Significance
HIV-1 is commonly assumed to have nine genes. However, in 1988 a 10th gene was suggested, overlapped by the env gene, but read on the antisense strand. The corresponding protein was named AntiSense Protein (ASP). Several pieces of evidence argue in favor of ASP expression in vivo, but its function is still unknown. We performed the first evolutionary study of ASP, using a very large number of HIV-1 and SIV (simian) sequences. Our results show that ASP is specific to group M of HIV-1, which is responsible for the pandemic. Moreover, we demonstrated that evolutionary forces act to maintain the asp gene within the M sequences and showed a striking correlation of asp with the spread of the pandemic.
Keywords: HIV-1, asp and env genes, overlapping genes, phylogenetic analyses, selection pressure
Abstract
Recent experiments provide sound arguments in favor of the in vivo expression of the AntiSense Protein (ASP) of HIV-1. This putative protein is encoded on the antisense strand of the provirus genome and entirely overlapped by the env gene with reading frame −2. The existence of ASP was suggested in 1988, but is still controversial, and its function has yet to be determined. We used a large dataset of ∼23,000 HIV-1 and SIV sequences to study the origin, evolution, and conservation of the asp gene. We found that the ASP ORF is specific to group M of HIV-1, which is responsible for the human pandemic. Moreover, the correlation between the presence of asp and the prevalence of HIV-1 groups and M subtypes appeared to be statistically significant. We then looked for evidence of selection pressure acting on asp. Using computer simulations, we showed that the conservation of the ASP ORF in the group M could not be due to chance. Standard methods were ineffective in disentangling the two selection pressures imposed by both the Env and ASP proteins—an expected outcome with overlaps in frame −2. We thus developed a method based on careful evolutionary analysis of the presence/absence of stop codons, revealing that ASP does impose significant selection pressure. All of these results support the idea that asp is the 10th gene of HIV-1 group M and indicate a correlation with the spread of the pandemic.
It is well established that retroviruses are able to perform antisense transcription from the 3′ long terminal repeat (LTR) of their proviral genome (1, 2). In 1988, the existence of an ORF on the antisense strand of the HIV type 1 (HIV-1) genome was suggested (3). This ORF encodes the putative AntiSense Protein (ASP). The existence of this ORF and of the encoded protein was controversial for many years, but now several pieces of evidence argue in favor of its expression (see ref. 4 for an extensive review): (i) several polyadenylated antisense transcripts capable of encoding ASP have been characterized within HIV-1–infected cells (1, 5, 6); (ii) it was demonstrated that the full-length ASP protein can be expressed ex vivo from the HIV-1 3′ LTR (7); (iii) ASP has been detected in freshly infected cells (2, 8, 9); and (iv) two recent independent clinical studies have shown the in vivo expression of ASP by detecting a cell-mediated immune response against several ASP epitopes within 30% of individuals infected with subtype B viruses (10, 11) [a percentage similar to those observed with other HIV-1 proteins, e.g., Tat and Pol (12)]. Moreover, experimental results suggested that ASP could form stable aggregates, be located partially at the plasma membrane, and be associated with autophagy (4, 7, 8). Despite this accumulation of evidence, the existence of ASP is still questioned because, for example, defective ribosome products with immune response have been reported for several viruses including HIV-1 (13, 14). Elucidating the function of ASP is thus a major goal, but studying the evolutionary forces acting on ASP is also crucial.
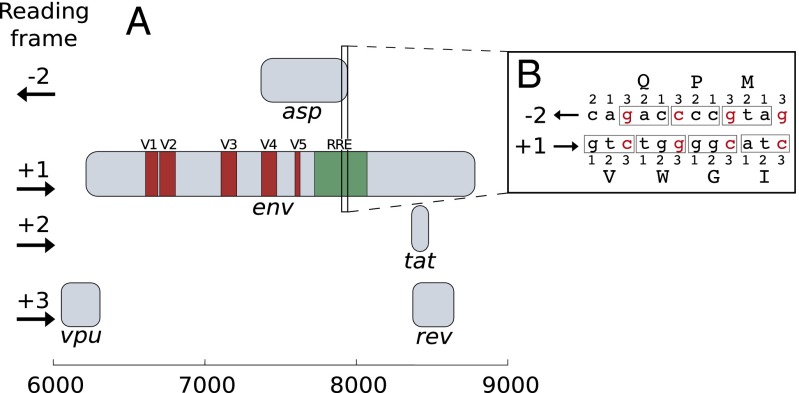
A striking fact with ASP ORF (and a challenge in terms of bioinformatics analyses) is its location on the provirus genome, as it overlaps the env (envelope) gene. Overlapping genes are a common feature of viruses to “compress” their genome (15). However, as the same portion of DNA encodes for several proteins, their adaptability is strongly lowered (16). Proteins encoded by overlapping genes are generally accessory proteins that play a role in viral pathogenicity or spreading (17). ASP ORF overlaps env on the frame −2: the codon positions 1, 2, and 3 in env face positions 2, 1, and 3 in asp, respectively (Fig. 1B). Because the two most important positions of env and asp codons are opposite each other, there is particularly little flexibility to encode amino acids (18).
Fig. 1.
Structure of the HIV-1 genome in the env gene region. (A) This region contains five overlapping ORFs: the env gene, the exon 2 of tat, the exon 2 of rev, the C-terminal extremity of vpu, and the ASP ORF. The env gene contains five variable regions (V1 to V5, reddish) and the RRE (greenish). (B) Overlapping sequences on the frames −2 and +1 [start of the asp region, HXB2 (20; GenBank accession no. K03455)].
The aims of this study were to assess the presence and conservation of the ASP ORF in the HIV-1 and SIVcpz/gor (chimpanzee and gorilla) groups and subtypes and to demonstrate the selection pressure induced by ASP to confirm its importance in some of the mechanisms of the virus.
Results
HIV-1 strains are classified into four phylogenetic groups: M, N, O, and P. These four groups resulted from four separate cross-species transmission events of Simian Immunodeficiency Virus (SIV) to humans (19). Group M is the pandemic group. It is divided into nine distinct subtypes, and more than 70 circulating recombinant forms (CRFs).
The ASP ORF is entirely overlapped by the env gene (Fig. 1A), which has several overlapping ORFs on different reading frames. The env gene contains five variable regions, separated by constant regions (21), and the Rev Response Element (RRE) (22), which is a highly structured RNA element that plays a role in the export of HIV-1 mRNAs.
Data.
