Significance
Single-stranded (ss) RNA viruses have ribonucleic acid as their genetic material and infect animals, plants, and bacteria. Here we used cryo-electron microscopy to reveal the genomic RNA (gRNA) of the ssRNA virus Qβ. The asymmetric gRNA adopts a single dominant structure in all virions and binds the capsid of Qβ at each coat protein. At the same time, we determined the structure of the maturation protein, A2, which functions both as the virion’s “tail” and its lysis protein. We see the gRNA is more ordered when interacting with A2. These results provide structural insights into gRNA packaging and host infection in ssRNA viruses.
Keywords: single-particle cryo-EM, Allolevivirus, ssRNA virus, genomic RNA, maturation protein
Abstract
Single-stranded (ss) RNA viruses infect all domains of life. To date, for most ssRNA virions, only the structures of the capsids and their associated protein components have been resolved to high resolution. Qβ, an ssRNA phage specific for the conjugative F-pilus, has a T = 3 icosahedral lattice of coat proteins assembled around its 4,217 nucleotides of genomic RNA (gRNA). In the mature virion, the maturation protein, A2, binds to the gRNA and is required for adsorption to the F-pilus. Here, we report the cryo-electron microscopy (cryo-EM) structures of Qβ with and without symmetry applied. The icosahedral structure, at 3.7-Å resolution, resolves loops not previously seen in the published X-ray structure, whereas the asymmetric structure, at 7-Å resolution, reveals A2 and the gRNA. A2 contains a bundle of α-helices and replaces one dimer of coat proteins at a twofold axis. The helix bundle binds gRNA, causing denser packing of RNA in its proximity, which asymmetrically expands the surrounding coat protein shell to potentially facilitate RNA release during infection. We observe a fixed pattern of gRNA organization among all viral particles, with the major and minor grooves of RNA helices clearly visible. A single layer of RNA directly contacts every copy of the coat protein, with one-third of the interactions occurring at operator-like RNA hairpins. These RNA–coat interactions stabilize the tertiary structure of gRNA within the virion, which could further provide a roadmap for capsid assembly.
Single-stranded (ss) RNA viruses are an abundant type of virus and infect all domains of life (1–4). One of the best-studied ssRNA virus systems is the Leviviridae, which infects Gram-negative bacteria via a variety of retractile pili (5); extensive genetic and biochemical studies have been performed on two of these phages: MS2 and Qβ (5–11). All of the Leviviridae have the same core genome, spanning 3.4–4.3 kb, encoding the maturation protein, the coat protein, and a subunit of RNA-dependent RNA replicase (SI Appendix, Fig. S1) (10). The MS2-like phages, designated true leviviruses, have a fourth gene that encodes the lysis protein, whereas the Qβ-like phages, designated alloleviviruses, have the lysis function as an additional feature of the maturation protein (called A2 in Qβ). Qβ also encodes a minor coat protein, called A1, arising from occasional read-through of the stop codon of the major coat protein; it has been estimated that the A1 protein replaces 3–10 copies of the major coat protein in the virion (11) and is required for infection (12). A1 consists of a coat domain and a read-through domain separated by a flexible linker (13). Unlike most dsDNA phages, which use specialized protein machinery to pump their genomic DNA into a capsid preassembled around a protein scaffold (14–17), ssRNA viruses, including the Leviviridae, assemble their coat proteins around the genomic RNA (gRNA), presumably because the extremely small genome does not allow for genes to encode proteins that help package the genetic material. Therefore, ssRNA viruses require direct gRNA–coat protein interactions, some of which are specific, to self-assemble the virion (18). This raises interesting questions, including how the viral RNA is selectively encapsidated over host RNAs. Addressing these questions will lead to a better understanding of the physiology of ssRNA viruses, and potentially novel therapeutics against them (19).
In Leviviridae, both Qβ and MS2 have capsids with T = 3 morphology, meaning each coat protein monomer adopts one of three conformers, termed A, B, and C (20, 21). Within the viral capsid, conformers A and B form an asymmetric A/B dimer and two C conformers form a symmetric C/C dimer. The gRNAs of Qβ and MS2 form secondary structures both in vitro (22, 23) and in vivo (7). There is a specific RNA hairpin near the start of the replicase gene called the “operator,” which has a high binding affinity for the coat protein (24, 25). Although both MS2 and Qβ operators form hairpins, the coat proteins for each phage are selectively attracted to their specific operator to the point at which, if purified RNAs from both phages are mixed with the coat protein of just one phage, only the cognate RNA is encapsidated (8). In MS2, the maturation protein binds to specific regions on the 5′ and 3′ of the gRNA during capsid assembly (26).
Neither the maturation protein nor the gRNA of Leviviridae was resolved in the previous high-resolution structural studies by X-ray crystallography, most likely because of the asymmetry of these two components in the stacking of symmetric particles within the crystals (20, 21). The first glimpse of RNA within the capsid came from a low-resolution structure of MS2 using single-particle cryo-EM (27). On the basis of another single-particle cryo-EM structure with icosahedral symmetry applied, it was proposed that MS2 has two concentric shells of RNA within the phage capsid (28). Subsequent work, using cryo-electron tomography and subtomogram averaging, yielded a 39-Å resolution symmetry-free density map of MS2 (29), suggesting the gRNA adopts the same conformation for each virus particle within the capsid and the maturation protein replaces a C/C dimer of the coat proteins. However, at this low resolution, it was not clear how the maturation protein, which contributes ∼1% of the molecular mass of the entire virion, interacts with the rest of the capsid or how the gRNA is organized inside the capsid shell.
