Significance
Antimicrobial drug resistance is a major threat to public health. Gram-negative bacteria are exceptionally resistant to antibiotics because of their outer-membrane barrier. Glycolipids called lipopolysaccharide (LPS) or lipooligosaccharide (LOS) fortify the outer membrane from many antimicrobials and biocides and were thought to be essential for Gram-negative bacterial survival. The last-resort treatment for multidrug-resistant Gram-negative infections is colistin, which targets the lipid A domain of LPS/LOS to disrupt the membrane, but the emerging pathogen Acinetobacter baumannii can develop colistin resistance by inactivating lipid A biosynthesis. This analysis advances our understanding of lipid A/LOS essentiality in A. baumannii and identifies antimicrobial targets.
Keywords: Acinetobacter, peptidoglycan, colistin, lipoprotein, lipopolysaccharide
Abstract
The Gram-negative bacterial outer membrane fortifies the cell against environmental toxins including antibiotics. Unique glycolipids called lipopolysaccharide/lipooligosaccharide (LPS/LOS) are enriched in the cell-surface monolayer of the outer membrane and promote antimicrobial resistance. Colistin, which targets the lipid A domain of LPS/LOS to lyse the cell, is the last-line treatment for multidrug-resistant Gram-negative infections. Lipid A is essential for the survival of most Gram-negative bacteria, but colistin-resistant Acinetobacter baumannii lacking lipid A were isolated after colistin exposure. Previously, strain ATCC 19606 was the only A. baumannii strain demonstrated to subsist without lipid A. Here, we show that other A. baumannii strains can also survive without lipid A, but some cannot, affording a unique model to study endotoxin essentiality. We assessed the capacity of 15 clinical A. baumannii isolates including 9 recent clinical isolates to develop colistin resistance through inactivation of the lipid A biosynthetic pathway, the products of which assemble the LOS precursor. Our investigation determined that expression of the well-conserved penicillin-binding protein (PBP) 1A, prevented LOS-deficient colony isolation. The glycosyltransferase activity of PBP1A, which aids in the polymerization of the peptidoglycan cell wall, was lethal to LOS-deficient A. baumannii. Global transcriptomic analysis of a PBP1A-deficient mutant and four LOS-deficient A. baumannii strains showed a concomitant increase in transcription of lipoproteins and their transporters. Examination of the LOS-deficient A. baumannii cell surface demonstrated that specific lipoproteins were overexpressed and decorated the cell surface, potentially compensating for LOS removal. This work expands our knowledge of lipid A essentiality and elucidates a drug resistance mechanism.
The bacterial cell envelope is a multifaceted structure that regulates uptake of essential nutrients and cofactors while protecting the cell from its often-hostile environment. The defining feature of Gram-negative bacteria is an essential second (outer) membrane in the cell envelope that encases the periplasm and peptidoglycan cell wall. The outer membrane forms a unique asymmetrical lipid bilayer barrier that enhances resistance to a variety of antibiotics and host immune mechanisms. Whereas the inner monolayer of the outer membrane is composed of glycerophospholipids, the cell surface-exposed monolayer is enriched in either lipopolysaccharide (LPS) or lipooligosaccharide (LOS), which are amphipathic glycolipids synthesized from the essential precursor, lipid A. The lipid A domain anchors LPS/LOS into the outer membrane, whereas the hydrophilic components including the core polysaccharides and O-polysaccharide(s) (in the case of LPS) extend into the extracellular milieu (1).
Host immune receptors recognize pathogen-associated molecular patterns (PAMPs) in the bacterial cell envelope during infection. The highly conserved lipid A (via TLR4/MD2 receptors) and peptidoglycan (via Nod receptors) chemical structures are PAMPs that stimulate immune pathways to clear the bacterial infection (2, 3). To circumvent detection by the host immune system, many Gram-negative pathogens alter structural motifs on the cell surface by adding chemical moieties to the lipid A domain of LPS/LOS. Not only does camouflaging PAMPs offer protection from host innate immune detection, but also chemical modification of lipid A enhances resistance to many antimicrobial agents (4). Consequently, bacterial remodeling of outer-membrane lipid A limits the effective antimicrobial arsenal to counteract Gram-negative bacterial infections.
Acinetobacter baumannii is an emerging Gram-negative pathogen that has quickly become pervasive throughout clinics and hospitals because it adapts to harsh environmental conditions and quickly develops resistance to antimicrobials. Multidrug resistant A. baumannii isolates, especially those resistant to carbapenems, have been isolated (5–7). Currently, colistin (polymyxin E) is prescribed as the last-resort antimicrobial to treat multidrug resistant A. baumannii infections. Colistin is a cationic lipopeptide that binds the negatively charged lipid A phosphate groups to perturb the membrane and kill the bacterial cell. However, A. baumannii has also evolved colistin resistance mechanisms (8–11).
A. baumannii amplifies colistin resistance through modification of the LOS lipid A anchor domain using three mechanisms. (i) Similar to lipid A modification strategies described in other Gram-negative pathogens (4), A. baumannii enzymatically adds phosphoethanolamine and/or galactosamine to lipid A to enhance antibiotic resistance (8–10). Incorporation of amine-containing chemical moieties neutralizes electrostatic interactions and reduces colistin binding to lipid A in the outer membrane. (ii) Whereas many bacterial pathogens respond to cellular assaults through acyl chain addition to the lipid A domain, A. baumannii encodes a mechanism that constitutively hyper-acylates the lipid A domain, presumably to alter host immune recognition and to strengthen the outer-membrane barrier (11). (iii) A. baumannii has also demonstrated a unique capacity to inactivate the lipid A biosynthetic pathway to develop resistance to colistin (12). In this work, we show that lipid A inactivation results not only in colistin resistance, but also in cross-resistance to other clinically important antibiotics.
A. baumannii is one of only three Gram-negative bacteria known to date that can survive after inactivation of lipid A. Whereas previous analyses indicated that lipid A was essential for survival of most Gram-negative bacteria (13), in vitro characterization of LOS-deficient isolates including A. baumannii, Neisseria meningitidis, and Moraxella catarrhalis demonstrate that some Gram-negative bacteria can persist without lipid A (12, 14, 15). Furthermore, LOS-deficient A. baumannii were isolated following colistin treatment from a patient in South Korea (16). We do not currently understand the mechanism that permits some Gram-negative bacterial species to survive without LOS whereas others cannot. Previous analysis suggested that LOS-deficient bacterial cells compensate for the absence of LOS through alterations to the cell envelope, but the molecular rearrangements have not been elucidated (17–19). Understanding how LOS-deficient pathogens remodel the cell envelope barrier without LOS will advance our understanding of lipid A essentiality and could identify targets to direct future therapeutic treatments.
To date, examination of LOS-deficient A. baumannii has been performed only in a single strain, designated ATCC 19606. Transcriptomic analysis (RNA-seq) of one isolate, which encoded a mutation that inactivated lipid A biosynthesis, suggested that LOS-deficient A. baumannii increased transcription of genes encoding cell-envelope biogenesis and transport proteins. Specifically, the maintenance of outer membrane lipid asymmetry (Mla) retrograde phospholipid transporter, localization of lipoproteins (Lol) lipoprotein transport, and poly-β-1,6-N-acetylglucosamine (PNAG) transcripts were increased relative to the wild-type parent strain (17). However, the contribution of each pathway to fitness after inactivation of lipid A biosynthesis and production of LOS was not determined.