We downloaded all available and complete HIV-1/SIVcpz/gor env sequences and data annotations from the Los Alamos HIV Sequence Database (www.hiv.lanl.gov/content/index). We also used GenBank to retrieve the original version of some of the sequences. After deleting problematic sequences, we obtained 22,992 env sequences belonging to 3,931 individuals. Codon-based, multiple alignments were performed on the env gene and on the frame −2 of this gene. To avoid counting several times sequences that are very close to each other and belong to the same individual, we used two strategies: (i) we used the complete multiple alignment, but weighted the sequences so that each individual had a total weight of 1 (we then obtained the “weighted” alignment); and (ii) we randomly selected one sequence per individual when it was required for computational reasons. Most of our results and statistics are based on weighted sequences, except where otherwise specified. Details are provided in SI Text.
Detection of the ASP ORF.
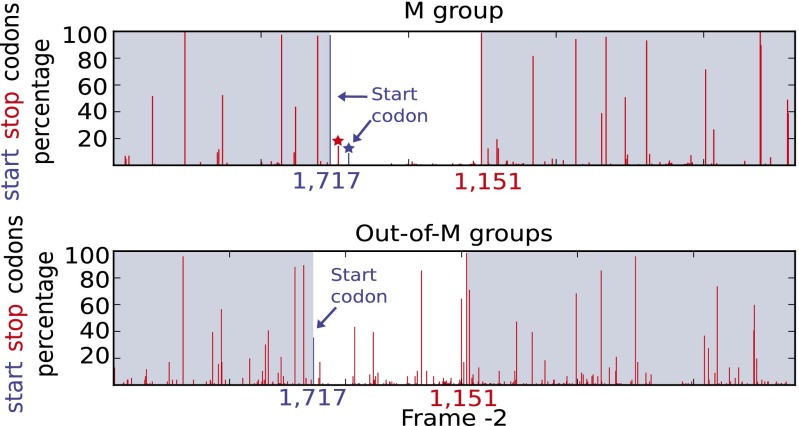
We based our analyses on the presence/absence of start and stop codons in frame −2 of the env region. The ASP ORF of the reference sequence HXB2 (20; GenBank accession no. K03455) has a length of 188 codons and is located between reference env positions 1,717 and 1,151. We thus searched all of our sequences for long DNA segments (>150 codons) with a start codon and no stop codon, read in frame −2 and located between these two reference positions. The analysis was carried out in the group M and an “out-of-M” dataset comprising all nonpandemic (N, O, P) HIV-1 and SIVcpz/gor sequences. For the sequences in group M and using the above criteria, we detected the ASP ORF for 77% of the (weighted) sequences. We clearly observed (Fig. 2) a region that is nearly free of stop codons and located between the reference positions of asp. At the beginning of the asp region, we note the presence of a stop codon for 14.5% of the sequences, located 12 codons after the start codon. However, most of these sequences (90%) belong to subtype A and its recombinants. One of the A recombinants in the asp region, namely CRF02_AG (112 individuals), has the early stop for ∼100% of the sequences, but only 7% of its sequences have the ASP ORF using our criteria. In contrast, in subtype A (240 sequences), ∼100% of sequences have the early stop, but a large percentage of them (∼90%) have an alternative start codon located 17 codons after the early stop; These sequences thus have a shorter version of ASP ORF, but still with a length of more than 150 codons.
Fig. 2.
Detection of the ASP ORF. Weighted percentages of start (blue) and stop (red) codons in frame −2, in the groups M and out-of-M. The asp region (white area) is located between the env positions 1,717 and 1,151 (HXB2 reference). The red star indicates an early stop codon that is specific to subtype A and A recombinants. This early stop codon is followed by an alternative start codon (blue star) in most of the A sequences and certain A recombinants.
In the out-of-M sequences, a number of stop codons are observed inside the asp region (Fig. 2). The start codon is not conserved (38% of sequences have a start/methionine codon), and less than 1.5% of out-of-M sequences have an ORF in asp region with length >150 codons.
Recent Emergence of the ASP ORF.
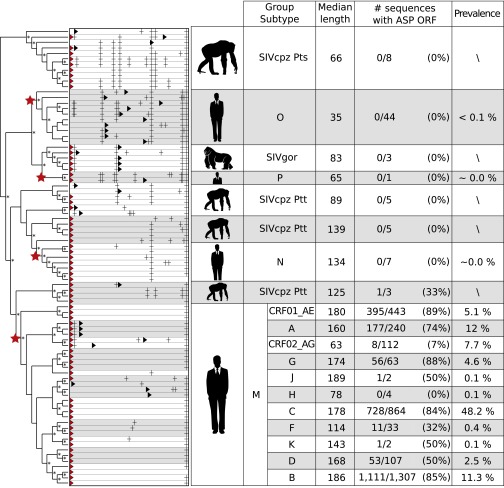
The contrasting results between the pandemic group M and the other groups (out-of-M) led us to study the emergence and evolution of the ASP ORF using a phylogenetic approach. For this purpose, we inferred a maximum-likelihood tree using PhyML (23) (GTR+Γ4+I model, 1,000 bootstrap replicates) on a selection of sequences extracted from our complete alignment. We used 33 reference sequences (24) of group M subtypes and CRFs (A, B, C, D, F, G, H, J, CRF01_AE, CRF02_AG), 10 randomly selected sequences from group O, and the 40 sequences of the other out-of-M groups. To complete this phylogeny, we computed statistics from the complete, weighted alignment for all groups, subtypes, and CRFs. By using the same detection criteria as above, we measured the length of the longest ORF in the asp region and the fraction of sequences that had the ASP ORF.
The phylogeny (Fig. 3) clearly shows the four introductions of HIV-1 in the human population, corresponding to the four groups O, P, N, and M. The start codon corresponding to asp is present in most of the studied sequences. However, group O sequences and some exceptions (e.g., subtype H, prevalence ∼0.1%) do not have this start codon. The median length of the ORF in the asp region of the out-of-M groups increases when approaching group M: there are 66 codons in SIVcpz_Pts (from Pan troglodytes schweinfurthii), which increases to 125 codons in SIVcpz_Ptt (from Pan troglodytes troglodytes) that is closest to the group M. For group M sequences, the ASP ORF is present in 77% of the sequences with a median length of 182 codons. All of this indicates that the ASP ORF was created recently and that its emergence in HIV-1 is concomitant with the emergence of the group M. This recent de novo creation is further supported by the fact that ASP does not have any known homologs (SI Text). Interestingly, among SIVcpz_Ptt sequences, there is one sequence that possesses the ASP ORF in its entirety. This simian ASP has the same structural features (4) as the human ones. However, it is phylogenetically remote, and we do not have enough SIVcpz_Ptt sequences to figure out whether ASP appeared in the HIV and SIV genomes independently or, rather, if ASP first appeared in the SIVcpz_Ptt genome and was maintained when the HIV-1 group M emerged from it (SI Text and Fig. S1).
