells, to counteract inflammation during IMQ treatment. Collectively, these data provide further support to a hierarchical model of events where Tnip1 acts upstream in IMQ (TLR7)-induced myeloid cells and downstream in IL-17–activated keratinocytes to counteract inflammation (Fig. 6K).
Fig. S6.
Tnip1 activity in hematopoietic cells counteracts IMQ-induced psoriasis-like inflammation. Tnip1+/+ mice were lethally irradiated and reconstituted with BM from Tnip1+/+ or Tnip1−/− mice, as indicated. (A) The back skin of reconstituted mice was treated with IMQ for 6 d (n = 5 mice per group), followed by microscopic analysis of H&E-stained sections from back skin. Representative images are shown. (Scale bars: 20 µm.) (B) Measurement of epidermal thickness (n = 5 mice per group). (C) qPCR analysis of mRNA from back skin (n = 5 mice per group). Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Results from one representative experiment of two independent experiments are shown.
Fig. 6.
K14-Cre-directed Tnip1-deletion in keratinocytes results in susceptibility against IMQ- and IL-17A-triggered psoriasis. (A–G) Tnip1f/f and Tnip1f/f K14-Cre mice (n = 6 mice per group) were treated with IMQ for 7 d and analyzed by macroscopic appearance (A), skin scaling by macroscopic severity score (B), microscopy of H&E-stained sections of back skin (C), microscopic measurement of epidermal thickness of back skin (D), flow cytometry-based quantification of CD45+ CD11b+ Ly6G+ neutrophils (E) and CD45+ CD3+ IL-17A+ T cells (F), and qPCR analysis of mRNA levels in back skin (G). (H–J) Tnip1f/f and Tnip1f/f K14-Cre mice (n = 7 mice per group) were challenged by intradermal injection with IL-17A for 4 d, followed by microscopic analysis of H&E-stained sections of ear skin (H), measurement of epidermal thickness (I), and qPCR analysis of mRNA from ear skin (J). (Scale bars: 20 µm.) Co, control. Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test. Results from one representative experiment of two independent experiments are shown. (K) Psoriasis model. A less well-defined stimulus, possibly containing TLR-activating nucleic acids, triggers myeloid cells to release IL-23, which mediates the expansion and activation of IL-17–producing T cells. IL-17 triggers keratinocytes and fibroblasts to release chemokines (and other factors), which attract myeloid cells and T17 cells. TNIP1/ABIN1 controls two critical processes involved: TLR-induced IL-23 production from myeloid cells and IL-17–induced chemokine production from keratinocytes and fibroblasts. Loss of TNIP1 function leads to increased production of IL-23 and, consequently, IL-17–producing T cells. In turn, IL-17 exposure of Tnip1-deficient keratinocytes and fibroblasts leads to exaggerated chemokine production, which fuels inflammation.
Loss of Tnip1 Function in Keratinocytes Promotes Psoriasis Inflammation.
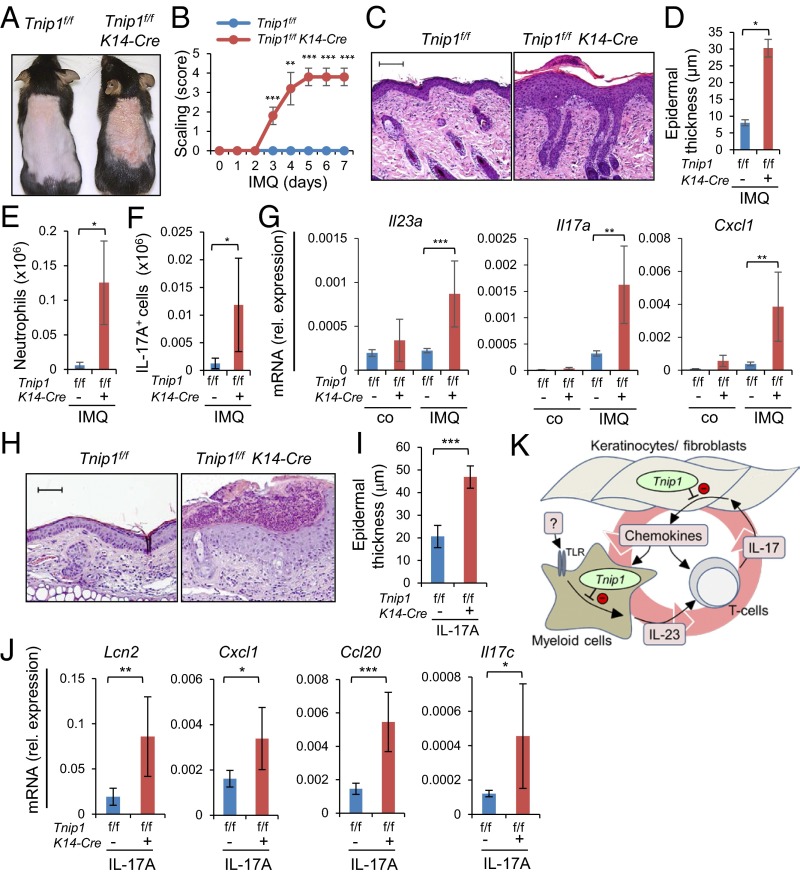
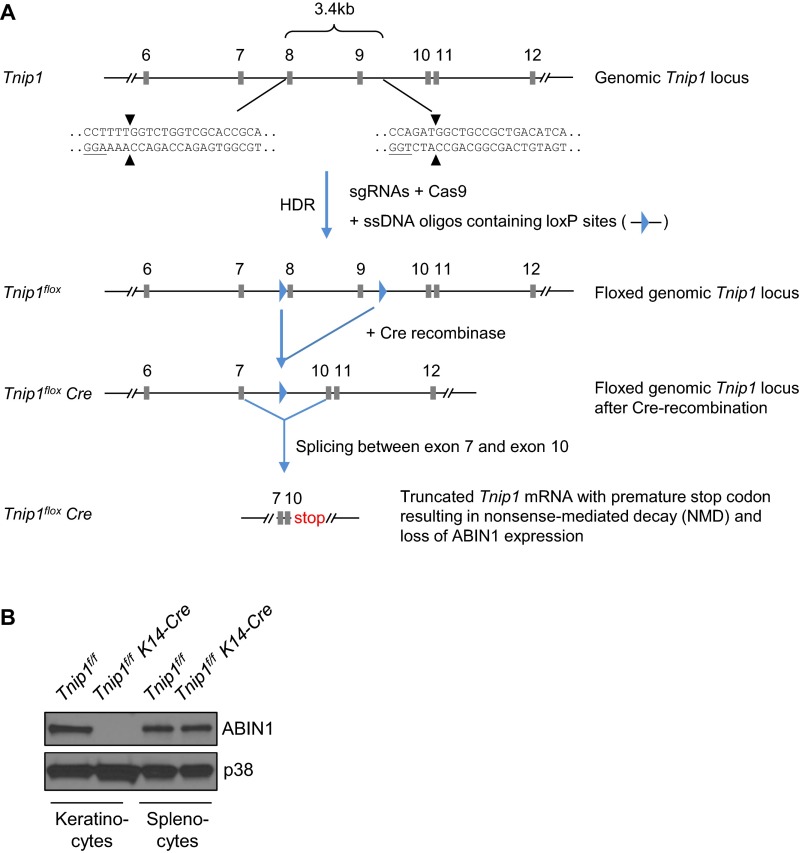