To understand the requirements for bacterial survival without lipid A/LOS, we examined 15 laboratory-adapted and clinical A. baumannii isolates. We confirmed that, although some clinical A. baumannii isolates including ATCC 19606 inactivated lipid A biosynthesis to develop colistin resistance, other isolates could not. We discovered that the penicillin-binding protein (PBP) 1A (encoded by ponA or mrcA), impeded isolation of LOS-deficient A. baumannii. Transcriptomic analysis of the PBP1A mutant and four LOS-deficient A. baumannii strains exemplified a significant conserved increase in transcripts encoding Lol lipoprotein transport proteins. Specific lipoproteins were overexpressed and enriched in the outer membrane to potentially compensate for the absence of lipid A/LOS. Here we show a link between the peptidoglycan assembly protein, PBP1A, and the survivability of LOS-deficient A. baumannii on the last-resort drug, colistin.
Results
Colistin Exposure Selects for LOS-Deficient A. baumannii.
In a previous report, in vitro selection of A. baumannii strain ATCC 19606 on colistin (10 µg/mL) yielded resistant mutants. Each isolate contained a single mutation in one of three genes including lpxA, lpxC, or lpxD, which encode the first three steps in the conserved lipid A biosynthetic pathway. Characterization of a single lpxA mutant confirmed that ATCC 19606 A. baumannii survived without LOS and its precursor, lipid A (12). To determine if survival without LOS is a characteristic of only strain ATCC 19606 (12) or all A. baumannii strains, 15 clinical isolates including four multidrug resistant strains were plated on colistin (10 µg/mL), and isolated colonies were analyzed to determine the frequency of LOS inactivation. As shown in Table 1, 9 of the 15 strains (60%) propagated LOS-deficient A. baumannii after colistin exposure (as determined by colistin resistance/vancomycin sensitivity assays described here) with a mutation frequency comparable to previously published analysis (12). Importantly, six strains did not yield LOS-deficient colistin-resistant colonies, demonstrating that some A. baumannii clinical isolates could not survive without LOS (Table 1).
Table 1.
Diverse A. baumannii strains develop resistance to colistin (polymyxin E) through inactivation of lipooligosaccharide biosynthesis
| A. baumannii strain | Isolation date | Isolation source | Drug resistance | Mutation frequency (LOS-deficient) |
| 5075 | 2008–2009 | Bone infection | Multidrug | 1.49 × 10−7 |
| ATCC 19606 | 1948 | Urine | Susceptible | 1.53 × 10−7 |
| ATCC 17978 | 1951 | Meninges | Susceptible | NA |
| AYE | 2003 | Blood | Multidrug | 5.09 × 10−8 |
| SDF | 2003 | Louse | Susceptible | 9.22 × 10−8 |
| ACICU | 2005 | Cerebrospinal fluid | Multidrug | NA |
| Recent clinical #1 | 1/1/10 | Blood | Susceptible | 4.23 × 10−8 |
| Recent clinical #2 | 3/15/10 | Vagina | Susceptible | NA |
| Recent clinical #3 | 4/30/12 | Lungs | Susceptible | 3.15 × 10−8 |
| Recent clinical #4 | 7/18/12 | Pustule (hand) | Susceptible | 3.13 × 10−8 |
| Recent clinical #5 | 8/19/12 | Blood | Susceptible | NA |
| Recent clinical #6 | 1/24/13 | Urine | Susceptible | NA |
| Recent clinical #7 | 2/26/13 | Blood | Multidrug | NA |
| Recent clinical #8 | 3/28/13 | Skin | Susceptible | 1.56 × 10−7 |
| Recent clinical #9 | 4/11/13 | Skin | Susceptible | 7.522 × 10−8 |
NA: indicates that no lipid A-deficient isolates were recovered.
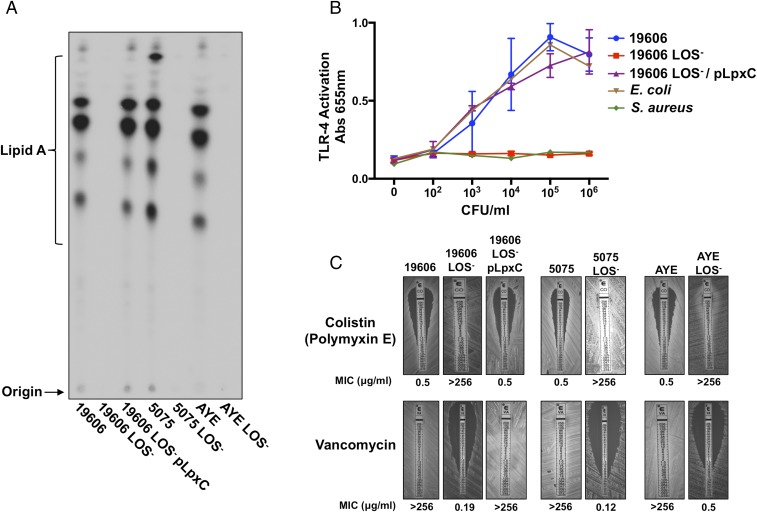
To ensure that colistin-resistant A. baumannii did not produce lipid A, three isolates were further characterized including the previously described ATCC 19606 and two multidrug-resistant A. baumannii strains, designated 5075 and AYE (12, 20, 21). Parent and colistin-resistant A. baumannii progeny were 32P-radiolabeled, and isolated lipid A was chromatographically separated based on hydrophobicity. Quantitative TLC demonstrated that parent A. baumannii strains produced equivalent lipid A, but the colistin-resistant progeny were defective in lipid A assembly (Fig. 1A). The genomes of parent and LOS-deficient isolates were sequenced and mutations were mapped to lpxC (SI Appendix, Table S1). The antibiotic-susceptible ATCC 19606 LOS-deficient strain that encoded a single-nucleotide polymorphism in lpxC was complemented with a plasmid expressing LpxC (pLpxC) to restore lipid A biosynthesis (Fig. 1A). To ensure that LOS-deficient A. baumannii did not produce an alternative lipid A glycoform, matrix-assisted laser desorption ionization (MALDI)–time of flight mass spectrometry (MS) analysis was performed on purified lipid A. Spectra collected from wild-type and complemented A. baumannii strains indicated major molecular ions at m/z 1728.0 and 1911.0 as previously described (8, 9, 11). Importantly, LOS-deficient A. baumannii lacked molecular ions indicative of lipid A (SI Appendix, Fig. S1A).
Fig. 1.
A. baumannii inactivates lipooligosaccharide biosynthesis to alter immune recognition and susceptibility to clinically relevant antibiotics. (A) 32P-radiolabeled lipid A was isolated from ATCC 19606, 5075, and AYE parent A. baumannii strains and their LOS-deficient progeny and separated based on hydrophobicity using TLC. (B) Stimulation of human TLR-4/MD2 complex following incubation of bacterial cells (cfu/mL) with HEK blue cells expressing the TLR-4 receptor complex is depicted. (C) MICs for parent and LOS-deficient A. baumannii strains.