Fig. 3.
Recent emergence of the ASP ORF using a phylogenetic approach. This phylogeny (* = bootstrap >80%) of the env gene contains reference sequences from HIV-1 and SIVcpz/gor groups, subtypes, and CRFs. The four distinct simian/human transmissions are indicated by red stars. For each of the sequences, we show the distribution of start codons (red triangle; black triangle for alternative start) and stop codons (black cross) in the asp region. For each group, subtype, and CRF, the table provides the median length of the ASP ORF, the fraction of sequences with the ASP ORF (length > 150 codons), and the prevalence in the human population (44).
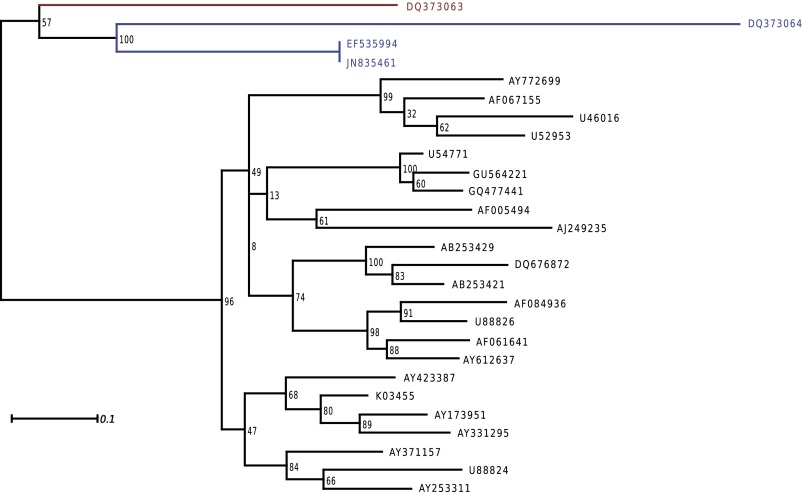
Fig. S1.
Phylogenetic analysis of the SIVcpz_Ptt sequence that has the ASP ORF. The phylogenetic tree was estimated using PhyML with a nucleic multiple alignment of the asp region, GTR+ Γ6, and 100 replicates. The DQ373063 sequence that has ASP is in red; the SIVcpz_Ptt sequences without ASP are in blue; the group M reference sequences that have ASP are in black.
However, the fraction of M sequences that have ASP ORF varies among subtypes and CRFs. The less prevalent subtypes (D, F, J, H, K, total prevalence ∼3%) have the ASP ORF for less than 45% of their sequences. As already mentioned, only a few sequences (7%) in CRF02_AG (prevalence 7.7%) have the ASP ORF. In contrast, the other prevalent subtypes and CRFs (A, B, C, G, and CRF01_AE, total prevalence = 81%) have ASP for 84% of their sequences. We thus see a clear correlation (P value = 0.003) (Materials and Methods): prevalent M subtypes and CRFs (except CRF02_AG) have the ASP ORF for a large majority of sequences, whereas low-prevalence subtypes and nonpandemic groups (N, O, P) have the ASP ORF in a minority of sequences (none in some groups/subtypes). This correlation is confirmed when accounting for phylogenetic correlation (Bayes factor = 3.8) (Materials and Methods).
The ASP ORF is present in 84% of sequences for prevalent subtypes (A, B, C, G) and CRF01_AE. This fraction is quite high and is not likely to be due to chance, as we shall see. However, 16% of sequences in these subtypes and CRF do not have the ASP ORF. This level of absence is similar to the one observed with nef [13.5% (25)], an accessory gene, and higher than the ∼5% that we found for env and pol (two obligatory genes) by scanning for the presence of stop codons [all available Los Alamos database sequences (December 2015)]. This nonnegligible fraction of stops in env and pol is explained by both sequencing errors (26) and the fact that some of the sequences are defective (27). The higher level of absence with ASP ORF is explained by the fact that asp is an accessory gene. As expected, ASP ORF was lost not only in some of the M subtypes and CRFs, but also in some of the individuals of prevalent subtypes and CRF01_AE, where 12% of individuals in our dataset do not have any sequence with the ASP ORF, whereas 81% of individuals have the ASP ORF for all of their sequences. These 12%, added to the 5% of sequencing errors and defective sequences observed with env and pol, roughly explain the 16% of asp absence in prevalent subtypes and CRF01_AE.
Conservation of the ASP ORF.
Previous analyses indicate that the ASP ORF is present in a large fraction of the group M sequences. We used computer simulations to demonstrate that there is a very low probability that this is due to chance.
We first estimated the probability of observing an ORF with ASP length overlapping the env gene in frame −2. For this purpose, we randomly generated sequences with the same length (856 codons) as the env gene of HXB2 and the same codon usage as HIV-1 (www.kazusa.or.jp/codon/). In this case, the probability of an ORF of length 180 in frame −2 is ∼3%. This is a low probability, but ASP is the longest overlapping ORF present in HXB2 in any reading frame (3), and one could argue that having such an ORF in the whole HIV-1 genome is quite likely. Using the same method as above, we thus generated sequences having the same length (3,239 codons) as HXB2 and searched for an ORF of length 180 in the five possible reading frames. The probability in this case is ∼19%. This is still a relatively low probability, but clearly we cannot reject that the presence of the ASP ORF at the root of HIV-1 M is merely due to chance. However, we observed the ASP ORF in 77% of our M sequences. The question then is: if we assume the ASP ORF presence at the root of HIV-1 M, would we have a significant chance of observing its presence in so many sequences at the phylogeny tips?
To answer this question, we simulated the evolution of the env gene along phylogenies inferred using 350 randomly selected strains. We used PhyML and codonPhyML (28) to infer 10 such phylogenies. For each one, we performed 100 codon-based simulations using Alf (29), starting from the env gene of HXB2 at the tree root (Materials and Methods). The maximum percentage of the tip sequences where the ASP ORF was still present (across 1,000 datasets) was equal to 67%, and on average, the ASP ORF was conserved in only 42% of sequences.
These results show that there is an extremely low probability that our observation of 77% on the conservation of the ASP ORF in the M group is due to chance, thus revealing a selection pressure that tends to conserve ASP. In the following section, we show that this selection pressure is also detected at the sequence level. We first used standard methods (evolutionary rate, nonsynonymous versus synonymous substitutions, codon usage), but none of these approaches provided a significant signal due to the specificity of frame −2 (30, 31) (SI Text). Thus, we developed a method dedicated to the frame −2.
