Significance
The bacterial envelope is a specialized compartment that plays a central role in energy production, cell–cell communication, and transport of nutrients into the cell and disposal of waste or toxic cell products. Despite this central role, however, only a few signal-specific envelope-sensing transduction systems are present in a typical bacterial genome. This may limit the cellular ability to mount the appropriate response to changes in its surroundings. In this work, we establish that the coordinated interaction between a broad envelope stress transduction system and a signal-specific cytoplasmic sensor integrates a precise signal- and compartment-restricted output. These types of concerted interactions contribute to expanding the spectra of monitored conditions within the bacterial envelope, favoring bacterial fitness and survival in a challenging environment.
Keywords: Salmonella, transcriptional codependence, periplasmic copper homeostasis, CpxR/CpxA, CueP
Abstract
Copper homeostasis is essential for bacterial pathogen fitness and infection, and has been the focus of a number of recent studies. In Salmonella, envelope protection against copper overload and macrophage survival depends on CueP, a major copper-binding protein in the periplasm. This protein is also required to deliver the metal ion to the Cu/Zn superoxide dismutase SodCII. The Salmonella-specific CueP-coding gene was originally identified as part of the Cue regulon under the transcriptional control of the cytoplasmic copper sensor CueR, but its expression differs from the rest of CueR-regulated genes. Here we show that cueP expression is controlled by the concerted action of CueR, which detects the presence of copper in the cytoplasm, and by CpxR/CpxA, which monitors envelope stress. Copper-activated CueR is necessary for the appropriate spatial arrangement of the −10 and −35 elements of the cueP promoter, and CpxR is essential to recruit the RNA polymerase. The integration of two ancestral sensory systems—CueR, which provides signal specificity, and CpxR/CpxA, which detects stress in the bacterial envelope—restricts the expression of this periplasmic copper resistance protein solely to cells encountering surplus copper that disturbs envelope homeostasis, emulating the role of the CusR/CusS regulatory system present in other enteric bacteria.
The bacterial envelope is a specialized compartment interacting with both the surrounding environment and the cytoplasm. It plays a central role in energy production and cell–cell communication, and actively controls the transport of nutrients and waste or toxic products (1). It is in this compartment where most of the perception of the bacterial surroundings takes place. Sensory devices, usually periplasmic-protruding inner-membrane histidine kinases or antisigma factors, detect changes in the environment and transduce this information to cytoplasmic effectors, usually transcriptional regulators (2, 3). This simple array provides an efficient and rapid response to modulate the expression of factors required to cope with a continuously challenging environment.
Dedicated signal-transduction systems are responsible for the homeostatic maintenance of specific components in this compartment (4, 5). One of these components is the transition redox copper ion. This essential ion participates in enzymatic reactions carried out by periplasmic cuproproteins, including cytochrome oxidases, NADH dehydrogenases, Cu,Zn-superoxide dismutases, laccases, and multicopper oxidases, among others (6). It is at the same time extremely reactive, causing damage to proteins, lipids, and other cellular components. Most enterobacterial species harbor a copper-responsive two-component system, CusR/CusS, to control copper levels in the cell envelope (7, 8). CusR/CusS responds to the surplus of periplasmic copper, inducing the expression of the CBA-type efflux complex, CusC(F)BA, that pumps the excess metal ion out of the cell (9). In contrast, the cytoplasm is not predicted to require copper (10, 11). This compartment is monitored by the copper-efflux regulator CueR, a MerR-like transcription factor that mounts the response to eliminate the toxic ion from the cytoplasm (12); thus, the independent monitoring of copper in each compartment provides a physiological advantage, allowing maintenance of the appropriate quota in the envelope and its exclusion from the cytoplasm.
Salmonella lacks both the genes coding for CusR/CusS and the operon encoding the CusC(F)BA efflux complex (8, 13). Different lines of evidence suggest that the CueR-induced periplasmic protein (CueP), product of the cueP gene, plays a major role in maintaining copper homeostasis in the Salmonella envelope. It participates in copper resistance, particularly in anaerobiosis (8, 14–16), and cueP is usually present in species lacking the cus locus (8).
Here we report that cueP expression requires both the presence of the toxic ion and the stress caused by the excess copper in the periplasm. We show that cueP induction requires the simultaneous activation of CueR and the CpxR/CpxA two-component system (17, 18). The absence of either CueR or CpxR abrogates cueP induction. We show that CueR is necessary for the appropriate spatial arrangement of the −10 and −35 elements in the cueP promoter, and that CpxR is essential for the recruitment of the RNA polymerase. This coregulation restricts the expression of CueP to cells encountering the copper excess that causes envelope stress. Given that the number of specific envelope-sensing signal transduction systems present in a typical bacterial genome is limited, coordinated interactions between an envelope stress detector and a signal-specific cytoplasmic sensor contribute to fine-tune the damage responses in the periplasmic compartment.
Results
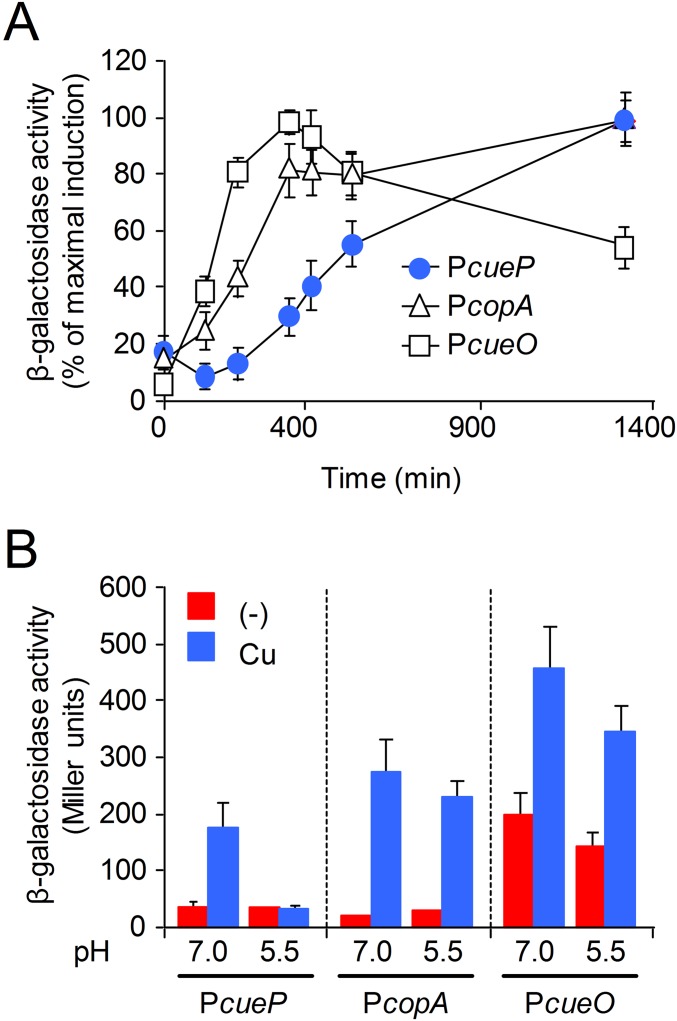
Differential Induction of cueP Within the Cue Regulon.
Several lines of evidence show that CueP parallels the enterobacterial Cus system. CueP is necessary for copper resistance in anaerobiosis, whereas its role in aerobic conditions is masked by other components of the Cue regulon (7, 8, 19). A mutant lacking CueP has an increased cellular copper load (15) and delayed copper-induced expression compared with the other Cue components (Fig. S1A), resembling the expression kinetics of the Escherichia coli cus locus (20). In addition, and unlike the other genes of the Cue regulon, its induction by copper is observed only in neutral conditions (pH 7.0), and not in acidic conditions (pH 5.5) (Fig. S1B).
Fig. S1.
Delayed expression and pH dependence of cueP. (A) Time course of CueR-controlled induction of cueP::lacZ, copA::lacZ, and cueO::lacZ transcriptional fusions in the presence of copper. Aliquots of overnight cells grown in LB were diluted in the same fresh medium containing 1 mM CuSO4 and incubated with shaking at 37 °C. At the indicated times, aliquots were withdrawn from the culture to determine OD630 and β-gal activity. The activity was normalized against the maximal response obtained for each transcriptional fusion. Data correspond to mean values of at least four independent experiments performed in duplicate. Error bars correspond to SD. (B) β-Gal activity from the transcriptional fusions shown in A. Cells were grown overnight in LB broth at pH 7.0 or pH 5.5 and without (−) or with the addition of 1 mM CuSO4 (Cu). The data correspond to mean values of three independent experiments performed in duplicate. Error bars represent SD.