The lipid A anchor of LPS/LOS is a PAMP that is bound with high affinity by the mammalian host toll-like receptor-4 myeloid differentiation protein-2 (TLR-4/MD-2) complex (2). Lipid A recognition by TLR-4/MD-2 activates MyD88- and TRIF-dependent pathways to initiate an inflammatory response and clear the bacterial infection. Wild-type, LOS-deficient, and complemented ATCC 19606 A. baumannii were incubated with a human embryonic kidney reporter cell line (HEK-blue) that expressed the human TLR-4/MD-2 receptor complex. Whereas the wild-type and complemented strains stimulated TLR-4–dependent activation comparable to Escherichia coli, the LOS-deficient A. baumannii failed to stimulate the TLR-4/MD-2 receptor complex similar to the Gram-positive Staphylococcus aureus, which does not produce lipid A (Fig. 1B). Analogous TLR-4/MD-2 activation was detected when colistin-resistant 5075 and AYE multidrug-resistant A. baumannii strains were analyzed (SI Appendix, Fig. S2). Together, TLC, MALDI-MS, and TLR-4/MD-2 activation assays demonstrate not only that A. baumannii strain ATCC 19606 can survive without lipid A, but also that other A. baumannii strains are capable of surviving, including multidrug-resistant isolates.
To understand the drug resistance profiles of wild-type and LOS-deficient A. baumannii isolates, we determined the minimum inhibitory concentration (MIC) of colistin and vancomycin, which are last-resort antimicrobials prescribed to treat either Gram-negative or Gram-positive infections, respectively. Following inactivation of lipid A biosynthesis, colistin resistance increased more than 500-fold, whereas vancomycin resistance decreased more than 500-fold relative to the wild-type parent A. baumannii (Fig. 1C). Our results demonstrated that removal of LOS through lipid A inactivation fostered colistin resistance, but also sensitized all strains including the multidrug-resistant strains to vancomycin, which is exclusively active against Gram-positive bacteria. The findings suggest that outer-membrane LOS is the major obstacle to vancomycin-dependent killing in A. baumannii, as previously shown in E. coli (22, 23). Furthermore, inactivation of lipid A biosynthesis in drug-susceptible A. baumannii ATCC 19606 resulted in multidrug resistance with increased resistance to ciprofloxacin and tigecycline, which are antibiotics prescribed to treat susceptible A. baumannii infections (SI Appendix, Fig. S3). These assays demonstrate that without acquiring antimicrobial resistance genes, drug-susceptible A. baumannii can develop resistance to clinically important antimicrobials. Notably, LOS-deficient A. baumannii demonstrated susceptibility to tobramycin, an aminoglycoside used to treat Pseudomonas aeruginosa infections (SI Appendix, Fig. S3).
Previous reports described alterations to the cellular glycerophospholipid composition after inactivation of lipid A biosynthesis (18, 19). To determine if phospholipids varied between wild-type and LOS-deficient A. baumannii, cells were 32P-radiolabeled and phospholipids were isolated. Quantitative TLC illustrated that wild-type and LOS-deficient A. baumannii produced equivalent glycerophospholipids including the major lipids phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL) (SI Appendix, Fig. S1B). Importantly, no discernible differences between primary phospholipid ratios of wild-type and LOS-deficient A. baumannii were observed. Although relative amounts did not vary, slight alterations to the acyl chains may not be obvious. Therefore, phospholipids were analyzed using normal-phase LC/MS/MS to discern subtle structural changes. One previous report described LOS-deficient A. baumannii producing phospholipids with shorter acyl chains (19), but our MS/MS analysis of two LOS-deficient A. baumannii strains did not indicate any notable differences in the ratio of phospholipid structures including PG (SI Appendix, Fig. S4A), CL (SI Appendix, Fig. S4B), PE (SI Appendix, Fig. S5A), or Lyso-PE and phosphatidic acid (SI Appendix, Fig. S5B) relative to the wild-type parent strains.
Inactivation of PBP1A Promotes Complete Loss of LOS After Exposure to Colistin.
Importantly, 6 of 15 strains in our screen did not yield LOS-deficient colonies after colistin exposure, suggesting a mechanism that inhibits LOS-deficient A. baumannii survival (Table 1). We constructed a random transposon (Tn) library of ∼250,000 mutants in A. baumannii strain ATCC 17978, a strain that did not produce LOS-deficient colonies after colistin exposure (Table 1). Selection of the ATCC 17978 Tn mutant library on colistin gave rise to colistin-resistant, vancomycin-sensitive isolates. Sequencing determined Tn insertion sites in the A1S_3196 and A1S_3197 (ponA or mrcA) genes, which encode the well-conserved penicillin-binding protein, PBP1A.
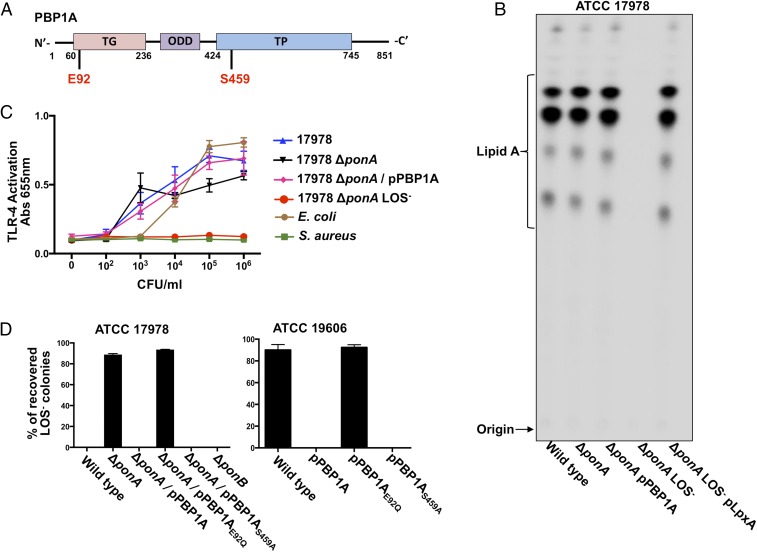
To confirm that the Tn-encoded isolates did not synthesize LOS, mutants were 32P-radiolabeled and lipid A was extracted. Quantitative TLC confirmed that two ponA::Tn isolates did not synthesize the LOS precursor, lipid A (SI Appendix, Fig. S6A). E-strip analysis established that the MIC of colistin increased 64-fold, whereas the MIC to vancomycin decreased more than 1,000-fold (SI Appendix, Fig. S6B). In addition, LOS-deficient ponA::Tn mutants did not stimulate the host TLR-4/MD2 receptor complex (SI Appendix, Fig. S6C). Collectively, these data indicated that PBP1A inhibited LOS-deficient ATCC 17978 A. baumannii survival after colistin exposure.
To validate that PBP1A inactivation enabled ATCC 17978 survival without lipid A, targeted mutagenesis removed the ponA-coding sequence from the A. baumannii chromosome. Deletion of ponA alone in the wild-type background did not alter lipid A assembly. However, colistin selection yielded ponA mutants that lacked LOS (SI Appendix, Fig. S9C) due to genetic mutation in the lipid A biosynthesis pathway (Fig. 2B). Genomic sequencing mapped the mutation of ATCC 17978 ∆ponA LOS-deficient strain to lpxA (SI Appendix, Table S1). LpxA expression in the LOS-deficient mutant restored lipid A production to wild-type levels (Fig. 2B), and ponA deletion alone did not alter TLR-4/MD-2 recognition (Fig. 2C). Together, these assays suggest that the ponA mutation alone does not impact LOS in any way before colistin treatment, but primes the cell to grow without LOS upon challenge with colistin.
Fig. 2.
