Significance
The mammalian circadian clock is controlled by a transcription-translation feedback loop consisting of transcriptional activators circadian locomotor output cycles kaput (CLOCK)–brain and muscle Arnt-like protein-1 (BMAL1), which function as a complex at E/E'-box elements, and repressors Cryptochrome 1 (CRY1)/CRY2 and PER1/PER2. CRYs repress upon binding as CRY–CLOCK–BMAL1–E-box complexes. Period proteins (PERs) repress by removing the heterotrimeric complexes from the E-box. We report here that in the Cry1 promoter, the CRY1–CLOCK–BMAL1–E-box complex represses a transcriptional activator acting in cis, and removal of the heterotrimeric complex by PER2 de-represses the transcriptional activator. ChIP-seq and RNA-seq experiments identified other genes also de-repressed by PER2. These data clarify the role of PER2 and reveal the level of complexity in regulation of Cry1 and other circadian-controlled genes.
Keywords: circadian, Period, transcription, activator, DNA binding
Abstract
The mammalian circadian clock is based on a transcription-translation feedback loop (TTFL) consolidated by secondary loops. In the primary TTFL, the circadian locomotor output cycles kaput (CLOCK)–brain and muscle Arnt-like protein-1 (BMAL1) heterodimer acts as the transcriptional activator, and Cryptochrome (CRY) and Period (PER) proteins function as repressors. PER represses by displacing CLOCK–BMAL1 from promoters in a CRY-dependent manner. Interestingly, genes with complex promoters may either be repressed or de-repressed by PER, depending on the particular promoter regulatory elements. Here, using mouse cell lines with defined knockout mutations in clock genes, RNA-seq, ChIP-seq, and reporter gene assays coupled with measurements of DNA–protein interactions in nuclear extracts, we elucidate the dual functions of PER as repressor and de-repressor in a context-dependent manner.
The circadian clock is the molecular system that confers a ∼24-h periodicity to many biological processes. In mammals, CLOCK, BMAL1, Cryptochrome (CRY), and Period (PER) proteins and their paralogs are essential for generating rhythmicity (1–4). Rhythmicity is the product of a transcription-translation feedback loop (TTFL): The circadian locomotor output cycles kaput (CLOCK)–brain and muscle Arnt-like protein-1 (BMAL1) heterodimer binds to E-boxes in the promoters of Cry1, Cry2, Per1, and Per2 genes and activates their transcription. The CRY and PER proteins, after a time lag, enter the nucleus and inhibit their own transcription (core clock circuit) (5–9), as well as transcription of other clock-controlled genes to maintain rhythmicity at the cellular and organismal levels. This core circuit is consolidated and stabilized by the nuclear receptors NR1D1 and NR1D2 (REV-ERBα/β), which bind to the retinoic acid response elements (RREs) in some of the clock gene promoters (10–14). Although the canonical model has provided a useful conceptual framework, it has been insufficient in constructing a mechanistic model for the clock. This is in part due to the multiple interactions among clock proteins and to the lack of an in vitro system for analyzing the clock using purified proteins and their target promoter elements along with an appropriate readout.
To simplify the analysis of clock protein functions, previously we reported the construction of cell lines that lack CRYs, PERs, and both CRYs and PERs and that express ectopic CRY or PER that can be targeted to the nucleus in a controllable manner (15). Using this system initially, we analyzed the canonical clock model by restricting our experiments to the Nr1d1 and D-site albumin promoter binding protein (Dbp) genes, which are controlled solely by the binding of CLOCK–BMAL1 to E-box regulatory elements. These experiments confirmed the repressor functions of CRY and PER but also revealed some features of their mechanisms: CRY alone can bind to the CLOCK–BMAL1–E-box complex and inhibit the transcription of cognate genes by forming a stable CRY–CLOCK–BMAL1 ternary complex in the promoter: “blocking-type” repression. In contrast, PER represses CLOCK–BMAL1–E-box-mediated transcription by removing the CLOCK–BMAL1 complex from the E-box in a CRY-dependent manner: “displacement-type” repression (15). Although these findings clarified the roles of PERs and CRYs in regulation of simple promoters exclusively controlled by E-box binding of transcription factors, they did not address the issue of the control of key clock genes with multiple cis-elements.
Here, we report the construction of cell lines with clock gene knockout mutations and expression systems to analyze the mechanisms of control of more complex promoters by PER2 protein. We show that the removal of CLOCK–BMAL1 from promoters affects gene expression in three different ways, depending on the type of regulatory elements present in the target promoters. First, in the case of a simple promoter controlled exclusively by an E-box, such as in the Nr1d1 gene, CLOCK–BMAL1 removal represses transcription. Second, in genes such as Bmal1, which is controlled exclusively by an RRE, CLOCK–BMAL1 removal from the Nr1d1 promoter indirectly activates transcription of Bmal1 by down-regulation of NR1D1/2, which represses by binding to the RRE in the Bmal1 promoter. Third, in the case of the Cry1 gene, removal of CLOCK–BMAL1 facilitates transactivation by other transcription factors that bind to the Cry1 promoter independently of NR1D1/2.
Results
Experimental System to Analyze the Role of PER in the TTFL.
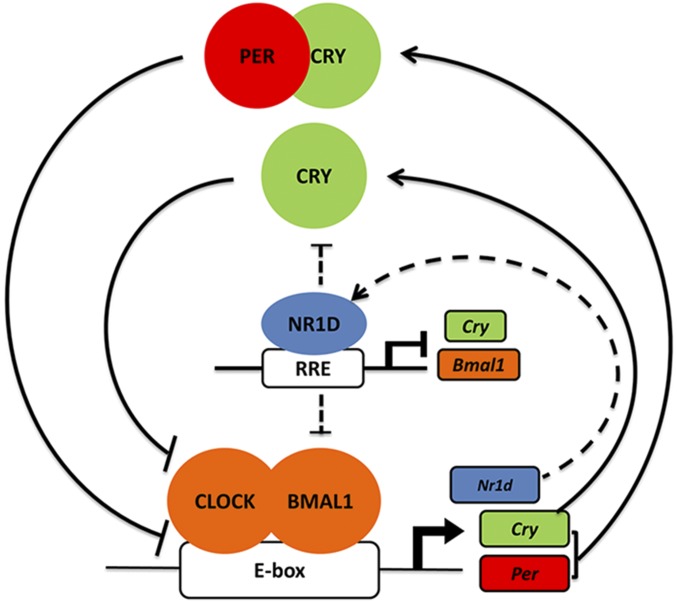
In Fig. 1 we present a highly idealized model for the mammalian circadian clock: The CLOCK–BMAL1 heterodimer activates transcription of Cry1/2, Per1/2, and Nr1d1/2 by binding to E-box (CACGTG/T) sequences in the promoters of these genes. The CRY and PER proteins inhibit transcriptional activation by CLOCK–BMAL1 to close the feedback loop, CRY by binding to the CLOCK–BMAL1–E-box and interfering with transactivation, and PER by causing the displacement of CLOCK–BMAL1 from E-boxes in a CRY-dependent manner. This primary circuit is consolidated by a stabilizing loop: The Nr1d1/2 nuclear receptor genes are also controlled by CLOCK–BMAL1, and NR1D1/2 proteins in turn repress Bmal1 and Cry1 transcription by binding to the RRE elements in the promoters of these genes (12–14, 16, 17). Some observations, such as the repressor activity of CLOCK–BMAL1 (18, 19) and the apparent transcriptional up-regulation by PER (20–23), do not readily fit into this model and require further experimentation.