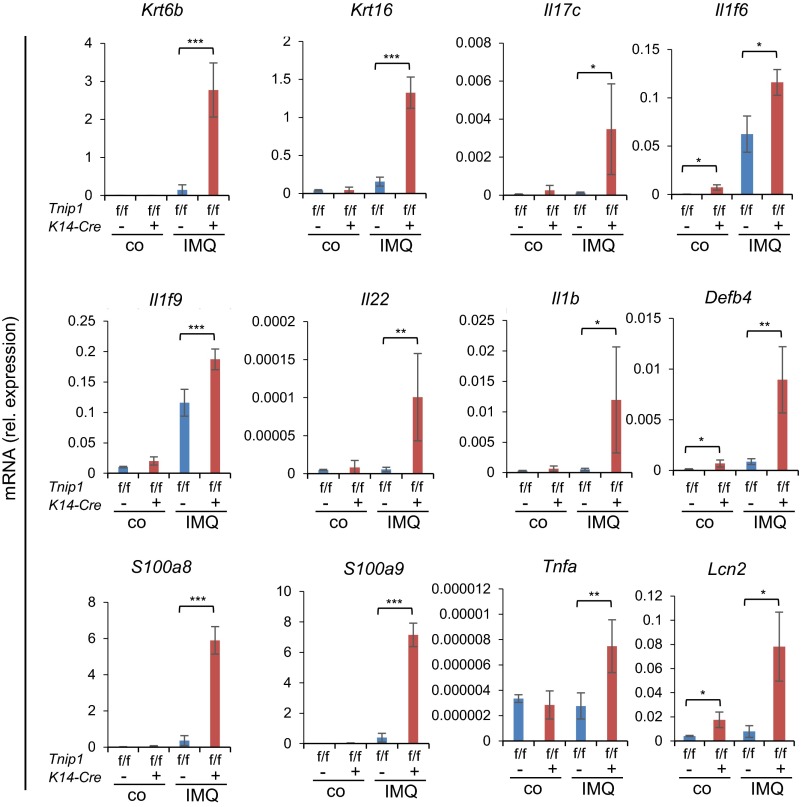
To confirm the foregoing data suggesting a critical role of Tnip1 in nonhematopoietic cells, and to directly assess a cell type-intrinsic function of Tnip1 in keratinocytes, we established a conditional Tnip1 mouse model based on site-specific recombination of LoxP sites by Cre-recombinase (Cre) (Tnip1flox/flox, also referred to as Tnip1f/f) (Fig. S7A). To delete Tnip1 in epidermal keratinocytes, Tnip1flox/flox mice were crossed to K14-Cre mice, which express Cre recombinase under control of the Keratin 14 (K14) promoter (37). Efficient, keratinocyte-specific deletion of Tnip1 was confirmed by immunoblotting based on keratinocytes and splenocytes isolated from Tnip1flox/flox K14-Cre and Tnip1flox/flox control mice (Fig. S7B). As expected, no spontaneous overt disease phenotype was observed in Tnip1flox/flox K14-Cre mice. These mice were treated with IMQ, and skin inflammation and gene expression were analyzed. As expected from BM chimeric mice, keratinocyte-specific Tnip1 deletion resulted in apparent disease symptoms, including redness and scaling and histopathological correlates of psoriasis, including epidermal thickness and microabscess formation (Fig. 6 A–D). Disease correlated with the appearance of CD11b+ myeloid cells, including Ly6Ghigh neutrophils, as well as IL-17A–producing γ/δ T cells in the skin, akin to mice with the germ-line Tnip1 deletion described above (Fig. 6 E and F). Accordingly, genes associated with psoriasis, including members of the IL-12, IL-17, IL-1, S100A, keratin, AMP, and chemokine families, were strongly deregulated (Fig. 6G and Fig. S8).
Fig. S7.
Generation of conditional Tnip1-deficient mice (Tnip1flox/flox). (A) LoxP sites were introduced into the Tnip1 genomic locus based on CRISPR/ CAS9 technology. Targeting sites were chosen strategically at 5′ of exon 8 and 3′ of exon 9. Cre-dependent recombination of loxP sites led to deletion of exon 8 and exon 9 and consequent splicing between exon 7 and exon 10, the latter of which is out of frame. This creates a premature stop codon in the truncated Tnip1 mRNA, resulting in nonsense-mediated decay and functional deletion of Tnip1/ABIN1 expression. Exons are annotated based on Tnip1 transcript variant 1 (NM_021327.4), and sequence of sgRNA homology region containing Cas9 cleavage sites and PAMs (underscored) are shown. (B) ABIN1 (TNIP1) protein expression in keratinocytes and splenocytes isolated from Tnip1f/f and Tnip1f/f K14-Cre mice and analyzed by immunoblotting.
Fig. S8.
K14-Cre-directed Tnip1-deletion in keratinocytes results in susceptibility against IMQ-induced psoriasis. Shown is qPCR analysis of mRNA levels in the back skin of Tnip1f/f and Tnip1f/f K14-Cre mice after 7 d of IMQ treatment (n = 6 mice per group). Data represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. Co, control. Results from one representative experiment of two independent experiments are shown.