Inactivation of ponA promotes complete loss of LOS in ATCC 17978 A. baumannii. (A) Illustration depicting PBP1A and its domains with respective catalytic residues (red). (B) 32P-radiolabeled lipid A isolated from ATCC 17978 wild-type and ponA mutant A. baumannii was separated based on hydrophobicity using TLC. (C) Stimulation of human TLR-4/MD-2 following incubation of bacterial cells (cfu/mL) with HEK blue cells expressing the TLR-4 receptor complex is depicted. (D) Percentage recovery of LOS-deficient A. baumannii after selection of plating either 109 cfu ATCC 17978 (Left) or ATCC19606 (Right) on colistin.
ATCC 19606 PBP1A is encoded by a single ponA gene and is orthologous to PBP1A in E. coli, which is a bifunctional enzyme with conserved glycosyltransferase and transpeptidase domains (SI Appendix, Fig. S7) (24, 25). However, the A. baumannii ATCC 17978 annotation predicted a guanine nucleotide insertion to result in a premature stop codon of ponA, resulting in two separate genes, annotated as A1S_3196 and A1S_3197 (21). The guanine nucleotide insertion was not identified by highly reliable Sanger sequencing analysis, which indicates that, like ATCC 19606, ATCC 17978 ponA is encoded by a single gene and produces a full-length PBP1A protein (SI Appendix, Fig. S8).
Deletion of PBP1A in E. coli had no significant effect on cell growth, but effected cell width and the timing of divisome assembly (26). Also, previous analysis performed on PBP1A distinguished two catalytic domains essential for glycosyltransferase and transpeptidase activities (25, 27). The glycosyltransferase activity catalyzes polymerization of peptidoglycan from lipid II, whereas the transpeptidase activity catalyzes peptide cross-links to form a rigid cell wall (Fig. 2A) (28). We were curious to see if either the glycosyltransferase and/or transpeptidase catalytic domains of PBP1A influenced isolation of LOS-deficient A. baumannii on colistin. Catalytic residues for either the glycosyltransferase (E92Q) or transpeptidase (S459A) activities were inactivated via point mutation (Fig. 2A). It is worthwhile to note that the E. coli glycosyltransferase mutant lacks both activities as the transpeptidase activity is dependent on ongoing glycosyltransferase reactions in the same enzyme molecule (25). Similar to the wild-type strain, expression of PBP1A or PBP1AS459A from a multicopy plasmid in ∆ponA did not yield LOS-deficient ATCC 17978 A. baumannii. In contrast, LOS-deficient ATCC 17978 A. baumannii were isolated on colistin when PBP1AE92Q was expressed in ∆ponA, analogous to the ∆ponA mutant. Thus, the glycosyltransferase activity of PBP1A is sufficient to inhibit LOS-deficient survival (Fig. 2D, Left). Using PBP1A specific antisera, we confirmed that PBP1A and PBP1AE92Q were expressed (Fig. 3B). Unlike ∆ponA, deletion of the closely related bifunctional PBP1B (encoded by ponB or mrcB) did not produce LOS-deficient survival following colistin exposure, illustrating that LOS-deficient survival is specific to PBP1A activity (Fig. 2D, Left).
Fig. 3.
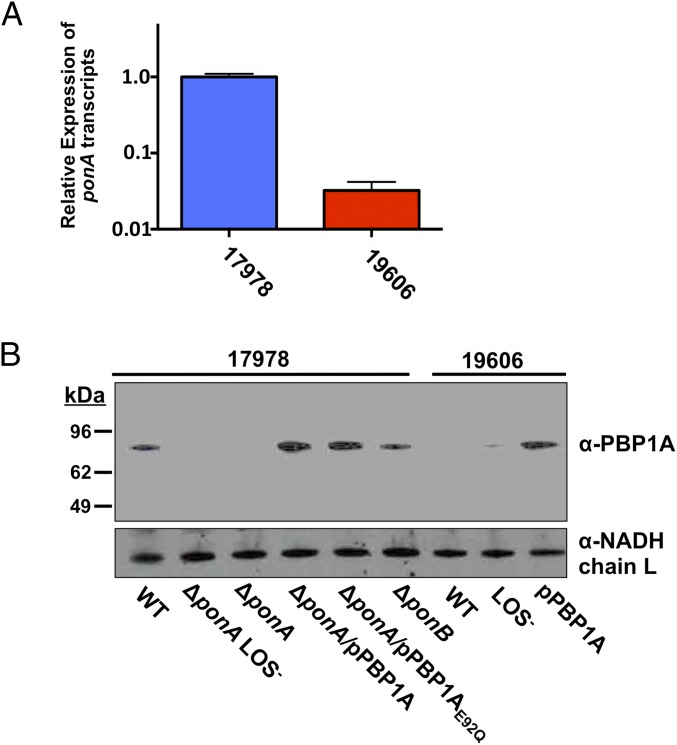
Differential expression of ponA and PBP1A in ATCC 17978 and ATCC 19606 A. baumannii. (A) Relative concentration of ponA mRNA in ATCC 17978 and ATCC 19606 wild-type A. baumannii. (B) Immunoblot analysis of PBP1A and NADH chain L proteins in whole-cell lysates from wild-type and mutant ATCC 17978 and ATCC 19606 A. baumannii strains. Each protein was detected using specific polyclonal antiserum.
To verify that mutation of ponA alone had no effect on antibiotic resistance or lipid A assembly, we assessed resistance to colistin and vancomycin in the wild-type, ∆ponA, and ∆ponA LOS-deficient ATCC 17978 strains. Consistent with analysis in the ponA::Tn mutant, colistin resistance and vancomycin sensitivity in ∆ponA were altered only after colistin selection (SI Appendix, Fig. S9A). Analysis confirmed that ∆ponA and strains expressing all complementation plasmids produced lipid A (SI Appendix, Fig. S9B) whereas a colistin-resistant, vancomycin-sensitive ∆ponA ATCC 17978 A. baumannii isolate did not (Fig. 2B). Finally, LOS staining demonstrated that ∆ponA ATCC 17978 synthesized LOS, but the ∆ponA LOS-deficient strain did not (SI Appendix, Fig. S9C).
Inactivation of PBP1A in A. baumannii Strain ATCC 17978 Alters Expression of Lipoprotein Transport Genes.
Expression of PBP1A prevented isolation of LOS-deficient ATCC 17978 A. baumannii (Fig. 2D, Left). However, we were curious to see if expression of native and mutant PBP1A proteins in ATCC 19606 A. baumannii, a strain known to survive without LOS, would block isolation of LOS-deficient colonies after colistin exposure. Selection of ATCC 19606 A. baumannii expressing PPB1A and PBP1AS459A from a multicopy plasmid yielded zero LOS-deficient isolates. Furthermore, PBP1AE92Q expression resulted in LOS-deficient isolates equivalent to wild-type levels (Fig. 2D, Right).
The ponA genes and respective promoters in ATCC 17978 and ATCC 19606 are nearly identical, indicating that each wild-type strain should produce similar levels of PBP1A. To understand why ATCC 19606, but not ATCC 17978, produces LOS-deficient isolates on colistin, we analyzed transcription of ponA in the two parent strains. Interestingly, an 80-fold lower abundance of ponA transcripts were detected in ATCC 19606 relative to ATCC 17978 A. baumannii (Fig. 3A). Furthermore, ATCC 17978 A. baumannii produced PBP1A, but ATCC 19606 A. baumannii did not (Fig. 3B).