Fig. 1.
TTFL model for the mammalian circadian clock. In the TTFL model, CLOCK–BMAL1 heterodimers bind to the E-boxes of Cry1/2, Per1/2, and Nr1d1/2 promoters and activate the transcription of these genes. Increased CRY protein then inhibits transcription through binding to the CLOCK–BMAL1 complex. PER protein inhibits transcription in a CRY-dependent manner by reducing CLOCK–BMAL1 binding to the E-box. This core circuit (solid lines) is stabilized by a secondary feedback loop (dashed lines) in which CLOCK–BMAL1-controlled NR1D1/2 inhibits the transcription of Bmal1 and Cry1 through binding to the RRE of their promoters.
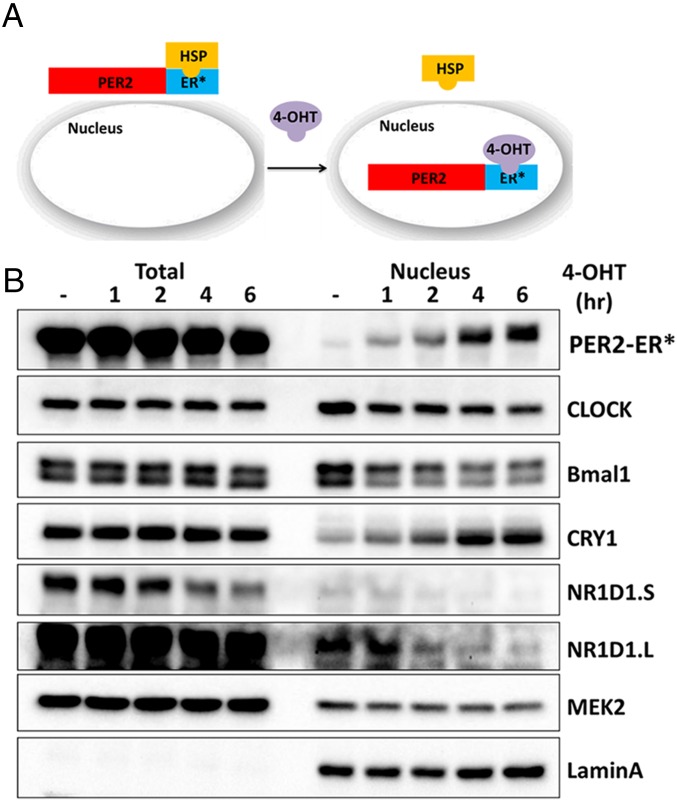
Because there is no in vitro system to analyze the roles of different clock proteins, individually or in various combinations, on transcription of cognate genes, we developed cell lines with various clock gene knockout mutations (Table S1) and with the controllable PER2 nuclear entry system shown in Fig. 2A for carrying out “in cellulo biochemistry” for better insight into the clock mechanism. These cell lines were isolated from knockout mouse strains or from mouse embryonic fibroblasts by using transcription activator-like effector nuclease (TALEN) and CRISPR technologies. The cell lines also ectopically express PER2-ER*, which is composed of PER2 protein fused to a mutant form of the ligand-binding domain of the estrogen receptor (ER*). PER2-ER* is trapped in the cytoplasm, and it enters the nucleus upon 4-hydroxytamoxifen (4-OHT) treatment. Thus, the effect of PER2 on gene expression can be analyzed in a controllable manner as shown in Fig. 2B. As previously reported (15), PER2 and CRY1 enter the nucleus together in this system.
Table S1.
Cell lines used in this study
| Name | Genotype | Generated by | Parental cell |
| P-P2ER | Per1−/−;Per2–/− PER2-ER* | Lentivirus | Per1−/−;Per2−/− |
| CP-P2ER | Per1−/−;Per2−/− | TALEN | Per1−/−;Per2−/− |
| Cry1−/−;Cry2−/− | Lentivirus | ||
| PER2-ER* | |||
| PN-P2ER | Per1−/−;Per2−/− Nr1d1−/−;Nr1d2−/− | CRISPR | Per1−/−;Per2−/− |
| PER2-ER* | Lentivirus | ||
| B-P2ER | Bmal1−/− | Lentivirus | Bmal1−/− |
| PER2-ER* |
Fig. 2.