In this study, we report the cryo-EM structures of the canonical Allolevivirus Qβ with and without symmetry applied, at 3.7- and 7-Å resolutions, respectively. Our structures reveal features never seen before for Qβ, such as the structure of A2, symmetry deviation of the coat proteins, organization of gRNA, and the interactions between them. These results are discussed in terms of a model for viral assembly and gRNA release in Qβ.
Results
Icosahedral Structure of Phage Qβ.
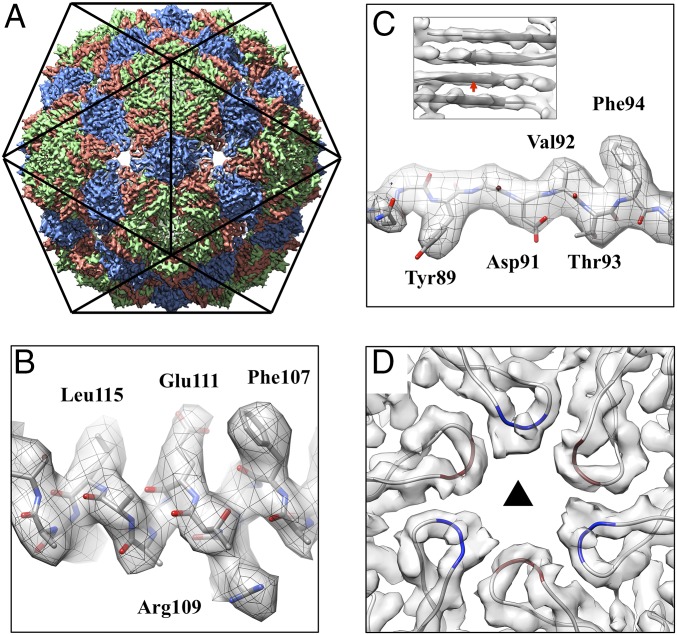
Ice-embedded Qβ phage particles were imaged using cryo-EM (SI Appendix, Fig. S2). A total of 712 image stacks were recorded on a direct detection camera in superresolution electron counting mode (30), yielding 51,815 high-quality phage particle images. Icosahedral symmetry was first applied to generate a structure of the coat protein shell at 3.7-Å resolution (SI Appendix, Fig. S3). Fig. 1 shows the icosahedral cryo-EM density map of Qβ with 180 copies of the 14-kDa coat protein. Each asymmetric unit within the capsid consists of three conformers: A, B, and C (Fig. 1A). Both types of dimers (A/B and C/C) are revealed. Protein α-helices and β-strands are clearly resolved throughout the cryo-EM density, with most of the bulky side-chains visible (Fig. 1 B and C). The cryo-EM structure is consistent with the previously solved crystal structure of the icosahedral Qβ phage capsid. PDB coordinates of the crystal structure fit into the cryo-EM density without the need for major adjustments (SI Appendix, Fig. S4). Two loops within the coat protein of note are the EF-loop, which protrudes into the interior of the capsid, and the FG-loop, which forms small pores at both the fivefold and threefold vertices of the capsid (SI Appendix, Fig. S5) (20). In our cryo-EM structure, the previously missing protein segment (residues 76–79) at the tip of the FG-loop of both conformers A and C is resolved (Fig. 1D), allowing us to build a more complete model for the capsid. The additional resolved residues are at the threefold vertices, with alternating FG-loops from conformers A and C. These residues form an ∼15-Å pore, in agreement with its predicted size (20). Residues 76–79 of conformer B are resolved both in our cryo-EM reconstruction and the previous crystal structure, mostly because of the location at the fivefold vertices, which allows smaller pores (∼8 Å in diameter) between neighboring subunits, restricting the movement of their FG-loops.
Fig. 1.
Icosahedral reconstruction of the Qβ coat protein shell at 3.7-Å resolution. (A) The map is oriented with the twofold axis pointing out of the figure. Coat protein conformers A, B, and C are colored salmon, green, and blue, respectively. (B) The α-helix density of the coat protein showing α-helical pitches and bulky amino acid side-chains. (C) The bulky side-chains in the β-strand. (Inset) Separation of four β-strands in a β-sheet formed within coat protein dimers, with the red arrow indicating the location and viewing angle for the C. (D) The cryo-EM densities and models of the FG-loops, located at the threefold axis, with residues 76–79 for conformers A and C colored red and blue, respectively. The black triangle indicates the threefold axis.
Asymmetric Structure of Phage Qβ.
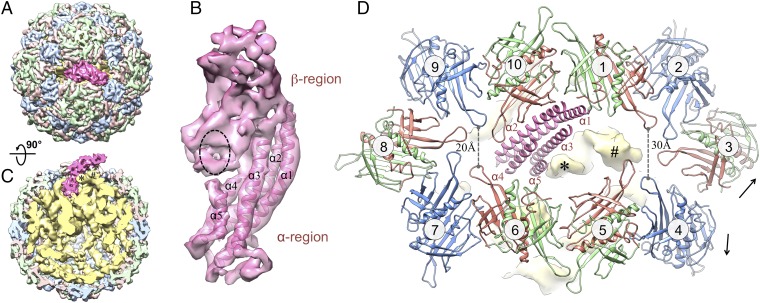
To determine the structures of the A2 protein and gRNA within the Qβ virion, we released the symmetry for our cryo-EM reconstruction. After 3D classification (Experimental Procedures), 12,975 particles were used to generate a symmetry-free reconstruction of Qβ at 7-Å resolution, with A2 and the gRNA clearly defined. The resolution for the asymmetric reconstruction of the coat proteins and A2 was further improved to 6.5 Å after masking out the RNA during map refinement (SI Appendix, Fig. S3 and S6). Fig. 2 and Movie S1 show the asymmetric cryo-EM structure of Qβ. The overall capsid stays similar to the icosahedral reconstruction of the coat proteins. The read-through domain of the A1 coat protein is not visible in our asymmetric reconstruction, possibly because of the flexible connector to the coat domain and/or the heterogeneity in the locations of A1 proteins among different virions. The most apparent difference in the capsid between our symmetric and asymmetric reconstructions is for A2 (colored hot pink in Fig. 2). A2 replaces one of the C/C dimers at a twofold axis (Fig. 2A), which leaves 178 copies of the coat protein in the virion. Much of the gRNA is clearly resolved, with major and minor grooves of the RNA helices discernible (SI Appendix, Fig. S7).