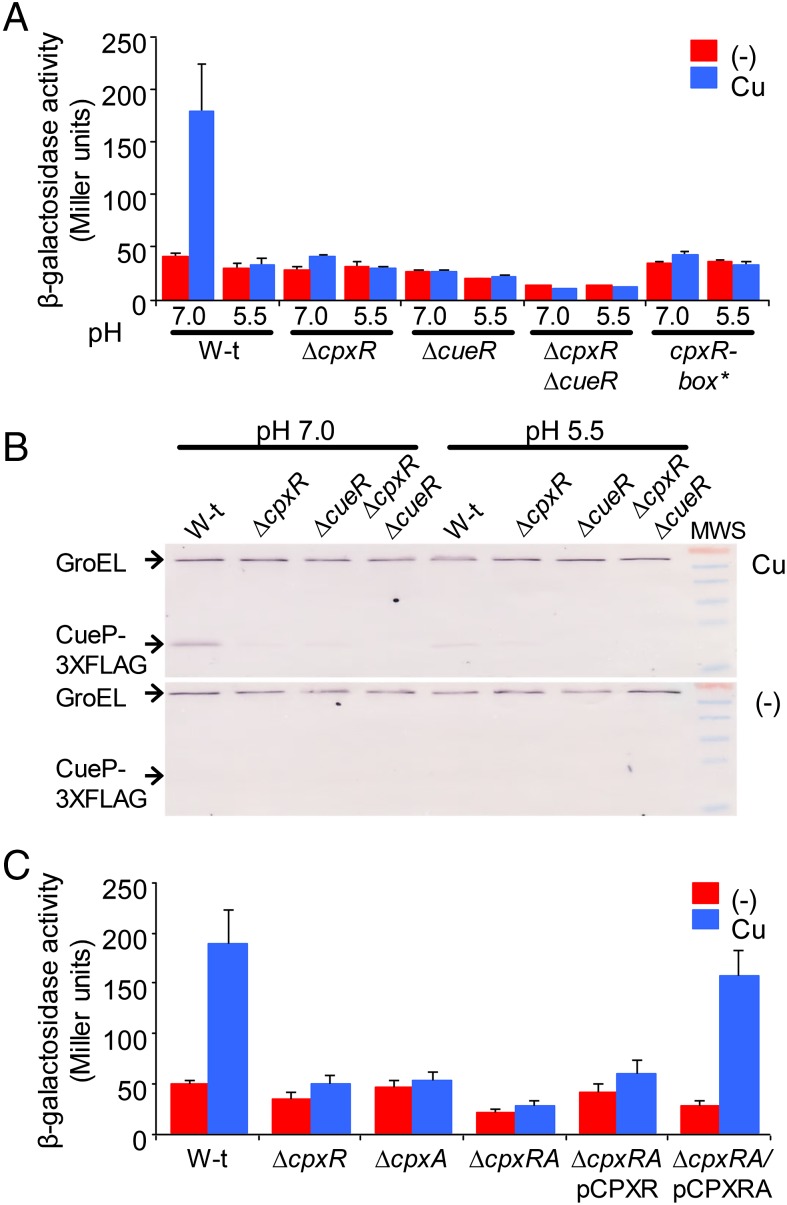
Copper-Induced cueP Expression Requires the Envelope-Stress Response CpxR/CpxA Regulatory System.
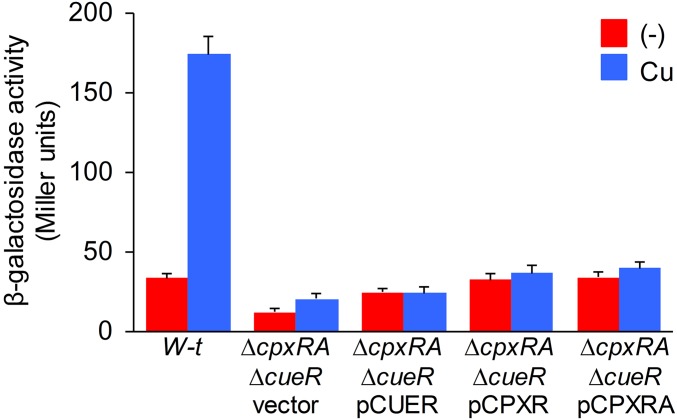
We and others have shown that copper can induce genes of the CpxR/CpxA regulon (21–23). This, along with the pH-dependence in cueP expression, another CpxR/CpxA-activating condition (24), prompted us to investigate the contribution of this regulatory system to cueP expression. Deletion of the response regulator gene cpxR and/or the sensor kinase coding gene cpxA abrogated the metal induction of cueP at pH 7.0 (Fig. 1). Remarkably, the absence of either CpxR or CueR hampered the metal induction of CueP (Fig. 1B), and overexpression of CueR or of CpxRA did not promote cueP expression in the ΔcueR ΔcpxRA double-mutant strain (Fig. S2), indicating that both factors are simultaneously required for the copper-promoted expression of this periplasmic protein.
Fig. 1.
Copper-induced expression of cueP depends on CpxR. (A) β-gal activity from a cueP::lacZ transcriptional fusion expressed on wild-type (W-t), ΔcpxR, ΔcueR, ΔcpxR ΔcueR, and cpxR-box* cells grown overnight in LB broth at pH 7.0 or pH 5.5 and without (−) or with the addition of 1 mM CuSO4 (Cu). The data correspond to mean values of four independent experiments performed in duplicate. Error bars represent SD. (B) Analysis of the expression of CueP-FLAG, using anti-FLAG antibodies. Here 20 μg of total crude extract protein of wild-type, ΔcpxR, ΔcueR, and ΔcpxR ΔcueR cells grown overnight in LB broth at pH 7.0 or pH 5.5 and without (−) or with (Cu) the addition of 1 mM CuSO4 was analyzed by SDS/PAGE, followed by transfer to nitrocellulose and development using monoclonal anti-FLAG antibodies. The PageRuler prestained protein ladder provided molecular weight standards. From top to bottom, bands of 70, 55, 40, 35, 25, and 15 kDa are shown. (C) β-Gal activity from the cueP::lacZ transcriptional fusion expressed on wild-type, ΔcpxR, ΔcpxA, ΔcpxRA, and cpxRA strains complemented with pCPXR (ΔcpxR/pCPXR) or with pCPXRA (ΔcpxR/pCPXRA) without (−) or with the addition of 1 mM CuSO4 (Cu). The data correspond to mean values of four independent experiments performed in duplicate. Error bars represent SD.
Fig. S2.
Copper-induced cueP expression requires the activation of both CueR and CpxR. Shown is β-gal activity from a cueP::lacZ transcriptional fusion expressed in the wild-type (W-t) and the ΔcpxRA ΔcueR strain complemented with pUHE21-2lacIq (vector), pCUER, pCPXR, or pCPXRA, without (−) or with the addition of 1 mM CuSO4 (Cu). The data correspond to mean values of four independent experiments performed in duplicate. Error bars represent SD.