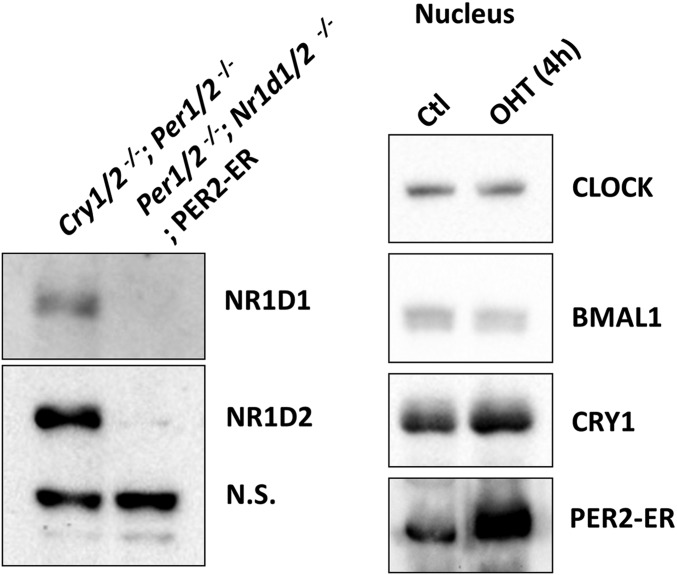
Cell-based system for study of mammalian clock regulation. (A) Schematic of the experimental system. PER2 fused to a mutant form of the estrogen receptor ligand-binding domain (ER*) is constrained to the cytosol when bound to endogenous heat-shock protein (HSP). Addition of 4-OHT to the cell culture displaces the HSP, and PER2-ER* then enters the nucleus. (B) Immunoblot analysis of clock proteins upon 4-OHT addition. Total and nuclear amounts of clock and control proteins (MEK2 and LaminA, which is a nuclear protein) up to 6 h following addition of 4-OHT to Per1/2−/−;PER2-ER* cells are shown. The blot shows that the total NR1D1 protein level (and thus nuclear NR1D1) decreases following incubation with 4-OHT. An increased level of nuclear CRY1 but not total CRY1 is associated with the increased nuclear PER2. This supports the idea that CRY and PER enter the nucleus together and that CRY-dependent PER2 activity is through the formation of a CRY–PER complex. NR1D1.s and NR1D1.L are short and long exposures of the same blot.
PER2 as a Repressor and Antirepressor.
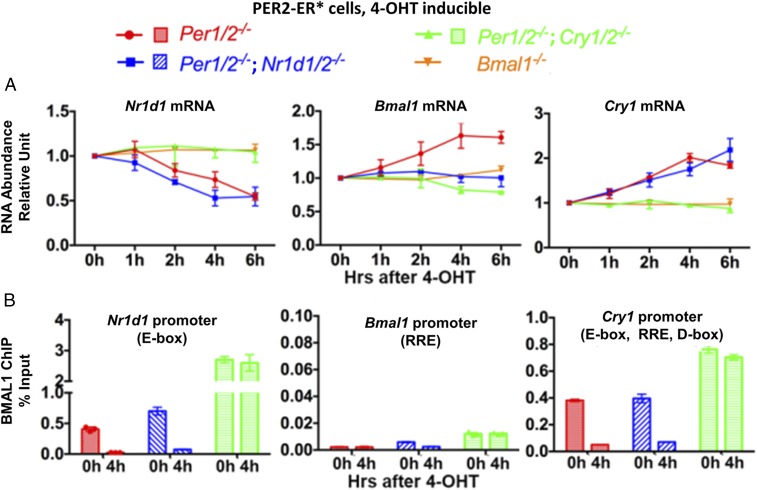
We addressed the regulatory roles of PER using a PER2-ER* ectopic expression system in cells with various clock gene knockout mutations. We analyzed transcription of three genes selected for their different promoter complexities, and reported differences in their regulation by PER2 (9–13). The plots in Fig. 3A show the levels of mRNA expressed from the Nr1d1, Bmal1, and Cry1 genes in Per1/2−/− cells following 0–6 h of treatment with 4-OHT, which induces translocation of PER2-ER* to the nucleus. The Nr1d1 promoter is controlled solely by an E-box, the Bmal1 promoter is controlled solely by the RRE element, and the Cry1 promoter is controlled by an E-box, an RRE element, and a D-box (14). As expected (15), the data in Fig. 3A (PER2-ER* enters the nucleus of Per1/2−/− cells) show that PER2 inhibits transcription of Nr1d1 (by removing CLOCK–BMAL1 from the E-box; Fig. 3A, Left, red); however, PER2 also induces transcription of Bmal1 and Cry1 (Fig. 3A, Middle and Right, red). Because PER2 inhibits the expression of Nr1d1 and because both Bmal1 and Cry1 have RRE elements in their promoters to which NR1D1/2 binds (12–14, 16, 24), it has been suggested that PER2 de-represses genes by down-regulating the NR1D1 repressor. To test this prediction, we investigated the effect of Nr1d1/2 deletions on PER2-ER*–mediated up-regulation of Bmal1 and Cry1 transcription. In the absence of NR1D1/2, there is no up-regulation of Bmal1 transcription by PER2-ER* (Fig. 3A, Middle, blue), supporting the view that PER2 up-regulates Bmal1 by down-regulating NR1D1/2. In contrast, and surprisingly, the NR1D1/2 deletions have no effect on PER2-mediated up-regulation of Cry1 (Fig. 3A, Right, blue). This agrees with the observation that NR1D1/2 is dispensable for the rhythmic expression of Cry1 (12) and indicates that PER2 up-regulates Cry1 transcription by a mechanism independent of NR1D1/2.
Fig. 3.
Effect of PER2 nuclear entry on transcription of Nr1d1, Bmal1, and Cry1 in Per1/2−/−, Per1/2−/−;Nr1d1/2−/−, Per1/2−/−;Cry1/2−/−, and Bmal1−/− cells. (A) Nuclear entry of PER2-ER* following addition of 4-OHT (red) represses Nr1d1 transcription (Left) but induces Bmal1 (Middle) and Cry1 (Right) transcription analyzed by RT-quantitative PCR. The effect of PER2 on these three genes is CRY-dependent (green). NR1D1/2 is required for PER2-induced Bmal1 transcription but is not required for the repression of Nr1d1 or for the induction of Cry1 transcription (blue). In Bmal1−/− cells (orange), nuclear entry of PER2-ER* has no effect on Nr1d1, Bmal1, or Cry1 transcription. Endogenous PER2 was not detected in Bmal1−/− cells (4). (B) PER2-dependent, CRY-dependent, NR1D1/2-independent displacement of CLOCK–BMAL1 from Nr1d1 and Cry1 E-boxes is analyzed by BMAL1–ChIP–quantitative PCR. Bmal1, which has no E-box and is not a primary target of CLOCK–BMAL1, is a negative control for ChIP. SE values are displayed in A and B.
PER2 Repressor and Antirepressor Activities Are BMAL1- and CRY-Dependent.