Given the foregoing findings demonstrating a critical function of Tnip1 in keratinocytes during IL-17 stimulation, we also tested the effect of IL-17 treatment in vivo in Tnip1flox/flox K14-Cre mice. As expected, ear injection of IL-17A led to significant psoriasis symptoms in Tnip1flox/flox K14-Cre mice, confirming the critical function of Tnip1 in keratinocytes during IL-17 activation in vivo (Fig. 6 H–J). In summary, the data demonstrate that Tnip1 is an essential regulator of IL-17 biology, and that loss of Tnip1 function in keratinocytes provides a critical contribution to the genetic susceptibility for psoriasis-like skin disease.
Discussion
Here we have characterized a mouse model for psoriasis based on the human susceptibility locus TNIP1, whose reduced expression in humans and apparent susceptibility in mice strongly suggest a hypomorphic or loss-of-function phenotype (16). The TNIP1 locus also has been linked genetically to the inflammatory disease systemic lupus erythematosus (SLE), and cells of humans with respective polymorphism have been found to express less TNIP1, also suggesting a loss-of-function phenotype (38–41). Consistent with this interpretation, Tnip1−/− mice were found to develop a progressive SLE-like disease with typical manifestations in parenchymatous organs, particularly the kidney glomeruli (19). Based on detailed pathology analysis, no lupus-like skin manifestations were observed in Tnip1−/− mice, neither constitutively nor on exposure to any of the triggers investigated, including IMQ, IL-23, or IL-17. As expected from previous experiments demonstrating that SLE-like disease is mediated by hematopoietic cells (19), mice with keratinocyte-specific deletion of Tnip1 did not show any lupus-like symptoms, but were susceptible to IMQ-induced psoriasis. As such, loss of Tnip1 function predisposes to two inflammatory diseases, psoriasis and lupus-like disease, which proceed independent of each other (in mouse and human) and involve different cell types.
In this work, we have focused on the role of Tnip1 in the skin and established a profound similarity between the mouse model and human psoriasis, based on detailed pathological, genomic, and therapeutic experiments. Pathology includes the typical epidermal symptoms reflecting changes in keratinocyte biology, as well as inflammation reflecting immune cell infiltrations with typical cell types, including myeloid cells, such as neutrophils, and T cells, particularly IL-17A–producing γ/δ T cells. The inflamed skin of mice is known to display an overall bias toward γ/δ T cells, whereas in humans α/β T cells prevail. More recently, however, IL-17–producing γ/δ T cells were also identified at high numbers in human psoriatic skin lesions, indicating a pathogenically relevant function (26). Interestingly, whereas γ/δ T cells were also identified as the primary cell type in a mouse model of IL-23–induced psoriasis, experiments based on BM chimeric mice and genetically deleted γ/δ T cells have suggested that α/β T cells can compensate for γ/δ T cells, indicating functional plasticity among IL-17–producing T-cell subsets (42). As such, the relative differences in the numbers of these T-cell subsets may be less relevant for disease development than the total number of IL-17–producing T cells.
The similarity between the Tnip1−/−/IMQ model and human disease is underscored by our analysis of gene regulation, which revealed several important points. First, there is a remarkable overall congruency of gene regulation between mouse and human skin, with 90% of genes being coherently deregulated in mouse model and human psoriasis (based on a cross-species comparison) (Fig. 2A and Table S2). Second, the genomic similarity between Tnip1−/− mice and psoriasis is sharply contrasted by the largely disparate pattern of gene regulation between Tnip1−/− mice and atopic eczema, strongly supporting the interpretation that the disease in Tnip1−/− mice replicates psoriasis, not skin inflammation in general. Third, the commonly deregulated genes include the major psoriasis signature genes and categories, as well as more recently identified factors, such as complement C3 and members of the IL-36 family, including the IL-36 antagonist Il36rn (42, 43). Even though it may be argued that the similarity between Tnip1−/− mice and human psoriasis does not come as a surprise, given their genetic relationship, it is this relationship that endows investigations related to the complex interplay of different cell types. Consistent with this interpretation, disease symptoms in Tnip1−/− mice are significantly ameliorated by genetic deletion of lymphocytes (including T cells) or therapeutic interference with IL-17, an established, T-cell–derived pathogenicity factor in humans, and, conversely, disease symptoms are exaggerated by injection with IL-23 and IL-17A. The roles of other factors with established relevance in human disease, including TNFα, remain to be investigated.
As mentioned earlier, the IMQ-driven inflammatory skin phenotype in Tnip1−/− mice was expected based on human case report studies and earlier reports demonstrating that Tnip1 deletion in myeloid cells resulted in up-regulation of TLR-induced IL-23 and increased inflammatory symptoms in IMQ-treated mice (20). In turn, the susceptibility against IL-17A described here was more surprising given the relative narrow cell tropism of IL-17 on largely nonhematopoietic cells, which indicated a hitherto undefined function of Tnip1. This hypothesis was confirmed by experiments based on BM chimeras and keratinocyte-specific Tnip1 deletion and, not least, analysis of chemokine release in vitro from IL-17–stimulated keratinocytes and fibroblasts. Collectively, these data and data from previous work demonstrate that TNIP1/ABIN1 controls proinflammatory factors acting at different stages of psoriatic inflammation, i.e., both myeloid cells and nonhematopoietic cells (19, 20). Whereas TLR-induced Tnip1-deficient myeloid cells produce increased amounts of IL-23, thereby promoting T-cell expansion and IL-17A production, Tnip1-deficient keratinocytes and fibroblast respond to IL-17 by increased production of chemokines (and other factors), promoting keratinocyte proliferation and further immune cell recruitment. These data contribute important information to the long-standing dispute about the pathogenic roles of immune cells vs. nonimmune cells in psoriasis and strongly favor a model in which defective anti-inflammatory mechanisms in different cell types allow for the reciprocal buildup of inflammation and disease (Fig. 6K).