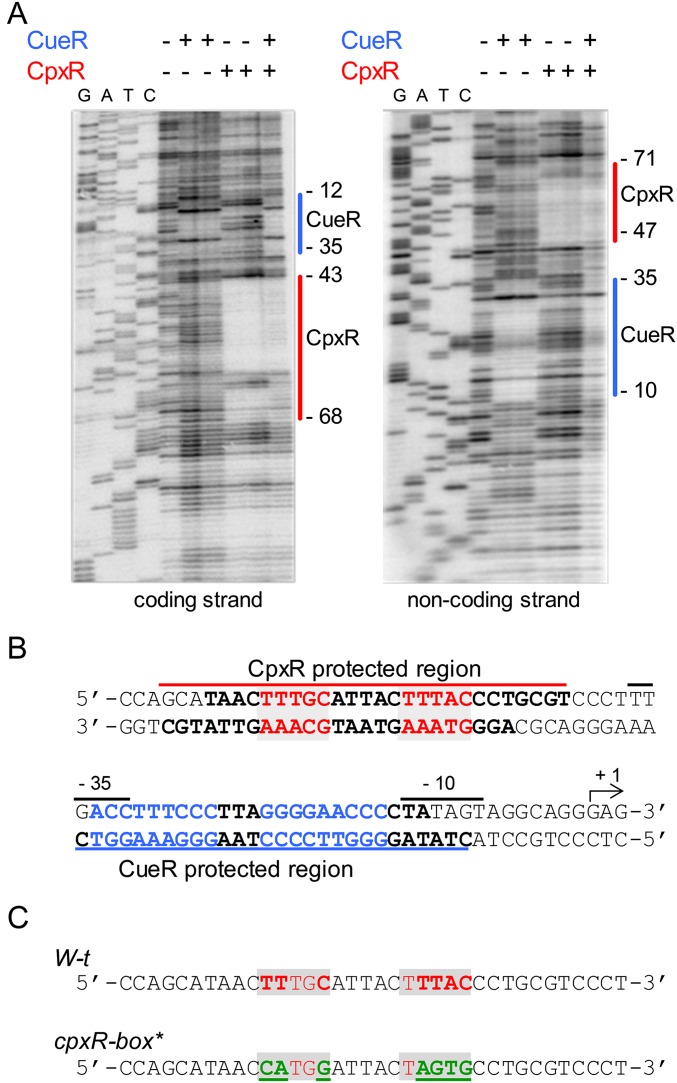
An in silico analysis identified a putative CpxR-binding site between nucleotides −64 and −50 relative to the cueP transcription start site. DNase I footprinting analysis (Fig. S3A) showed that CpxR protected from nucleotide −43 to nucleotide −68 relative to the transcription start site of the promoter in the coding strand, and from nucleotide −47 to nucleotide −71 in the noncoding strand, with an overlap of 22 bp between the two strands in which the predicted Cpx-binding box was centered (Fig. S3B). The CpxR-protected region was 6 nt upstream of the region protected by CueR, and there was no interference in the simultaneous interaction of both regulators with the promoter fragment (Fig. S3A). A chromosomal cpxR-box* mutant strain was generated, replacing the predicted CpxR-binding motif TTtgC-n5-tTTAC with the CAtgG-n5-tAGTG sequence (Fig. S3C). Copper-induced expression of cueP was abrogated in this mutant strain (Fig. 1A), confirming the requirement of this direct-repeat region for CpxR recognition.
Fig. S3.
Mapping the CpxR- and CueR-binding sites by DNA footprinting analysis of the cueP promoter region. (A) DNA footprint analysis of the cueP promoter region performed on both end-labeled coding and noncoding strands. The DNA fragments were incubated with purified CueR at final concentrations of 0.25 and 0.5 μM, or with purified and acetyl phosphate-preincubated CpxR at final concentrations of 0.5 and 1 μM. Solid lines indicate the CueR- and CpxR-protected regions. (B) DNA sequence of the cueP promoter region shown in the CpxR- and CueR-protected regions. Lines above or below the sequence indicate DNase I-protected regions; in each strand, the protected sequence is in bold type. The predicted CpxR direct repeat is shown in red and shaded; the CueR operator is indicated in blue. (C) Comparison of DNA sequences of the WT cueP promoter and the generated promoter with mutations in the predicted CpxR-binding site (CpxR-box*). The mutated bases are depicted in bold green characters and underscored.
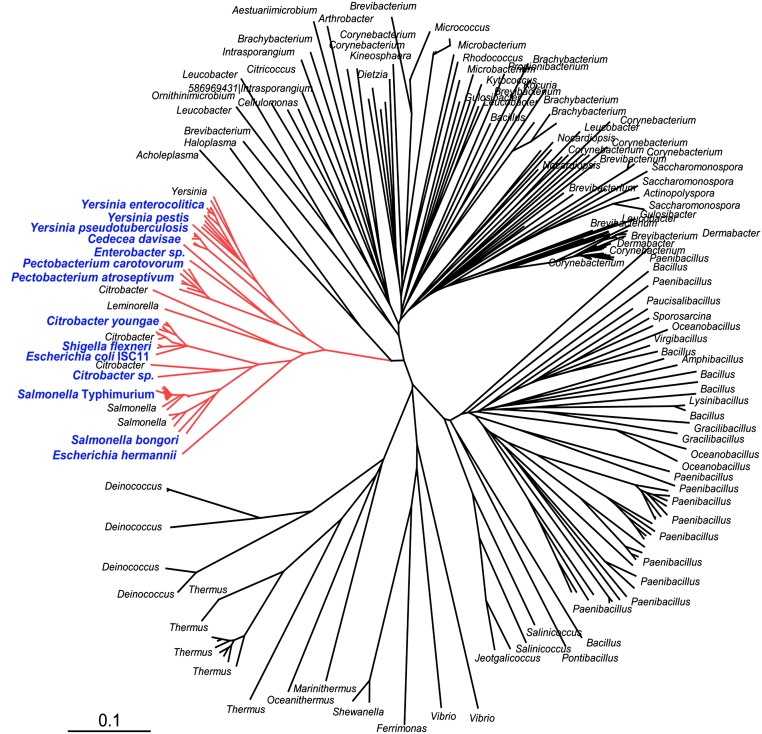
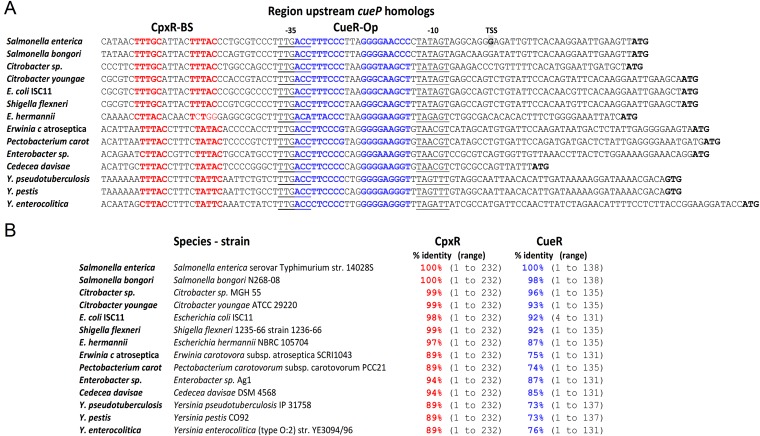
Although characterized only in Salmonella, genes coding for CueP homologs are present in species of Actinobacteria, Deinococcus-Thermus, Firmicutes, Tenericutes, and Proteobacteria (Fig. S4). The presence of CueP homologs in Proteobacteria is restricted to a few species of the Gammaproteobacterial class and within this, mostly to enterobacterial species, including Salmonella. The region upstream of cueP in this cluster contains a highly conserved CueR operator, an almost identical −35 promoter element, and a CpxR-binding site (Fig. S5), reflecting a conserved selective pressure to maintain the coordinated expression control of these xenolog proteins during evolution.
Fig. S4.
Phylogenetic tree obtained by comparison of CueP-homologs. The tree was constructed by Bayesian inference as described previously (33, 34). For simplicity, only reference species are shown. The proteobacterial branch is depicted in red. Blue letters indicate the upstream sequences of cueP homologs selected for the alignments shown in Fig. S5A.
Fig. S5.
Identification of CueR operators and CpxR-binding sites at the promoter regions of cueP-encoded enterobacterial species. (A) Alignment of the predicted promoter regions of enterobacterial cueP homologs. Both the predicted CueR operator (bold/blue) and CpxR-binding sites (bold/red) are highlighted. (B) The enterobacterial strain chosen at random to analyze the cueP promoter, as well as the presence of CpxR and CueR homologs. The identity percentage against Salmonella Typhimuriun 14028s CpxR and CueR, as well as the range of amino acid identities, are indicated in each case. Sequences were obtained from the following strain genomes: CP001363.1, Salmonella enterica subsp. enterica serovar Typhimurium strain 14028S; NC_021870.1, Salmonella bongori N268-08; NZ_JMU.K.00000000.1, Citrobacter sp. MGH 55; NZ_ABWL00000000.2, Citrobacter youngae ATCC 29220; E. coli ISC11 WGS project CBWP000000000 data, contig c7; AKNF01001182.1, Shigella flexneri 1235–66 strain 1236–66; BAFF01000002.1, Escherichia hermannii NBRC 105704; NC_004547.2, Erwinia carotovora subsp. atroseptica SCRI1043; NC_018525.1, Pectobacterium carotovorum subsp. carotovorum PCC21; NZ_AKXM01000034.1, Enterobacter sp. Ag1 Contig35; NZ_KE161030.1, Cedecea davisae DSM 4568; NC_009708.1, Yersinia pseudotuberculosis IP 31758; CP009973.1, Yersinia pestis CO92; NZ_CAQH00000000.1, and Yersinia enterocolitica (type O:2) strain YE3094/96. Both the predicted CueR operator (bold/blue) and CpxR-binding sites (bold/red) are highlighted.
Coordinated Regulation of cueP Is Required for Copper Resistance.