Fig. 2.
Asymmetric structure of the Qβ virus at 7-Å resolution. (A) Top view of Qβ with coat protein conformers A, B, and C colored light red, light green, and light blue, respectively. The maturation protein A2 is hot pink. (B) Segmented density of A2 showing the top β-region and bottom α-region. Five α-helices were modeled into A2. They are labeled from α1 to α5 according to their lengths, with the longest as α1. Black dashed oval indicates the location where one RNA stem-loop contacts. (C) Side view of Qβ rotated 90° from the top view, with A2 pointing up. The virion is sliced down the middle to reveal the interaction of A2 with the RNA (yellow). The RNA density is low-pass–filtered to 10-Å resolution. Densities of two RNA stem-loops, which are close to A2, are labeled with star and pound signs. (D) Zoom-in top view around A2 with the hot pink helix models shown for the α-helix bundle of α1 to α5. Ten coat protein dimers around A2 are labeled from 1 to 10 clockwise and colored salmon, green, and blue for conformers A, B, and C, respectively. Black arrows label the disruption of the interface between dimers 3 and 4. The yellow densities of two RNA stem-loops are labeled with star and pound signs as in C.
Structure of the Maturation Protein A2.
The Qβ maturation protein, A2, consists of two structural regions positioned at an angle of 120° (Fig. 2). The top structural region, named the β-region, consists mostly of sheet-like densities from β-strands, whereas the bottom segment, termed the α-region, is mostly composed of rod-like densities from α-helices. Five α-helices were identified in the α-region and labeled α1 to α5, from the longest to shortest helix lengths (Fig. 2B), with α1 being ∼60 amino acids. Sequence-based secondary structure analysis predicted several α-helices in A2 (SI Appendix, Fig. S8). Remarkably, one predicted α-helix (from residues 130–190) has a high confidence score and is consistent in length with α1. The α-region connects the gRNA through the capsid, the β-region, in contrast, is exposed on the surface of the phage with a tilt angle of 30° from the capsid shell (Fig. 2C). The dimers around A2 are numbered clockwise from 1 to 10 (Fig. 2D). The top halves of helices α1 and α2 interact with two A/B dimers (labeled 1 and 10) of the coat proteins. The bottom halves of helices α1 and α2, along with α3–α5, insert into the gRNA. Although most of the A2–gRNA interactions occur at the bottom of the α-helix bundle, one RNA stem-loop, labeled with an asterisk in Fig. 2, leans toward the top of the helix α5, forming an interaction between RNA with the middle of A2 (seen as the black dashed oval in Fig. 2B). Another RNA stem-loop, labeled with a pound sign in Fig. 2, displaces the FG-loop on conformer A of dimer 5 (SI Appendix, Fig. S9 and Movie S2). The presence of A2 and the gRNA forces the coat proteins to deviate from the perfect icosahedral arrangement seen in our symmetric reconstruction (SI Appendix, Fig. S9 and Movie S3), especially for the coat proteins around A2. Notably, the deviations are larger for dimers 1–5 compared with dimers 6–10. The distance between the protein backbones of opposite FG-loops increases from 20 Å for dimers 6–10 to 30 Å for dimers 1–5. The break in symmetry around A2 causes a fissure between dimers 3 and 4, disrupting the interactions that hold them together (Fig. 2D, SI Appendix, Fig. S9, and Movies S2 and S3), leaving more of an opening on the right toward which the β-region of A2 is angled. These disrupted coat protein interactions may allow for an easier detachment of A2, along with the gRNA, from the capsid upon infection. Furthermore, although A2 interacts with neighboring coat proteins, it does not make up for all of the interactions that would have taken place with the two coat proteins for which it has substituted. As a result, the weakened coat protein interactions around A2 may cause less obstruction during release of the gRNA from the capsid.
Organization of the gRNA.