From a mechanistic standpoint, the function of TNIP1/ABIN1 in IL-17 biology is not entirely unexpected. We have identified ABIN1 as part of the TLR signaling complex, and more detailed analyses revealed that both the ubiquitin-binding motif contained in ABIN1 and the ubiquitin ligase TRAF6, a core component of the TLR pathway, are required for ABIN1 engagement (19, 44). Thus, it seems possible that TRAF6-mediated synthesis of polyubiquitin chains is the basis for (and possibly the target of) ABIN1-mediated anti-inflammatory activity. IL-17R signaling also has been shown to involve TRAF6, providing a common denominator in both TLR and IL-17R signal transduction pathways (45). Moreover, ABIN1 was originally identified as interacting protein of A20, an anti-inflammatory protein with deubiquitinating activity that is also linked by GWAS to psoriasis (16, 18). Overexpression of A20 along with ABIN1 revealed synergistic negative regulatory activity in the TNFR pathway, indicating that ABIN1 may serve as an adaptor protein to target A20 to ubiquitinated proteins, including IKKγ/NEMO, which controls NF-κB activation (18, 46). Interestingly, A20 has been shown to counteract IL-17R–mediated signaling, at least in cell lines in vitro (47). Assembling all of these pieces of information into a unifying model, it seems possible that ABIN1 and A20 proteins act together in different major inflammatory signaling pathways, including the TNFRI, TLRs, and IL-17R, with ABIN1 providing substrate specificity for A20 via its ubiquitin-binding moiety. It should be noted, however, that several aspects of this model are not yet supported by experimental data, and some that observations suggest a more complex scenario. For example, A20, but not ABIN1, has been linked genetically to rheumatoid arthritis, suggesting nonoverlapping (independent) functions of the two proteins (48, 49). Moreover, keratinocyte-specific deletion of A20 (Tnfaip3) was found to result in increased keratinocyte proliferation and various ectodermal organ abnormalities, which are not observed in Tnip1−/− mice (50). Not least, TLR activation of A20-deficient myeloid cells has been shown to lead to deregulation of the NF-κB pathway, whereas ABIN1-deficient cells do not recapitulate this phenotype, but rather exhibit a select increase in C/EBPβ activity (19, 51). The latter observation is particularly interesting in the context of IL-17–induced gene regulation, given that C/EBP family members appear to be important regulators of IL-17–mediated gene activation (52, 53). Taken together, genetic and limited biochemical evidence support a model in which ABIN1 controls part of the spectrum of A20-mediated activities, although the possibility of independent activities cannot be dismissed. In any case, the data shown here establish TNIP1/ABIN1as a critical factor controlling IL-17 biology in nonhematopoietic cells (i.e., keratinocytes and fibroblasts) both in vivo and in vitro, providing an important piece of information about which cells contribute causally to psoriasis.
Materials and Methods
Mice.
Tnip1−/− mice, based on the ES cell clone E059E05 (obtained from the German Gene Trap Consortium), were backcrossed to a C57BL/6 background for more than 10 generations and have been described previously (19). All mouse strains used were on a C57BL/6 background. Rag1tm1Mom (Rag1−/−) mice and Il17atm1.1(icre)Stck (Il17a−/−) mice were purchased from The Jackson Laboratories (30, 54). Tnip1flox/flox mice were generated as detailed below and crossed to K14-Cre mice established by Dussule et al. (37) and obtained from The Jackson Laboratory [stock Tg (KRT14-cre) 1Amc/J]. Age- and sex-matched littermates were used for all experiments at 8–12 wk of age. All mouse studies were carried out in accordance with protocols approved by the Institutional Animal Care and Use Committee at St. Jude Children's Research Hospital.
Generation of Conditional Tnip1 (Tnip1flox) Mice.
Tnip1flox mice were generated using CRISPR-Cas9 technology (55, 56), as detailed in SI Materials and Methods.
Generation of BM Chimeric Mice and Cytokine Treatment.
BM cells were obtained from flushing femurs and tibias of donor Tnip1+/+ and Tnip1−/− mice. BM cells were collected by centrifugation and transferred in 200 µL of PBS containing 1% FBS via tail vain injection into recipient mice that had been lethally irradiated at 950 rad 1 d before cell transfer. Recipient mice were treated with antibiotics (2% SulfaTrim; Hi-Tech Pharmacal) in the drinking water for 5–6 wk, starting 1 wk before irradiation. At 9 wk after reconstitution, the ear skin of these mice was injected intradermally with 500 ng of recombinant murine IL-17A daily for 4 d (days 0, 1, 2, and 3) or treated topically with IMQ for 6 d as detailed below.
IMQ Treatment, Cytokine Injection, and Neutralization of IL-17A and IL-17RA.
Mice at 8–12 wk of age received a daily topical dose of 32 mg of 5% IMQ cream (Perrigo; corresponding to 1.6 mg of the active compound), unless otherwise stated, for 5–6 consecutive days on the Nair crème-treated and shaved back. The severity of the scaling was scored on a scale of 0–4 (0, none; 1, slight; 2, moderate; 3, marked; 4, very marked). Intradermal injections in the ears were performed every other day for 14 d with either recombinant mouse IL-23 (100 ng; R&D Systems) or recombinant mouse IL-17A (100 ng; R&D Systems) in a total volume of 20 μL using a 30G needle under isoflurane anesthesia. Ear thickness was measured using a digital micrometer (Mitutoyo) by an observer blinded to treatment conditions and mouse genotype.
For the antibody-blocking experiments with anti–IL-17A, the ear skin of Tnip1−/− mice was treated intradermally with 200 ng/d of IL-23 for 4 d. Then 100 μg of anti–IL-17A (eBioscience) or 100 μg of mouse IgG1 control antibody (eBioscience) was injected i.p. at 1 h before cytokine injection on days 0 and 2. For the antibody-blocking experiments with anti–IL-17RA, the depilated back skin of Tnip1−/− mice was treated with 20 mg/d of IMQ cream for 5 d, and 125 μg of anti–IL-17RA (R&D Systems) or 125 μg of IgG2A control antibody (R&D Systems) was injected i.p. 1 h before IMQ treatment on days 0 and 2.
In Vitro Culture and Functional Assays of Keratinocytes and Fibroblasts.
Keratinocytes and dermal fibroblasts were prepared as described previously (57) and detailed in SI Materials and Methods.
Antibodies and Flow Cytometry.