Measuring Selection Pressure.
This method is based on a careful analysis of the presence/absence of start and stop codons. Let us first discuss the start codon (included in the RRE) (Fig. 1). Having a start codon (atg) on frame −2 implies the presence of the two codons on (env) frame +1: xxc followed by atx (x represents any nucleotide) (Fig. 1B). Any mutation on the third base of the first codon (xxc) leads to the disappearance of the start codon on frame −2. However, for all amino acids that are encoded by a codon ending with c, a mutation of c into t leaves the amino acid unchanged. Thus, on frame −2, the start codon is never imposed by the sense gene and may appear/disappear synonymously. In fact, we observe (Fig. 2) that the start codon is highly frequent (97%) among the sequences of HIV-1 M. This is a first indication of selection pressure acting on asp. This selection effect is not found in out-of-M sequences, where the start codon is present in only 38% of sequences. In other words, this selection effect is most likely attributable to the maintenance of the ASP ORF in HIV-1 M, and not to some other cause (e.g., RRE structural constraints), which would impact both M and out-of-M sequences. However, the start codon is not included in further statistical significance calculations, as the RRE secondary structure could differ between M and out-of-M sequences.
Moreover, ∼90% of A sequences (240 individuals) with an early stop codon have an alternative start/methionine codon (Fig. 2), which is observed in only 4.8% of the sequences that do not have this early stop. This is another indication of the pressure to maintain the ASP ORF, acting specifically in subtype A and some A recombinants.
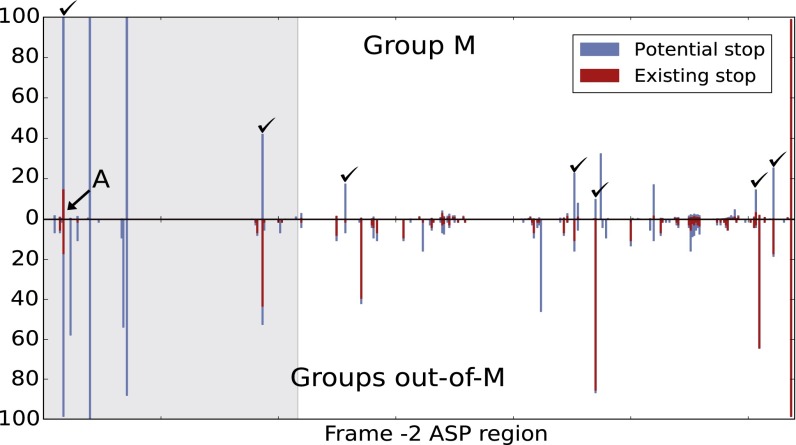
We used the same type of reasoning with the stop codons. There are actually two types of stop codons on frame −2 (see Materials and Methods for details). Some are imposed by the coding of Env, as is the case for the terminal stop. This explains why nearly 99% of our sequences do have the terminal stop codon, even when they do not have the ASP ORF. The other type of stops may appear/disappear without modifying Env, just as with the start codon. When they are absent, we call them “potential” stops, meaning that they can mutate into a stop codon synonymously for Env. Fig. 4 displays the frequency of the potential/existing stop codons in the asp region of both M and out-of-M sequences. For 11 sites, we observe potential stops for more than 10% of M sequences, but, on average, these sites contain stops for only 0.5% sequences (disregarding A and A recombinant sequences with early stop and alternative start). Moreover, for seven of these sites (labeled in Fig. 4), stop codons are actually observed in out-of-M sequences. When we remove the RRE region, which clearly imposes strong structural constraints, five of these seven sites remain, with (on average) <0.7% and >23% stops in M and out-of-M sequences, respectively. We thus have a second, strong indication of the selection pressure to maintain ASP: stop codons could be observed and are actually observed in out-of-M sequences, but not in M sequences. Note, however, that we cannot exclude the existence of some yet-unknown constraint (e.g., large-scale RNA structure), differing in M and out-of-M groups and inducing such an effect.
Fig. 4.
Percentage of potential (blue) and existing (red) stop codons in the asp region. Shaded area = RRE region. In the top panel, the stop codon that is conserved in 14.5% of sequences is characteristic of subtype A and A recombinants. The labeled sites contain potential stop codons for more than 10% of M sequences and existing stop codons in some of the out-of-M sequences.
To measure the statistical significance of these findings, we used a method inspired by Firth (32), but dedicated to frame −2 and the analysis of potential stop codons. The original principle is to count the number of synonymous mutations on the reference gene and check whether this number is significantly less than expected. However, in frame −2 most synonymous mutations in the reference frame are also synonymous in the overlapping frame due to the fact that the third codon positions face each other (Fig. 1B). This makes the original method unable to detect any global pressure induced by ASP, although it performs well with other reading frames (Fig. S2). Our adaptation involves focusing solely on the stop codons and counting the synonymous mutations corresponding to the change of potential stops into existing stops. We compute a statistic that is equal to the expected number of such mutations minus the observed number of mutations; then we perform a Z-score–like test to derive a P value (Materials and Methods).
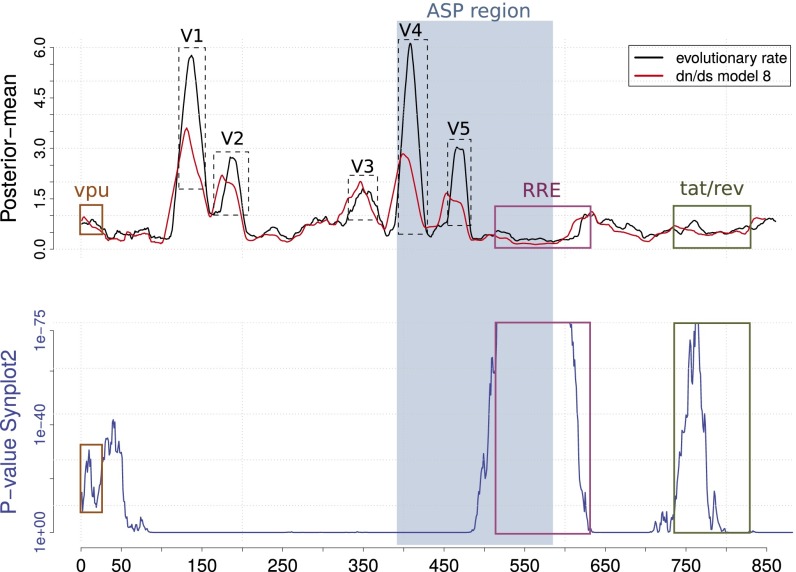
Fig. S2.