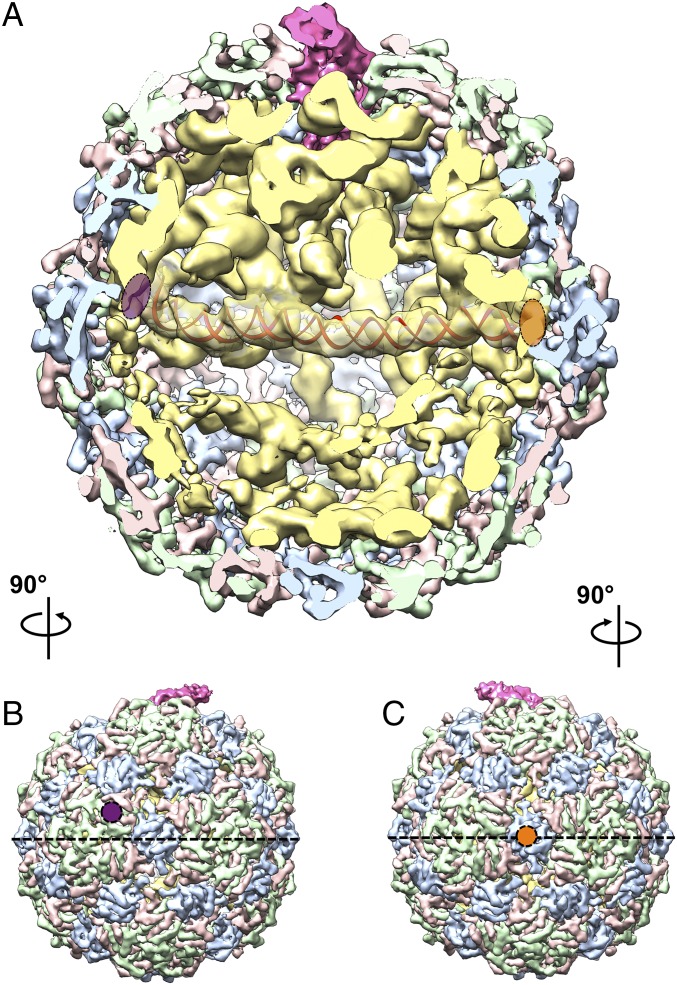
In the asymmetric structure, the gRNA exhibits well-defined secondary and tertiary structures within the ∼30-Å-thick coat protein shell. Shown in Fig. 3 is a cutaway view of the asymmetric cryo-EM density map of Qβ oriented with A2 at the “North Pole” and its β-region, pointing out of the figure. The RNA elements (yellow density in Fig. 3A) show clear RNA helices and junctions. Only one shell of RNA lies beneath the capsid, with RNA further inside the virion showing distinct asymmetric tertiary structures. Several long RNA helices are observed to traverse the interior of the capsid (Movie S1). One striking feature is a 200-Å-long RNA helix (density fitted with a red RNA helix model in Fig. 3A) spanning the middle of the capsid close to its equator. One end of this long helix touches the inner surface of an A/B dimer that is slightly above the equator (marked with a purple oval in Fig. 3 A and B); the other end of the long helix touches the inner surface of a C/C dimer on the equator at the opposite side (marked with an orange oval in Fig. 3 A and C). In our asymmetric reconstruction, ∼60% of the RNA density is located above the equator, and ∼40% below. South of the equator, the local resolution of the RNA is lower compared with the RNA in contact with the capsid or proximal to A2 (SI Appendix, Fig. S6). This indicates that the RNA is more flexible in the interior of the southern hemisphere, possibly because of it being less packed in this region.
Fig. 3.
gRNA organization inside the Qβ capsid. (A) Cutaway front view of Qβ with A2 in the North Pole and pointing out of the paper. Proteins and RNA are colored as in Fig. 2. The density of a long RNA helix, ∼200 Å in length, is docked with a red RNA duplex model and spans the two ends of the equator. Purple and orange ovals indicate the left and right of the helix, respectively. (B) Left outside view of the Qβ map, with the purple circle annotating the A/B dimer at the left end of the long helix in A. (C) Right outside view of the Qβ map, with the orange circle annotating the C/C dimer at the right end of the long helix in A. The black dashed lines, in B and C, label the equator.
Interactions Between the Capsid and gRNA.
The EF-loop of each coat protein thrusts inside the capsid, toward the RNA (Fig. 4A and SI Appendix, Fig. S5). Close inspection of the coat protein sequence of Qβ reveals that there are several positively charged residues at the tip of the EF-loop: residues 56–60 with the amino acid sequence SRNRK. These positively charged side-chains of Arginine and Lysine, at the tip of its EF-loop, may help anchor the coat proteins of Qβ to the negatively charged gRNA backbone during assembly. Moreover, it was previously proposed that, in many ssRNA viruses, the capsid proteins consist of highly basic semiflexible peptide arms, which provide nonspecific electrostatic interactions to control both the length and conformations of the gRNA (31).
Fig. 4.
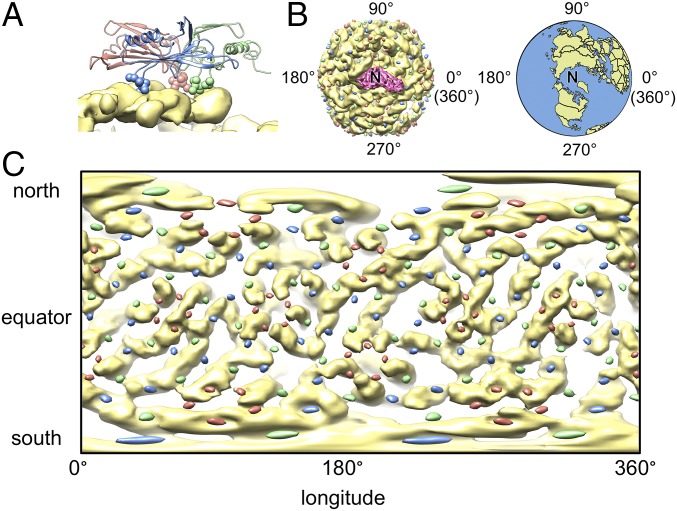
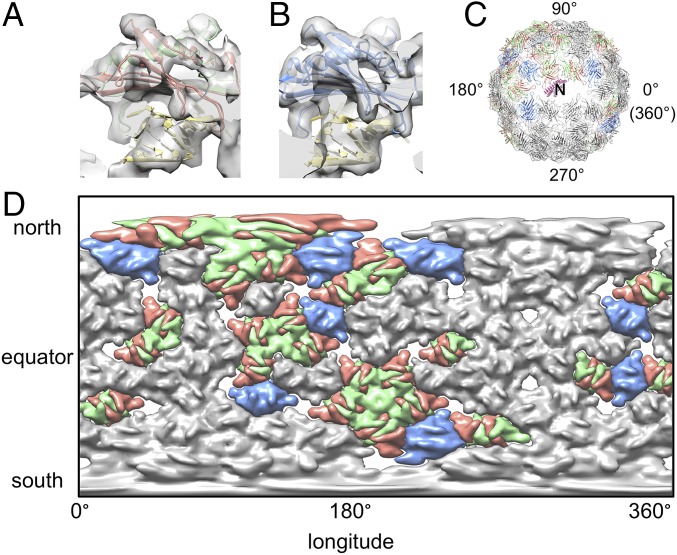
EF-loops anchor the coat proteins onto the outmost shell of gRNA. (A) Interaction between the gRNA and an asymmetric unit of the coat protein with the residues at tip of the EF-loop shown as sphere models. The density of gRNA is in yellow, whereas the models of the coat proteins are colored salmon, green, and blue for conformers A, B, and C, respectively. (B, Left) A2 (hot pink) and Qβ gRNA (yellow) are viewed with A2 as the North Pole (labeled “N”). Tips of the EF-loop are presented as colored dots (with color conventions as in A) around the Qβ gRNA globe. (Right) Earth in the same orientation. (C) Mercator projection of the outmost shell of the Qβ gRNA (yellow) and the locations of EF-loops (salmon, green, and blue ellipsoids).