To understand how peptidoglycan was altered in LOS-deficient A. baumannii, we isolated muropeptides from wild-type ATCC 17978, ∆ponA, the ∆ponA LOS-deficient strain, and ∆ponA expressing either PBP1A or PBP1AE92Q from a multicopy plasmid. The extracted muropeptide pool was separated using high-performance liquid chromatography to quantify the differences in the chemical composition (28). The peptidoglycan structure of wild-type A. baumannii contained significantly more peptide cross-links (∼60%) than either E. coli (∼40–50%) (28) or Helicobacter pylori (∼40%) (29) with an average chain length of 30 disaccharides (SI Appendix, Table S2). Neither the average peptidoglycan cross-links nor the chain lengths were significantly altered in the ∆ponA or the LOS-deficient mutant compared with wild-type ATCC 17978 A. baumannii (SI Appendix, Table S2).
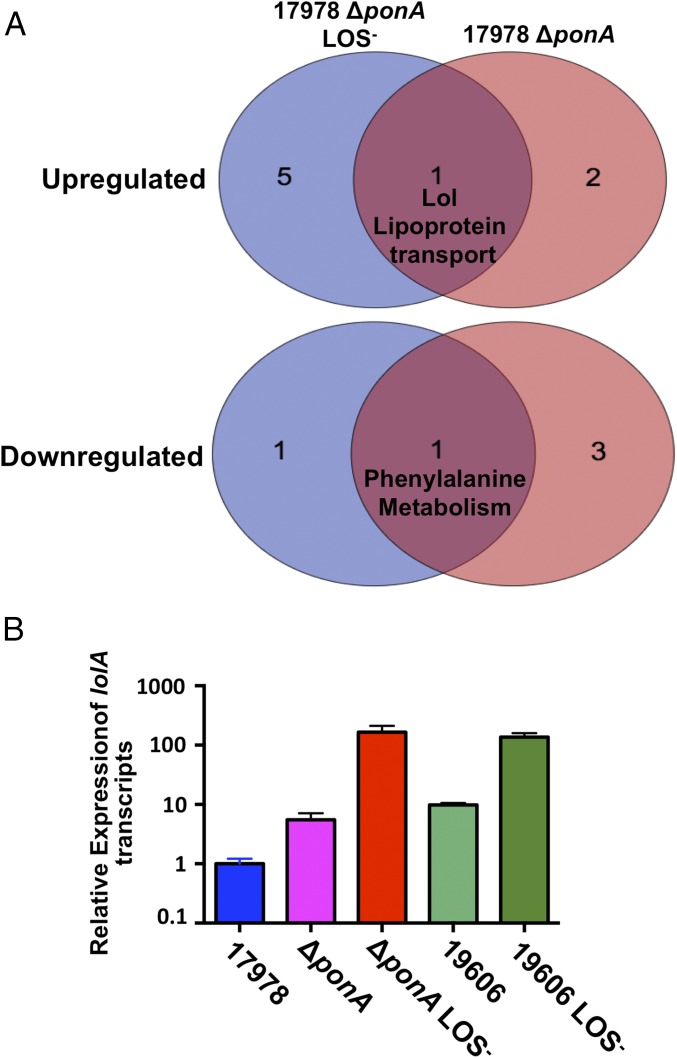
To understand how PBP1A hindered survival of some A. baumannii strains without LOS, we isolated total RNA transcripts from wild-type, ∆ponA, and ∆ponA LOS-deficient ATCC 17978 A. baumannii strains. RNA sequencing detected altered transcription of genes encoding important cell envelope pathways relative to the wild-type parent (Datasets S1 and S2). Pathway analysis determined by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations (P value ≤0.05) specified increased transcription of three pathways in ∆ponA and six pathways in ∆ponA LOS-deficient A. baumannii. Only increased transcription of genes encoding the Lol transport pathway was conserved, which included overexpression of lolA (Fig. 4A), where transcripts relative to the wild-type parent increased >4-fold in ∆ponA and >56-fold in the ∆ponA LOS-deficient strain (Datasets S1 and S2). In addition, analysis indicated reduced transcription of four pathways using a false discovery rate adjusted P value ≤0.05 in the ∆ponA strain and two pathways in the ∆ponA LOS-deficient strain relative to the parent ATCC 17978. Although not the focus of this article, transcripts in the phenylalanine biosynthesis pathway were reduced in both strains (Fig. 4A).
Fig. 4.
Inactivation of ponA in ATCC 17978 A. baumannii increases expression of lipoprotein transport genes. (A) Venn diagram showing altered pathway expression in ATCC 17978 ΔponA LOS-deficient and ATCC 17978 ΔponA A. baumannii strains (P value ≤0.05). Gene expression was calculated relative to the wild-type parent ATCC 17978 A. baumannii. (B) Relative concentration of lolA mRNA in wild-type and mutant ATCC 17978 and ATCC 19606 A. baumannii strains.
The Lol transport system is an essential pathway in Gram-negative bacteria (reviewed in ref. 27). LolA is a periplasmic chaperone that transports lipoproteins from LolCDE at the inner membrane to LolB at the outer membrane, where the lipoproteins are anchored. Consistent with global transcriptome analysis, qPCR analysis confirmed that lolA transcription increased ∼8-fold in ∆ponA and ∼110-fold in the ∆ponA LOS-deficient strain ATCC 17978 relative to wild type. Furthermore, lolA transcription in wild-type ATCC 19606 was ∼10-fold higher relative to ATCC 17978 and >100-fold higher in LOS-deficient ATCC 19606 (Fig. 4B).
Global RNA-seq Analysis in Multiple A. baumannii Strains Highlights Pathways Important for Loss of LOS.
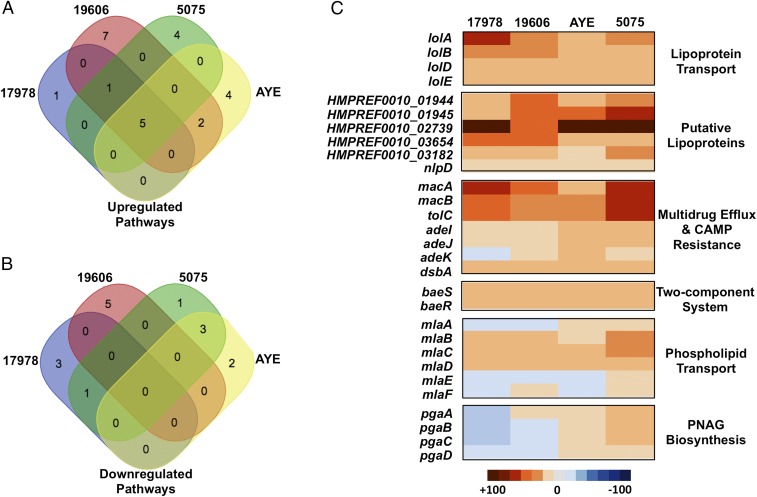
Previously published analysis on a single LOS-deficient strain, a 19606 A. baumannii lpxA mutant, showed increased transcription of genes encoding Lol-lipoprotein transporters, Mla-retrograde phospholipid transporters, and carbohydrate transporters, but reduced transcription of genes encoding surface structures (17). Congruent with our multistrain analysis, we sought to understand if the transcriptional alterations in LOS-deficient A. baumannii were conserved. RNA was isolated from four LOS-deficient A. baumannii strains and their wild-type parent strains including ATCC 17978 ∆ponA, ATCC 19606, 5075, and AYE. The RNA transcripts were sequenced to ascertain the altered transcriptional profile of each isolate relative to the wild-type parent strain (Datasets S2–S5). Pathway analysis (P value ≤ 0.05) indicated that increased transcription of five pathways was conserved among the LOS-deficient strains (Fig. 5A), whereas no down-regulated pathways were conserved (Fig. 5B). Conserved up-regulated pathways that shared significant increased transcription among all LOS-deficient strains included Lol lipoprotein transport genes, putative lipoproteins, retrograde phospholipid transport genes, genes involved in drug efflux and CAMP resistance, and genes encoding the BaeSR two-component system. A heat map illustrates the fold change in expression for each gene transcript in the conserved pathways relative to the respective wild-type A. baumannii strain (Fig. 5C). Transcription of PNAG biosynthetic genes was not conserved in our analysis, but the genes were included in the heat map because a previous report suggested that these pathways could contribute to A. baumannii strain ATCC 19606 survival without LOS (17).