For flow cytometry testing of skin T cells and neutrophils, the back skin of mice was cut into small pieces and digested with 0.35% collagenase type I (Worthington) for 30 min at 37 °C, followed by treatment with 20 µg/mL DNase (Sigma-Aldrich) for 15 min at 37 °C. Flow cytometry of skin DCs and monocytes was performed following the extraction procedure and gating strategy described by Malosse et al. (58). In brief, the back skin of mice was cut into small pieces and digested with 0.15% collagenase 4 and 0.05% DNase for 90 min at 37 °C. Single-cell suspensions were obtained by passing cell preparation through 100-μm and 40-μm cell strainers. DCs were identified as lineage-negative (CD19−, CD3−, CD161c−, Ly6G−) CD45+ MHC II+ cells (excluding CD11b+ CD24high Langerhans and Ly6C+ CD64+ monocytes), and monocytes/monocyte-derived DCs were identified as lineage-negative (CD19−, CD3−, CD161c−, Ly6G−) CD45+CD11b+ cells (excluding Ly6C− CD64− DCs), as described previously (58). Antibodies used for staining are described in SI Materials and Methods.
For intracellular cytokine staining, isolated cells were stimulated with phorbol 12-myristate 13-acetate (5 ng/mL; Sigma-Aldrich) and ionomycin (500 ng/mL; Sigma-Aldrich). GolgiStop (BD Biosciences) was added 1 h later, followed by incubation for another 4 h. Stimulated cells were stained for extracellular markers, followed by fixation and permeabilization using the Cytofix/Cytoperm Kit (BD Biosciences). Samples were analyzed with a FACSCanto II flow cytometer (BD Bioscience) and FlowJo 7.6.5 software.
Histology and Immunofluorescence.
Standard procedures were used as detailed in SI Materials and Methods.
RNA Analysis by RNAseq and qPCR.
RNA-based analyses, including computational analyses, were based on total RNA obtained from snap-frozen skin biopsy specimens, as described in detail in SI Materials and Methods (59–62). RNAseq data of mouse samples have been deposited (GSE85891) in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo). The genes and primer sequences used for qPCR are listed in Table S3.
ELISA.
CXCL1 and CCL20 levels in the cell culture supernatant were determined using the DuoSet ELISA Development Kit (R&D Systems) according to the manufacturer's instructions.
Statistics.
All data are expressed as mean ± SD. Statistically significant differences were assessed by the two-tailed Student's t test for two groups.
SI Materials and Methods
Generation of Conditional Tnip1 (Tnip1flox) Mice.
Tnip1flox mice were generated using CRISPR-Cas9 technology (55). In brief, loxP sites were inserted in exons 7 and 9 of the Tnip1 gene together with an EcoRI restriction site in 3′ of the 5′ loxP site and 5′ of the 3′ loxP site by injecting C57BL/6 zygotes with two single guide RNAs (5′ guide sequence: 5′-TGCGGTGCGACCAGACCAAA-3′; 3′ guide sequence: 5′-TGATGTCAGCGGCAGCCATC-3′), along with Cas9 mRNA transcripts and two single-stranded DNA oligonucleotides that served as homology-directed repair (HDR) templates. The LoxP sites (underscored) and EcoRI sites (italic) were inserted 3 bp upstream of the protospacer adjacent motif (PAM) sequences (bold; on reverse complement), thereby disrupting the sgRNA-binding sites in the targeted allele. (5′ loxP HDR: 5′-CCTACAGATGGGGGAAATTCTGCTGTTAAGGATTCTCATGCAGCCTGCACACCTTCTGGGTGGCTTACAGATGCCCTTTTATAACTTCGTATAATGTATGCTATACGAAGTTATGAATTCGGTCTGGTCGCACCGCAGCAGTGCCTTTGACAGCAGATCCTTGGCTTTGCAAGTGTCTTAACCCAGGGCTGACATTGTGA-3′; 3′ loxP HDR: 5′- CATTCATATTCTAGATTGACTCATCTCTCAGAAGGAAAAGAATGTTCATTATGTAATGAACCAAGAGTCTCCCTCCAGATGAATTCATAACTTCGTATAATGTATGCTATACGAAGTTATGGCTGCCGCTGACATCAGCTCACTGTGTTCCCTTGGGGTTGTTTGGTTTTCCTTTTTTCTGTGGCACTAGGAACTGAACC-3′). The target sites for each sgRNA are unique in the genome and have no potential off-target sites with fewer than three mismatches as determined using the Cas-OFFinder algorithm, minimizing the risk of off-target mutations of Cas9/sgRNA injection in zygotes (55, 56). Genetically modified zygotes were generated by mating superovulated 12-wk-old C57BL/6J females to C57BL/6J males (The Jackson Laboratory). ssDNA (2 pmol/μL), sgRNA (50 ng/μL each), and Cas9 mRNA (100 ng/μL) were mixed in water and injected into the cytoplasm of pronuclear stage zygotes (Transgenic/Gene Knockout Shared Resource, St. Jude Children’s Research Hospital). Injected embryos were surgically transplanted into oviducts of pseudo-pregnant CD1 females. The genomic loci were amplified by PCR using the primer pair Tnip1-i7-R1 (5′-AGGACTGGTCATACCACGGA-3′) and Tnip1-i7-F1 (5′-AGAAGGTCTCACCCAGAGCA-3′) for amplification of the 5′ intronic loxP site (i7) and primer pair Tnip1-i9-R1 (CGGGAGAACTGGCTTATGCT) and Tnip1-i9-F1 (AGTCAGGACAGTCGGCATTG) for amplification of the 3′ intronic loxP site (i9). Amplicons were then subjected to restriction digestion using EcoRI and analyzed by electrophoresis and Sanger sequencing using the same primers. Mice harboring the two loxP sites on the same allele were identified by long-range PCR based on the primers Tnip1-i7-F1 and Tnip1-i9-R1, whose amplicon-analysis by Sanger sequencing confirmed the correct sequence of the loxP-flanked Tnip1 locus.
In Vitro Culture and Functional Assays of Keratinocytes and Fibroblasts.