We highlighted the tip of the EF-loops as spheres on our ribbon model of the coat proteins with colors salmon, green, and blue for conformers A, B, and C, respectively (Fig. 4A). Fig. 4B shows the density of gRNA and A2 with the locations of EF-loops labeled as dots in corresponding colors. After defining A2 as the North Pole, similar to that of Earth, we can unwrap the outmost spherical shell of the gRNA such that it would be like a map of the world. This is referred to as a Mercator projection, where the poles of the globe appear much larger in the projection than parts at the equator (Fig. 4C). What is readily seen is that each colored dot, representing EF-loops from the different-colored coat protein conformers, is immediately proximal to RNA, with most of the EF-loops appearing to be within the grooves of the RNA helices or at the tips of stem-loops. Interestingly, at some of the fivefold vertices, surrounded by the salmon ellipsoids (EF-loop of conformer A), the RNA also adopts a pseudo fivefold arrangement (Fig. 4C), which provides evidence that the structure of the outmost shell of the RNA depends on steric contributions from the capsid inner surface topography (32).
The interactions between coat proteins and gRNA in Qβ were first seen through a crystal structure of the complex between the operator and a coat protein dimer (24). We can fit this crystal structure (PDB ID code 4L8H) as a template into our asymmetric density map to look for interactions that are similar to the one between operator and coat protein dimers. To our surprise, 31 of the 89 coat protein dimers interact with operator-like hairpins, suggesting they may share a similar mode of interaction with high binding affinity. The operator-like hairpins interact with both types of dimer, A/B and C/C (Fig. 5 A and B), with a preference of A/B dimers (23 of the 31 dimers). When visualized over the whole capsid in a Mercator projection, the operator-like hairpin-coat protein interactions are asymmetrically distributed within the capsid, mostly between longitudes 90° and 180° in the northern hemisphere, close to A2 (Fig. 5 C and D). The coat protein dimers also interact with the gRNA in other locations, such as in the middle of RNA helices or on RNA junctions (SI Appendix, Fig. S10).
Fig. 5.
Interaction between coat protein dimers and the operator-like hairpins of gRNA. (A) Operator-like hairpin interacting with an A/B dimer. (B) Operator-like hairpin interacting with a C/C dimer. (C) Model of the coat protein dimers interacting with operator-like hairpins are in salmon, green, and blue for their conformers A, B, and C, respectively. Dimers not interacting with operator-like hairpins are in gray. The α-helix bundle of A2 is in hot pink and shown as the North Pole (labeled “N”). (D) Mercator projection of the capsid density with coat protein dimers colored as in C.
Discussion
Role of A2 as the Maturation and Lysis Protein.
The results presented here may have implications for the mechanism of host infection and of gRNA packaging in ssRNA phages. The Qβ gRNA is surrounded by a coat protein shell, whose symmetry is broken by the maturation protein, A2. A2 replaces a C/C dimer with its β-region tilting at an angle of ∼30° from the virus surface, just barely rising above the capsid. As in all Leviviridae, the maturation protein A2 is required for infection. The β-region of A2, outside the capsid, may carry out the role of binding to the receptor, the F-pilus, whereas the α-region binds to the gRNA and hangs on until the RNA is inside the cell. In Qβ and Qβ-like phages, the portion of A2 outside the capsid has the expanded role of causing lysis. At the time of lysis, it has been shown that A2, either free in the cytosol or as a single protein on the virion, can both bind and inhibit MurA (33), a universally conserved enzyme that catalyzes the first committed step in cell wall biosynthesis. From our asymmetric structure of Qβ, the MurA-binding site of A2 is likely to be exposed and around the β-region.
Structural Implication for gRNA Release.