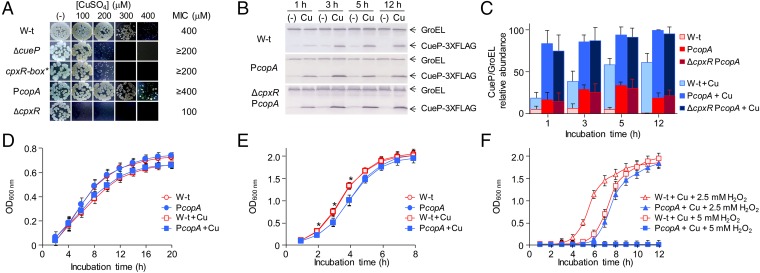
CueP is a copper-binding protein (14, 16) required for resistance to the metal ion (8). Its presence alleviates copper stress (25) and reduces the cellular content of the ion (15). As expected for its reduced expression, the cpxR-box* mutant showed a decreased resistance to copper under anaerobic conditions (Fig. 2A), indicating that its CpxR activation is essential for copper resistance. Furthermore, a ΔcpxR mutant displayed a marked sensitivity to the metal ion (Fig. 2A) (26), suggesting that other factors under CpxR control are also involved in the resistance to copper.
Fig. 2.
Coordinated regulation of cueP is required for optimal growth. (A) Comparative Cu MIC values in wild-type (W-t) Salmonella, ΔcueP, cpxR-box*, PcopA, and ΔcpxR mutant strains on LB plates containing increasing amounts of CuSO4 in anaerobic conditions. (B) Detection of CueP in whole-cell extracts obtained from the cueP-3×FLAG (WT), the PcopA-cueP-3×FLAG mutant strain (PcopA), and the PcopA-cueP-3×FLAG in the ΔcpxR mutant strain (ΔcpxR PcopA), grown in LB without or with the addition of 1 mM CuSO4. Here, 20 µg of total protein cell extracts was analyzed by SDS/PAGE, followed by transfer to nitrocellulose and development using monoclonal anti-FLAG antibodies. CueP relative levels were normalized to GroEL. (C) Mean value of three biological replicates analyzed in duplicate in each case. (D and E) Wild-type Salmonella and the PcopA-cueP mutant strain growth in static/microaerobic (D) or in aerobic conditions (E) in LB without or with the addition of sublethal concentrations of CuSO4. The data correspond to mean values of at least four independent experiments performed in duplicate. *Significant differences in growth (P < 0.005) from the wild-type to the PcopA mutant strains grown aerobically in either the absence or the presence of Cu. (F) Wild-type Salmonella and the PcopA-cueP mutant strain growth in LB with sublethal concentrations of CuSO4 and with the addition of 2.5 or 5 mM H2O2. The data correspond to mean values of at least four independent experiments performed in duplicate.
Because CueP is important for copper resistance under anaerobiosis but dispensable under aerobiosis (25), we examined whether its unregulated expression could affect growth under aerobic or even more oxidative conditions. For this, we constructed a mutant strain with cueP under control of the CpxR-independent copA promoter (PcopA), a promoter that responds to Cu-CueR and has similar levels of metal induction as the native cueP promoter at neutral pH (Fig. S1B). Earlier and increased CueP production was attained when expressed from the PcopA promoter during the early exponential growth phase (Fig. 2 B and C). We detected no differences in growth between these strains in LB under static, microaerobic conditions (Fig. 2D). Interestingly, under aerobic conditions, the PcopA strain showed a growth defect during exponential phase compared with the wild-type strain in both the absence and the presence of a sublethal (1 mM) copper concentration (Fig. 2E), and increased oxidative stress caused by the addition of H2O2 (Fig. 2F) further exacerbated the growth difference between these two strains. These results indicate that tight transcriptional control of CueP is necessary both to allow its expression under envelope copper overload and to prevent its presence when it is not required.
Productive Interaction of the RNA Polymerase with PcueP Requires Both Cu-CueR and Phosphorylated CpxR.
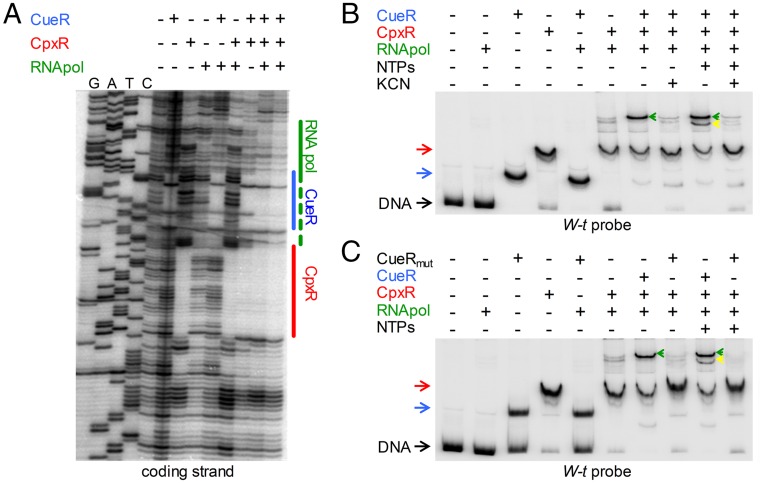
Incubation of a cueP promoter fragment containing the CpxR- and CueR-binding sites with RNA polymerase alone did not show any protection against DNase I digestion (Fig. 3A), supporting the need for additional factors to favor this fragment’s interaction with PcueP. Addition of the RNA polymerase with either Cu-CueR or phosphorylated CpxR (CpxR∼P) did not extend the protected region generated by the regulators alone. Only the simultaneous incubation of the three factors produced an extension of the protected region toward the transcription start site (Fig. 3A, last two columns), suggesting that both Cu-CueR and CpxR∼P are simultaneously required for the productive interaction of the RNA polymerase with the DNA.
Fig. 3.
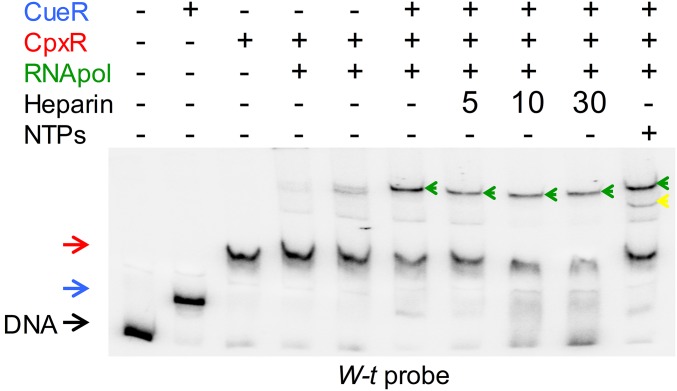
CpxR recruits the RNA polymerase to the cueP promoter. (A) DNA footprinting analysis of the cueP promoter region performed on the end-labeled coding strand. The DNA fragment was incubated when indicated with 0.5 μM of purified CueR, 1 μM of acetyl phosphate-preincubated CpxR, and 0.1 or 0.2 μM RNA polymerase (last two lines). Solid lines indicate the protected region detected in the presence of each transcription factor or the combination of CueR and CpxR. The blue, red, and green lines indicate the regions protected by CueR, CpxR, and the RNA pol, respectively. (This latter protection was observed only with the simultaneous addition of the three proteins.) (B and C) EMSA analysis performed using 6 fmol of a 32P 3′ end-labeled PCR fragment from the cueP promoter region with the addition of 0.5 μM of purified CueR (pretreated with KCN as indicated) or purified CueRC120S, 1 μM acetyl phosphate-preincubated CpxR, 0.2 μM RNA polymerase, and 250 μM NTPs, as indicated. DNA represents the unshifted PCR fragment; CueR and the CpxR shifted bands are indicated with blue and red arrows, respectively. Green and yellow arrows indicate the supershifts observed by the presence of CpxR, CueR, and the RNA polymerase or by the presence of CpxR, CueR, the RNA polymerase, and NTPs, respectively.
Confirming the foregoing observations, the RNA polymerase did not affect the electrophoretic migration of the DNA fragment in EMSAs (Fig. 3 B and C). Shifted bands were observed when either CueR or CpxR was incubated with the DNA probe. The addition of CpxR∼P, but not of Cu-CueR, together with the RNA polymerase produced a supershift of the probe (Fig. 3 B and C).
