Significance
Ca2+ is one of the most important second messengers regulating numerous cellular functions; therefore, the regulation of Ca2+ release from and its uptake into the endoplasmic reticulum (ER) are both critical for calcium signaling. The activity of sarco/endoplasmic reticulum Ca2+-ATPase isoform 2b (SERCA2b), a calcium pump on the ER membrane, was reported to be negatively regulated by the oxidation of two cysteines in its ER-luminal portion, and it is expected to be activated by its reduction. However, no molecules responsible for this reduction have been identified. Here, we showed for the first time that ERdj5, the reductase in the ER of mammalian cells, activates SERCA2b by reducing its disulfide bonds in a [Ca2+]ER-dependent manner.
Keywords: ERdj5, SERCA2, endoplasmic reticulum, calcium homeostasis, redox regulation
Abstract
Calcium ion (Ca2+) is an important second messenger that regulates numerous cellular functions. Intracellular Ca2+ concentration ([Ca2+]i) is strictly controlled by Ca2+ channels and pumps on the endoplasmic reticulum (ER) and plasma membranes. The ER calcium pump, sarco/endoplasmic reticulum calcium ATPase (SERCA), imports Ca2+ from the cytosol into the ER in an ATPase activity-dependent manner. The activity of SERCA2b, the ubiquitous isoform of SERCA, is negatively regulated by disulfide bond formation between two luminal cysteines. Here, we show that ERdj5, a mammalian ER disulfide reductase, which we reported to be involved in the ER-associated degradation of misfolded proteins, activates the pump function of SERCA2b by reducing its luminal disulfide bond. Notably, ERdj5 activated SERCA2b at a lower ER luminal [Ca2+] ([Ca2+]ER), whereas a higher [Ca2+]ER induced ERdj5 to form oligomers that were no longer able to interact with the pump, suggesting [Ca2+]ER-dependent regulation. Binding Ig protein, an ER-resident molecular chaperone, exerted a regulatory role in the oligomerization by binding to the J domain of ERdj5. These results identify ERdj5 as one of the master regulators of ER calcium homeostasis and thus shed light on the importance of cross talk among redox, Ca2+, and protein homeostasis in the ER.
Intracellular Ca2+ acts as one of the most important signaling molecules in the cytosol and regulates numerous cellular functions including muscle contraction, cellular motility, and vesicular transport through the function of calcium-binding proteins including calmodulin in the cytosol (1, 2). Thus, maintenance of the intracellular Ca2+ concentration ([Ca2+]i) is critical for cellular signaling. Calcium homeostasis in the cytosol is maintained by release and influx of Ca2+ through calcium channels and pumps, respectively, in the plasma and endoplasmic reticulum (ER) membranes (1, 2). Calcium release from the ER into the cytosol is mediated by the inositol 1,4,5-trisphosphate receptor channel (3) and ryanodine receptor (4, 5) localized on the ER membrane. On the other hand, intracellular Ca2+ is taken up by the ER via the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) in an ATPase activity-dependent manner (2, 6, 7).
Subtypes of SERCAs (SERCAs 1–3) are variously expressed in eukaryotic cells. SERCA1a and -1b are expressed in fast skeletal muscles, whereas SERCA2a is expressed in cardiac, smooth, and slow skeletal muscles. There are five isoforms of SERCA3, which are expressed in various nonmuscle cells in a tissue-dependent manner. Among subtypes of SERCAs, SERCA2b is a housekeeping isoform that is ubiquitously and abundantly expressed in nonmuscle cells and smooth muscles. SERCA2b consists of 11 transmembrane domains, and its activity is negatively regulated by the oxidation of two cysteines in its ER-luminal domain (8). In the resting state of SERCA2b under the high ER luminal [Ca2+] ([Ca2+]ER) condition, the N domain of calreticulin, a molecular chaperone in the ER, interacts with the C-terminal sequence of SERCA2b and recruits ERp57, a ubiquitous ER thiol-dependent oxidoreductase that promotes the formation of disulfide bonds, to target the intraluminal loop 4 of SERCA2b (8). Intramolecular disulfide bond formation between two cysteines in loop 4 inhibits the pump activity of SERCA2b. When [Ca2+]ER decreases to lower than 50 µM, ERp57 dissociates from SERCA2b to reactivate the pump function (8). Considering the oxidative condition in the ER lumen, the reduction of this disulfide bond should require a molecule(s) with reductase activity, which has not been identified.
Newly synthesized secretory proteins are cotranslationally translocated into the ER, where they are correctly folded with the aid of various molecular chaperones and enzymes. The major chaperones in the ER, including calnexin, calreticulin, BiP, and some protein disulfide isomerases (PDIs), require Ca2+ for their functions (6, 9). Inhibition of Ca2+ uptake into the ER by the SERCA inhibitor thapsigargin causes ER stress due to the accumulation of misfolded proteins. Consequently, maintenance of calcium homeostasis in the ER by the SERCA calcium pump is critically important for the functional integrity of the ER.
Proteins that harbor genetic mutations or are terminally misfolded must be eliminated to prevent formation of toxic aggregates. Terminally misfolded glycoproteins in the ER are transferred from calnexin/calreticulin to ER degradation-enhancing α-mannosidase-like protein 1 (EDEM1) in an N-glycan trimming-dependent manner (10, 11) and are subsequently transferred to ERdj5, the first disulfide reductase identified in the mammalian ER, which contains a J domain at its N terminus and four thioredoxin-like domains with redox-active motifs (Cys-X-X-Cys, CXXC) (12). ERdj5 cleaves disulfide bonds within the misfolded proteins to facilitate their passage through the retro-translocation channel in the ER membrane. Misfolded substrates are transferred from ERdj5 to Binding Ig Protein (BiP), a major molecular chaperone in the ER, which binds to ERdj5 through the HPD (Hys-Pro-Asp) motif in the J domain. BiP recruits the substrates to the retro-translocation channel to promote their ER-associated degradation (ERAD) pathway (13, 14).
ERdj5 is the first identified ER reductase; therefore, we postulated and examined whether ERdj5 regulates SERCA2b activity by cleaving the disulfide bond in its intraluminal loop. Here, we show that ERdj5 activates the pump function of SERCA2b by reducing its luminal disulfide bond. Remarkably, we also found that ERdj5 activates SERCA2b in a [Ca2+]ER-dependent manner. These results have established that ERdj5, in addition to ERp57, works as one of the master regulators of SERCA2b and thus for ER calcium homeostasis. ERdj5 is a PDI family member involved in redox homeostasis in the ER; therefore, our findings shed light on the importance of cross talk among redox, Ca2+, and protein homeostasis in the ER.
Results
ER-Resident Reductase ERdj5 Interacts with SERCA2b.
Although dissociation of ERp57 from the intraluminal loop 4 of SERCA2b was reported to reactivate its ATPase activity under the lower [Ca2+]ER condition, the fate of the disulfide bond after release of ERp57 was not examined. The redox condition in the ER lumen is oxidizing; therefore, an oxidoreductase(s) with reducing activity is postulated to be required for reduction of this disulfide bond. We hypothesized and examined whether ERdj5 can activate SERCA2b by cleaving the disulfide bond.