Standard analyses. Moving average of evolutionary rate (black) and dN/dS (red) with a sliding window of 20 codons. Results of Synplot2 (blue) with a sliding window of 100 codons.
For sequences of group M (3,855 sequences), we observe a positive result (Z = 6.47, P value = 10−10) in the asp region. In other words, there are fewer stops than expected if the null model was true, and we clearly see the presence of selection pressure. When excluding the RRE region, which induces strong structural constraints (Fig. 4), the difference is still highly significant, with fewer stops than expected (Z = 2.9, P value = 0.006).
As a negative control, we applied the same method to the rest of the env gene (i.e., the env-asp region). In this case, the number of observed stops in group M was greater than expected (Z = −1.9, P value = 0.06), which further supports the significance of our observations in the asp region. Moreover, for out-of-M groups (76 sequences) we did not detect any significant difference between the observed and expected number of stops, both in the asp region (Z = 1.53, P value = 0.13) and in the env-asp region (Z = 1.04, P value = 0.3). When excluding the RRE, we obtained a negative score (Z = −0.17, P value = 0.71), meaning that the positive score (Z = 1.53) observed in the whole asp region was induced by the RRE.
To summarize, these results indicate that the ASP ORF in group M is maintained selectively by conserving the start codon and avoiding stop codons. This finding is highly significant and explains, at the sequence level, the computer simulation results presented in the previous section. Moreover, the same results were not observed in either the env-asp region of M sequences or in the asp and env-asp regions of out-of-M sequences.
Discussion
By looking at the presence/absence of start and stop codons, we showed the existence of the ASP ORF in most of the prevalent subtypes and recombinant forms of HIV-1 M (with the notable exception of the CRF02_AG recombinant). In contrast, the ASP ORF appears to be absent in the other nonpandemic subtypes and human groups. These results indicate that the ASP ORF was created recently, concomitantly with the emergence of HIV-1 M, and is specific to this group, which is responsible for the human pandemic.
However, a relatively large fraction (16%) of M sequences do not have the ASP ORF, even in the prevalent subtypes and recombinant forms. This level of absence, and the fact that asp is a recent de novo creation, indicate that asp is an accessory gene that can be lost without compromising the viability of the virus. This could explain why finding the function of ASP has proven to be so difficult since its discovery in the 1990s.
It has been observed that viral de novo genes often play a role in pathogenicity or spreading, rather than being central to viral replication or structure (17, 33, 34). This scheme most likely applies to asp. The striking correlation between the presence of asp in nonpandemic groups and M subtypes and CRFs, and their prevalence, strongly supports the idea that ASP could play a role in spreading. This contradicts a common argument that the difference among HIV-1 groups in terms of prevalence and impact in human populations has no molecular basis, but is mostly due to social changes in ∼1960 in central Africa, where the group M was already well established (35).
Our simulations and careful analyses of start and stop codons all indicate the presence of selection pressure to maintain the ASP ORF. However, we have not been able to reveal selection pressure acting at the amino acid level, which could be related to the structure and function of ASP protein. Similar difficulties were encountered in other studies on overlapping genes (e.g., ref. 36). Although such pressure possibly exists, one hypothesis could be that the function of ASP is essentially related to the expression of its ORF, for example, by interfering with the regulation of env. Alternatively, ASP could be involved in some mechanisms that alter the cellular functioning of specific cells, such as dendritic cells (2), for example, by forming aggregates (7). An interaction is possible with the recently discovered HIV antisense long noncoding RNA, which was suggested to modulate viral transcription (5, 37). However, the transcripts seem to be different (38), and the selection pressure acting on ASP ORF to maintain the start codon and avoid stop codons is clearly in favor of a coding part. Such mechanisms could be sufficient to improve the fitness of viral strains that have ASP and produce the observations reported in this study. Deciphering the function of ASP is now a major goal for further research, as all of our results support the idea that asp is the 10th gene of HIV-1 group M and indicate a correlation with the spread of the pandemic.
Materials and Methods
Correlation Between Prevalence and Presence of ASP.
We used an exact Spearman rank correlation test to demonstrate the statistical significance of the correlation between the presence of ASP in N, O, and P groups and M subtypes and CRFs and their prevalence (data in Fig. 3). Only the two prevalent CRFs (i.e., CRF01_AE and CRF02_AG, with prevalence 5.1% and 7.7%, respectively) were considered because the other CRFs appeared recently in most cases (hence their low prevalence) and conform with the original subtype from which they derive [e.g., all but one of the CRFs deriving from subtype B in the asp region (19) have ASP, just like the subtype B]. Using the rankor function (“Spearman,” “exact,” and “greater” options) of the pvrank R package, we obtained a rho value of 0.705 and a one-sided P value of 0.003.
To account for phylogenetic correlation, we used BayesTraits V2 (39). As there is no consensus on the phylogeny of the HIV-1 groups and subtypes, we computed 1,000 PhyML trees (GTR+Γ6) with one randomly selected sequence per subtype, prevalent CRF, and nonpandemic group. We then launched BayesTraits with this set of trees, and the same presence and prevalence values as in previous test. The mean log of the Bayes Factor (correlation model versus independence assumption) was equal to 3.8, that is, again, a strong evidence for correlation.
Statistical Significance of ASP ORF Conservation Using Simulations.
Our statistics on ASP ORF conservation are based on computer simulations. The basic principle is to assume that the ASP ORF was present at the phylogeny root and then simulate the evolution of sequences (read in env frame +1) along the tree and count the number of tip sequences that still have the ASP ORF. For this purpose, we selected 350 env sequences of group M at random with at most one sequence per individual. To root the tree, we added one env sequence of the closest SIVcpz_ptt group (GenBank entry: DQ373064). Ten samples of 350+1 sequences were obtained in this manner. For each sample, we estimated a phylogenetic tree using PhyML (GTR+FreeRate, six rate categories). Branch lengths were re-estimated for the asp region, using CodonPhyML with empirical codon model (40) and env codon usage. Having rooted the tree with the SIVcpz_ptt sequence, we ran simulation using Alf (same model options as CodonPhyML) with sequences evolving along this tree, starting with the asp region of HXB2 at the tree root. Moreover, specific codon rates were used to account for the variability of rates in the asp region. We used three codon categories, corresponding to the RRE, the variable regions V4 and V5, and the remaining codons. The rate of each category was estimated using the tree length estimated for that category by CodonPhyML (same options), divided by the tree length for the whole asp region. Finally, we calculated the percentage of tip sequences that have the ASP ORF in frame −2 (ORF length > 150 codons).
Obligatory and Potential stop Codons.