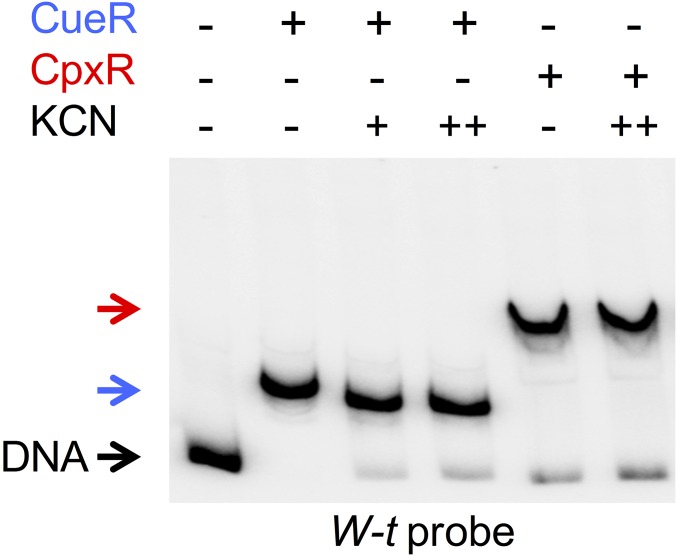
The simultaneous addition of the three factors increased the intensity of a supershifted band, with the disappearance of the remnant unshifted DNA. This supershifted band matched a transcriptional open complex as the addition of NTPs generated a faster electrophoresis migrating initiating complex (Fig. 3B), and, in contrast to the CpxR-DNA complex, remained stable after treatment with heparin (Fig. S6). Cu-activated CueR is required, because the presence of the Cu chelator KCN abrogated both the open and the initiating complexes, whereas KCN did not affect the binding capacity of either CueR or CpxR (Fig. S7). Furthermore, a mutant protein, CueRC120S, which cannot interact with Cu (27), produced a similar shift of the probe as the wild-type regulator, but was unable to generate either the open or the initiating complexes (Fig. 3C).
Fig. S6.
The CueR-CpxR-RNA polymerase-DNA complex is resistant to heparin. EMSA analysis was performed using 6 fmol of a 32P 3′ end-labeled PCR fragment from the cueP promoter region with the addition of 0.5 μM purified CueR or 25 mM acetyl phosphate-incubated CpxR, 0.2 μM RNA polymerase, and 250 μM NTPs, and treatment with a final concentration of 200 µg/mL heparin at the indicated time. DNA stands for the unshifted PCR fragment; CueR and the CpxR shifted bands, are indicated with a blue and red arrow, respectively. Green and yellow arrows indicate the supershifts observed by the presence of CpxR, CueR, and the RNA polymerase or by the presence of CpxR, CueR, the RNA polymerase, and NTPs, respectively.
Fig. S7.
KCN does not affect either CueR or CpxR DNA binding. Shown are the results of EMSA analyses performed using 6 fmol of a 32P 3′ end-labeled PCR fragment from the cueP promoter region with the addition of 0.5 μM of purified CueR or 25 mM of acetyl phosphate-incubated CpxR, and without or with a 30-min preincubation with 100 (+) or 300 µM (++) KCN. CueR and CpxR shifted bands are indicated by a blue arrow and a red arrow, respectively.
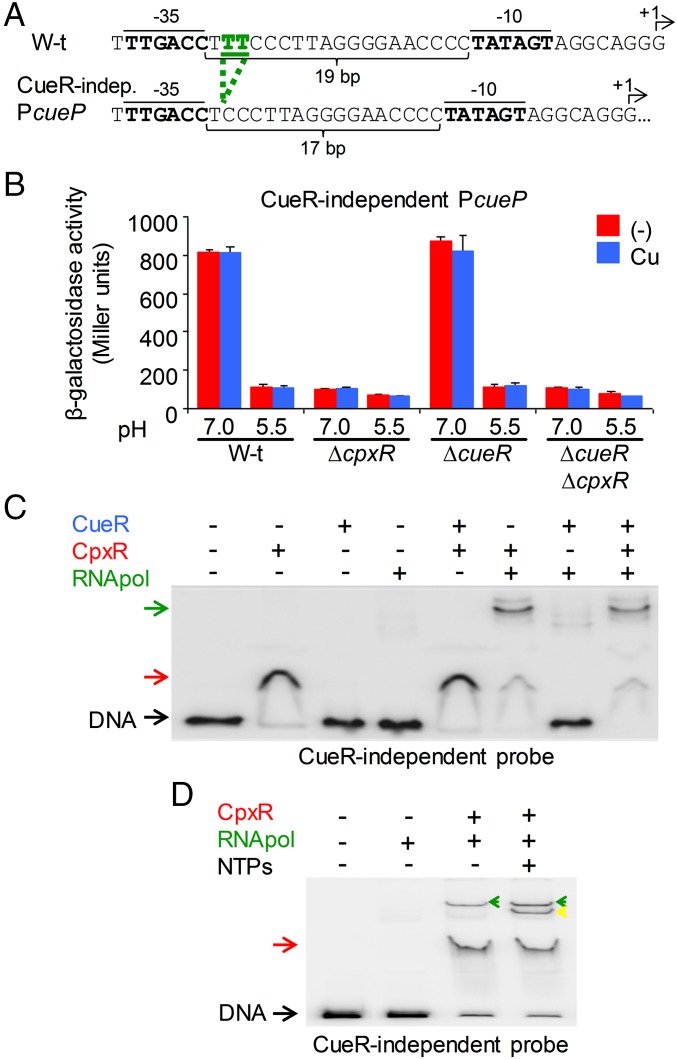
The foregoing results strongly suggest that CpxR is required for recruitment of the RNA polymerase, whereas CueR, as a typical MerR-like regulator, facilitates the spatial orientation of the −35 and −10 elements. In a CueR-dependent promoter, these elements are separated by 19 bp, a longer distance than the 17 ± 1 bp present in a typical σ70 promoter. Distortion of the DNA by copper-activated CueR is necessary for the RNA polymerase promoter recognition (12); however, in the cueP promoter, the DNA distortion may be necessary but not sufficient for its transcription. To test this, we constructed a CueR-independent promoter by shortening the distance between the −35 and −10 elements to the ideal of 17 bp (Fig. 4A). As predicted, CueR was no longer required for activation of the mutant promoter, which remained dependent on CpxR (Fig. 4B). CueR did not interact with the mutated promoter, whereas CpxR was able to recognize it (Fig. 4C). Moreover, CpxR was necessary and sufficient for the productive interaction of the RNA polymerase and the mutated promoter, promoting formation of the open and initiating complexes (Fig. 4D).
Fig. 4.
CueR facilitates the spatial orientation of the cueP promoter elements for its productive interaction with the RNA polymerase. (A) DNA sequence of the WT and CueR-independent promoter region of cueP. The two bases deleted within the spacer between the −10 and −35 RNA-polymerase elements are shown in green. The indicated 10 and −35 elements (bold) and the transcription start sites (+1 and arrows) are indicated. (B) β-Gal activity from a CueR-independent cueP::lacZ transcriptional fusion was determined on overnight cultures in LB broth at pH 7.0 or pH 5.5 and without (−) or with the addition of 1 mM CuSO4 (Cu). The data correspond to mean values of three independent experiments performed in duplicate. Error bars represent SD. (C and D) EMSA analysis performed essentially as indicated in Fig. 3B using 6 fmol of a 32P 3′ end-labeled PCR fragment from the CueR-independent cueP promoter region (C) and with the addition of NTPs (D) as indicated. Green and yellow arrows indicate the supershifts observed by the presence of CpxR and the RNA polymerase or by the presence of CpxR, the RNA polymerase, and NTPs, respectively.
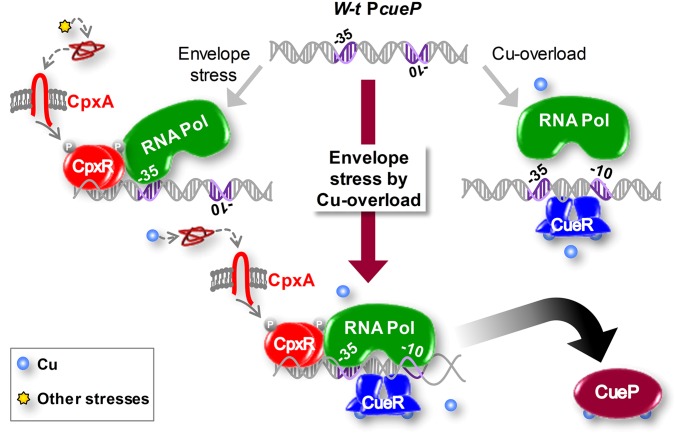
Taken together, the foregoing results indicate that the expression of cueP requires the simultaneous activation of CpxR, which recruits the RNA polymerase to the cueP promoter, and of CueR, which is necessary for the appropriate spatial arrangement of the −10 and −35 elements to initiate transcription (Fig. 5).
Fig. 5.