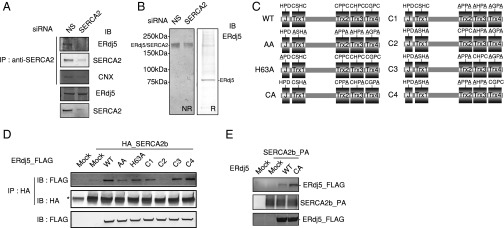
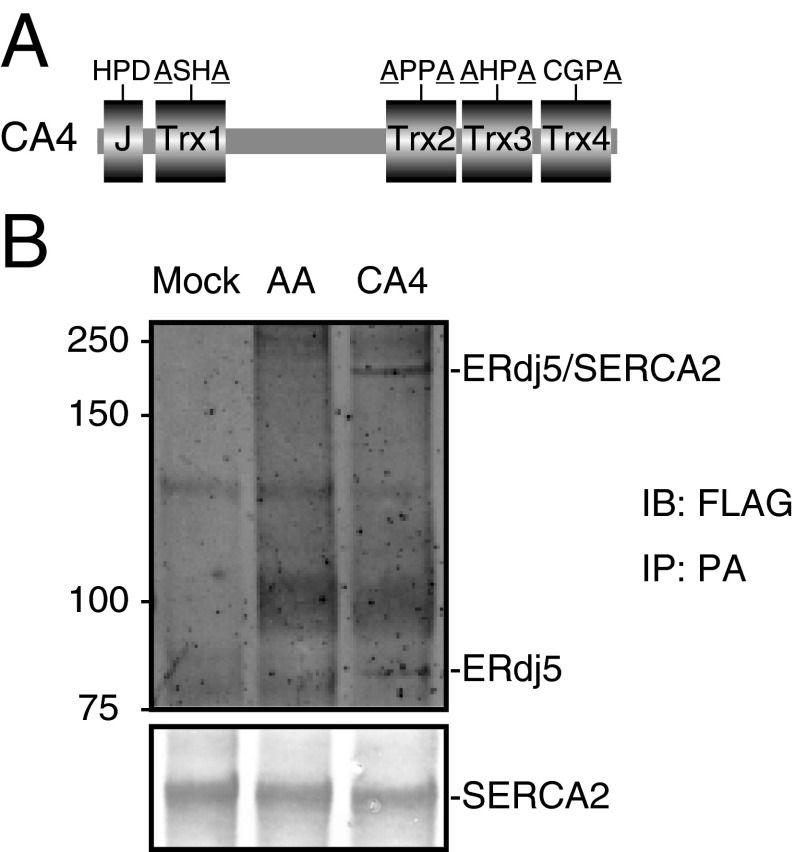
Immunoprecipitation and immunoblot analysis clearly showed that endogenous ERdj5 interacted with endogenous SERCA2b. The specificity of this interaction was confirmed by small interfering RNA (siRNA) knockdown of SERCA2b (Fig. 1A). Calnexin, a SERCA2b-binding protein (15), was also coprecipitated in this complex. In nonreducing gels, the band containing ERdj5 was shifted to around 200 kDa because cysteines of CXXC motifs of ERdj5 make mixed disulfide bonds with SERCA2b (Fig. 1B). When all CXXC motifs were converted to Ala-X-X-Ala (AXXA) (Fig. 1C), this ERdj5/AA mutant lost its reducing activity (12, 13). The ERdj5/AA mutant barely bound to SERCA2b (Fig. 1D). These results suggest that this complex forms via the mixed disulfide bond(s) between ERdj5 and SERCA2b. Consistent with this, the ERdj5/CA mutant, in which all CXXC motifs were converted to CXXA (Fig. 1C), bound much more strongly to SERCA2b because the CA mutant acts as a trapping mutant of redox proteins (Fig. 1E). Using a set of ERdj5 mutants in which only one CXXC motif remained intact, we found that SERCA2b preferentially bound to ERdj5 via Trx3 and Trx4 (Fig. 1D), which have significant reductase activity and are hence involved in ERAD (13). This was more directly shown using the ERdj5/CA4 mutant, in which the CXXC motif in the Trx4 domain was mutated to CXXA and other CXXCs to AXXA. The complex of ERdj5/CA4 mutant and SERCA2b was shown as an around 200-kDa band in the presence of divinylsulfone (DVSF), which is a chemical cross-linker for disulfide bridge (Fig. S1).
Fig. 1.
Interaction between ERdj5 and SERCA2. (A and B) Forty-eight hours after transfection of nonspecific (NS) or SERCA2-specific siRNA into HeLa cells, cell lysates were prepared for immunoprecipitation with an anti-SERCA2 antibody. Immunoprecipitates were subjected to (A) reducing or (B) nonreducing (Left) and reducing (Right) SDS/PAGE for the analysis by immunoblotting with the indicated antibodies. (C) The series of ERdj5 mutants constructed in this work. (D and E) Twenty-four hours after cotransfection of (D) HA-tagged or (E) PA-tagged SERCA2b and FLAG-tagged ERdj5/WT or the indicated ERdj5 mutants into HEK293 cells, cell lysates were prepared for immunoprecipitation with (D) anti-HA or (E) anti-PA antibodies. (D and E) All immunoprecipitates were subjected to reducing SDS/PAGE and analyzed by immunoblotting with the indicated antibodies. The asterisk in D indicates an NS band. CNX, calnexin.
Fig. S1.
ERdj5/CA4 interacts with SERCA2 through the disulfide bond. (A) Schematic representation of ERdj5/CA4. (B) Twenty-four hours after cotransfection of PA-tagged SERCA2b and FLAG-tagged ERdj5/AA or ERdj5/CA4 mutant into HEK293 cells, cells were treated with DVSF, which is the cross-linker specific for disulfide bond. The lysates were added with 1% SDS to dissociate the noncovalent interaction. After the dilution of SDS to 0.1%, the cell lyates were immunoprecipitated with anti-PA antibodies and analyzed by SDS/PAGE under reducing condition.
A mutation in the J domain of ERdj5, in which histidine 63 in the HPD motif was replaced with alanine (ERdj5/H63A), weakened the interaction with SERCA2b (Fig. 1D). This suggests that BiP contributes to efficient formation of the SERCA2b–ERdj5 complex.
ERdj5 Regulates SERCA2b Pump Function Through Its Reductase Activity.
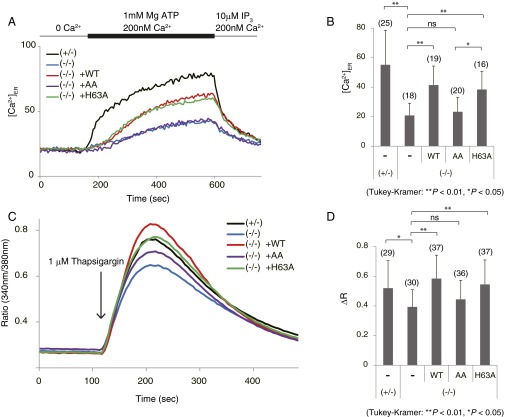
Next, we examined the effect of ERdj5 binding to SERCA2b on Ca2+ uptake into the ER from the cytosol using ERdj5-disrupted mouse embryonic fibroblasts (MEFs) semipermeabilized with digitonin. Very little Ca2+ was imported into the ER of ERdj5 (−/−) cells (blue line), whereas it was imported rapidly into the ER of ERdj5 (+/−) cells (black line) (Fig. 2 A and B). Ca2+ uptake into the ER of ERdj5 (−/−) cells was rescued by overexpression of wild-type (WT) ERdj5 (red line), whereas the ERdj5/AA mutant (purple line) had no effect. Overexpression of the ERdj5/H63A mutant (green line) had almost the same effect on Ca2+ uptake as ERdj5/WT in semipermeabilized cells. In these experiments, the expression levels of SERCA2b were confirmed not to be changed among these cells (Fig. S2). These results clearly indicate that redox-active ERdj5 is required for the efficient uptake of Ca2+ into the ER.
Fig. 2.
Deficiency of ERdj5 suppresses Ca2+ uptake into the ER through SERCA2b. (A and B) Cells were loaded with Mag-Fura-2 to estimate [Ca2+]ER. Semipermeabilized MEFs were treated with EGTA for 20 min to remove Ca2+. After depletion of Ca2+, ATP and Ca2+ were added to stimulate SERCA2 pump functions. Mag-Fura-2 fluorescence was measured as [Ca2+]ER. The quantifications of each Ca2+ uptake are as shown in B. (C and D) After 1 µM thapsigargin treatment, [Ca2+]i was measured using Mag-Fura-2. Peak amplitudes are shown as a bar graph in D.
Fig. S2.
The expression levels of SERCA2b were unaffected by the overexpression of ERdj5s. Twenty-four hours after transfection of ERdj5 WT, AA mutant, or H63A mutant into ERdj5 (−/−) MEF cells, cell lysates were prepared for immunoblotting with anti-SERCA antibody (Upper) or anti–β-actin antibody (Lower).