Fig. 5.
Global transcriptional analysis from multiple A. baumannii strains highlights conserved pathways important for loss of LOS. Venn diagrams showing up-regulated (A) and down-regulated (B) pathways in four LOS-deficient A. baumannii strains (P value ≤ 0.05). Gene expression was calculated relative to the parent wild-type A. baumannii strains. The five conserved significantly up-regulated pathways in LOS-deficient A. baumannii included lipoprotein transport, putative lipoproteins, retrograde phospholipid transport, multidrug efflux pumps/CAMP resistance proteins, and the BaeSR two-component system. (C) Heat map illustrating the altered expression of each gene in the five conserved pathways and for PNAG biosynthesis. Our statistical analysis did not highlight the PNAG biosynthesis as conserved responses to loss of LOS, but genes were included in the heat map because this pathway was previously thought to be important for A. baumannii survival without LOS.
Lipoproteins Are Overexpressed and Surface-Displayed in LOS-Deficient A. baumannii.
Together, the transcriptomic analysis performed on four LOS-deficient A. baumannii strains (Fig. 5C) and the previous analysis performed on ATCC 19606 (17) indicated that transcripts encoding lipoprotein transporters and putative lipoproteins are increased relative to the wild-type strain. Specifically, transcription of HMPREF0010_01944 (1944), HMPREF0010_01945 (1945), and HMPREF0010_02739 (2739), as annotated in ATCC 19606 A. baumannii, exhibited the largest conserved increased fold-change in the RNA-seq analysis among the four LOS-deficient isolates (Fig. 5C).
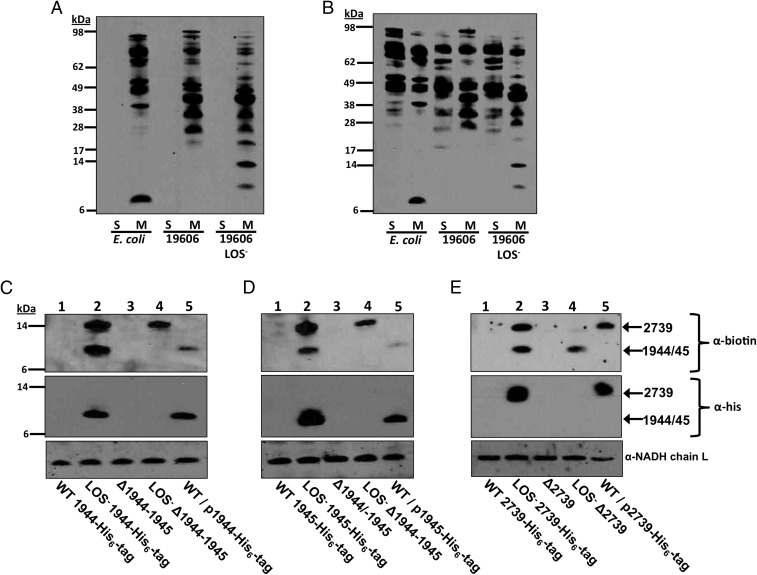
To evaluate lipoprotein localization, wild-type and LOS-deficient A. baumannii were treated with sulfo-NHS-LC-LC-biotin, which reacts with primary amines to conjugate a biotin molecule onto surface-exposed proteins. Previously, a derivative of this molecule (NHS-LC-LC-biotin) identified surface proteins of E. coli (30) and Vibrio cholerae (31). This compound does not permeate the outer membrane because of its size and the charge added by the sulfo-NHS groups. To confirm that the sulfo-NHS-LC-LC-biotin molecule does not enter A. baumannii, whole cells from wild-type E. coli, A. baumannii, and LOS-deficient A. baumannii were labeled. Western blotting with streptavidin-HRP showed biotinylation of membrane proteins, but no soluble proteins, illustrating that the molecule does not enter the cell (Fig. 6A). In contrast, when the cells were lysed before the addition of sulfo-NHS-LC-LC-biotin, both membrane and soluble proteins were biotinylated (Fig. 6B).
Fig. 6.
Lipoproteins are overexpressed and surface-displayed in LOS-deficient A. baumannii. Intact whole cells (A) or cell lysates (B) were incubated with sulfo-NHS-LC-LC-biotin to biotinylate accessible proteins. After labeling, proteins from the soluble (S) and membrane (M) fractions were subjected to SDS/PAGE and Western blotted using streptavidin-HRP. Intact whole cells containing chromosomal His6-tag fusions including (C) HMPREF0010_1944 (1944-His6) (11.52 kDa), (D) HMPREF0010_1945 (1945-His6) (11.83 kDa), and (E) HMPREF0010_2739 (2739-His6) (14.76 kDa) were treated with sulfo-NHS-LC-LC-biotin, and proteins were separated using SDS/PAGE. Wild-type, LOS-deficient for each respective mutant, and overexpression strains were blotted using a streptavidin-HRP conjugate (Top), anti-his antibody (Middle), or a polyclonal NADH chain L antibody (Bottom).
A number of biotinylated proteins detected in the LOS-deficient cell surface were absent in wild-type cells (Fig. 6 A and B), especially lower-molecular-weight proteins (<17 kDa). To determine if the lipoproteins 1944, 1945, and 2739 were enriched on the surface of LOS-deficient A. baumannii, we generated three separate strains where each lipoprotein was fused to a C-terminal His6-tag. The theoretical mass of 1944-His6 and 1945-His6 is 11.52 and 11.83 kDa, respectively, whereas the theoretical mass of 2739-His6 is 14.76 kDa. Intact whole cells were treated with sulfo-NHS-LC-LC-biotin and lysed. The proteins in each lysate were separated using SDS/PAGE. Streptavidin-HRP blotting indicated that protein(s) at the indicated theoretical masses of each lipoprotein were biotinylated in LOS-deficient cells, but not the wild-type A. baumannii (Fig. 6 C–E, lanes 1 and 2, Top). To determine if the biotinylated proteins represented the target lipoproteins, blots were probed with an anti-his antibody. Biotinylated and His6-tagged proteins had equivalent masses, suggesting that the biotinylated proteins were the target lipoproteins (Fig. 6 C–E, lanes 1 and 2, Top and Middle). Furthermore, each respective lipoprotein mutant (∆2739 and ∆1944–45) did not conjugate biotin in the LOS-deficient mutant, confirming that the three lipoproteins are enriched in the outer membrane of LOS-deficient A. baumannii (Fig. 6 C–E, lanes 3 and 4). Surprisingly, the three lipoproteins (1944, 1945, and 2739) were not detected in the wild-type A. baumannii strain, suggesting that expression and/or outer membrane localization transpires only in the LOS-deficient mutant (Fig. 6 C–E, lane 1). Next, we questioned if lipoprotein overexpression would result in outer-membrane accumulation in wild-type A. baumannii. Each overexpressed His6-tagged lipoprotein was produced (Fig. 6 C–E, lane 5, Middle), and localized to the outer membrane in the wild-type strain (Fig. 6 C–E, lane 5, Top). Therefore, lipoprotein overexpression in the LOS-deficient strain does not redirect lipoprotein transport. Instead, overexpressed lipoproteins likely accumulate at the surface-exposed face of the outer membrane to potentially compensate for the absence of LOS. Importantly, LOS-deficient A. baumannii was isolated in each lipoprotein mutant, suggesting that no individual lipoprotein was essential for survival without LOS.