Proposed model for the dual control of cueP by CueR and CpxR. Activation of the Cpx signaling pathway caused by a stressor other than copper will recruit the RNA polymerase (RNA Pol) to the promoter region of cueP. Nevertheless, transcription will not occur, because of the unfavorable spatial distribution of the −10 and −35 promoter elements. Moreover, the sole activation of CueR by the presence of copper in the cytoplasm reorientates the −35 and −10 elements, but transcription is not initiated because the RNA polymerase does not recognize the promoter. Only the simultaneous activation of CpxR, by copper-induced periplasmic stress, and CueR, by cytoplasmic copper, will induce transcription of cueP. Further details are provided in the text.
Discussion
Copper is an essential metal ion, but it can be toxic even at low levels, especially when its local concentration or intracellular distribution is not properly controlled (28). The essential as well as toxic nature of copper makes its active handling a vital skill for most organisms (6, 11). Recently reported evidence indicates that intracellular copper actively contributes to the host innate immune response against bacterial infections, and that pathogens have acquired specific traits to deal with this intoxicant (29–31).
Most known bacterial cuproproteins localize to the envelope, making this compartment the main target for copper homeostasis, whereas there is no reported requirement for this metal in the bacterial cytoplasm (10, 11). In fact, it has become increasingly evident that both Gram-positive and Gram-negative bacteria make a great physiological effort to tightly control the envelope copper concentration (32). Whereas in most enteric species, periplasmic copper homeostasis is maintained by the CusR/CusS-controlled CusC(F)BA RND system, this periplasmic sensor/efflux system is absent in Salmonella (8, 19). Instead, Salmonella uses the periplasmic CueP protein to cope with copper toxicity (8). This protein is required for macrophage survival (33), coinciding with an increased copper content in the Salmonella phagosome (15). Despite its function in envelope copper handling, CueP protein expression relies on the cytoplasmic MerR-like regulator CueR (8, 34, 35); thus, why a major player of periplasmic copper homeostasis is controlled by a cytoplasmic copper sensor was not clear.
Here we have demonstrated that CueP expression is controlled not only by CueR, which is activated by the surge in cytoplasmic copper (36), but also, and simultaneously, by CpxR/CpxA, which senses alterations in envelope homeostasis, particularly in the periplasm and the inner membrane (17, 18). Whereas CpxR∼P is necessary to recruit the RNA polymerase to PcueP (Fig. 3 B and C), Cu-CueR introduces torsional stresses that kink and undertwist the promoter, reshaping the −35 and −10 promoter elements for optimal recognition by the σ70-dependent RNA polymerase (12). Indeed, shortening the distance between the −35 and −10 elements renders a promoter independent of CueR-regulation (Fig. 4B). This mutant promoter remains dependent on CpxR, facilitating the RNA polymerase–promoter interaction (Fig. 4 C and D). We show that this simultaneous regulation is required for copper resistance in anaerobiosis.
In addition, earlier expression of CueP in the exponential phase (Fig. 2 B and C) by the CopA promoter mutant (PcopA) shows a growth defect in aerobiosis (Fig. 2E), This defect was exacerbated under oxidative stress caused by H2O2 (Fig. 2F), indicating that surplus CueP can be toxic in these conditions. Furthermore, codependent transcriptional control of cueP by CueR and CpxR/CpxA is conserved among all cueP-harboring Gammaproteobacteria species (Fig. S5A), indicating a selective pressure kept in evolution to coordinate the expression of the periplasmic copper-homeostasis protein.
The growth defect in the strain expressing CueP independently of CpxR/CpxA could be caused by subtle differences in the metallation of envelope cuproproteins. As a Cu-binding protein (14, 16), the unregulated expression of CueP would restrict the availability of the metal ion for other factors. Nevertheless, attempts to determine differences in the copper content of periplasmic cuproproteins like SodCII at each growth time point were unsuccessful, likely owing to a lack of sensitivity.
A cooperative or even synergistic effect between transcription factors is a common regulatory mechanism controlling the expression of bacterial promoters. Examples include PhoP and SlyA, which countersilence the H-NS repression of horizontally acquired genes in Salmonella (37); the Escherichia coli melAB promoter, which requires the AraC/XylS-regulator MelR for positive activation by the cAMP receptor protein CRP (38); and the multiple factors acting to control expression of the master regulator of HilA, the SPI1 pathogenicity island (39). Each factor converges to enhance or reduce the transcriptional level of the target gene in response to different conditions. Remarkably, we show here the transcriptional codependence of the cueP promoter on CpxR/CpxA and CueR. By itself, neither the induction by Cu-activated CueR nor the activation of the CpxR/CpxA by a pH shift or NlpE overexpression (Fig. 1 and Fig. S8) was able to promote transcription in the absence of the other factor.
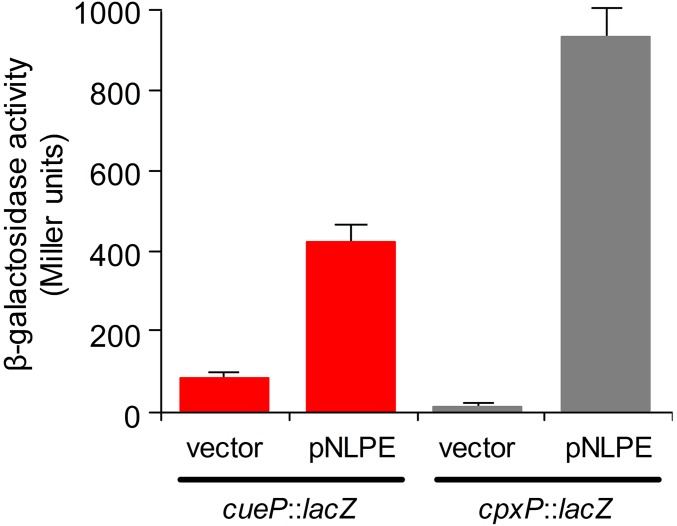
Fig. S8.
NlpE overexpression induces cueP transcription from the CpxR-dependent/CueR-independent promoter. β-gal activity from the cueP::lacZ transcriptional fusion expressed on the CueR-independent PcueP strain complemented with pUHE21-2lacIq (vector) or pNLPE at pH 5.5, with the addition of 100 µM IPTG. The CpxR-dependent cpxP::lacZ transcriptional fusion served as a positive control. The data correspond to mean values of three independent experiments performed in duplicate. Error bars represent SD.
A small amount of CueP is produced in wild-type as well as in ΔcpxR or ΔcueR strains, both in the presence and the absence of copper (Fig. 1B). This basal expression is likely required for CueP’s role as a copper chaperone to deliver the metal ion to SodCII, one of the Salmonella periplasmic Cu/Zn superoxide dismutases in unstressed low-copper conditions (40). Nevertheless, CueP’s stress induction, the sensitivity to copper of the ΔcueP mutant (8), the growth deficiency of the strain in which cueP is expressed from a CpxR-independent promoter (Fig. 2), and the observation that a cueP-deleted mutant accumulates threefold more copper than the WT (15) cumulatively support the role for this protein in copper resistance.
Envelope-sensing two-component systems constitute <10% of the total number of signal-dependent transcription factors present in a bacterial chromosome. Thus, the integration of these ancestral signal- and envelope-specific sensory systems contributes to broadening the spectra of monitored conditions that can affect this bacterial cell compartment and that require a particular response.
Experimental Procedures
Bacterial Strains and Growth Conditions.
The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Tables S1 and S2. Cells were grown aerobically (41) or microaerobically (42) at 37 °C in Luria broth (LB) or LB-agar plates, except for the metal induction assays, in which pH was adjusted to either 5.5 or 7.0 by the addition of 100 mM MOPS-MES or on LB agar plates treated in the same way. Ampicillin, kanamycin, and chloramphenicol were used at 100, 25, and 10 µg mL−1 respectively. Next, 50 µM isopropyl-β-thiogalactopyranoside (IPTG) was added to induce the expression of cpxR from plasmid. Cell culture medium reagents, chemicals, and oligonucleotides were obtained from Sigma-Aldrich, except the LB culture medium, which was obtained from Difco. CuSO4 salt was of at least analytical grade (>95%). Statistical analysis was performed by one-way analysis of variance and the Holm–Sidak test. Minimum inhibitory concentration (MIC) values were determined on LB plates containing increasing amounts of CuSO4 under anaerobic conditions, as reported previously (8).