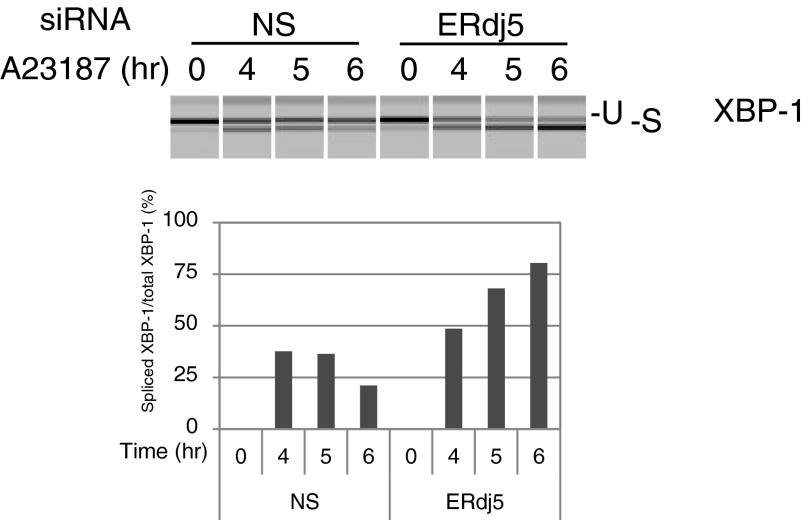
We next investigated calcium storage in the ER by treating cells with thapsigargin, an inhibitor of SERCA ATPase activity (Fig. 2 C and D). Ca2+ release from the ER under inhibition of SERCA2b was lower in ERdj5 (−/−) MEFs than in ERdj5 (+/−) MEFs, which suggests that the inactive state of SERCA2b in the absence of ERdj5 caused low calcium storage in the ER. Overexpression of ERdj5/WT and the ERdj5/H63A mutant recovered Ca2+ storage in the ER, but the ERdj5/AA mutant failed to rescue Ca2+ storage in the ER, which is consistent with the results shown in Fig. 2B. Taken together, these observations strongly suggest that ERdj5 stimulates SERCA2b activity and that, in the absence of ERdj5, SERCA2b is maintained in an inactive state. Indeed, ERdj5 knockdown decreased cellular tolerance to ER stress induced by A23187, a Ca2+ ionophore that decreases [Ca2+]ER (Fig. S3).
Fig. S3.
ERdj5-knockdown cells are sensitive to ER stress induced by the Ca2+ ionophore A23187. Unfolded protein response induction was examined by detecting the spliced form of XBP1 mRNA. Total RNA was prepared from HEK293T cells 48 h after transfection. Cells treated with 100 nM A23187 for the indicated amounts of time were used as positive controls. Unspliced (U) and spliced (S) forms of hXBP1 mRNA amplified by RT-PCR are shown.
ERdj5 Enhances the ATPase Activity of SERCA2b by Reducing the Luminal Disulfide Bond.
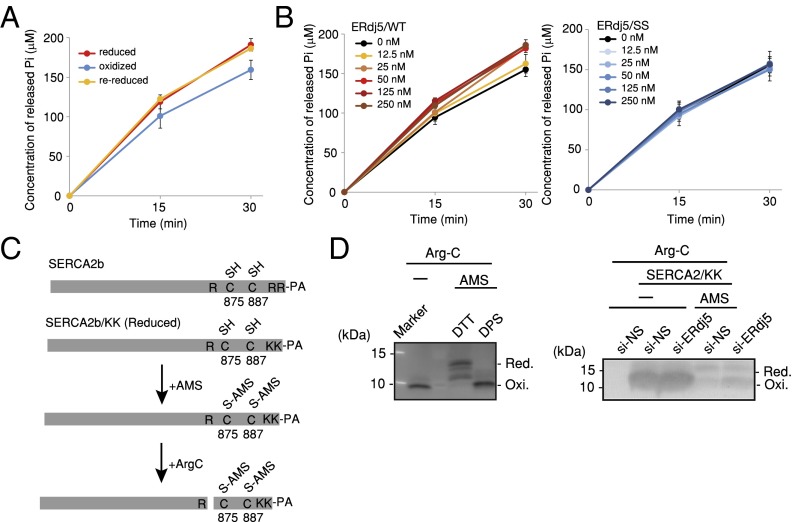
SERCA2b is a member of the P-type ion-transport ATPase family, and its ATPase activity is indispensable for Ca2+ transport into the ER (16, 17). We examined the effect of oxidation/reduction of Cys875 and Cys887 on ATPase activity using purified recombinant SERCA2b/WT (Fig. S4). The reduced form of SERCA2b had significantly higher ATPase activity than the oxidized form (Fig. 3A). The rereduction of oxidized SERCA2b restored its ATPase activity to that of the reduced form, indicating that the redox states of the Cys875–Cys887 pair reversibly regulate the ATPase activity of SERCA2b. The ATPase activity of oxidized SERCA2b was increased by incubation with the reduced form of recombinant ERdj5/WT, whereas the ERdj5/SS mutant, in which all of the CXXC motifs of ERdj5 were mutated to Ser-X-X-Ser motifs, had little effect (Fig. 3B).
Fig. S4.
Preparation of recombinant PA-SERCA2b, ERdj5/WT-FLAG, and ERdj5/SS-FLAG. Purified PA-SERCA2b, ERdj5/WT-FLAG, and ERdj5/SS-FLAG were subjected to reducing SDS/PAGE and stained with Coomassie brilliant blue (CBB).
Fig. 3.
ERdj5 cleaves the disulfide bond of SERCA2b and activates its function. (A) Comparison of ATPase activity between the oxidized and reduced forms of SERCA2b. The ATPase activities of reduced SERCA2b (red line), oxidized SERCA2b (blue line), and rereduced SERCA2b (orange line) were assessed by quantifying released Pi. Results presented in the graphs represent means ± SD of three independent experiments. (B) Activation of the SERCA2b ATPase by ERdj5/WT and the ERdj5/SS mutant. The concentration of Pi released by 50 nM SERCA2b in the presence of 0, 12.5, 25, 50, 125, or 250 nM ERdj5/WT or ERdj5/SS was quantified and plotted as a function of the reaction time. Results presented in the graphs represent means ± SD of three independent experiments. (C) To accurately estimate the redox states of SERCA2b, SERCA2b/KK was constructed, in which two arginines in the C terminus were converted to lysines. After SERCA2b/KK was transfected into HEK293T cells, the cells were precipitated with 10% TCA, and free thiol groups were modified with 150 mM AMS. The C-terminal region containing C875, C887, and the PA tag was excised using Arg-C endopeptidase, which selectively cleaves C-terminal to arginine residues. (D) Forty-eight hours after transfection of nonspecific (NS) or ERdj5-specific siRNA into HeLa cells, cell lysates were prepared for Arg-C treatment as described in C. After immunoprecipitation with anti-PA antibody, a concentrated C-terminal region was detected by Western blotting with an anti-PA antibody.
The addition of reduced ERdj5 caused activation of SERCA2b; therefore, we directly assessed the redox state of the luminal disulfide bond of SERCA2b. Because SERCA2b is too large to analyze its redox state by the modification method of two cysteines, and also because it is necessary to exclude the effect of the many cysteine residues on the cytosolic domain of SERCA2b, we tried to detect the peptide fragment of SERCA2b containing these two cysteines. PA-tagged SERCA2b/KK, in which two arginines (Arg923 and Arg988) in the C-terminal region were mutated to lysines to abolish the cleavage sites by Arg-C endopeptidase, was transfected into HEK293 cells and modified with 4-acetamido-4-maleimidylstilbene-2–2-disulfonic acid (AMS) after the redox state was frozen by treatment of the cells with 10% trichloroacetic acid (TCA) (Fig. 3C). The C-terminal portion of SERCA2b, including the two luminal cysteines, was detected with an anti-PA antibody after cleavage by Arg-C endopeptidase. The reduced and oxidized states of SERCA2b could be clearly discriminated by this method (Fig. 3D, Left). In ERdj5-knockdown cells, the two cysteines existed predominantly in the oxidized form, whereas in WT cells, significant fraction was in the reduced form (Fig. 3D, Right). These observations suggested that ERdj5 is required to maintain Cys875 and Cys887 of SERCA2b in their reduced forms.
[Ca2+]ER Influences the Interaction of ERdj5 with SERCA2b.