In all HIV-1 groups, the final stop codon is highly conserved. This stop codon is obligatory in most sequences. It is induced by Env, which contains a phenylalanine followed by a tyrosine at that position for 99% of sequences. At the nucleotide level, we thus have one of the four possibilities: tt{c,t} ta{c,t} (the overlap is in boldface), and on the opposite strand in frame −2, we necessarily have one of the two stop codons ta{g,a}.
Potential stop codons correspond to particular configurations, easily derived from the genetic code. For example, let us consider tga, one of the three stop codons. Having tga in frame −2 imposes having the two codons xxt cax in frame +1. The second codon (cax) encodes for histidine or glutamine and cannot be mutated synonymously on the first and second overlapping positions. In contrast, the first codon (xxt) can be mutated in a number of ways on the third position without changing the corresponding amino acid. For example, aac (asparagine) ↔ aat (asparagine), whereas in frame −2 we have tgg (tryptophan) ↔ tga (stop codon). Thus, aac cax defines a potential stop, whereas aat cax corresponds to an existing stop. Other potential stops are similar, and their mutations into existing stops always involve the third position of the first codon (hence our restriction to the third codon position; see below).
To measure the statistical significance of findings in Fig. 4, we used some ideas from ref. 32. Our adaptation involves focusing solely on the frame −2 and on synonymous sites corresponding to potential/existing stops. For an aligned sequence pair, we analyzed synonymous sites (i.e., the same Env amino acid pair is encoded in both sequences) for which, in the first sequence (denoted as sequence 1), there is a potential stop in frame −2. For each of these sites, we compared the number (0 or 1) of stop codons present in the second sequence (denoted as sequence 2) and the expected number of stop codons, assuming no selection pressure. For this purpose, we used DNADIST (41) with the F84 substitution model to estimate the evolutionary distance (δ) between both sequences. We restricted ourselves to third codon positions and synonymous sites, using both the asp and env-asp regions. We thus estimated the expected number of substitutions per site being synonymous regarding Env. The transition/transversion ratio (κ) and nucleotide frequencies (required by DNADIST) were estimated globally from all sequence pairs assuming HKY substitution model (nearly identical to F84, easy formulae). We then estimated, for each synonymous site with a potential stop, the expected number of stop codons in sequence 2 in frame −2. This estimation was achieved assuming HKY, using the previously estimated evolutionary distance (δ) and parameters (κ, nucleotide frequencies). The difference between this expected number and the observed number of stop codons in sequence 2 in frame −2 formed the statistics that we used to assess the significance of our observations (Fig. 4). Assuming no selection pressure, both expected and observed numbers of stop codons should be nearly equal. Conversely, assuming that ASP imposes some selection pressure, the expected number of stop codons should be larger than the actual number of stop codons, which is close to zero in group M (Fig. 4). Assuming a Poisson process (42), the variance of this statistic is equal to its mean. The alignment of the sequence pair being studied was scanned for sites corresponding to this pattern, summing for this pair the expectations, observations, and variances of all occurrences.
To select the list of sequence pairs, we used an unrooted phylogenetic tree [FastTree with GTR+CAT (43); one-per-individual multiple alignment; group M: 3,855 sequences; groups out-of-M: 76 sequences], tracing around the outside of a two-dimensional drawing of the tree. A sequence pair corresponded to two neighboring leaves in the tree. This procedure (derived from ref. 32) was used to account for the evolutionary dependency of the sequences: as every branch in the tree was run twice, we multiplied by 2 (worst case analysis) the variance computed under the independence assumption. We then summed our statistics over all sites and all sequences in the tree and computed a Z-score from which a P value was computed under the assumption of a normal distribution. A positive Z-score indicated that we had fewer stop codons than expected, assuming no selection pressure. Because the sequence pairs depended on the two-dimensional tree drawing, we randomly rotated subtrees and obtained a new set of sequence pairs from which the same statistics were computed. We obtained 10 replicates in this manner, providing nearly the same results that were averaged (e.g., Z-score of the whole asp region = 6.47 ± 0.23 and 1.53 ± 0.21 for groups M and out-of-M, respectively).
SI Text
Data Sets and Multiple Alignments.
Available nucleotide and protein sequences with metadata of the env gene were downloaded from the Los Alamos HIV Sequence Database (HIV-1 groups, SIVcpz, and SIVgor). Because sequences of HIV-1 P and SIVgor were modified in this database, we retrieved the original version from GenBank. We then identified problematic sequences that showed differences between the translation of the nucleotide sequence and the corresponding protein sequence available in the database. These sequences were discarded, as were incomplete sequences (more than 10 gaps/unknowns at the beginning and/or at the end), multiple-copy sequences, in which case only one sequence was conserved, and HIV-1 M sequences without patient identifiers. After this filtering, we obtained 22,992 env sequences of HIV-1/SIVcpz/gor (22,900 M, 92 out-of-M), corresponding to 3,855 M individuals, 68 out-of-M individuals, and 8 out-of-M sequences without individual identifiers, which were kept because of the small number of out-of-M sequences. The number of sequences per individual was between 1 and 285.
The Los Alamos database provides nucleotide and protein multiple alignments of HIV-1 sequences. To obtain an alignment with all of our sequences, we added unaligned protein sequences of HIV-1 P and SIV groups into the Los Alamos HIV-1 protein alignment (of better quality than the nucleotide alignment). We used MAFFT version 7.123b (45) with the option–add. We used the obtained protein multiple alignment as a template to produce the corresponding codon alignment. Some of our analyses were performed on a codon-based multiple alignment in frame −2. To obtain this antisense alignment, we generated the complementary strand of the frame +1 codon alignment and then shifted the codon bounds to match the frame −2.
Depending on the computational burden, these very large multiple alignments (22,992 sequences) were used either in a weighted manner so that each individual had a total weight of 1 (i.e., each sequence belonging to an individual with n sequences had a weight of 1/n) or by randomly selecting one sequence per individual. Except where otherwise specified, all of our statistics are based on weighted sequences.
Searching for Homologs of ASP.
We used PSI-BLAST (46) (default parameters) with the ASP versions of the M-reference sequences without stop codons (23 sequences). We searched all nonredundant GenBank coding region (CDS) translations + Protein Data Bank (PDB) + Protein Information Resource (PIR) + Protein Research Foundation (PRF), excluding environmental samples from Whole Genome Shotgun (WGS). The PSI-BLAST first iteration uses a scoring matrix and performs a BLASTp search. We obtained four sequences above the 0.005 threshold, but all were ASP protein sequences. At the second iteration, no new sequence was found. A search in the Hidden Markov Model databases was also performed using HMMScan (47) with the HXB2 reference sequence. No hit was found in any of the tested databases [Pfam, TIGRFAM, Gene3D, Superfamily, Protein Information Resource SuperFamily (PIRSF)]]. These results confirm (if needed) that ASP is a new gene created de novo.