Table S1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source |
| S. typhimurium strain | ||
| 14028s | Wild-type | ATCC |
| PB6127 | ∆cueP::CmR | (8) |
| PB5449 | ∆cueR | (20) |
| PB3167 | cuiD::MudJ | (47) |
| PB5948 | cueP::lacZ::CmR | (8) |
| PB8214 | cueP::lacZ | (8) |
| PB6028 | cueP::3xFLAG KmR | (8) |
| PB5949 | ∆cueR cueP::lacZ::CmR | (8) |
| PB10123 | ∆cpxR cueP::lacZ::CmR | Present study |
| PB10029 | ∆cueR::KmR ∆cpxR cueP::lacZ::CmR | Present study |
| PB10400 | ∆cueR ∆cpxR cueP::3xFLAG KmR | Present study |
| PB10326 | ∆cpxR::CmR cueP::3xFLAG KmR | Present study |
| PB10399 | ∆cueR cueP::3xFLAG KmR | Present study |
| PB10009 | CueR-independent cueP::lacZ::CmR | Present study |
| PB10010 | ∆cpxR::KmR CueR-independent cueP::lacZ::CmR | Present study |
| PB10424 | ∆cueR::KmR CueR-independent cueP::lacZ::CmR | Present study |
| PB10423 | ∆cueR::KmR ∆cpxR CueR-independent cueP::lacZ::CmR | Present study |
| PB9958 | cueP::lacZ cpxR-box*::CmR | Present study |
| PB9662 | ∆cpxRA::CmR | Present study |
| PB5292 | copA::lacZY+ | (47) |
| PB10009 | CueR-independent cueP::lacZ::CmR | Present study |
| PB10864 | cpxP::lacZ | Present study |
| PB11609 | PcopAcueP | Present study |
| PB11893 | PcopA::CmR cueP::3xFLAG KmR | Present study |
| PB12519 | cpxR-box*-CmR | Present study |
| PB12526 | ∆cpxA::CmR | Present study |
| PB12527 | ∆cpxA::CmR cueP::lacZ | Present study |
| PB12531 | ∆cpxR PcopA::CmR cueP::3xFLAG KmR | Present study |
| Plasmids | ||
| pUH21–2lacIq | reppMB1 ApR lacIq | (47) |
| pPB1205 | pUH::cueR (pCUER) | (20) |
| pPB1466 | pUH::cpxR (pCPXR) | Present study |
| pPB1467 | pQE32::cpxR | Present study |
| pPB1468 | pUH::cpxRA (pCPXRA) | Present study |
| pPB1474 | pUH::nlpE (pNLPE) | Present study |
ATCC, American Type Culture Collection.
Table S2.
Oligonucleotides used in this study
| Primer name | Sequence (5′→3′) | Purpose |
| cpxR-BamHI-Fwd | ACGGGATCCATATGAATAAAATCCTGTTAG | To clone cpxR |
| cpxR-HindIII-Rv | AGCAAGCTTTCATGAAGCGGAAACCATC | To clone cpxR |
| cueP-prext-Rv | TCTGAGGATGCCGCAAGCGCGGAGCC | DNase I footprinting/EMSA |
| cueP-fwd-ftprin | GCATTTTGAATCCCTGCCTGATGG | DNase I footprinting/EMSA |
| P1-Rv | CGAAGCAGCTCCAGCCTACAC | EMSA |
| P1-CueP-constit | GTCCCTTTGACCTCCCTTAGGGGAACCCCTATAGTAGGCAGGGAGATTGTTCACAAGGAATTGAAGTTGTGTAGGCTGGAGCTGCTTCG | Construction of CueR-independent cueP::lacZ transcriptional fusion |
| P2-CueP-constit | TATAGGCCCGATAACCCATTATGTTATCGGGCATTTTTCATATGAATATCCTCCTTA | Construction of CueR-independent cueP::lacZ transcriptional fusion |
| cpxR-P1-Fwd | CCTGATGACGTAATTTCTGCCTCGGAGGTACGTAAACAGTGTAGGCTGGAGCTGCTTCG | λ Red deletion of cpxR |
| cpxR-int-P2-Rv | CGGCGTCAGGCGCTTACCCAGCACTTCCTGACTTAAATGCATATGAATATCCTCCTTA | λ Red deletion of cpxR |
| cpxR P2 Rv | CCAGAAGATGGCGAAGATGCGCGCGGTTAAACTTCCTACATATGAATATCCTCCTTA | λ Red deletion of cpxRA |
| P1-site-cpxR | CTGATGGCGGGGATTTTTTTTATTCCAATTCCCCCCTCGTGTAGGCTGGAGCTGCTTCG | Mutagenesis of the CpxR box in PcueP |
| P2-cpxR-mut | GGACGCAGGCACTAGTAATCCACAGTTATGCTGGGGGACATATGAATATCCTCCTTA | Mutagenesis of the CpxR box in PcueP |
| Cpx-Mut-Kpn-Fwd | TCCCCCAGCATAACCATGGATTACTAGTGCCTGCGTCC | Mutagenesis of the CpxR box in PcueP |
| P-int-cueP-Rv | ACTTCAATTCCTTGTGAACAATC | Mutagenesis of the CpxR box in PcueP |
| cueP-prom-copA-Fwd | GTAATGGCGGCGTCACCAGC | Construction of PcopA-cueP strain |
| P2-rv-cueP-prom-CopA | CGGTTTTATGAAGAGAAAGGGCTGGTGACGCCGCCATTACCATATGAATATCCTCCTTA | Construction of PcopA-cueP strain |
| P1-Fwd-cueP-prom-copA | CCAGCATAACTTTGCATTACTTTACCCTGCGTCCCTTTGAGTGTAGGCTGGAGCTGCTTCG | Construction of PcopA-cueP strain |
| cpxA-int-P1-Fwd | TAGGAAGTTTAACCGCGCGCATCTTCGCCATCTTCTGGGTGTAGGCTGGAGCTGCTTCG | λ Red deletion of cpxA |
| cpxA-int-P2-Rv | GAGATAAAAAATCGGCCTGCATTCGCAGGCCGATGGTTTCATATGAATATCCTCCTATA | λ Red deletion of cpxA |
Bacterial Genetic and Molecular Biology Techniques.
All S. enterica serovar Typhimurium strains used in this study were derived from wild-type 14028s and were constructed by phage P22-mediated transduction as described previously (43). Strains carrying deletions or lacZ reporter fusion to promoters on the chromosome were carried out using lambda Red-mediated recombination (44, 45). When necessary, the antibiotic-resistance cassette inserted at the deletion point was removed using the temperature-sensitive plasmid pCP20 carrying the FLP recombinase. The CueR-independent cueP promoter with the lacZ reporter fusion was built by lambda Red-mediated recombination. Mutation of the the CpxR box in strain PB9958 was constructed by the gene splicing by overlap extension PCR method (46). Strain PB11609 with the copA promoter driving the expression of cueP was obtained in the same manner. All constructs were verified by DNA sequencing. DNA was introduced into bacterial strains by electroporation using a Bio-Rad E. coli Pulser Electroporator following the manufacturer’s recommendations.
Cu-Induction Assays.
The β-gal assays were carried out essentially as described previously (47), using overnight cultures on LB 100 mM MOPS-MES buffer (Sigma-Aldrich) to adjust the pH value to 7.0 or 5.5, in the presence of 1 mM CuSO4 or without metal added.
Protein Purification and Western Blot Analysis.
The CpxR-His–tagged fusion protein was purified by Ni2+-NTA-agarose affinity chromatography. CueR and CueRC120S were purified essentially as described previously (19, 27), with the addition of 1 mM CuSO4 in the LB culture. (One equivalent of CuSO4 per CueR monomer was added to the purified protein before storage.) All procedures were carried out at 4 °C. The purified proteins were analyzed by SDS/PAGE, and their concentrations were calculated using the Bradford assay.
Western blot analyses of 3× FLAG-tagged proteins or GroEL were carried out as described previously (8, 35), with mouse anti-FLAG monoclonal (Sigma-Aldrich) or rabbit polyclonal anti-GroEL antibodies.
Protein–DNA Interaction Analysis.
EMSAs were performed as described previously (34) using purified CueR and CueRC120S (27, 34), CpxR (preincubated with 25 mM acetyl phosphate), and RNA polymerase (Epicentre). DNase I protection assays were done for both DNA strands (34). For KCN treatment, CueR was incubated with KCN to reach a final concentration of 100 or 300 µM in the protein/DNA-binding assay (36). Heparin and NTPs were used at a final concentration of 200 µg/mL and 250 µM, respectively, as indicated previously (48).
Acknowledgments
We thank Eleonora García Véscovi and Susana K. Checa for advice. This study was supported by the Agencia Nacional de Promoción Científica y Tecnológica (Grant PICT-2013-1513). A.J., L.B.P., and C.L. are fellows of the National Scientific and Technical Research Council (CONICET). F.C.S. is a career investigator of CONICET and of the Rosario National University Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603192113/-/DCSupplemental.