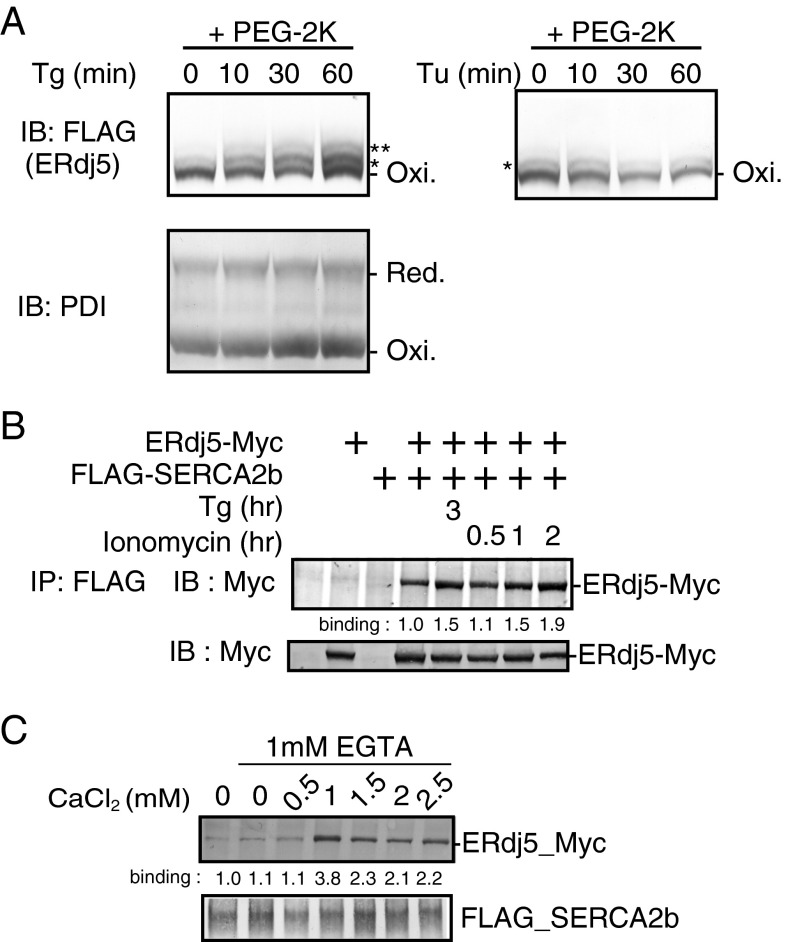
Next, we sought to determine whether the activation of SERCA2b by ERdj5 is constitutive or regulative. To maintain Ca2+ homeostasis in the ER, ERdj5 should activate SERCA2b in a [Ca2+]ER-dependent manner. Although ERdj5 was present in its oxidized form in normal cells, treatment with thapsigargin converted it to the reduced form, which is capable of reducing SERCA2b (Fig. 4A). The redox state of another ER-resident oxidoreductase, PDI, was not changed by thapsigargin treatment, suggesting that Ca2+ deficiency in the ER specifically converted ERdj5 to the reduced form. This was not simply due to ER stress because tunicamycin, an inhibitor of N-glycosylation, had no effect on the redox state of ERdj5 (Fig. 4A, Right).
Fig. 4.
Interaction between ERdj5 and SERCA2 depending on [Ca2+]. (A) Redox states of ERdj5 under treatment with thapsigargin (Tg) or tunicamycin (Tu). Transfected FLAG-tagged ERdj5 was modified with mPEG2000-mal in HEK293T cells treated with Tg or Tu for the indicated amount of time. Double or single asterisks denote modified ERdj5 bands. (B) Twenty-four hours after cotransfection of FLAG-tagged SERCA2b and Myc-tagged ERdj5/WT into HEK293 cells, the cells were treated with Tg or ionomycin for the indicated amount of time. The percentage binding of ERdj5 was normalized to that in untreated cells. (C) FLAG-SERCA2b was captured with FLAG-conjugated beads in lysates of cells transfected with FLAG-SERCA2b. Lysates of cells transfected with Myc-tagged ERdj5 were adjusted to contain the indicated concentration of CaCl2. The adjusted cell lysates were incubated with FLAG-SERCA2b-bound beads for the pull-down assay. ERdj5-Myc bound to SERCA2b was detected by immunoblotting with an anti-Myc antibody. Binding of ERdj5 with SERCA2b was quantified and shown below the panel.
The interaction of ERdj5 with SERCA2b was strengthened when cells were treated with thapsigargin or ionomycin, both of which decrease [Ca2+]ER (Fig. 4B). When [Ca2+]i was titrated in the presence of 1 mM ethylene glycol tetraacetic acid (EGTA), the ERdj5/SERCA2b interaction was maximal at submillimolar [Ca2+] (Fig. 4C).
ERdj5 Forms Oligomers at High [Ca2+]ER.
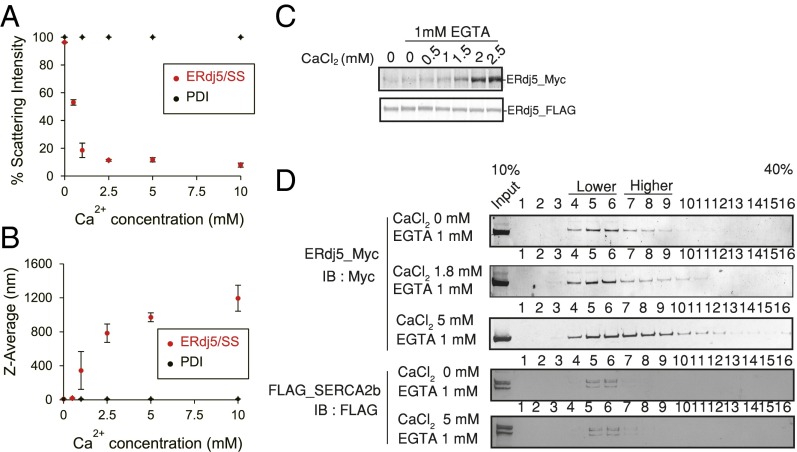
To address why ERdj5 is incapable of binding to SERCA2b at high [Ca2+], we referred to previous studies of calsequestrin, a regulator of the ryanodine receptor (18, 19). Calsequestrin consists of three tandem repeats of thioredoxin-like domains, which are similar to a part of ERdj5, and was reported to convert its molecular conformation from a monomer to an oligomer depending on [Ca2+], and this conversion affects the interaction with the ryanodine receptor (20, 21). Thus, we analyzed the size distribution and hydrodynamic diameter of ERdj5 in solution by dynamic light scattering (DLS) measurement of recombinant ERdj5 at various [Ca2+]. Upon addition of submillimolar [Ca2+], the scattering intensity derived from the monomer species rapidly decreased (Fig. 5A), whereas the average diameter rapidly increased (Fig. 5B). These observations indicate that ERdj5 is converted to homo-oligomers at high [Ca2+], in sharp contrast to the behavior of PDI.
Fig. 5.
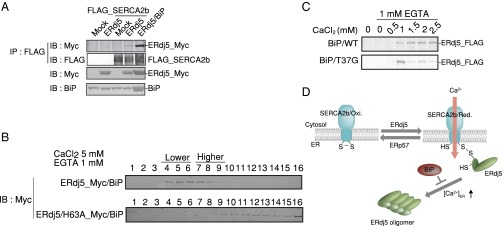
ERdj5 regulates [Ca2+]ER by oligomer formation. (A) Relative scattering intensity derived from the monomeric component of ERdj5/SS or PDI at various calcium concentrations. (B) The Z-average, the mean diameter of ensemble particles in solution, of ERdj5/SS or PDI under various calcium concentrations. Values are the means ± SD of five independent experiments. (C) ERdj5-FLAG was captured with FLAG-conjugated beads in lysates of cells transfected with ERdj5-FLAG. Lysates of cells cotransfected with Myc-tagged ERdj5 and BiP/WT or the T37G mutant were adjusted to contain the indicated concentration of CaCl2. The adjusted cell lysates were incubated with ERdj5-FLAG-bound beads for the pull-down assay. After pull down with ERdj5-FLAG-bound beads, Myc-tagged ERdj5 bound to FLAG-tagged ERdj5 was detected by immunoblotting with an anti-Myc antibody. (D) Twenty-four hours after transfection of the indicated constructs into HEK293 cells, cells lysates containing 1 mM EGTA and titrated CaCl2 were prepared. Cell lysates were applied to a 10–40% sucrose density gradient and centrifuged. Each fraction was separated by SDS/PAGE for immunoblotting with the indicated antibodies.
The [Ca2+] dependency of oligomerization was examined by coprecipitation of ERdj5-Myc and ERdj5-FLAG. ERdj5 started to form oligomers in the presence of 1.5 mM CaCl2 and 1 mM EGTA (Fig. 5C). Sucrose density gradient centrifugation of cells revealed that ERdj5 sedimented in lighter fractions (corresponding to a smaller molecular size) when [Ca2+] was submillimolar or lower. However, at high [Ca2+] (5 mM), ERdj5 tended to sediment in denser fractions (Fig. 5D, Upper). SERCA2b cosedimented with ERdj5 in lighter fractions (Fig. 5D, Lower), suggesting that ERdj5 molecules that do not form higher-order oligomers can interact with SERCA2b.