Keratinocytes and dermal fibroblasts were prepared as described previously (57). In brief, for preparation of epidermal keratinocytes and dermal fibroblasts, adult mouse tail skin was incubated in 1% Trypsin without EDTA (Thermo Fisher Scientific) at 37 °C for 60 min. The epidermis was separated from the dermis and placed in keratinocyte growth medium (s-MEM; Sigma-Aldrich) supplemented with 8% (vol/vol) Chelex (Sigma-Aldrich)-treated FBS (HyClone), 1% penicillin-streptomycin (Thermo Fisher Scientific), 2 nM glutamate (Thermo Fisher Scientific), 1 mM sodium pyruvate (Thermo Fisher Scientific), and 0.2 mM calcium chloride. The Chelex resin (pH 7.0) had been stirred with two volumes of FBS for 30 min at room temperature, followed by filtering through 0.22-μm filters. Single-cell suspensions were prepared by repeated pipetting with a 10-mL pipette and filtering through 100-μm cell strainers. Keratinocytes were plated on tissue culture plastic (precoated with bovine plasma fibronectin (0.01 mg/mL; Thermo Fisher Scientific) and bovine type I collagen (3 mg/mL; Thermo Fisher Scientific) and incubated overnight at 37 °C in an atmosphere of 5% CO2. Loosely attached cells were removed by washing with PBS before feeding the attached cells with keratinocyte growth medium containing 0.05 mM CaCl. Medium was replaced with fresh medium every 2 d. Keratinocytes were used for experiments on day 3 after the initial plating.
To isolate fibroblasts, the dermis was cut into small pieces and digested with 0.35% collagenase type I (Worthington) in fibroblast growth medium (DMEM; Thermo Fisher Scientific) supplemented with 10% FBS (HyClone), 1% penicillin-streptomycin (Thermo Fisher Scientific), 2 mM glutamate (Thermo Fisher Scientific), and 1 mM sodium pyruvate (Thermo Fisher Scientific) for 30 min at 37 °C, followed by treatment with 20 µg/mL NDase (Worthington) for 15 min at 37 °C. Single-cell suspensions were prepared as described for keratinocytes and collected by centrifugation at 430 × g for 5 min at room temperature. The cells were suspended in fibroblast growth medium and cultured at 37 °C in an atmosphere of 5% CO2. The fibroblasts were trypsinized (0.25% trypsin-EDTA; Thermo Fisher Scientific) and passaged to new cell culture dishes every other day, and were used in the experiments after two passages. Two-hundred ng/mL of recombinant murine IL-17A (rmIL-17A; R&D Systems) was used for stimulation.
Antibodies Used for Flow Cytometry.
The following antibodies were used for flow cytometry staining: FITC-conjugated antibodies to mouse MHC-II (M5/114.15.2; eBioscience; 1:300 dilution) and CD3e (17A2; eBioscience; 1:100), PE-conjugated antibodies to mouse IL-17A (TC11-18H10; BD Bioscience; 1:200) and CCR2 (475301; R&D Systems; 1:50), PE-Cy7–conjugated antibodies to mouse Ly-6G (1A8; BD Bioscience; 1:200) and CD45 (30-F11; Biolegend; 1:250), allophycocyanin (APC)-conjugated antibodies to mouse γ/δ TCRs (GL3; eBioscience; 1:200) and CD64 (X54-5/7.1; Biolegend; 1:100), APC/Cy7-conjugated antibodies to mouse Ly6C (HK1.4; Biolegend; 1:200) and CD19 (1D3; BD Bioscience; 1:200), Brilliant Violet (BV) 421-conjugated antibodies to mouse CD11b (M1/70; Biolegend; 1:200), PerCP-Cy5.5–conjugated antibodies to mouse CD24 (M1/69; eBioscience; 1:200) and CD45 (30-F11; BD Bioscience; 1:100), and biotin-conjugated antibodies to mouse CD19 (1D3; eBioscience; 1:300), CD3e (145-2C11; BD Bioscience; 1:100), NK1.1 (PK136; TONBO Bioscience; 1:300), Ly6G (1A8; Biolegend; 1:200), and BV 570-conjugated streptavidin (Biolegend; 1:100). Specificity of all antibodies was controlled by respective antibody isotypes.
Histology and Immunofluorescence.
Skin biopsy specimens were fixed with 10% phosphate-buffered formalin and embedded in paraffin. Deparaffinized 5-μm sections were stained with H&E and analyzed by light microscopy using a Zeiss Axio Scope.A1 microscope equipped with an AxioCam MRc digital camera and Axiovision software. Epidermal thickness (excluding stratum corneum and “rete-like” ridges) was determined by taking measurements on 12 individual sections 150 µm apart from one another.
For immunofluorescence analysis, mouse skin was immersed in OCT compound (BD Biosciences) and frozen in liquid nitrogen. Cryosections of skin (6 μm thick) were fixed in 3% paraformaldehyde. Sections were permeabilized with 0.2% Triton X-100 in PBS for 10 min, blocked in 4% nonfat milk in TBST (20 mM Tris pH 7.2, 150 mM NaCl, and 0.5% Tween-20), and then incubated for 1 h with primary antibodies (FITC-conjugated anti-CD3; 17A2; eBioscience). DAPI was used to counterstain nuclei. Images were acquired using a Zeiss LSM 780 confocal microscope, and images from x–y sections (1 μm) were collected using Zen software. Images were stacked and processed using ImageJ version 1.48 and Adobe Photoshop CS6. All images of tissue samples from different groups of mice were collected and processed under identical conditions.
RNA Analysis by RNAseq and qPCR.
Skin biopsy specimens (10 × 5 mm) were snap-frozen in liquid nitrogen and pulverized using a BioPulverizer (BioSpec) that was precooled in liquid nitrogen. Total RNA was isolated using TRIzol (Invitrogen) and quantified by measuring the optical density at 260 nm using a NanoDrop spectrophotometer. For RNAseq, libraries were generated from ∼1 µg of total RNA using the Illumina TruSeq Stranded Total RNA Prep Kit with the Ribo-Zero Gold Human/Mouse/Rat Ribosomal Reduction Kit. Libraries were sequenced on an Illumina HiSeq 2500 Sequencing system using paired-end 100-bp sequencing chemistry. For qPCR, cDNA was prepared from 1 μg of total RNA by reverse transcription using SuperScript II reverse transcriptase (Thermo Fisher Scientific) and oligo(dT) primers. Real-time PCR was performed using SYBR Green Master Mix (Thermo Fisher Scientific) and gene-specific primers in a 7300 Sequence Detector System (Thermo Fisher Scientific). Data were normalized by the level of GAPDH expression in each individual sample. The genes and primer sequences used for qPCR are listed in Table S3.