Based on indirect evidence, it has been previously reported that PER2 can up-regulate gene transcription by interacting with another transcription factor (25) or by removing CRY from CLOCK–BMAL1 (20). We have shown that PER2 represses transcription by removing the CLOCK–BMAL1 complex from cognate promoters in a CRY-dependent manner (15). We wished to know if the transcriptional up-regulation effect of PER was also mediated through removal of the CLOCK–BMAL1–CRY complex. To this end, we tested the effect of PER2-ER* nuclear entry on transcription of the three sentinel genes in cells lacking BMAL1. In these Bmal1 knockout cells, endogenous PER2 is undetectable (4), and thus any effect that might be seen upon 4-OHT addition can be ascribed to PER2-ER*. The results (Fig. 3A, orange) show that in the absence of BMAL1 there is no repression or de-repression of these genes upon nuclear entry of PER2-ER*. This excluded the possibility that PER2 facilitates a transcriptional activator in a BMAL1-independent manner to activate Cry1 transcription. To examine whether PER2-induced up-regulation is dependent on CRY proteins, we tested the effect of PER2-ER* on expression of all three sentinel genes in cells lacking CRY1/2. As seen in Fig. 3A, the effect of PER2 on all these three genes (red) is abolished in the Cry1/2 knockout background (green), indicating that PER2 exerts its up-regulation activity only through a CRY-mediated mechanism as it does its repressor activity.
Gene Up-Regulation by PER2 Is Associated with Removal of CLOCK–BMAL1.
We then used ChIP to directly examine the binding of BMAL1 to the sentinel gene promoters in response to PER2. In Fig. 3B, Middle, there is no BMAL1 binding to the Bmal1 promoter, as it lacks an E-box (1, 2, 4). Fig. 3B, Left shows that, as expected, nuclear entry of PER2 following 4-OHT treatment causes displacement of BMAL1 from the (PER2-repressed) Nr1d1 promoter (red), and this displacement is NR1D1/2-independent (blue) but CRY-dependent (green). Importantly, the positive regulation of the (PER2-activated) Cry1 promoter shown in Fig. 3A, Right coincides with NR1D1/2-independent but CRY-dependent displacement of BMAL1 (Fig. 3B, Right). Taken together, these data indicate that BMAL1 and CRY are required for both PER-mediated transcriptional repression and up-regulation in a context-dependent manner. Indeed, the idea of repression by CRY in a CLOCK–BMAL1-dependent manner was suggested in a previous study, although there was no direct experimental evidence (19). We suggest that in the Cry1 promoter, CLOCK–BMAL1 binding to an E-box represses another activator, which is the predominant transcription activator for this gene. Here, we show that the PER2-mediated up-regulation of Cry1 gene transcription is accompanied with the removal of the CLOCK–BMAL1–CRY complex from the Cry1 promoter and thus present direct evidence for these counterintuitive roles of CLOCK–BMAL1 in gene repression and PER2 (with CRY) in gene de-repression.
Genome-Wide Effect of PER on Regulation and Promoter Binding by the CLOCK–BMAL1 Complex.
The model that emerges from the results presented here as well as our previous studies is that PER displaces CLOCK–BMAL1 from cognate gene promoters and in doing so, depending on the complement of regulatory elements in the target promoter, it either inhibits or up-regulates (de-represses) transcription in a CRY-dependent manner (15). Because there are several thousands of CLOCK–BMAL1 binding sites in the genome (26, 27), we wished to find out whether PER-mediated CLOCK–BMAL1 removal and both the repression and de-repression functions of PER are observed on a global scale. To this end, we performed ChIP-seq experiments using an anti-BMAL1 antibody to probe CLOCK–BMAL1 binding sites and RNA-seq experiments to analyze RNA levels before and after nuclear entry of PER2 in Per1/2−/−;Nr1d1/2−/−;PER2-ER* cells so as to avoid complications arising from second-order clock-controlled genes regulated by NR1D1/2 (such as Bmal1).
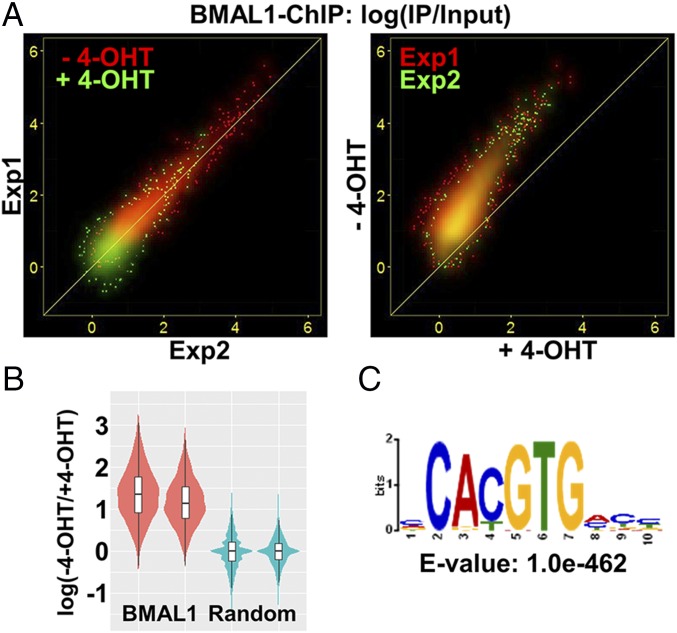
ChIP-seq experiments were conducted in duplicate with cells not treated with 4-OHT. Specific BMAL1 binding sites were identified based on 10–15 million input and precipitated DNA reads (Materials and Methods). There were 4,789 and 8,615 binding sites in the two experiments and 4,740 common binding sites seen in both experiments. Induction of PER2 with 4-OHT reduced the number of common BMAL1 binding sites to 483, and PER2 induction produced no novel BMAL1 binding sites. Thus, PER2 eliminated detectable binding of BMAL1 to most of the regulatory regions to which it binds. To further assess BMAL1 binding, we analyzed reads mapped to the 4,740 common BMAL1 binding sites as relative BMAL1 binding strength, expressed as “IP/input.” We observed that relative BMAL1 binding strength was negatively associated with PER2 induction and was highly correlated between the two independent experiments (Fig. 4A, Left). BMAL1 binding was stronger without 4-OHT than with 4-OHT on almost all regions (Fig. 4A, Right). To determine whether the observed decrease in BMAL1 binding was due to experiment-specific technical factors such as sequencing depth, we compared binding differences from these BMAL1 binding sites to reads randomly selected from the genome. Relative to the BMAL1 binding sites, there is no difference of BMAL1 binding between without 4-OHT and with 4-OHT in the random areas (Fig. 4B). Results of motif analysis of the BMAL1 binding sites showed high enrichment of the E-box sequence (Fig. 4C), which further supports the finding that PER2 removes CLOCK–BMAL1 from the E-box sequence of the promoters.