Analysis of the SIVcpz_Ptt Sequence Having ASP.
The ASP ORF is present in one SIVcpz_Ptt sequence (GenBank: DQ373063). Thus, the following question emerges: Did ASP appear in group M and SIVcpz_Ptt genomes independently, or rather did this ORF first appear in the SIVcpz_Ptt genome and then was maintained when HIV-1 group M emerged from it?
We first checked that this SIVcpz_Ptt sequence possesses the key structural features of the ASP protein (4): a double cysteine triplet separated by nine amino acids, a ProXXProXXPro motif, and two transmembrane domains [as predicted using TOPCONS (50)]. Because the DQ373063 sequence does possess these key features, we can assume that it is functional. Note, however, that some of the SIV sequences show the same features when read in frame −2, although they contain stop codons in the asp region and do not have the ASP ORF.
We then built a phylogeny with PhyML (GTR+Γ6, asp region), using the 4 sequences available for the basal SIVcpz_Ptt group containing the DQ373063 sequence, and the 24 group M reference sequences (24) having ASP. The tree (Fig. S1) shows that the DQ373063 sequence is in basal position regarding the M sequences, thus supporting the idea that the ASP ORF first appeared in the SIVcpz_Ptt genome and was then transmitted to humans. However, this basal position is moderately supported (57% bootstrap support), the DQ373063 sequence is remote from the group M sequences, and our sample of SIVcpz_Ptt sequences is clearly too small to reject two independent emergences.
Use of Standard Methods to Measure the Selection Pressure Imposed by ASP.
To measure the selection pressure induced by the ASP protein, we first used several standard methods, which were applied to the env gene (read in frame +1): (i) estimation of the evolutionary rate; (ii) estimation of the dN/dS ratio (dN: rate of nonsynonymous substitutions; dS: rate of synonymous substitutions); (iii) and a recently developed method for the detection of overlapping genes, which is based on the existence of biases in the codon usage (32). Indeed:
-
i)
HIV-1 is one of the fastest evolving viruses, and most mathematical models show that the presence of overlapping ORFs increases the stability of the genome and thus decreases the evolutionary rate. In accordance with this prediction, a reduction of the evolutionary rate in overlapping regions was observed for several retroviruses (16).
-
ii)
The dN/dS ratio determines the type of selection pressure induced by the protein and its residues: the ratio is close to 0 if the residue(s) is subjected to purifying selection (which is the most common observation); it is close to 1 when the evolution is neutral; and it is higher than 1 when positive selection occurs (rarely observed, but typically seen in epitopes). With overlapping genes, a synonymous substitution for one gene is generally nonsynonymous for the other overlapping gene. Assuming that the overlapping gene (e.g., asp or tat) is under purifying selection, as is common with most genes, then we should observe that the reference gene (e.g., env) has a high dN/dS in the corresponding region with nonsynonymous substitutions corresponding to synonymous substitutions in the overlapping gene. This prediction was observed for tat (frame +2) and vpr (frame +3) (48).
-
iii)
Firth’s method Synplot2 (32) analyzes multiple alignments of protein-coding virus sequences to identify regions where there is a statistically significant reduction in the degree of variability at synonymous sites—a characteristic signature of overlapping functional elements.
To estimate the evolutionary rate of the nucleic sites of the env gene, we used PhyML (23) with a “FreeRate” model of rates across sites and eight rate categories. Ten samples of 500 sequences were randomly drawn from our complete multiple alignment with at most one sequence per individual, and the results were averaged over all samples. To estimate the dN/dS ratio of the env codons, we used PAML with the M8 model (49). Thirty samples of 100 sequences were randomly drawn from our complete multiple alignment with at most one sequence per individual, and the results were averaged over all samples. Synplot2 software (32) was launched with the one-per-individual env alignment, using a phylogenetic tree estimated by FastTree (43) with a GTR+CAT model.
Results are shown in Fig. S2. They are all interpretable by the known structure of the env gene and are seemingly not impacted by ASP:
Evolutionary rate: In the asp region, we observe the two—very fast—variable regions V4 and V5, followed by the—very slow—RRE region, which is a highly structured RNA element with multiple stems and loops. In the env-asp region, we clearly see the other (fast) variable regions, whereas the overlapping regions corresponding to vpu, tat, and rev are rather slow, as expected. Globally, the asp region is not any slower than env-asp (1.3 versus 0.9, respectively).
dN/dS ratio: We observe a strong correlation between the dN/dS ratio and the evolutionary rate, which is a common observation, and thus no clear signal for asp (dN/dS = 0.9) versus env-asp (dN/dS = 0.8). Actually, computer simulations (31) showed that it is difficult to detect a selection pressure difference for overlapping genes on the frame −2. Considering the overlap of the third codon positions (which is the most flexible codon position), the presence of a selection pressure on the reference reading frame (+1), whether it is a positive or purifying selection pressure, will result in an identical selection pressure on frame −2.
Synplot2: This program provides a clear signal for the overlapping genes in the sense strand (vpu, tat, and rev) and the RRE (obeying to structural constraints, which may impact the codon usage). However, as with the other methods, asp is not detected. Hence, the development of our method, which uses some of the ideas of Synplot2 [i.e., synonymous substitutions regarding the reference gene (env), P value computation], but is dedicated to the frame −2 and uses potential stops exclusively.
Acknowledgments
We thank M. Jung, S. Laverdure, S. Lèbre, and V. Lefort for their help initiating this study and T. Stadler, E. Simon-Loriere, and three anonymous referees for their comments on the manuscript. This project and a PhD grant (to E.C.) were supported by the “Projet Exploratoire Premier Soutien (PEPS) de Site CNRS/Université de Montpellier: Comprendre les maladies émergentes et les épidémies.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All of our data, alignments, methods, and detailed results are available from FigShare at https://figshare.com/s/9668ef62e84488d4787a.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605739113/-/DCSupplemental.