Finally, we examined the effect of BiP binding on ERdj5 oligomerization using the ERdj5/H63A mutant, which is incapable of BiP binding, and the BiP/T37G mutant, which lacks ATPase activity (22). First, to examine the effect of BiP on the interaction between ERdj5 and SERCA2b, BiP was cotransfected with ERdj5 and SERCA2b for immunoprecipitation. Overexpressed BiP facilitated ERdj5 binding to SERCA2b (Fig. 6A). Meanwhile, H63A mutation of ERdj5 prevented the interaction between ERdj5 and SERCA2b (Fig. 1D). In the presence of BiP, ERdj5 sedimented in lighter fractions even at high [Ca2+] (Fig. 6B), which is in contrast to the previous observation without BiP (Fig. 5D). However, the ERdj5/H63A mutant sedimented in denser fractions under the same conditions (Fig. 6B). The association of ERdj5 with SERCA2b at high [Ca2+] was promoted by cotransfection with BiP and ERdj5 (Fig. 6C), whereas it was weakened in the absence of BiP (Fig. 4C). The BiP mutant lacking ATPase activity had little effect on binding of ERdj5 to SERCA2b at high [Ca2+] (Fig. 6C). These observations suggest that BiP prevents ERdj5 from forming higher-order oligomers, thereby maintaining its ability to bind to and activate SERCA2b (Fig. 6D).
Fig. 6.
BiP binding to ERdj5 prevents ERdj5 oligomer formation. (A) Twenty-four hours after cotransfection of FLAG-tagged SERCA2b, FLAG-tagged ERdj5/WT, and BiP into HEK293 cells, cell lysates were prepared for immunoprecipitation with an anti-FLAG antibody. Myc-tagged ERdj5 bound to FLAG-tagged SERCA2b was detected by immunoblotting with an anti-Myc antibody. (B) Twenty-four hours after cotransfection of BiP and ERdj5/WT or the H63A mutant into HEK293 cells, cell lysates containing 1 mM EGTA and 5 mM CaCl2 were prepared. Cell lysates were applied to a 10–40% sucrose density gradient and centrifuged. Each fraction was separated by SDS/PAGE for immunoblotting with an anti-Myc antibody. (C and D) SERCA2b activation mechanism via the reducing activity of ERdj5 depending on [Ca2+]ER. ERdj5 activates SERCA2b only at a lower [Ca2+]ER, whereas a higher [Ca2+]ER induces ERdj5 to form oligomers that are no longer able to interact with the pump. BiP exerts a regulatory role in the oligomerization of ERdj5 by binding to its J domain.
Discussion
In this report, we showed that ERdj5 activated the pump function of SERCA2b by reducing the intraluminal disulfide bond of SERCA2b. This activation by ERdj5 was [Ca2+]ER-dependent, and at a higher [Ca2+]ER, ERdj5 was converted to an oligomer form, which was no longer effective for the activation of SERCA2b.
A possible regulatory mechanism is shown in Fig. 6D. The oxidized (low-activity state) form of SERCA2b can be converted into the high-active form by reduction of the disulfide bond in the ER-luminal portion, which activates Ca2+ uptake into the ER. When [Ca2+]ER becomes higher than 1 mM, it causes oligomerization of ERdj5, which loses the ability to bind to SERCA2b. This feedback regulation by oligomerization of ERdj5 serves to maintain Ca2+ homeostasis in the ER lumen. When [Ca2+] ER is lowered by treatment of cells with thapsigargin, the decrease in [Ca2+]ER shifts the lumen to a reducing state (23), providing favorable conditions for the reducing activity of ERdj5 and activation of SERCA2b. Treatment with thapsigargin also causes inactivation of PDI, a major oxidoreductase in the ER, and contributes to the shift of the ER redox condition to a reducing state (24). The decrease in [Ca2+]ER induces ER stress, resulting in higher expression levels of ER molecular chaperones such as BiP, which prevents the oligomerization of ERdj5 to counteract the decrease in [Ca2+]ER by activating SERCA2b. Molecular chaperones induced by ER stresses including BiP require Ca2+, which is provided by activation of SERCA2b (6). Thus, regulation of SERCA2b by ERdj5 and BiP provides a calcium-mediated regulatory mechanism that maintains organelle homeostasis of the ER.
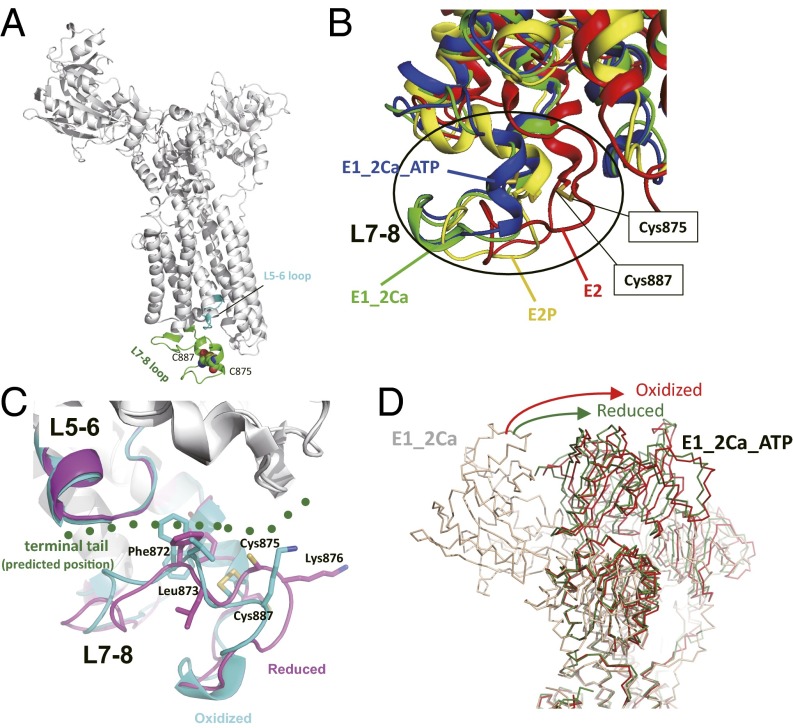
One important question is how SERCA2b activity is regulated by oxidation/reduction of the Cys875–Cys887 pair located in L7–L8. To obtain insights into this question, homology models of SERCA2 in several catalytic and redox states were built based on the previously reported crystal structures of SERCA1 (25–27) (Fig.7 A–D and Fig. S5). In these homology models, L7–L8 of SERCA2 is predicted to consist of two short α-helices with loops. Cys875 and Cys887 are located in each of the two α-helices, respectively (Fig. 7 A and B). In the oxidized state, Cys875 and Cys887 form a disulfide bridge, stabilizing the L7–L8 conformation (Fig. 7C). Comparison of the oxidized and reduced states suggests that reduction of the Cys875–Cys887 disulfide bond alters the backbone structure and orientation of the side chains of several residues, leading to significant conformational change of L7–L8. On the other hand, the C-terminal tail characteristic of SERCA2b is predicted to interact with the cleft between L7–L8 and L5–L6 and stabilize the Ca2+-bound E1 state (Fig. 7C), thereby preventing Ca2+ release from SERCA2b (28). Therefore, reduction of the Cys875–Cys887 disulfide bond will change the L7–L8 conformation and affect the interaction between the loop and the C-terminal tail, thus destabilizing the E1 state. Consequently, Ca2+ release is likely facilitated in the reduced form of SERCA2b.
Fig. 7.
Homology models of SERCA2 provide a likely mechanism of the redox-dependent regulation of SERCA2b activity. (A) The Cys875–Cys887 pair and L5–L6 and L7–L8 loops in a SERCA2 homology model in the E1-2Ca state are highlighted. (B) Superposition of homology models of SERCA2 in the E2 (red), E1-2Ca (green), E1-2Ca-ATP (blue), and E2P (yellow) states. The homology model of SERCA2 was constructed for each state with SWISS-Model (39) using previously reported crystal structures of SERCA1a (Protein Data Bank ID: 3W5C for E2, 1SU4 for E1-2Ca, 3AR2 for E1-2Ca-ATP, and 3B9B for E2P) as templates. The transmembrane regions of these four SERCA2 models were superposed to minimize the root-mean-square deviation of their Cα atoms. Comparison of the models in different catalytic states suggests that the L7–L8 region of SERCA2 undergoes significant conformational changes during its catalytic cycle. (C) Conformational changes in the L7–L8 region upon reduction of the luminal Cys875–Cys887 disulfide bond. The homology models in the oxidized (cyan) and reduced (magenta) forms were constructed based on the crystal structures of both of the oxidized and the reduced forms of SERCA1a in the E1-2Ca-ATP state. The predicted position of the C-terminal tail of SERCA2b is shown in green dots. (D) Comparison of the relative position of the SERCA2 N domain in the E1-2Ca-ATP state. The homology models of the oxidized and reduced forms in the E1-2Ca-ATP state are shown in red and green, respectively. The model in the E1-2Ca form is shown in light brown.