We see that A2 coordinates denser packing of gRNA around it, locally expanding the capsid and causing a fissure between coat dimers (SI Appendix, Fig. S9 and Movie S3). Upon attachment to the F-pilus, the β-region of A2 may have weakened interactions with the neighboring coat proteins, easing the detachment of A2-bound gRNA. Upon infection, most of the RNA tertiary interactions, as well as the gRNA–coat interactions, need to be abolished for the gRNA to exit through the narrow slot (∼60 × 40 Å), left on the capsid after A2 is detached. As ssRNA viruses of Leviviridae do not have a pressurized capsid, the gRNA cannot be mechanically injected into the host from the capsid. The retractile character of the F-pilus may provide some or all of the driving force to pull the A2-bound gRNA out of the capsid. Notably, the loose interactions of A2 with the coat proteins and the disrupted interfaces between coat protein dimers around the exit may facilitate the gRNA release. The fissure between coat protein dimers 3 and 4 may further expand during gRNA release and come together when the capsid is emptied. The loose packing in the southern hemisphere of the capsid may enable the gRNA to sample different conformations to unfold during release through the narrow exit.
Structural Mechanism for Capsid Assembly.
For Qβ and other ssRNA viruses, there is clearly an evolutionary mandate for efficient and accurate virion assembly (34). After the gRNA is replicated and coat proteins are produced, the rate-limiting step in virion production is encapsidation of the RNA (35). It was first proposed that the assembly pathway was initiated when the first coat protein dimer bound the operator hairpin, serving as a nucleation event for other coat protein dimers to “walk” down a “Hamiltonian path” binding neighboring RNAs until the virion is complete (36). To accommodate this perspective, it was later proposed that there are many sites within the gRNA, termed “packaging signals,” that can interact with the coat protein (6). These packaging signals were thought to be related to the consensus sequence of the operator. In our reconstruction of Qβ, we see the outmost layer of the gRNA coming in contact with almost every coat protein, presenting 31 operator-like hairpins for the binding of coat protein dimers. Many of the gRNA secondary structures involve an extensive array of long-range tertiary interactions, which have been shown to play roles in gene regulation and capsid assembly (37). These tertiary interactions may contract the gRNA to a rigid structure in which the operator-like hairpins, which may be far apart in the genome, are located near each other. As seen in our structure, the operator-like hairpins are asymmetrically distributed on the outmost shell of the gRNA, possibly presenting a roadmap to sequentially recruit more coat protein dimers. The recruited coat protein dimers strengthen their interactions with neighboring coat protein dimers, which in turn stabilize the gRNA underneath. Such a mutual chaperoning of both RNA and coat protein conformations (38) may facilitate the fast and accurate assembly of these viruses in vivo.
Experimental Procedures
Sample and Grid Preparation.
Phage Qβ was purified as previously described (9). The final protein concentration of Qβ was 5 mg/mL, ∼1012 plaque-forming units. For imaging, 3 µL of the sample solution was first applied to a C-Flat 1.2/1.3 400 mesh holey carbon grid at 20 °C with 100% relative humidity and vitrified using a Vitrobot Mark III (FEI Company).
Cryo-EM Imaging.
The thin-ice areas that showed clearly visible and monodispersed particles were imaged under a JEM3200FSC cryo-electron microscope with a field emission gun (JEOL) operated at 300 kV. Data were collected using the manual mode of SerialEM (39) on a Gatan K2 Summit direct detection camera (Gatan) in the superresolution electron counting mode. A nominal magnification of 30,000× was used, yielding a subpixel size of 0.6 Å. The beam intensity was adjusted to a dose rate of 7.2 e−/pixel/s on the camera. A 50-frame movie stack was recorded for each picture, with 0.2 s per frame, for a total exposure time of 10 s. Each frame in the movie stack had an exposure dose of 1 e−/Å2 per frame. An in-column energy filter was used with a slit width of 29 eV.
Data Preprocessing.
A total of 712 superresolution movie stacks were collected and first binned by 2 to yield a pixel size of 1.2 Å. Using Unblur (40), these stacks were aligned, filtered according to electron dose, and summed to generate two sets of sum images, with one set from frames 1–30 and the other set from frames 2–12, respectively.
These sum images were visually screened, and 582 of each set with strong power spectra were selected for further processing. Contrast transfer functions of the micrographs were estimated using CTFFind4 (41). Batchboxer in EMAN (42) was first used to automatically pick all the particles from these 582 sum images with a box size of 320 × 320 pixel2, yielding 111,507 particles. The automatically picked particles were then shrunk by eight times to a pixel size of 9.6 Å and screened for high-contrast particles for three rounds of the reference-free 2D classification in Relion (43), with each round having 18, 16, and 25 iterations, respectively. The coordinates of the 53,999 screened particles were then imported into the e2boxer.py in EMAN2 (44) to manually screen for missed false positives, leaving only 51,815 particles.
Icosahedral Refinement for the Coat Proteins.
The 51,815 clean particles were then refined in Relion, with the icosahedral symmetry applied. Initial refinement was run using the sum images from frame 1–30 (total dose of 31 e−/Å2 with dose filter on in Unblur). The starting map was generated from EMAN2 and low-pass-filtered to 60-Å resolution. The refinement first yielded a map at 4.0-Å resolution. This was followed by a continuing map refinement with sum images of only frame 2–12 (total dose of 11 e−/Å2 with dose filter on) and the local contrast transfer functions for each particle reestimated using Gctf (45). The final refined icosahedral map yielded a resolution of 3.7 Å.
Asymmetric Refinement for the Entire Virion.