Discussion
Gram-negative bacteria are defined by a second membrane bilayer in the cell envelope that encases the peptidoglycan and acts as a fortified barrier to protect the cell from environmental toxins. Unlike the cytoplasmic membrane, the outer membrane is asymmetric with the surface-exposed monolayer enriched in either LPS or LOS glycolipids, which prevent entry of many antibiotics. Whereas the LOS/LPS precursor, lipid A, was thought to be essential for survival of most Gram-negative bacteria (13), recent reports confirm that A. baumannii also persists after inactivation of lipid A biosynthesis (12, 14, 15). Importantly, colistin selection in vitro enables isolation of LOS-deficient A. baumannii that are colistin resistant due to inactivation of lipid A biosynthesis (12). As previously reported, a growth defect is associated with LOS-deficient A. baumannii under standard growth conditions, and cells are smaller and round during exponential growth (32). We observed a similar growth phenotype in our study. In addition, LOS-deficient A. baumannii displayed a virulence defect in a murine model (32), but, importantly, LOS-deficient A. baumannii were isolated from a patient after colistin treatment (16). A more detailed understanding how A. baumannii modifies its outer membrane to persist in the host without LOS will elucidate a novel drug resistance mechanism.
We have confirmed not only that selection of drug-susceptible A. baumannii strain ATCC 19606 on colistin results in isolation of resistant LOS-deficient colonies, but also have demonstrated the resistance of the strain to other clinically relevant antibiotics including ciprofloxacin and tigecycline in the ATCC 19606 LOS-deficient isolates (SI Appendix, Fig. S3). Resistance acquisition to multiple antimicrobials following colistin treatment has not been described and is extremely worrisome because drug-susceptible cells acquired multidrug resistance after only one exposure to colistin without acquisition of new genes. Although characterization of these drug resistance mechanisms is outside the scope of this work, overexpression of resistance pumps and transporters reported in our transcriptomic analysis could indicate resistance mechanisms (Datasets S1–S5). Interestingly, this study also showed that removal of lipid A/LOS molecules from the outer membrane increased A. baumannii sensitivity to vancomycin and tobramycin (an aminoglycoside) (Fig. 1B and SI Appendix, Fig. S3). This important finding demonstrates that alterations to the outer-membrane permeability barrier sensitize otherwise drug-resistant bacteria to specific classes of antibiotics, which could be useful for antimicrobial synergism to treat bacterial infections.
To date, bacterial survival after inactivation of the lipid A biosynthetic pathway is limited to N. meningitidis, M. catarrhalis, and A. baumannii, which are all Gram-negative mucosal pathogens that synthesize the LOS glycoform (12, 14, 15). We discovered that, although some A. baumannii strains can survive without lipid A, other strains cannot, providing a model system to explore lipid A essentiality. Screening of an A. baumannii ATCC 17978 Tn mutant library directed us to examine PBP1A, a major cell-wall synthase. Confirming our mutagenesis screen, removal of the ponA (which encodes PBP1A) coding sequence permitted A. baumannii ATCC 17978 to develop colistin resistance through inactivation of the lipid biosynthesis pathway. Furthermore, expression of PBP1A from a multicopy plasmid in A. baumannii ATCC 19606 impeded isolation of LOS-deficient colonies, further supporting a role for PBP1A. Unexpectedly, our results link two fundamental cell-envelope processes, lipid A biosynthesis and biogenesis of the peptidoglycan cell wall. Interestingly, pathways involved in cell-wall maintenance/biogenesis and the steps in lipid biosynthesis do not directly impact each other, but previous analysis showed that mutations resulted in convergence of the two distinct biogenesis pathways (23).
Curiously, A. baumannii strains such as ATCC 17978, which produce PBP1A, and ATCC 19606, which does not (Fig. 3 A and B), encode almost identical ponA genetic coding sequences and promoters. The varied ponA expression between the two wild-type strains suggests complex genetic regulation among A. baumannii isolates. Although we cannot conclude why PBP1A is detrimental to LOS-deficient A. baumannii, we hypothesize that small changes in the cell wall that we could not detect act as a signal to initiate increased transcription of the gene encoding the lipoprotein transporter, LolA. Our analysis demonstrated that increased transcription of lipoprotein transporters and lipoproteins in LOS-deficient A. baumannii resulted in outer-membrane remodeling that could promote A. baumannii survival without lipid A/LOS. Regulation of lolA in E. coli is controlled by the Rcs phosphorelay system, which detects cell-envelope stress (33). However, A. baumannii does not encode an orthologous system, and regulatory mechanisms controlling lolA gene expression in A. baumannii are not known. It is within reason to speculate that increased LolA-dependent lipoprotein transport could result in accumulation of lipoproteins in the outer membrane where they assemble onto the cell surface of A. baumannii. Increased localization of lipoproteins at the cell surface could compensate for the absence of LOS. Although it is unknown how lipoproteins in Gram-negative bacteria flip from the inner leaflet to the cell-surface leaflet of the outer membrane, a recent report demonstrated that a protein termed SLAM is required for surface localization of key lipoproteins in N. meningitidis, and a similar mechanism may exist in A. baumannii (34).
The pathogenic spirochetes Treponema pallidum and Borrelia burgdorferi do not encode genes to synthesize lipid A (35, 36). Instead, the surface-exposed monolayer of their outer membrane is enriched in lipoproteins and devoid of outer-membrane proteins (37). Although T. pallidum is an obligate intracellular human pathogen (38), B. burgdorferi adapts to dramatic environmental fluctuations (39), suggesting that Gram-negative bacteria lacking surface-exposed LPS/LOS, such as A. baumannii, can potentially survive in a variety of environments including the human host.
Furthermore, A. baumannii encodes an O-linked glycosylation system that adds glycans to many proteins (40). If the surface-exposed lipoproteins were glycosylated in A. baumannii, they could partially mimic the biophysical properties of LPS/LOS in LOS-deficient A. baumannii to maintain the outer-membrane barrier. Although previous analysis determined the O-glycosylated proteome of A. baumannii strain ATCC 17978 (40), the lipoproteins in this analysis are not expressed in wild-type A. baumannii and could have been overlooked. Although we have shown that three putative lipoproteins, including 1944, 1945, and 2739, are overexpressed in the outer membrane and are potentially surface-localized, no single lipoprotein was required for isolation of LOS-deficient A. baumannii (Fig. 6 A–C). Interestingly, higher-molecular-weight (>17 kDa) proteins were biotin-labeled in the LOS-deficient A. baumannii cells (Fig. 6 A and B), indicating that other outer membrane proteins could contribute to survival of LOS-deficient A. baumannii.