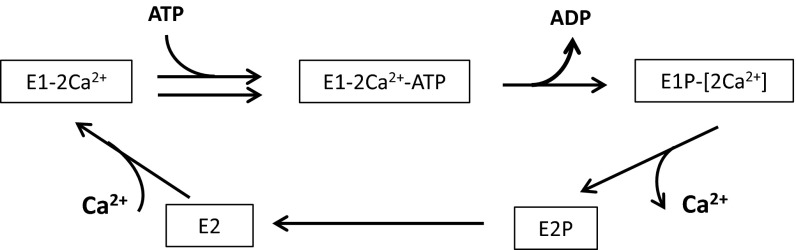
Fig. S5.
Simplified reaction scheme during the Ca2+ pump cycle of SERCA.
In addition to the redox-dependent conformational changes of L7–L8, homology models in the E1-2Ca-ATP state suggest that the cytosolic N domain is also involved in the regulation of the catalytic cycle in a redox-dependent manner. The relative position of the N domain differs significantly between the reduced (green) and oxidized (red) forms of SERCA2 in the E1-2Ca-ATP state (Fig. 7D). Thus, compared with the reduced state, the larger movement of the N domain is expected in the oxidized state during the transition from the E1-2Ca to the E1-2Ca-ATP state. These observations suggest that the reduced state requires lower activation energy than the oxidized state for the transition from the E1-2Ca to the E1-2Ca-ATP states, resulting in acceleration of the catalytic cycle.
In addition to the reducing activity of ERdj5, the interaction of ERdj5 with BiP through its J domain is also noted. The interaction of BiP with ERdj5 is not necessary for the activation of Ca2+ uptake under the low [Ca2+] condition because the ERdj5/H63A mutant and ERdj5/WT had an equivalent activity in the semipermeabilized assay system (Fig. 2 A and B). On the contrary, BiP binding to ERdj5 was necessary to prevent oligomerization of ERdj5 and thus for the interaction of ERdj5 with SERCA2b under high [Ca2+] (Fig. 6 A–C). The involvement of BiP in the activation of SERCA2b by ERdj5 may be particularly relevant to the maintenance of [Ca2+] homeostasis in the ER under the stress conditions as mentioned above.
We previously showed that the interplay of ERdj5 with BiP is indispensable for the elimination of misfolded proteins from the ER through ERAD. Glycoproteins misfolded in the ER are recognized by EDEM1 and are recruited to ERdj5, which reduces the intramolecular or intermolecular disulfide bonds of misfolded proteins to facilitate retro-translocation through the dislocon channel. When nonglycoproteins are misfolded in the ER or when EDEM1 is overwhelmed with misfolded glycoproteins, BiP recruits these misfolded proteins to ERdj5 (14). After reduction of disulfide bonds, substrates to be degraded are handed to BiP and recruited to the dislocon channel. Thus, ERdj5 together with BiP have critical roles for both ERAD and calcium regulation. At present, it is not clear how this interplay of ERdj5 with BiP is regulated to induce either ERAD or SERCA2b activation. However, our findings shed light on coordinated cross talk among protein, redox, and calcium homeostasis, which are three major types of homeostasis in the ER.
Darier’s disease, an autosomal dominant inherited disorder of the skin, was reported to be caused by an anomaly of Ca2+ signaling due to mutations of SERCA2b (29–32). Because of loss-of-function mutations in SERCA2b, keratinocytes in Darier’s disease patients were reported to have a reduced pool of Ca2+ in the ER. It is worth noting that mutation of one of two cysteines in loops 7 and 8 also causes the phenotype of Darier’s disease, which suggests the importance of these cysteines in the regulation of pump function by SERCA2b. On the other hand, disruption of the ERdj5 gene in mice was reported to enhance sensitivity to ER stresses in the salivary gland (33). The salivary gland is one of the major secretory organs in which calcium signaling plays critical roles for the secretion of various proteins including α-amylase into saliva. The possibility that disruption of ERdj5 affecting SERCA2b activity in salivary glands is the cause of ER stress is worth examining in the future.
Materials and Methods
Cell Culture and Transfections.
HEK293T cells, HeLa cells, and MEFs were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco 11995) supplemented with 10% (vol/vol) inactivated FCS. Plasmids were transfected using Lipofectamine 2000 (Life Technologies) for HEK293T or HeLa cells and Lipofectamine LTX (Life Technologies) for MEFs. Cells were transfected with siRNA using RNAiMAX (Life Technologies) reagents. Stealth RNA Negative Control Low GC and Stealth siRNAs specific to human SERCA2 and ERdj5 were obtained from Life Technologies.
Plasmid Construction.
Human SERCA2b cDNA was amplified by PCR from the Matchmaker Pretransformed Human HeLa library (Clontech) and subcloned into pcDNA3.1. Mouse ERdj5/WT and other ERdj5 mutants (AA, CA, H63A, C1, C2, C3, and C4) were constructed as described previously (12, 13). The SERCA2b C875AC887A (SERCA2b/AA) and R923KR988K (SERCA2b/KK) double mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene). pIRES2-NLS-EGFP was constructed as follows: the internal ribosome entry site (IRES) sequence was PCR-amplified from pIRES2-AcGFP1 (Invitrogen) using a primer set (5′-CACCAAAATCAACGGGACTTTCC-3′ and 5′-ATCTCTAGATGGCCATATTATCATCGTG-3′) and then digested with EcoRI and XbaI. EGFP cDNA was PCR-amplified using a primer set (5′-TCTCTAGAATGTCTCCCAAGAAGAAGCGCAAGGTGGGGCCAGTATGGTGAGCAAGGGCGAGGAG-3′ and 5′-GCTGGCAACTAGAAGGCACAG-3′), which includes a nuclear localization signal (NLS), and digested with XbaI and NotI. Restriction fragments containing the IRES and NLS-EGFP cDNA were inserted into the EcoRI-NotI sites of pIRES2-AcGFP1. To construct ERdj5s/pIRES2-NLS-EGFP, ERdj5s cDNA was isolated by digestion with BamHI and EcoRI and then inserted into the BglII-EcoRI sites of pIRES2-NLS-EGFP. To construct purified recombinant SERCA2b, DNA encoding human SERCA2b with a PA tag (GVAMPGAEDDVV) at the N terminus (34) (PA-SERCA2b) was inserted into the PiggyBac Cumate Switch Inducible Vector (System Bioscience). Hamster BiP/WT and BiP/T37G were a gift of Linda Hendershot, Department of Tumor Cell Biology, St. Jude Children’s Research Hospital, Memphis, TN.
Antibodies.
Mouse monoclonal anti-FLAG M2, anti-SERCA2, and anti-BiP antibodies were purchased from Sigma, Calbiochem, and BD Biosciences, respectively. Rabbit polyclonal antibodies against HA and calnexin were obtained from Santa Cruz Biotechnology and Enzo Life Sciences, respectively. A mouse polyclonal antibody against ERdj5 was purchased from Abnova. A rat monoclonal antibody NZ-1 against the PA tag was purchased from Wako Pure Chemical Co. and coupled to CNBr-activated Sepharose 4 Fast Flow as described previously (34).
Cell Lysis and Immunoprecipitation.
Cells were washed with PBS[−], incubated on ice for 20 min in lysis buffer [50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 10 mM N-ethylmaleimide, and 1% Nonidet P-40 or 1% CHAPS] supplemented with protease inhibitors, and then immunoprecipitated with specific antibodies. Immunoprecipitants were separated by SDS/PAGE. Immunoblotting was conducted under reducing or nonreducing conditions with the antibodies indicated in the text (Figs. 1 and 3–6). To detect the mixed disulfide complex (Fig. S1), cells were treated with 100 μM DVSF for 1 h. After the cell lysis, the cell lysates are incubated with 1% SDS and diluted with lysis buffer by 10 times for immunoprecipitation.
Imaging.
ERdj5 (+/−) and ERdj5 (−/−) MEFs were transfected with pIRES2-NLS-EGFP or ERdj5s/pIRES2-NLS-EGFP using TransIT-LT1 (Mirus Bio). To measure [Ca2+]ER, after 24–30 h of transfection, transfected MEFs were loaded with Mag-Fura-2:00 AM (Life Technologies) (5 μM) for 60 min at room temperature and perfused with cytosolic-like medium (CLM) containing 125 mM KCl, 19 mM NaCl, 10 mM 4-(2-hydroxyethyl)-1-piperazinethanesulfonic acid (Hepes), and 1 mM EGTA (pH adjusted to 7.3 with KOH). The cells were then permeabilized by exposure to CLM containing 20 μM digitonin (Calbiochem) for 2–3 min. After incubation for 15 min in CLM, MEFs were perfused with CLM containing 1 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) (Abcam) to prevent Ca2+ uptake into mitochondria (35). The ER was Ca2+-loaded by switching to CLM in which the free [Ca2+] was adjusted to 200 nM containing 1 μM FCCP and 1 mM MgATP. To induce Ca2+ release, cells were incubated in CLM containing the same concentration of Ca2+ and 10 μM inositol trisphosphate (Dojindo) but lacking MgATP to prevent Ca2+ reuptake into the ER.