Fig. 4.
Effect of PER2 nuclear entry on BMAL1 promoter binding by BMAL1–ChIP-Seq in Per1/2−/−;Nr1d1/2−/− cells. (A) Strength of BMAL1 binding (ChIP/Input) to 4,740 common binding sites is shown as a scatter plot. (Left) Results with (green) and without (red) 4-OHT show high correlation between the two experiments. The results obtained with 4-OHT are closest to the origin, indicating weak or no binding of BMAL1 following PER2 induction. (Right) The strength of BMAL1 binding in the condition with (x axis) and without (y axis) 4-OHT is plotted. The distributions largely overlap in the two experiments and are above the line representing y = x, indicating reduced BMAL1 binding following PER2 induction. (B) Distribution of relative strength of BMAL1 binding –4-OHT/+4-OHT was plotted by violin and box plot. Stronger binding of BMAL1 without 4-OHT was detected on the BMAL1 binding sites (red) but not with reads randomly selected from the genome (blue). (C) Result from motif analysis of BMAL1 binding sites shows strong enrichment of the E-box sequence.
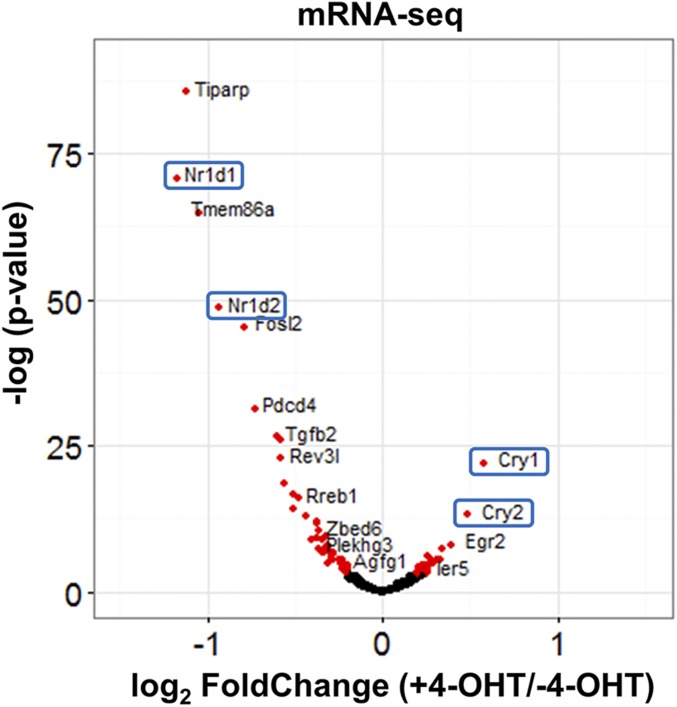
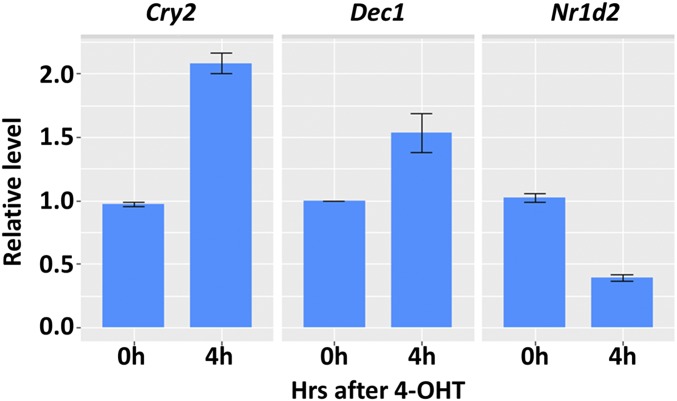
To analyze the effect of PER2 on CLOCK–BMAL1-controlled gene expression, we first defined CLOCK–BMAL1-controlled genes as genes with transcription start sites within ±5 kb from BMAL1 binding sites. Based on our ChIP-seq data, 3,317 genes were selected for the analysis. As apparent from the volcano plot in Fig. 5, nuclear entry of PER2-ER* changes the expression pattern, with 53 transcripts going down, 28 going up, and most remaining unchanged (Table S2). Interestingly, Cry1 and Cry2 are increased and Nr1d1 and Nr1d2 are decreased when PER2 enters the nucleus. To ascertain the quality of the data from BMAL1 ChIP-seq and RNA-seq, in Fig. 6 we focus on Cry1 and Nr1d1 loci, on chromosomes 10 and 11, respectively: In agreement with the RT-quantitative PCR and ChIP-quantitative PCR data in Fig. 3, nuclear entry of PER2 up-regulates Cry1 expression and represses the Nr1d1 transcript level and both are associated with a decrease in BMAL1 binding on the promoter. Similarly, increased Cry2, Dec1 (Bhlhe40), and decreased Nr1d2 transcription (Table S2) are confirmed by RT-quantitative PCR (Fig. S1).
Fig. 5.
Effect of PER2 nuclear entry on transcriptome by mRNA-Seq in Per1/2−/−;Nr1d1/2−/− cells. Effect of PER2 nuclear entry on transcription of individual genes is shown by Volcano plot with the x axis representing level of difference (+4-OHT vs. –4-OHT) and the y axis representing the level of statistical significance. Genes with a statistically significant difference between with and without 4-OHT are shown in red.
Table S2.