References
- 1.Ruiz N, Silhavy TJ. Sensing external stress: Watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol. 2005;8(2):122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Guo MS, Gross CA. Stress-induced remodeling of the bacterial proteome. Curr Biol. 2014;24(10):R424–R434. doi: 10.1016/j.cub.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacRitchie DM, Buelow DR, Price NL, Raivio TL. Two-component signaling and gram-negative envelope stress response systems. Adv Exp Med Biol. 2008;631:80–110. doi: 10.1007/978-0-387-78885-2_6. [DOI] [PubMed] [Google Scholar]
- 4.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raivio TL. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol. 2005;56(5):1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 6.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460(7257):823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 7.Outten FW, Huffman DL, Hale JA, O’Halloran TV. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem. 2001;276(33):30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 8.Pontel LB, Soncini FC. Alternative periplasmic copper-resistance mechanisms in Gram-negative bacteria. Mol Microbiol. 2009;73(2):212–225. doi: 10.1111/j.1365-2958.2009.06763.x. [DOI] [PubMed] [Google Scholar]
- 9.Su CC, et al. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature. 2011;470(7335):558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argüello JM, Raimunda D, Padilla-Benavides T. Mechanisms of copper homeostasis in bacteria. Front Cell Infect Microbiol. 2013;3:73. doi: 10.3389/fcimb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philips SJ, et al. TRANSCRIPTION. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science. 2015;349(6250):877–881. doi: 10.1126/science.aaa9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClelland M, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413(6858):852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 14.Abriata LA, Pontel LB, Vila AJ, Dal Peraro M, Soncini FC. A dimerization interface mediated by functionally critical residues creates interfacial disulfide bonds and copper sites in CueP. J Inorg Biochem. 2014;140:199–201. doi: 10.1016/j.jinorgbio.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Osman D, et al. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J Biol Chem. 2010;285(33):25259–25268. doi: 10.1074/jbc.M110.145953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoon BY, et al. Structure of the periplasmic copper-binding protein CueP from Salmonella enterica serovar Typhimurium. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 10):1867–1875. doi: 10.1107/S090744491301531X. [DOI] [PubMed] [Google Scholar]
- 17.Hunke S, Keller R, Müller VS. Signal integration by the Cpx-envelope stress system. FEMS Microbiol Lett. 2012;326(1):12–22. doi: 10.1111/j.1574-6968.2011.02436.x. [DOI] [PubMed] [Google Scholar]
- 18.Vogt SL, Raivio TL. Just scratching the surface: An expanding view of the Cpx envelope stress response. FEMS Microbiol Lett. 2012;326(1):2–11. doi: 10.1111/j.1574-6968.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- 19.Espariz M, Checa SK, Audero ME, Pontel LB, Soncini FC. Dissecting the Salmonella response to copper. Microbiology. 2007;153(Pt 9):2989–2997. doi: 10.1099/mic.0.2007/006536-0. [DOI] [PubMed] [Google Scholar]
- 20.Thieme D, Neubauer P, Nies DH, Grass G. Sandwich hybridization assay for sensitive detection of dynamic changes in mRNA transcript levels in crude Escherichia coli cell extracts in response to copper ions. Appl Environ Microbiol. 2008;74(24):7463–7470. doi: 10.1128/AEM.01370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external copper. Mol Microbiol. 2005;56(1):215–227. doi: 10.1111/j.1365-2958.2005.04532.x. [DOI] [PubMed] [Google Scholar]
- 22.Kershaw CJ, Brown NL, Constantinidou C, Patel MD, Hobman JL. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology. 2005;151(Pt 4):1187–1198. doi: 10.1099/mic.0.27650-0. [DOI] [PubMed] [Google Scholar]
- 23.Pontel LB, et al. Identification of a Salmonella ancillary copper detoxification mechanism by a comparative analysis of the genome-wide transcriptional response to copper and zinc excess. Microbiology. 2014;160(Pt 8):1659–1669. doi: 10.1099/mic.0.080473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raivio TL. Everything old is new again: An update on current research on the Cpx envelope stress response. Biochim Biophys Acta. 2014;1843(8):1529–1541. doi: 10.1016/j.bbamcr.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Pontel LB, Pezza A, Soncini FC. Copper stress targets the rcs system to induce multiaggregative behavior in a copper-sensitive Salmonella strain. J Bacteriol. 2010;192(23):6287–6290. doi: 10.1128/JB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto K, Ishihama A. Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci Biotechnol Biochem. 2006;70(7):1688–1695. doi: 10.1271/bbb.60024. [DOI] [PubMed] [Google Scholar]
- 27.Ibáñez MM, Checa SK, Soncini FC. A single serine residue determines selectivity to monovalent metal ions in metalloregulators of the MerR family. J Bacteriol. 2015;197(9):1606–1613. doi: 10.1128/JB.02565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braymer JJ, Giedroc DP. Recent developments in copper and zinc homeostasis in bacterial pathogens. Curr Opin Chem Biol. 2014;19:59–66. doi: 10.1016/j.cbpa.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pontel LB, Checa SK, Soncini FC. Bacterial copper resistance and virulence. In: Saffarini DA, editor. Bacteria–Metal Interactions. Springer International; Cham, Switzerland: 2015. pp. 1–19. [Google Scholar]
- 30.Djoko KY, Ong CL, Walker MJ, McEwan AG. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem. 2015;290(31):18954–18961. doi: 10.1074/jbc.R115.647099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samanovic MI, Ding C, Thiele DJ, Darwin KH. Copper in microbial pathogenesis: Meddling with the metal. Cell Host Microbe. 2012;11(2):106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nies DH, Herzberg M. A fresh view of the cell biology of copper in enterobacteria. Mol Microbiol. 2013;87(3):447–454. doi: 10.1111/mmi.12123. [DOI] [PubMed] [Google Scholar]
- 33.Yoon BY, et al. Direct ROS scavenging activity of CueP from Salmonella enterica serovar Typhimurium. Mol Cells. 2014;37(2):100–108. doi: 10.14348/molcells.2014.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humbert MV, Rasia RM, Checa SK, Soncini FC. Protein signatures that promote operator selectivity among paralog MerR monovalent metal ion regulators. J Biol Chem. 2013;288(28):20510–20519. doi: 10.1074/jbc.M113.452797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez Audero ME, et al. Target transcription binding sites differentiate two groups of MerR-monovalent metal ion sensors. Mol Microbiol. 2010;78(4):853–865. doi: 10.1111/j.1365-2958.2010.07370.x. [DOI] [PubMed] [Google Scholar]
- 36.Changela A, et al. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301(5638):1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 37.Will WR, Bale DH, Reid PJ, Libby SJ, Fang FC. Evolutionary expansion of a regulatory network by counter-silencing. Nat Commun. 2014;5:5270. doi: 10.1038/ncomms6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wade JT, Belyaeva TA, Hyde EI, Busby SJ. A simple mechanism for co-dependence on two activators at an Escherichia coli promoter. EMBO J. 2001;20(24):7160–7167. doi: 10.1093/emboj/20.24.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellermeier JR, Slauch JM. Adaptation to the host environment: Regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol. 2007;10(1):24–29. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Osman D, et al. The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Mol Microbiol. 2013;87(3):466–477. doi: 10.1111/mmi.12107. [DOI] [PubMed] [Google Scholar]
- 41.Pontel LB, Audero ME, Espariz M, Checa SK, Soncini FC. GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol Microbiol. 2007;66(3):814–825. doi: 10.1111/j.1365-2958.2007.05963.x. [DOI] [PubMed] [Google Scholar]
- 42.Sevcík M, Sebková A, Volf J, Rychlík I. Transcription of arcA and rpoS during growth of Salmonella typhimurium under aerobic and microaerobic conditions. Microbiology. 2001;147(Pt 3):701–708. doi: 10.1099/00221287-147-3-701. [DOI] [PubMed] [Google Scholar]
- 43.Davis RW, Bolstein D, Roth JR. Advanced Bacterial Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1980. [Google Scholar]
- 44.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellermeier CD, Janakiraman A, Slauch JM. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene. 2002;290(1-2):153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 46.Horton RM, et al. Gene splicing by overlap extension. Methods Enzymol. 1993;217:270–279. doi: 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- 47.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 48.Boulanger A, et al. Bordetella pertussis fim3 gene regulation by BvgA: Phosphorylation controls the formation of inactive vs. active transcription complexes. Proc Natl Acad Sci USA. 2015;112(6):E526–E535. doi: 10.1073/pnas.1421045112. [DOI] [PMC free article] [PubMed] [Google Scholar]