To measure [Ca2+]i, transfected MEFs were loaded with Fura-2:00 AM (Life Technologies) (5 μM) for 60 min at room temperature in balanced salt solution (BSS) containing 20 mM Hepes (pH-adjusted to 7.4 with NaOH), 115 mM NaCl, 5 mM KCl, 1 mM MgCl2, 10 mM glucose, and 2 mM CaCl2. To induce Ca2+ release from the ER, cells were stimulated with 1 μM thapsigargin (Sigma-Aldrich) in BSS lacking CaCl2. Imaging data analysis was performed using MetaMorph (Molecular Devices) and Igor Pro (WaveMetrics) software.
Expression and Purification of Recombinant SERCA2b and ERdj5.
The PA-SERCA2b vector was transfected into HEK293T cells to establish a cell line stably expressing PA-SERCA2b. High expression of PA-SERCA2b was induced with cumate, 50 ng/mL phorbol 12-myristate 13-acetate (PMA), and 1 mM sodium butyrate (36, 37). The cells were solubilized with 1% n-dodecyl-β-d-maltoside (DDM), 50 mM Hepes (pH 7.0), 100 mM NaCl, 20% (vol/vol) glycerol, 1 mM CaCl2, and 1 mM MgCl2. PA-SERCA2b was bound to anti-PA tag NZ-1-Sepharose and eluted with 0.2 mg/mL PA14 peptide, as described previously (34). The eluted PA-SERCA2b sample was treated with 1 mM diamide or 1 mM DTT for 1 h at 4 °C for full oxidation or reduction of the Cys875–Cys887 pair, respectively, and then loaded onto a Superose6 10/300 GL column (GE Healthcare) pre-equilibrated with 50 mM Hepes (pH 7.0), 100 mM NaCl, 20% (vol/vol) glycerol, 1 mM CaCl2, 1 mM MgCl2, and 0.1% DDM.
C-terminally FLAG-tagged ERdj5 and its cysteine mutant (ERdj5/SS) (12, 13) were inserted into the PiggyBac Cumate Switch Inducible Vector (System Biosciences). The resultant vector was transfected into HEK293T cells to generate cell lines stably overexpressing ERdj5-FLAG or ERdj5-FLAG/SS. The cells were induced with 10× cumate and 50 ng/mL PMA, and the harvested cells were solubilized with 2% (wt/vol) DDM, 50 mM Tris⋅HCl (pH 8.0), 500 mM NaCl, 10% (vol/vol) glycerol, 1 mM EDTA, 1 mM DTT, and 1 mM PMSF. ERdj5-FLAG was purified with anti-FLAG agarose (Medical & Biological Laboratories). The eluted sample was concentrated in the presence of 1 M Non-Detergents Sulfobetaines-201 (NDSB-201) and then loaded onto a Superdex200 10/300 GL column (GE Healthcare) pre-equilibrated with 20 mM Hepes (pH 8.0), 0.05% Tween-20, 10% (vol/vol) glycerol, and 1 M NDSB. The purity of the resultant samples (reduced and oxidized PA-SERCA2b, ERdj5-FLAG, and ERdj5-FLAG/SS) was assessed by reducing SDS/PAGE (Fig. S4).
ATPase Activity Measurement of SERCA2b With/Without ERdj5.
The purified PA-SERCA2b samples were incubated in buffer containing 1 mM MgCl2, 10 μM CaCl2, and 0.1% DDM for 10 min at 37 °C. After incubation, 1 mM ATP was added to initiate the SERCA2b ATPase cycle. The rereduced SERCA2b was prepared by incubation of oxidized SERCA2b with 1 mM DTT for 1 h at 4 °C. Reduced forms of ERdj5-FLAG and ERdj5-FLAG/SS were prepared by treatment with 1 mM DTT for 1 h at 4 °C. Oxidized PA-SERCA2b (50 nM) was incubated with 0, 12.5, 25, 50, 125, and 250 nM of the reduced form of ERdj5-FLAG or ERdj5-FLAG/SS for 10 min at 37 °C. After incubation, 1 mM ATP was added to initiate the SERCA2b ATPase cycle. The phosphate group released during this reaction cycle was quantified using the EnzCheck Phosphate Assay Kit (Life Technologies). Briefly, free phosphate groups generated upon ATP hydrolysis by SERCA2b reacted with 2-amino-6-mercapto-7-methylpurine riboside (MESG), resulting in a prominent absorbance peak of the MESG product at 360 nm, which was recorded on a U3310 spectrophotometer (Hitachi). The concentration of the released phosphate groups was calculated using an absorbance at 360 nm of 3.4 × 103/M.
DLS Measurement.
His-ERdj5/SS was overexpressed in Escherichia coli and purified as described previously (13) and used for DLS measurement. The size distributions of ERdj5/SS were determined using a Malvern Zetasizer Nano ZS (Malvern Instruments) essentially as described previously (38). Briefly, the polydispersity/heterogeneity of Brownian movements was calculated by the SD value of the distribution at each calcium concentration. The Z-average diameter, which is the mean diameter of an ensemble of particles in solution, was calculated by the slope of the linearized form of the correlation function. The hydrodynamic diameter was converted by the Stokes–Einstein equation. ERdj5/SS or PDI (50 μM of each protein) dissolved in 20 mM Hepes (pH 7.5) containing 10% (wt/vol) glycerol, 0.5% (wt/vol) Tween-20, and 1 M NDSB was centrifuged at 15,000 × g for 15 min at 4 °C. The supernatants were filtered through membranes with a pore size of 0.22 mm. Calcium chloride (0.5, 1, 2.5, 5, and 10 mM) was titrated into the sample, and DLS measurements were performed at 20 °C after incubation for 10 min.
Detection of the Redox States of SERCA2b and ERdj5.
Cells in suspension were either untreated or treated with 10 mM DTT or 1 mM dipyridyl disulfide and then precipitated with 10% (vol/vol) TCA on ice for 20 min. After centrifugation at 12,000 × g, cell pellets were washed twice with acetone, washed again with dimethyl ether, and dried completely. To determine the protein redox state in vivo, proteins in pellets were lysed and modified by incubation for 30 min at room temperature in buffer containing 30 mM AMS (Life Technologies) or mPEG2000-mal (Nichiyu) and 1% SDS. In the case of SERCA2b, the lysates were precipitated again with 10% (vol/vol) TCA and washed with acetone and dimethyl ether, as described above. To concentrate the PA-tagged SERCA2b, dried pellets are resuspended with lysis buffer and immunoprecipitated with PA antibody-conjugated beads. The immunoprecipitants were incubated overnight at 37 °C in buffer containing Arg-C protease (Promega). Samples were adjusted to contain 1× Laemmli buffer for SDS/PAGE and subjected to immunoblotting with the anti-PA tag antibody NZ-1.
Pull-Down Assay.
FLAG-tagged SERCA2b or ERdj5 transiently expressed in HEK293T cells was immunoprecipitated with Dynabeads Protein G (Thermo Fisher) conjugated with the anti-FLAG M2 antibody. Lysates of cells transfected with Myc-tagged ERdj5 were incubated with titrated CaCl2 and 1 mM EGTA for 1 h at 4 °C. The conjugated beads were mixed with cell lysates and incubated with rotation for 10 min at room temperature. Coprecipitated ERdj5-Myc was resolved by SDS/PAGE and immunoblotted with an anti-Myc antibody.
Sucrose Density Gradient Centrifugation.
Cell lysates were applied to a 10–40% sucrose density gradient created using a Gradient Master (BioComp). Lysates were centrifuged at 36,000 × g for 16 h. Each fraction of 250 μL collected from the top was adjusted to contain 1× Laemmli buffer for separation by SDS/PAGE.
Acknowledgments
We thank Takao Iwawaki (Advanced Scientific Research Leaders Development Unit, Gunma University) and Linda Hendershot (Department of Tumor Cell Biology, St. Jude Children’s Research Hospital) for sharing ERdj5-deficient MEFs and BiP constructs, respectively. R.U. is supported by the Japan Society for the Promotion of Science (JSPS), a Grant-in-Aid for Young Scientists (B) (Grant Number 25840079), and Grants-in-Aid for Scientific Research on Innovative Areas (Grants 15H01545, 26111521, and 24121725). K.N. was supported by the JSPS and a Grant-in-Aid for Scientific Research (S) (Grant 24227009). This research was partially supported by the Platform Project for Supporting in Drug Discovery and Life Science Research from the Ministry of Education, Culture, Sports, Science and Technology and the Japan Agency for Medical Research and Development (AMED).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605818113/-/DCSupplemental.