Computational Biology Analyses.
RNAseq reads were mapped to the mm9 genome using an internal pipeline that uses STAR and Tophat2 (59, 60) Counting and FPKM (fragments per kilobase of transcript per million mapped reads) calculations were performed using HTSEQ (61). Log2 transformation, file merging, correlations, and visualization were done using STATA/MP SE 11.2 (College Station). The density plots were created by dividing the data in 10K bins by first dividing the range of the x and y logratios by 100 and counting membership for each (x, y). Cross-species analyses were joined by case-insensitive gene symbol matching. Statistical analyses, including unequal variance tests, were performed using Partek Genomic Suite 6.6. The heat map was generated using Spotfire DecisionSite 9.1.2 (TIBCO). GO category enrichment was done using DAVID (62).
Public data for cross-species comparison were downloaded from the GEO database as matrices. For psoriasis, RNAseq data from 82 normal skin samples and 92 psoriasis skin samples (accession no. GSE54456) were downloaded, log 2 transformed, and compared with the mouse model data. For atopic eczema, 30 human skin samples (15 from subjects with actopic dermatitis and 15 from healthy subjects) were compared from arrays (accession no. GSE12511).
Supplementary Material
Acknowledgments
We thank Hartmut Berns for the generation of Tnip1flox mice; the staff of the Transgenic/Gene Knockout Shared Resource, the Animal Resource Center, and the Veterinary Pathology Core at St. Jude Children’s Research Hospital; and Doug Green for generously providing resources of the St. Jude Immunology Department. This work was supported by the American Lebanese Syrian Associated Charities.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE85891).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606996113/-/DCSupplemental.
References
- 1.Perera GK, Di Meglio P, Nestle FO. Psoriasis. Annu Rev Pathol. 2012;7:385–422. doi: 10.1146/annurev-pathol-011811-132448. [DOI] [PubMed] [Google Scholar]
- 2.Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227–255. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee E, et al. Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. J Exp Med. 2004;199(1):125–130. doi: 10.1084/jem.20030451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGeachy MJ, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10(3):314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuentes-Duculan J, et al. A subpopulation of CD163-positive macrophages is classically activated in psoriasis. J Invest Dermatol. 2010;130(10):2412–2422. doi: 10.1038/jid.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillitzer R, Ritter U, Spandau U, Goebeler M, Bröcker EB. Differential expression of GRO-alpha and IL-8 mRNA in psoriasis: A model for neutrophil migration and accumulation in vivo. J Invest Dermatol. 1996;107(5):778–782. doi: 10.1111/1523-1747.ep12371803. [DOI] [PubMed] [Google Scholar]
- 7.Kulke R, et al. Co-localized overexpression of GRO-alpha and IL-8 mRNA is restricted to the suprapapillary layers of psoriatic lesions. J Invest Dermatol. 1996;106(3):526–530. doi: 10.1111/1523-1747.ep12343916. [DOI] [PubMed] [Google Scholar]
- 8.Nograles KE, et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol. 2008;159(5):1092–1102. doi: 10.1111/j.1365-2133.2008.08769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedrick MN, et al. CCR6 is required for IL-23-induced psoriasis-like inflammation in mice. J Clin Invest. 2009;119(8):2317–2329. doi: 10.1172/JCI37378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha HL, et al. IL-17 drives psoriatic inflammation via distinct, target cell-specific mechanisms. Proc Natl Acad Sci USA. 2014;111(33):E3422–E3431. doi: 10.1073/pnas.1400513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krueger JG, et al. IL-17A is essential for cell activation and inflammatory gene circuits in subjects with psoriasis. J Allergy Clin Immunol. 2012;130(1):145–154. doi: 10.1016/j.jaci.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopp T, et al. Clinical improvement in psoriasis with specific targeting of interleukin-23. Nature. 2015;521(7551):222–226. doi: 10.1038/nature14175. [DOI] [PubMed] [Google Scholar]
- 13.Jullien D, Prinz JC, Nestle FO. Immunogenicity of biotherapy used in psoriasis: The science behind the scenes. J Invest Dermatol. 2015;135(1):31–38. doi: 10.1038/jid.2014.295. [DOI] [PubMed] [Google Scholar]
- 14.Wagner EF, Schonthaler HB, Guinea-Viniegra J, Tschachler E. Psoriasis: What we have learned from mouse models. Nat Rev Rheumatol. 2010;6(12):704–714. doi: 10.1038/nrrheum.2010.157. [DOI] [PubMed] [Google Scholar]
- 15.Swindell WR, et al. Genome-wide expression profiling of five mouse models identifies similarities and differences with human psoriasis. PLoS One. 2011;6(4):e18266. doi: 10.1371/journal.pone.0018266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair RP, et al. Collaborative Association Study of Psoriasis Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41(2):199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harden JL, Krueger JG, Bowcock AM. The immunogenetics of psoriasis: A comprehensive review. J Autoimmun. 2015;64:66–73. doi: 10.1016/j.jaut.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyninck K, et al. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol. 1999;145(7):1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, et al. A20-binding inhibitor of NF-κB (ABIN1) controls Toll-like receptor-mediated CCAAT/enhancer-binding protein β activation and protects from inflammatory disease. Proc Natl Acad Sci USA. 2011;108(44):E998–E1006. doi: 10.1073/pnas.1106232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callahan JA, et al. Cutting edge: ABIN-1 protects against psoriasis by restricting MyD88 signals in dendritic cells. J Immunol. 2013;191(2):535–539. doi: 10.4049/jimmunol.1203335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilliet M, et al. Psoriasis triggered by Toll-like receptor 7 agonist imiquimod in the presence of dermal plasmacytoid dendritic cell precursors. Arch Dermatol. 2004;140(12):1490–1495. doi: 10.1001/archderm.140.12.1490. [DOI] [PubMed] [Google Scholar]
- 22.Wu JK, Siller G, Strutton G. Psoriasis induced by topical imiquimod. Australas J Dermatol. 2004;45(1):47–50. doi: 10.1111/j.1440-0960.2004.00030.x. [DOI] [PubMed] [Google Scholar]
- 23.Rajan N, Langtry JA. Generalized exacerbation of psoriasis associated with imiquimod cream treatment of superficial basal cell carcinomas. Clin Exp Dermatol. 2006;31(1):140–141. doi: 10.1111/j.1365-2230.2005.01938.x. [DOI] [PubMed] [Google Scholar]