Genes repressed (left) and enhanced (right) following PER2 nuclear entry after 4-OHT treatment were identified by change levels (log2FoldChange)
| Repressed symbol | log2FoldChange | padj | Enhanced symbol | log2FoldChange | padj |
| Nr1d1 | –1.171079188 | 9.54E-68 | Cry1 | 0.582509459 | 7.01E-20 |
| Tiparp | –1.116491793 | 3.53E-82 | Cry2 | 0.488970366 | 2.39E-11 |
| Tmem86a | –1.048065647 | 6.27E-62 | Egr2 | 0.390135015 | 1.55E-06 |
| Nr1d2 | –0.929677858 | 5.60E-46 | Eif5a2 | 0.336959307 | 4.65E-06 |
| Fosl2 | –0.792265666 | 7.92E-43 | Ier5l | 0.326283989 | 0.000286143 |
| Pdcd4 | –0.727566796 | 4.29E-29 | Ccdc85b | 0.307035599 | 0.000334602 |
| Tgfb2 | –0.606135515 | 3.10E-24 | Scx | 0.286860505 | 0.000894672 |
| Rev3l | –0.583470454 | 1.25E-20 | Irf2bp2 | 0.26441073 | 0.000225802 |
| Tgfbr3 | –0.582549054 | 7.42E-24 | Junb | 0.262316777 | 0.002386822 |
| Rybp | –0.559187374 | 1.88E-16 | Hnrnpa0 | 0.259303336 | 0.000103812 |
| Blnk | –0.51000592 | 1.90E-12 | Maml3 | 0.25522041 | 0.004340804 |
| Fabp5 | –0.50465813 | 9.26E-15 | Socs1 | 0.252669957 | 0.026963439 |
| Rreb1 | –0.479442855 | 3.90E-14 | Egln3 | 0.252133043 | 0.011534232 |
| Mef2a | –0.436003054 | 4.39E-11 | Bhlhe41 | 0.244322867 | 0.039279442 |
| Gadd45g | –0.402989654 | 2.06E-07 | Hes1 | 0.243902722 | 0.033322159 |
| Errfi1 | –0.378623151 | 2.97E-10 | Klf10 | 0.243786348 | 0.002680085 |
| Rai1 | –0.377851524 | 1.15E-07 | Bhlhe40 | 0.241411878 | 0.003330418 |
| Pgrmc2 | –0.371340375 | 5.17E-10 | Enpp1 | 0.228086522 | 0.002704116 |
| Il6ra | –0.370987194 | 9.63E-08 | Lfng | 0.223648315 | 0.037798981 |
| Larp1b | –0.368270213 | 5.36E-06 | Hopx | 0.221850293 | 0.04072607 |
| Zbed6 | –0.359685857 | 9.21E-09 | Nrep | 0.221701588 | 0.003621797 |
| Aspa | –0.355603725 | 1.32E-05 | Tubb2a | 0.216466109 | 0.03395037 |
| Per3 | –0.341887931 | 2.29E-05 | Cited2 | 0.213776972 | 0.007570276 |
| Atl2 | –0.341145571 | 3.07E-07 | Txnip | 0.200820508 | 0.004317338 |
| Gclc | –0.337416771 | 1.90E-05 | Ier5 | 0.199428583 | 0.022612445 |
| Plekhg3 | –0.329756366 | 4.89E-06 | Mthfd1l | 0.192298143 | 0.020957662 |
| Ywhaq | –0.320749749 | 6.68E-08 | Samd1 | 0.190782997 | 0.046340934 |
| Ttc39b | –0.317267083 | 5.40E-06 | Tef | 0.182625049 | 0.037649738 |
| Sybu | –0.3120783 | 0.00132 | |||
| Ppp3ca | –0.309866297 | 1.89E-06 | |||
| Lrig1 | –0.299946889 | 2.20E-05 | |||
| Znhit6 | –0.289347349 | 0.00017 | |||
| Stambpl1 | –0.286900666 | 0.00026 | |||
| 1110057K04Rik | –0.284080917 | 4.87E-05 | |||
| Samd4 | –0.279540221 | 2.62E-05 | |||
| Ccdc86 | –0.278351595 | 0.000389 | |||
| Hat1 | –0.250139791 | 0.000241 | |||
| Zc3h11a | –0.239148722 | 0.000251 | |||
| Zmiz1 | –0.236867525 | 0.001491 | |||
| Cyr61 | –0.232502284 | 0.000332 | |||
| 5031439G07Rik | –0.231286348 | 0.002064 | |||
| Nfat5 | –0.229662991 | 0.000452 | |||
| Gpam | –0.22907332 | 0.007249 | |||
| Gemin5 | –0.223552359 | 0.007229 | |||
| Fnip2 | –0.218089956 | 0.00414 | |||
| Loxl4 | –0.21716325 | 0.002321 | |||
| Agfg1 | –0.216934213 | 0.002398 | |||
| Arhgap42 | –0.216566318 | 0.019797 | |||
| Serpine1 | –0.208667754 | 0.025378 | |||
| Nov | –0.202948844 | 0.004056 | |||
| Gm15408 | –0.202621692 | 0.003095 | |||
| Fbxo30 | –0.199338968 | 0.024817 | |||
| Bmp4 | –0.196890312 | 0.04087 |
Statistical significance was shown by adjusted P value (padj).
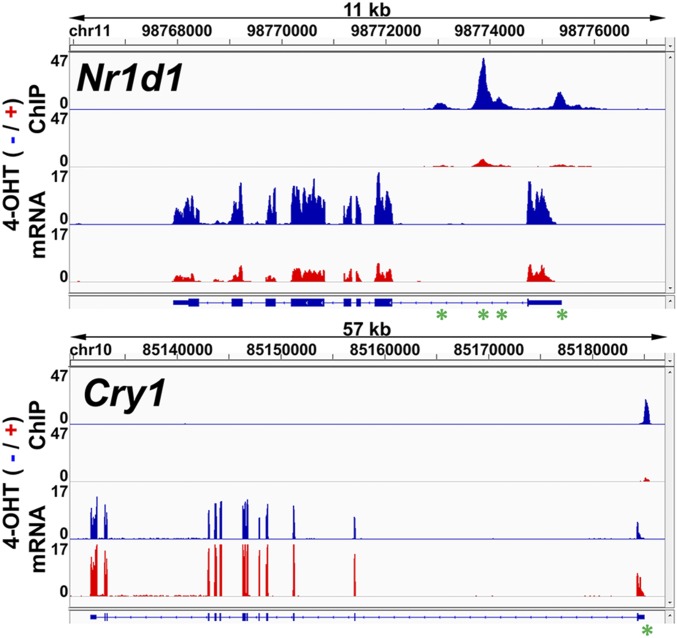
Fig. 6.
Visualization of BMAL1–ChIP-seq and RNA-seq on Nr1d1 (Top) and Cry1 (Bottom) gene loci. (Top) An 11 kbp region of chromosome 11 containing the Nr1d1 gene. (Bottom) A 57 kbp region of chromosome 10 containing the Cry1 gene. Gene structures are illustrated along the x axis beneath each panel, with E/E’-box regulatory elements indicated with green asterisks. Within each panel, the magnitude of the ChIP-seq signal (y axis) is shown in the top half, and the magnitude of the RNA-seq signal (y axis) is shown in the bottom half. Signals observed with 4-OHT are shown in red, and signals without 4-OHT are shown in blue. With both genes, lower ChIP-seq signals were observed with 4-OHT (red) than without 4-OHT (blue). However, following induction of PER2 with 4-OHT, RNA-seq data show a weaker signal in the Nr1d1 gene and a stronger signal in the Cry1 gene. Data were visualized using IGV software.