To better understand how the outer-membrane glycolipids LPS or LOS protect Gram-negative bacteria from various environmental niches, we focused on A. baumannii not only because it is one of only three bacterial pathogens that survives after lipid A inactivation, but also because it shuts off LOS biosynthesis to promote colistin resistance. This study illustrates the plasticity of the Gram-negative outer membrane, which tolerates dramatic alterations in cell-envelope composition to protect the cell in the presence of toxic antibiotics. Furthermore, our work demonstrates that inactivation of lipid A increases expression of genes encoding lipoproteins and their transporters and that the lipoproteins accumulate on the cell surface when LOS is not produced, likely to maintain the outer-membrane barrier. Altogether our work indicates that the outer membrane of Gram-negative bacteria is a dynamic barrier that is modified to protect the cell from detrimental environmental conditions.
Materials and Methods
Description of all strains, plasmids, and primers can be found in SI Appendix, Materials and Methods, for all experiments including strain construction and isolation of LOS-deficient A. baumannii, lipid extractions, MS analysis, TLR-4–signaling assays, RNA and genomic sequencing, surface biotin labeling, and MICs can be found in SI Appendix. The data discussed in this publication have been deposited in the National Center for Biotechnology’s Gene Expression Omnibus (41) (accession no. GSE84282).
Supplementary Material
Acknowledgments
We thank Alan Hauser for providing plasmid pJNW684 used in the transposon mutagenesis screen and Andrew Camilli for providing the E. coli MFD (mutation frequency decline) pir donor strain. Funding from the NIH (Grants AI119879, AI064184, and AI076322 to M.S.T. and Grant F32GM113488 to J.M.B.); the Army Research Office (Grant W911NF-12-1-0390 to M.S.T.); and the Wellcome Trust (Grant 101824/Z/13/Z to W.V.) is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data discussed in this publication have been deposited in the National Center for Biotechnology’s Gene Expression Omnibus (accession no. GSE84282).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611594113/-/DCSupplemental.
References
- 1.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 2.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458(7242):1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 3.Girardin SE, et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003;278(43):41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 4.Needham BD, Trent MS. Fortifying the barrier: The impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol. 2013;11(7):467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56(3):395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaara M. Polymyxins and their novel derivatives. Curr Opin Microbiol. 2010;13(5):574–581. doi: 10.1016/j.mib.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Kassamali Z, Rotschafer JC, Jones RN, Prince RA, Danziger LH. Polymyxins: Wisdom does not always come with age. Clin Infect Dis. 2013;57(6):877–883. doi: 10.1093/cid/cit367. [DOI] [PubMed] [Google Scholar]
- 8.Beceiro A, et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother. 2011;55(7):3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arroyo LA, et al. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother. 2011;55(8):3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelletier MR, et al. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother. 2013;57(10):4831–4840. doi: 10.1128/AAC.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boll JM, et al. Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. MBio. 2015;6(3):e00478-15. doi: 10.1128/mBio.00478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffatt JH, et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. 2010;54(12):4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steeghs L, et al. Meningitis bacterium is viable without endotoxin. Nature. 1998;392(6675):449–450. doi: 10.1038/33046. [DOI] [PubMed] [Google Scholar]
- 15.Peng D, Hong W, Choudhury BP, Carlson RW, Gu X-X. Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infect Immun. 2005;73(11):7569–7577. doi: 10.1128/IAI.73.11.7569-7577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park YK, et al. Independent emergence of colistin-resistant Acinetobacter spp. isolates from Korea. Diagn Microbiol Infect Dis. 2009;64(1):43–51. doi: 10.1016/j.diagmicrobio.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Henry R, et al. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrob Agents Chemother. 2012;56(1):59–69. doi: 10.1128/AAC.05191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steeghs L, et al. Outer membrane composition of a lipopolysaccharide-deficient Neisseria meningitidis mutant. EMBO J. 2001;20(24):6937–6945. doi: 10.1093/emboj/20.24.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maifiah MH, et al. Global metabolic analyses identify key differences in metabolite levels between polymyxin-susceptible and polymyxin-resistant Acinetobacter baumannii. Sci Rep. 2016;6:22287. doi: 10.1038/srep22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs AC, et al. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio. 2014;5(3):e01076-14. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams MD, et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol. 2008;190(24):8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell. 2005;121(2):307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Grabowicz M, et al. A mutant Escherichia coli that attaches peptidoglycan to lipopolysaccharide and displays cell wall on its surface. eLife. 2014;3:e05334. doi: 10.7554/eLife.05334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishino F, Mitsui K, Tamaki S, Matsuhashi M. Dual enzyme activities of cell wall peptidoglycan synthesis, peptidoglycan transglycosylase and penicillin-sensitive transpeptidase, in purified preparations of Escherichia coli penicillin-binding protein 1A. Biochem Biophys Res Commun. 1980;97(1):287–293. doi: 10.1016/s0006-291x(80)80166-5. [DOI] [PubMed] [Google Scholar]
- 25.Born P, Breukink E, Vollmer W. In vitro synthesis of cross-linked murein and its attachment to sacculi by PBP1A from Escherichia coli. J Biol Chem. 2006;281(37):26985–26993. doi: 10.1074/jbc.M604083200. [DOI] [PubMed] [Google Scholar]
- 26.Banzhaf M, et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol. 2012;85(1):179–194. doi: 10.1111/j.1365-2958.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- 27.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32(2):234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 28.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32(2):149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 29.Sycuro LK, et al. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141(5):822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowles CE, Li Y, Semmelhack MF, Cristea IM, Silhavy TJ. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol Microbiol. 2011;79(5):1168–1181. doi: 10.1111/j.1365-2958.2011.07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pride AC, Herrera CM, Guan Z, Giles DK, Trent MS. The outer surface lipoprotein VolA mediates utilization of exogenous lipids by Vibrio cholerae. MBio. 2013;4(3):e00305–e00313. doi: 10.1128/mBio.00305-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beceiro A, et al. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother. 2014;58(1):518–526. doi: 10.1128/AAC.01597-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao K, Narita S, Tokuda H. Defective lipoprotein sorting induces lolA expression through the Rcs stress response phosphorelay system. J Bacteriol. 2012;194(14):3643–3650. doi: 10.1128/JB.00553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hooda Y, et al. Slam is an outer membrane protein that is required for the surface display of lipidated virulence factors in Neisseria. Nature Microbiology. 2016;1(4):16009. doi: 10.1038/nmicrobiol.2016.9. [DOI] [PubMed] [Google Scholar]
- 35.Fraser CM, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281(5375):375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 36.Fraser CM, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390(6660):580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 37.Cullen PA, Haake DA, Adler B. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol Rev. 2004;28(3):291–318. doi: 10.1016/j.femsre.2003.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norris SJ. Treponema Pallidum Polypeptide Research Group Polypeptides of Treponema pallidum: Progress toward understanding their structural, functional, and immunologic roles. Microbiol Rev. 1993;57(3):750–779. doi: 10.1128/mr.57.3.750-779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coburn J, Leong J, Chaconas G. Illuminating the roles of the Borrelia burgdorferi adhesins. Trends Microbiol. 2013;21(8):372–379. doi: 10.1016/j.tim.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwashkiw JA, et al. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 2012;8(6):e1002758. doi: 10.1371/journal.ppat.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.