References
- 1.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1(1):11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: Dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4(7):517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 3.Miyawaki A, Furuichi T, Maeda N, Mikoshiba K. Expressed cerebellar-type inositol 1,4,5-trisphosphate receptor, P400, has calcium release activity in a fibroblast L cell line. Neuron. 1990;5(1):11–18. doi: 10.1016/0896-6273(90)90029-f. [DOI] [PubMed] [Google Scholar]
- 4.Lai FA, Erickson HP, Rousseau E, Liu QY, Meissner G. Purification and reconstitution of the calcium release channel from skeletal muscle. Nature. 1988;331(6154):315–319. doi: 10.1038/331315a0. [DOI] [PubMed] [Google Scholar]
- 5.Inui M, Saito A, Fleischer S. Purification of the ryanodine receptor and identity with feet structures of junctional terminal cisternae of sarcoplasmic reticulum from fast skeletal muscle. J Biol Chem. 1987;262(4):1740–1747. [PubMed] [Google Scholar]
- 6.Burdakov D, Petersen OH, Verkhratsky A. Intraluminal calcium as a primary regulator of endoplasmic reticulum function. Cell Calcium. 2005;38(3-4):303–310. doi: 10.1016/j.ceca.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Verkhratsky A. Physiology and pathophysiology of the calcium store in the endoplasmic reticulum of neurons. Physiol Rev. 2005;85(1):201–279. doi: 10.1152/physrev.00004.2004. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Camacho P. Ca2+-dependent redox modulation of SERCA 2b by ERp57. J Cell Biol. 2004;164(1):35–46. doi: 10.1083/jcb.200307010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michalak M, Robert Parker JM, Opas M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium. 2002;32(5-6):269–278. doi: 10.1016/s0143416002001884. [DOI] [PubMed] [Google Scholar]
- 10.Oda Y, Hosokawa N, Wada I, Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299(5611):1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- 11.Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299(5611):1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- 12.Ushioda R, et al. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321(5888):569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 13.Hagiwara M, et al. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol Cell. 2011;41(4):432–444. doi: 10.1016/j.molcel.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Ushioda R, Hoseki J, Nagata K. Glycosylation-independent ERAD pathway serves as a backup system under ER stress. Mol Biol Cell. 2013;24(20):3155–3163. doi: 10.1091/mbc.E13-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roderick HL, Lechleiter JD, Camacho P. Cytosolic phosphorylation of calnexin controls intracellular Ca(2+) oscillations via an interaction with SERCA2b. J Cell Biol. 2000;149(6):1235–1248. doi: 10.1083/jcb.149.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyoshima C. Structural aspects of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. Arch Biochem Biophys. 2008;476(1):3–11. doi: 10.1016/j.abb.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 17.Toyoshima C. How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim Biophys Acta. 2009;1793(6):941–946. doi: 10.1016/j.bbamcr.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Beard NA, Sakowska MM, Dulhunty AF, Laver DR. Calsequestrin is an inhibitor of skeletal muscle ryanodine receptor calcium release channels. Biophys J. 2002;82(1 Pt 1):310–320. doi: 10.1016/S0006-3495(02)75396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Kelley J, Schmeisser G, Kobayashi YM, Jones LR. Complex formation between junctin, triadin, calsequestrin, and the ryanodine receptor. Proteins of the cardiac junctional sarcoplasmic reticulum membrane. J Biol Chem. 1997;272(37):23389–23397. doi: 10.1074/jbc.272.37.23389. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, et al. Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum. Nat Struct Biol. 1998;5(6):476–483. doi: 10.1038/nsb0698-476. [DOI] [PubMed] [Google Scholar]
- 21.Fryer MW, Stephenson DG. Total and sarcoplasmic reticulum calcium contents of skinned fibres from rat skeletal muscle. J Physiol. 1996;493(Pt 2):357–370. doi: 10.1113/jphysiol.1996.sp021388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellman R, Vanhove M, Lejeune A, Stevens FJ, Hendershot LM. The in vivo association of BiP with newly synthesized proteins is dependent on the rate and stability of folding and not simply on the presence of sequences that can bind to BiP. J Cell Biol. 1999;144(1):21–30. doi: 10.1083/jcb.144.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avezov E, et al. Lifetime imaging of a fluorescent protein sensor reveals surprising stability of ER thiol redox. J Cell Biol. 2013;201(2):337–349. doi: 10.1083/jcb.201211155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avezov E, et al. Retarded PDI diffusion and a reductive shift in poise of the calcium depleted endoplasmic reticulum. BMC Biol. 2015;13:2. doi: 10.1186/s12915-014-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toyoshima C, Cornelius F. New crystal structures of PII-type ATPases: Excitement continues. Curr Opin Struct Biol. 2013;23(4):507–514. doi: 10.1016/j.sbi.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Toyoshima C, Yonekura S, Tsueda J, Iwasawa S. Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+ Proc Natl Acad Sci USA. 2011;108(5):1833–1838. doi: 10.1073/pnas.1017659108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bublitz M, et al. Ion pathways in the sarcoplasmic reticulum Ca2+-ATPase. J Biol Chem. 2013;288(15):10759–10765. doi: 10.1074/jbc.R112.436550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandecaetsbeek I, et al. Structural basis for the high Ca2+ affinity of the ubiquitous SERCA2b Ca2+ pump. Proc Natl Acad Sci USA. 2009;106(44):18533–18538. doi: 10.1073/pnas.0906797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakuntabhai A, et al. Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nat Genet. 1999;21(3):271–277. doi: 10.1038/6784. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Perez VL, et al. ATP2A2 mutations in Darier’s disease: Variant cutaneous phenotypes are associated with missense mutations, but neuropsychiatric features are independent of mutation class. Hum Mol Genet. 1999;8(9):1621–1630. doi: 10.1093/hmg/8.9.1621. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi H, et al. Novel mutations of ATP2A2 gene in Japanese patients of Darier’s disease. J Dermatol Sci. 2001;26(3):169–172. doi: 10.1016/s0923-1811(00)00173-0. [DOI] [PubMed] [Google Scholar]
- 32.Wada T, et al. A Japanese case of segmental Darier’s disease caused by mosaicism for the ATP2A2 mutation. Br J Dermatol. 2003;149(1):185–188. doi: 10.1046/j.1365-2133.2003.05412.x. [DOI] [PubMed] [Google Scholar]
- 33.Hosoda A, Tokuda M, Akai R, Kohno K, Iwawaki T. Positive contribution of ERdj5/JPDI to endoplasmic reticulum protein quality control in the salivary gland. Biochem J. 2009;425(1):117–125. doi: 10.1042/BJ20091269. [DOI] [PubMed] [Google Scholar]
- 34.Fujii Y, et al. PA tag: A versatile protein tagging system using a super high affinity antibody against a dodecapeptide derived from human podoplanin. Protein Expr Purif. 2014;95:240–247. doi: 10.1016/j.pep.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Gurney AM, Drummond RM, Fay FS. Calcium signalling in sarcoplasmic reticulum, cytoplasm and mitochondria during activation of rabbit aorta myocytes. Cell Calcium. 2000;27(6):339–351. doi: 10.1054/ceca.2000.0124. [DOI] [PubMed] [Google Scholar]
- 36.Hao QL, et al. Expression of biologically active human factor IX in human hematopoietic cells after retroviral vector-mediated gene transduction. Hum Gene Ther. 1995;6(7):873–880. doi: 10.1089/hum.1995.6.7-873. [DOI] [PubMed] [Google Scholar]
- 37.Laubach VE, Garvey EP, Sherman PA. High-level expression of human inducible nitric oxide synthase in Chinese hamster ovary cells and characterization of the purified enzyme. Biochem Biophys Res Commun. 1996;218(3):802–807. doi: 10.1006/bbrc.1996.0143. [DOI] [PubMed] [Google Scholar]
- 38.Okumura M, et al. Inhibition of the functional interplay between endoplasmic reticulum (ER) oxidoreduclin-1α (Ero1α) and protein-disulfide isomerase (PDI) by the endocrine disruptor bisphenol A. J Biol Chem. 2014;289(39):27004–27018. doi: 10.1074/jbc.M114.564104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biasini M, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]