Fig. S1.
Nuclear entry of PER2-ER* following addition of 4-OHT (4 h) induces Cry2 (Left) and Dec1 (Middle) transcription but represses Nr1d2 (Right) transcription in Per1/2−/−;Nr1d1/2−/−;PER2-ER* cells analyzed by RT-quantitative PCR. SEs are plotted.
Decreased CLOCK–BMAL1 Binding on the Cry1 Promoter Increases Cry1 Transcription.
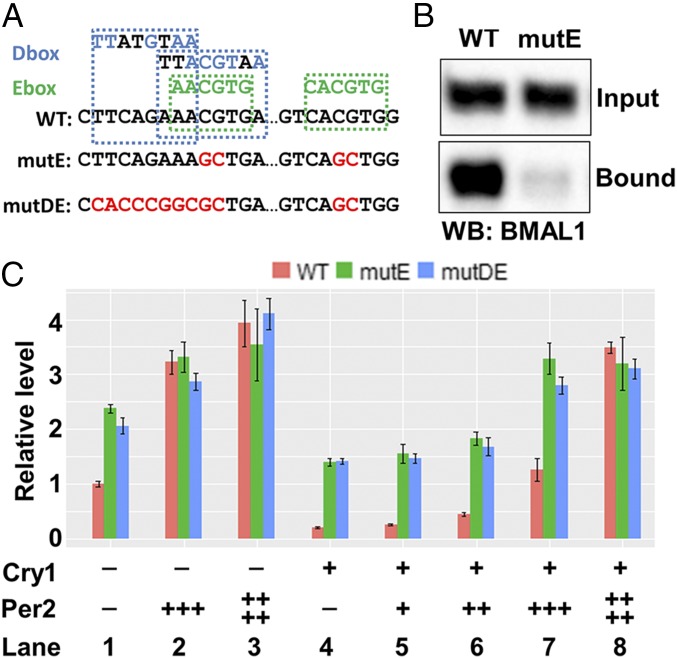
Finally, the experiments described so far were carried out with 4-OHT–induced nuclear entry of PER2-ER*, and it could be argued that in this model system, the observed removal of CLOCK–BMAL1 from promoters and the accompanying up-regulation of Cry1 might be due to some unique feature of our system and that the CLOCK–BMAL1 removal from the E-box upon PER2-ER* nuclear entry and Cry1 up-regulation might not be physiologically relevant. To address this issue, we conducted reporter gene assays using a 1.5 kbp promoter fragment of Cry1, which can sustain rhythmic transcription (14). This fragment possesses two D-box and two E-box sequences that partially overlap, as shown in Fig. 7A. To characterize the effect of CLOCK–BMAL1 binding on Cry1 transcription, we first compared the reporter activity of Cry1 promoters with either wild-type (WT) or mutant E-box (mutE) sequences (Fig. 7A). In a preliminary control experiment to ascertain the specificity of CLOCK–BMAL1 binding to the E-box in the Cry1 promoter, the two 1.5 kbp promoter fragments, end-labeled with biotin, were incubated with NIH 3T3 cell-free extract. The fragments were pulled down and the bound BMAL1 was visualized in the Western blot shown in Fig. 7B. BMAL1 bound to the WT promoter but only weakly to the mutant promoter.
Fig. 7.
Decreased CLOCK–BMAL1 binding increases reporter gene expression driven from the Cry1 promoter. (A) Sequences of the Cry1 promoter region including WT, mutant E-boxes, and D-boxes are shown. Nucleotides conserved in the E-box and D-box sequences are colored in green and blue, respectively. Nucleotides mutated in mutE (E-box mutation) and mutDE (D-box and E-box mutation) are colored in red. (B) CLOCK–BMAL1 binding affinity to Cry1 promoters with WT or mutE sequences. BMAL1 was pulled down from nuclear extract from NIH 3T3 cells by immobilized biotin–DNA and analyzed by Western blot. (C) Reporter gene assay of the Cry1 promoters with the WT or mutant promoter elements is shown. Reporter signals were first normalized to the signals from cotransfected control plasmid and then were normalized to the condition without Cry1 and Per2 plasmids. Note that reporter gene expression driven by the mutant Cry1 promoter is higher than expression driven by the WT Cry1 promoter. SEs are plotted in C.
Reporter assays were then performed following cotransfection of the Cry1 promoter/reporter constructs with and without PER2 and/or CRY1-expressing plasmids, which is a commonly used circadian experimental system. Fig. 7C, lane 1 shows that in the absence of exogenous PER2 or CRY1, there was much more expression from the mutE (green) than the WT (red) promoter. This increase is consistent with strong transcription from another element on the 1.5 kbp fragment that is inhibited by CLOCK–BMAL1–CRY1 binding to the WT E-box. Expression of PER2 led to substantially increased transcription from the WT promoter fragment (lanes 2 and 3). Apparently the basal level of CRY1/2 in these cells is sufficient to enable the removal of CLOCK–BMAL1 by exogenous PER2 so as to reveal strong transcription. A modest increase in transcription of the mutE promoter by exogenous PER2 is consistent with the removal of residual CLOCK–BMAL1 bound weakly to the mutant promoter (green, lane 1 vs. lanes 2 and 3). Comparing lane 4 versus lane 1 (red) shows that exogenous CRY1 inhibited transcription mediated by CLOCK–BMAL1. Increasing the amount of exogenous PER2, in the presence of exogenous CRY1 (lanes 5–8), resulted in increased transcription, as PER2 removed CLOCK–BMAL1 from the E-boxes in a CRY1-dependent manner. These data from cellular oscillators support our conclusion that up-regulation of Cry1 by PER2 is not simply due to removal of CRY1 to facilitate CLOCK–BMAL1-mediated transcription but in fact is due to the removal of the entire CRY1–CLOCK–BMAL1 “repressor complex” to facilitate the activation of transcription by another sequence element in the 1.5 kbp Cry1 promoter region.