References
- 1.Landry S, et al. Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology. 2007;4:71. doi: 10.1186/1742-4690-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laverdure S, et al. HIV-1 antisense transcription is preferentially activated in primary monocyte-derived cells. J Virol. 2012;86(24):13785–13789. doi: 10.1128/JVI.01723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller RH. Human immunodeficiency virus may encode a novel protein on the genomic DNA plus strand. Science. 1988;239(4846):1420–1422. doi: 10.1126/science.3347840. [DOI] [PubMed] [Google Scholar]
- 4.Torresilla C, Mesnard JM, Barbeau B. Reviving an old HIV-1 gene: The HIV-1 antisense protein. Curr HIV Res. 2015;13(2):117–124. doi: 10.2174/1570162x12666141202125943. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi-Ishihara M, et al. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology. 2012;9:38. doi: 10.1186/1742-4690-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbagallo MS, Birch KE, Deacon NJ, Mosse JA. Potential control of human immunodeficiency virus type 1 asp expression by alternative splicing in the upstream untranslated region. DNA Cell Biol. 2012;31(7):1303–1313. doi: 10.1089/dna.2011.1585. [DOI] [PubMed] [Google Scholar]
- 7.Torresilla C, et al. Detection of the HIV-1 minus-strand-encoded antisense protein and its association with autophagy. J Virol. 2013;87(9):5089–5105. doi: 10.1128/JVI.00225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clerc I, et al. Polarized expression of the membrane ASP protein derived from HIV-1 antisense transcription in T cells. Retrovirology. 2011;8:74. doi: 10.1186/1742-4690-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briquet S, Vaquero C. Immunolocalization studies of an antisense protein in HIV-1-infected cells and viral particles. Virology. 2002;292(2):177–184. doi: 10.1006/viro.2001.1224. [DOI] [PubMed] [Google Scholar]
- 10.Bet A, et al. The HIV-1 antisense protein (ASP) induces CD8 T cell responses during chronic infection. Retrovirology. 2015;12:15. doi: 10.1186/s12977-015-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger CT, et al. Immune screening identifies novel T cell targets encoded by antisense reading frames of HIV-1. J Virol. 2015;89(7):4015–4019. doi: 10.1128/JVI.03435-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frahm N, et al. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol. 2004;78(5):2187–2200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardinaud S, et al. Identification of cryptic MHC I-restricted epitopes encoded by HIV-1 alternative reading frames. J Exp Med. 2004;199(8):1053–1063. doi: 10.1084/jem.20031869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maness NJ, et al. CD8+ T cell recognition of cryptic epitopes is a ubiquitous feature of AIDS virus infection. J Virol. 2010;84(21):11569–11574. doi: 10.1128/JVI.01419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chirico N, Vianelli A, Belshaw R. Why genes overlap in viruses. Proc Biol Sci. 2010;277(1701):3809–3817. doi: 10.1098/rspb.2010.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon-Loriere E, Holmes EC, Pagán I. The effect of gene overlapping on the rate of RNA virus evolution. Mol Biol Evol. 2013;30(8):1916–1928. doi: 10.1093/molbev/mst094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol. 2009;83(20):10719–10736. doi: 10.1128/JVI.00595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith TF, Waterman MS. Protein constraints induced by multiframe encoding. Math Biosci. 1980;49(1):17–26. [Google Scholar]
- 19.Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med. 2012;18(3):182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Ratner L, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 21.Willey RL, et al. Identification of conserved and divergent domains within the envelope gene of the acquired immunodeficiency syndrome retrovirus. Proc Natl Acad Sci USA. 1986;83(14):5038–5042. doi: 10.1073/pnas.83.14.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cullen BR. Nuclear mRNA export: Insights from virology. Trends Biochem Sci. 2003;28(8):419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 23.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 24.Leitner T, Korber B, Daniels M, Calef C, Foley B. HIV-1 subtype and circulating recombinant form (CRF) reference sequences, 2005. HIV Seq Compend. 2005;2005:41–48. [Google Scholar]
- 25.Pushker R, Jacqué JM, Shields DC. Meta-analysis to test the association of HIV-1 nef amino acid differences and deletions with disease progression. J Virol. 2010;84(7):3644–3653. doi: 10.1128/JVI.01959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halin M, et al. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2-mediated transcription. Blood. 2009;114(12):2427–2438. doi: 10.1182/blood-2008-09-179879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malim MH, Emerman M. HIV-1 accessory proteins: Ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3(6):388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Gil M, Zanetti MS, Zoller S, Anisimova M. CodonPhyML: Fast maximum likelihood phylogeny estimation under codon substitution models. Mol Biol Evol. 2013;30(6):1270–1280. doi: 10.1093/molbev/mst034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalquen DA, Anisimova M, Gonnet GH, Dessimoz C. ALF: A simulation framework for genome evolution. Mol Biol Evol. 2012;29(4):1115–1123. doi: 10.1093/molbev/msr268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabath N, Landan G, Graur D. A method for the simultaneous estimation of selection intensities in overlapping genes. PLoS One. 2008;3(12):e3996. doi: 10.1371/journal.pone.0003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mir K, Schober S. Selection pressure in alternative reading frames. PLoS One. 2014;9(10):e108768. doi: 10.1371/journal.pone.0108768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Firth AE. Mapping overlapping functional elements embedded within the protein-coding regions of RNA viruses. Nucleic Acids Res. 2014;42(20):12425–12439. doi: 10.1093/nar/gku981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F, Ding SW. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabath N, Wagner A, Karlin D. Evolution of viral proteins originated de novo by overprinting. Mol Biol Evol. 2012;29(12):3767–3780. doi: 10.1093/molbev/mss179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faria NR, et al. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346(6205):56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi M, et al. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J Virol. 2012;86(22):12411–12413. doi: 10.1128/JVI.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saayman S, et al. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther. 2014;22(6):1164–1175. doi: 10.1038/mt.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbeau B, Mesnard JM. Does chronic infection in retroviruses have a sense? Trends Microbiol. 2015;23(6):367–375. doi: 10.1016/j.tim.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401(6756):877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 40.Kosiol C, Holmes I, Goldman N. An empirical codon model for protein sequence evolution. Mol Biol Evol. 2007;24(7):1464–1479. doi: 10.1093/molbev/msm064. [DOI] [PubMed] [Google Scholar]
- 41.Felsenstein J. PHYLIP: Phylogeny inference package (version 3.2) Cladistics. 1989;5:163–166. [Google Scholar]
- 42.Bulmer M. Use of the method of generalized least squares in reconstructing phylogenies from sequence data. Mol Biol Evol. 1991;8(6):868. [Google Scholar]
- 43.Price MN, Dehal PS, Arkin AP. FastTree 2: Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25(5):679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(W1):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes AL, Westover K, da Silva J, O’Connor DH, Watkins DI. Simultaneous positive and purifying selection on overlapping reading frames of the tat and vpr genes of simian immunodeficiency virus. J Virol. 2001;75(17):7966–7972. doi: 10.1128/JVI.75.17.7966-7972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 50.Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015;43(W1):W401-7. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]