The same 51,815 clean particles were binned by 2 to a pixel size of 2.4 Å for the asymmetric reconstruction of the entire virion. The icosahedral map of the coat proteins was low-pass-filtered to 60 Å as an initial model. Unsupervised 3D classification in Relion was first performed with C1 symmetry, and 12,975 particles went into a class, which showed clear density of the gRNA inside the capsid. These 12,975 particles were further refined in Relion with C1 symmetry to yield a map at 7.0-Å resolution, where the protein secondary structures were clearly visible. To further improve the resolution of the coat proteins and A2, a mask was applied to focus the refinement on the outer shell of the particle, which yielded a resolution of 6.5 Å. Further refinement of the asymmetric structure using all of the 51,815 particles did not improve the resolvability of the map, potentially because the 12,975 particles, selected from the 3D classification in Relion, have the best contrast and are most homogeneous in conformations.
Resolution Estimation.
The overall resolutions of all these reconstructed maps were assessed using the gold-standard criterion of Fourier Shell Correlation, with a cutoff at 0.143, between two half maps from two independent half-sets of data (46). Local resolutions were estimated using Blocres (47).
Modeling, Map Segmentation, and Visualization.
To build the model for the coat protein from the icosahedral reconstruction, the crystal structure (PDB ID code 1QBE) was used as the initial model. The missing loop in the crystal structure was built in COOT (48) and then further refined in the real-space refinement in Phenix (49). The refined model showed good model geometry from Molprobity (50) (SI Appendix, Table S1). The Fourier Shell Correlation between the icosahedral map and the refined PDB model agreed at 3.9-Å resolution at 0.5 threshold (SI Appendix, Fig. S3). This more complete model for the coat protein was then used to fit into the asymmetric map of the Qβ virion to guide the segmentation of the gRNA and A2 protein with Segger (51) as a plugin in University of California, San Francisco (UCSF) Chimera (52). The α-helices of the A2 protein were identified in Helixhunter (53). Secondary structure prediction of A2 was performed with I-TASSER (54). The 200-Å long RNA helix that spanned the equator of the gRNA was first built in UCSF Chimera with a straight A-form RNA helix with 70 GC base pairs and then refined in Molecular Dynamics Flexible Fitting to improve the fit to the density (55). All of the figures and movies were made in UCSF chimera and VMD (56). Unwrapping of the gRNA into Mercator projection was done in EMAN2 with the tool “e2unwrap3d.py” in two steps for the longitude and latitude, respectively.
Supplementary Material
Acknowledgments
We thank Jeng-Yih Chang for performing the molecular dynamics flexible fitting of the long RNA helix model. We thank the Microscopy and Imaging Center at Texas A&M University for providing instrumentation for the initial screening of the phage particles; the Texas A&M High Performance Research Computing Center for providing the computational resources for the data processing; and the National Center for Macromolecular Imaging (NCMI) at Baylor College of Medicine for data collection. NCMI is supported by NIH Grants P41GM103832 and U24GM116787. J.Z. is supported by start-up funding from the Department of Biochemistry and Biophysics at Texas A&M University. R.Y. is supported by Public Health Service Grant GM27099. J.Z. and R.Y. acknowledge the support of the Center for Phage Technology, jointly sponsored by Texas AgriLife and Texas A&M University. K.V.G. is partly supported by the Welch Foundation Grant A-1863 (to J.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the EMDataBank database, www.emdatabank.org (accession nos. EMD-8253, EMD-8254, and EMD-8255). The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5KIP).
See Commentary on page 11390.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609482113/-/DCSupplemental.
References
- 1.Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology. 2015;479-480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 3.Bidet K, Garcia-Blanco MA. Flaviviral RNAs: Weapons and targets in the war between virus and host. Biochem J. 2014;462(2):215–230. doi: 10.1042/BJ20140456. [DOI] [PubMed] [Google Scholar]
- 4.Kolakofsky D. A short biased history of RNA viruses. RNA. 2015;21(4):667–669. doi: 10.1261/rna.049916.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valentine RC, Strand M. Complexes of F-Pili and RNA bacteriophage. Science. 1965;148(3669):511–513. doi: 10.1126/science.148.3669.511. [DOI] [PubMed] [Google Scholar]
- 6.Rolfsson Ó, et al. Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J Mol Biol. 2016;428(2 Pt B):431–448. doi: 10.1016/j.jmb.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed CA, Langlais C, Wang IN, Young R. A2 expression and assembly regulates lysis in Qβ infections. Microbiology. 2013;159(Pt 3):507–514. doi: 10.1099/mic.0.064790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling CM, Hung PP, Overby LR. Independent assembly of Qβ and MS2 phages in doubly infected Escherichia coli. Virology. 1970;40(4):920–929. doi: 10.1016/0042-6822(70)90138-8. [DOI] [PubMed] [Google Scholar]
- 9.Strauss JH, Jr, Sinsheimer RL. Purification and properties of bacteriophage MS2 and of its ribonucleic acid. J Mol Biol. 1963;7:43–54. doi: 10.1016/s0022-2836(63)80017-0. [DOI] [PubMed] [Google Scholar]
- 10.Reed CA, Langlais C, Kuznetsov V, Young R. Inhibitory mechanism of the Qβ lysis protein A2. Mol Microbiol. 2012;86(4):836–844. doi: 10.1111/mmi.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber K, Konigsberg W. Proteins of the RNA phages. In: Zinder ND, editor. RNA Phages. Cold Spring Harbor Lab Press; Cold Spring Harbor, New York: 1975. pp. 51–84. [Google Scholar]
- 12.Hofstetter H, Monstein HJ, Weissmann C. The readthrough protein A1 is essential for the formation of viable Qβ particles. Biochim Biophys Acta. 1974;374(2):238–251. doi: 10.1016/0005-2787(74)90366-9. [DOI] [PubMed] [Google Scholar]