- 24.Fanti PA, Dika E, Vaccari S, Miscial C, Varotti C. Generalized psoriasis induced by topical treatment of actinic keratosis with imiquimod. Int J Dermatol. 2006;45(12):1464–1465. doi: 10.1111/j.1365-4632.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- 25.van der Fits L, et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol. 2009;182(9):5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- 26.Cai Y, et al. Pivotal role of dermal IL-17-producing γδ T cells in skin inflammation. Immunity. 2011;35(4):596–610. doi: 10.1016/j.immuni.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray EE, Suzuki K, Cyster JG. Cutting edge: Identification of a motile IL-17-producing gammadelta T cell population in the dermis. J Immunol. 2011;186(11):6091–6095. doi: 10.4049/jimmunol.1100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sumaria N, et al. Cutaneous immunosurveillance by self-renewing dermal gammadelta T cells. J Exp Med. 2011;208(3):505–518. doi: 10.1084/jem.20101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai Y, Fleming C, Yan J. New insights of T cells in the pathogenesis of psoriasis. Cell Mol Immunol. 2012;9(4):302–309. doi: 10.1038/cmi.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68(5):869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 31.Ramirez-Carrozzi V, et al. IL-17C regulates the innate immune function of epithelial cells in an autocrine manner. Nat Immunol. 2011;12(12):1159–1166. doi: 10.1038/ni.2156. [DOI] [PubMed] [Google Scholar]
- 32.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278(3):1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 33.Ghilardi N, et al. Compromised humoral and delayed-type hypersensitivity responses in IL-23-deficient mice. J Immunol. 2004;172(5):2827–2833. doi: 10.4049/jimmunol.172.5.2827. [DOI] [PubMed] [Google Scholar]
- 34.Lindroos J, et al. IL-23-mediated epidermal hyperplasia is dependent on IL-6. J Invest Dermatol. 2011;131(5):1110–1118. doi: 10.1038/jid.2010.432. [DOI] [PubMed] [Google Scholar]
- 35.Rizzo HL, et al. IL-23-mediated psoriasis-like epidermal hyperplasia is dependent on IL-17A. J Immunol. 2011;186(3):1495–1502. doi: 10.4049/jimmunol.1001001. [DOI] [PubMed] [Google Scholar]
- 36.Friedrich M, Tillack C, Wollenberg A, Schauber J, Brand S. IL-36γ sustains a proinflammatory self-amplifying loop with IL-17C in anti-TNF-induced psoriasiform skin lesions of patients with Crohn’s disease. Inflamm Bowel Dis. 2014;20(11):1891–1901. doi: 10.1097/MIB.0000000000000198. [DOI] [PubMed] [Google Scholar]
- 37.Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127(22):4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- 38.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 39.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawasaki A, et al. Association of TNFAIP3 interacting protein 1, TNIP1 with systemic lupus erythematosus in a Japanese population: A case-control association study. Arthritis Res Ther. 2010;12(5):R174. doi: 10.1186/ar3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adrianto I, et al. BIOLUPUS and GENLES Networks Association of two independent functional risk haplotypes in TNIP1 with systemic lupus erythematosus. Arthritis Rheum. 2012;64(11):3695–3705. doi: 10.1002/art.34642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tortola L, et al. Psoriasiform dermatitis is driven by IL-36-mediated DC-keratinocyte crosstalk. J Clin Invest. 2012;122(11):3965–3976. doi: 10.1172/JCI63451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schonthaler HB, et al. S100A8-S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity. 2013;39(6):1171–1181. doi: 10.1016/j.immuni.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Häcker H, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439(7073):204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 45.Schwandner R, Yamaguchi K, Cao Z. Requirement of tumor necrosis factor receptor-associated factor (TRAF)6 in interleukin 17 signal transduction. J Exp Med. 2000;191(7):1233–1240. doi: 10.1084/jem.191.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mauro C, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281(27):18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 47.Garg AV, Ahmed M, Vallejo AN, Ma A, Gaffen SL. The deubiquitinase A20 mediates feedback inhibition of interleukin-17 receptor signaling. Sci Signal. 2013;6(278):ra44. doi: 10.1126/scisignal.2003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomson W, et al. Wellcome Trust Case Control Consortium YEAR Consortium Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39(12):1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plenge RM, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39(12):1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lippens S, et al. Keratinocyte-specific ablation of the NF-κB regulatory protein A20 (TNFAIP3) reveals a role in the control of epidermal homeostasis. Cell Death Differ. 2011;18(12):1845–1853. doi: 10.1038/cdd.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289(5488):2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruddy MJ, et al. Functional cooperation between interleukin-17 and tumor necrosis factor-alpha is mediated by CCAAT/enhancer-binding protein family members. J Biol Chem. 2004;279(4):2559–2567. doi: 10.1074/jbc.M308809200. [DOI] [PubMed] [Google Scholar]
- 53.Shen F, Hu Z, Goswami J, Gaffen SL. Identification of common transcriptional regulatory elements in interleukin-17 target genes. J Biol Chem. 2006;281(34):24138–24148. doi: 10.1074/jbc.M604597200. [DOI] [PubMed] [Google Scholar]
- 54.Hirota K, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12(3):255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pelletier S, Gingras S, Green DR. Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity. 2015;42(1):18–27. doi: 10.1016/j.immuni.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bae S, Park J, Kim JS. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30(10):1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lichti U, Anders J, Yuspa SH. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc. 2008;3(5):799–810. doi: 10.1038/nprot.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malosse C, Henri S. Isolation of mouse dendritic cell subsets and macrophages from the skin. Methods Mol Biol. 2016;1423:129–137. doi: 10.1007/978-1-4939-3606-9_9. [DOI] [PubMed] [Google Scholar]
- 59.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders S, Pyl PT, Huber W. HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.