- 13.Rumnieks J, Tars K. Crystal structure of the read-through domain from bacteriophage Qβ A1 protein. Protein Sci. 2011;20(10):1707–1712. doi: 10.1002/pro.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lander GC, et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312(5781):1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 15.Jiang W, et al. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439(7076):612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen DH, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci USA. 2011;108(4):1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yap ML, et al. Role of bacteriophage T4 baseplate in regulating assembly and infection. Proc Natl Acad Sci USA. 2016;113(10):2654–2659. doi: 10.1073/pnas.1601654113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stockley PG, et al. Bacteriophage MS2 genomic RNA encodes an assembly instruction manual for its capsid. Bacteriophage. 2016;6(1):e1157666. doi: 10.1080/21597081.2016.1157666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKnight KL, Heinz BA. RNA as a target for developing antivirals. Antivir Chem Chemother. 2003;14(2):61–73. doi: 10.1177/095632020301400201. [DOI] [PubMed] [Google Scholar]
- 20.Golmohammadi R, Fridborg K, Bundule M, Valegård K, Liljas L. The crystal structure of bacteriophage Qβ at 3.5 Å resolution. Structure. 1996;4(5):543–554. doi: 10.1016/s0969-2126(96)00060-3. [DOI] [PubMed] [Google Scholar]
- 21.Golmohammadi R, Valegård K, Fridborg K, Liljas L. The refined structure of bacteriophage MS2 at 2.8 A resolution. J Mol Biol. 1993;234(3):620–639. doi: 10.1006/jmbi.1993.1616. [DOI] [PubMed] [Google Scholar]
- 22.Beekwilder J, Nieuwenhuizen R, Poot R, van Duin J. Secondary structure model for the first three domains of Qβ RNA. Control of A-protein synthesis. J Mol Biol. 1996;256(1):8–19. doi: 10.1006/jmbi.1996.0064. [DOI] [PubMed] [Google Scholar]
- 23.Beekwilder MJ, Nieuwenhuizen R, van Duin J. Secondary structure model for the last two domains of single-stranded RNA phage Qβ. J Mol Biol. 1995;247(5):903–917. doi: 10.1006/jmbi.1995.0189. [DOI] [PubMed] [Google Scholar]
- 24.Rumnieks J, Tars K. Crystal structure of the bacteriophage Qβ coat protein in complex with the RNA operator of the replicase gene. J Mol Biol. 2014;426(5):1039–1049. doi: 10.1016/j.jmb.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 25.Valegård K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature. 1994;371(6498):623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 26.Shiba T, Suzuki Y. Localization of A protein in the RNA-A protein complex of RNA phage MS2. Biochim Biophys Acta. 1981;654(2):249–255. doi: 10.1016/0005-2787(81)90179-9. [DOI] [PubMed] [Google Scholar]
- 27.Koning R, et al. Visualization by cryo-electron microscopy of genomic RNA that binds to the protein capsid inside bacteriophage MS2. J Mol Biol. 2003;332(2):415–422. doi: 10.1016/s0022-2836(03)00846-5. [DOI] [PubMed] [Google Scholar]
- 28.Toropova K, Basnak G, Twarock R, Stockley PG, Ranson NA. The three-dimensional structure of genomic RNA in bacteriophage MS2: Implications for assembly. J Mol Biol. 2008;375(3):824–836. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 29.Dent KC, et al. The asymmetric structure of an icosahedral virus bound to its receptor suggests a mechanism for genome release. Structure. 2013;21(7):1225–1234. doi: 10.1016/j.str.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belyi VA, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci USA. 2006;103(46):17174–17178. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ElSawy KM, Caves LS, Twarock R. On the origin of order in the genome organization of ssRNA viruses. Biophys J. 2011;101(4):774–780. doi: 10.1016/j.bpj.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernhardt TG, Wang IN, Struck DK, Young R. A protein antibiotic in the phage Qbeta virion: Diversity in lysis targets. Science. 2001;292(5525):2326–2329. doi: 10.1126/science.1058289. [DOI] [PubMed] [Google Scholar]
- 34.Dykeman EC, Stockley PG, Twarock R. Solving a Levinthal’s paradox for virus assembly identifies a unique antiviral strategy. Proc Natl Acad Sci USA. 2014;111(14):5361–5366. doi: 10.1073/pnas.1319479111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heldt FS, Frensing T, Reichl U. Modeling the intracellular dynamics of influenza virus replication to understand the control of viral RNA synthesis. J Virol. 2012;86(15):7806–7817. doi: 10.1128/JVI.00080-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dykeman EC, et al. Simple rules for efficient assembly predict the layout of a packaged viral RNA. J Mol Biol. 2011;408(3):399–407. doi: 10.1016/j.jmb.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 37.Klovins J, Berzins V, van Duin J. A long-range interaction in Qβ RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA. 1998;4(8):948–957. doi: 10.1017/s1355838298980177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rolfsson O, Toropova K, Ranson NA, Stockley PG. Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid. J Mol Biol. 2010;401(2):309–322. doi: 10.1016/j.jmb.2010.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192(2):216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 43.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193(1):1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9(9):853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157(1):3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pintilie G, Zhang J, Chiu W, Gossard D. 2009. Identifying Components in 3D Density Maps of Protein Nanomachines by Multi-scale Segmentation. IEEE NIH Life Sci Syst Appl Workshop 2009 2009:44–47. [DOI] [PMC free article] [PubMed]
- 52.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Jiang W, Baker ML, Ludtke SJ, Chiu W. Bridging the information gap: Computational tools for intermediate resolution structure interpretation. J Mol Biol. 2001;308(5):1033–1044. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- 54.Yang J, et al. The I-TASSER suite: Protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49(2):174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.