It has been shown that Cry1 transcription could be regulated through the D-box in the promoter (14). It is possible that removal of CLOCK–BMAL1 from the E-boxes by PER facilitates the transcription of Cry1 through the nearby and overlapping D-boxes (Fig. 7A). We considered that D-box binding proteins, such as DBP, TEF, and HLF, would activate the transcription of Cry1 when not blocked by the binding of CLOCK–BMAL1–CRY to the E-boxes. To test this, we measured the reporter activity of a Cry1 promoter with mutated D-box and E-box (mutDE, Fig. 7A). As seen with the mutE promoter, mutDE promoter activity in the absence of exogenous PER2 or CRY1 was much higher than WT (lane 1, blue) and furthermore was very similar to mutE in all conditions tested. Thus, the increase in Cry1 transcription mediated by PER as a result of CLOCK–BMAL1 dissociation is not by removal of a repressor complex interfering with D-box–mediated activation. It is most likely that another transcriptional activator with a binding site in the 1.5 kbp Cry1 promoter fragment is responsible for increased Cry1 expression following removal of CLOCK–BMAL1. For purposes of discussion, this possible transcription factor will be referred to as factor T, which binds to a postulated T element in the Cry1 promoter (Fig. 8).
Fig. 8.
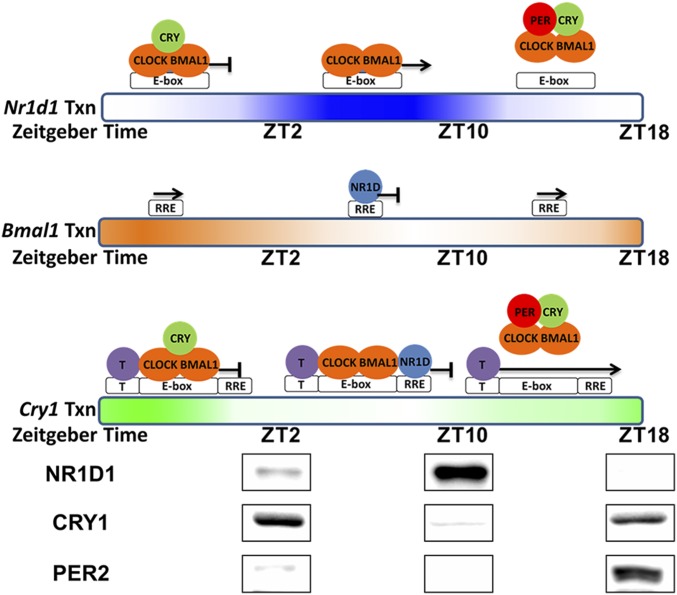
Model of PER-mediated regulation of Nr1d1, Bmal1, and Cry1 genes. This model represents transcription and regulation patterns of the Nr1d1, Bmal1, and Cry1 genes individually as a function of the circadian time. Gradient-colored bars represent transcription of Nr1d1 (blue), Bmal1 (orange), and Cry1 (green) genes based on nascent RNA-seq data from mouse liver (27). Above the bars are the proposed status of promoter occupation and transcription. Spacing of the T element, E-box, and RRE in this representation of the Cry1 promoter is for illustrative purposes only. The RRE element is in intron 1 of Cry1 ∼23 kbp downstream from the E-box. At the bottom of the model are the levels of NR1D1, CRY1, and PER2 in mouse liver nuclei. The levels of these and additional proteins from different times were cropped from a single image (Fig. S2). For Nr1d1, which is controlled solely by an E-box, transcription is inhibited by either binding of CRY on CLOCK–BMAL1 or removal of CRY1–CLOCK–BMAL1 from the E-box by PER. For Bmal1, which is controlled solely by an RRE, transcription is inversely correlated with the NR1D1 level. For Cry1, which is controlled by an E-box, RRE, and a putative factor T, binding of either the CRY–CLOCK–BMAL1 complex on the E-box or NR1D1 on the RRE represses the transcription of Cry1 from another element (T). PER eliminates both pathways of inhibition by removing CLOCK–BMAL1 from the E-box.
Fig. S2.
Original Western blot image used in Fig. 8 shows the changes of different circadian proteins in the nucleus of mouse liver at different Zeitgeber Times (ZT).
Discussion
The mammalian molecular circadian clock consists of a TTFL generated by four genes (core clock genes) and their paralogs. Research over the past two decades has supported the TTFL model and identified a secondary loop involving nuclear receptors Nr1d1/2. Furthermore, identification of kinases (28, 29) and ubiquitin ligases (30, 31) that modulate the activity and stability of the core clock proteins has provided insights into the mechanisms that are necessary to establish a daily rhythm of high amplitude, precise period, and flexibility for phase resetting in response to stimuli.
Despite these important developments, the negative arm of the TTFL model has remained poorly defined. In particular, the respective roles of CRY and PER in repression have been unclear. There are several reasons for this uncertainty. First, the two proteins interact strongly and aid in one another’s stability and nuclear entry (32–34). Second, removal of one affects the posttranslational modification and stability of the other. Also, although CRY has been shown to be capable of repressing target genes on its own, there is no convincing evidence that PER affects the TTFL in the absence of CRY (15). Moreover, depending on the clock gene tested, PER has been reported to function as either a repressor (for Nr1d1) or an up-regulator (for Cry1 and Bmal1).
To examine the clock model, we have developed an “in cellulo biochemical” system consisting of mouse cell lines with knockout mutations in one or more of the core clock genes and that have a controllable delivery system for CRY or PER proteins (15). In this study, using the PER2 delivery system, we have obtained RT-quantitative PCR, RNA-seq, ChIP, and ChIP-seq data that have enabled us to revise the consensus mammalian clock model so as to reconcile various views regarding the roles of CRY and PER in the negative arm of the TTFL and eliminate the “internal inconsistencies” of the conventional model (3).
We previously reported that PER causes the removal of CLOCK–BMAL1 from the cognate promoters in a CRY-dependent manner and that it inhibits the transcription of certain genes with simple promoters (promoters with only an E-box control element) by this mechanism. In this study, we investigated the effect of PER2 on transcription of genes with more complex promoters. In aggregate, our work shows that PER2 exerts three types of effect on clock gene transcription, depending upon the type of promoter. Furthermore, we show that all of these effects are CRY-dependent, and moreover our data reconcile some previous seemingly contradictory reports on PER function in the circadian clock. We explain the three different effects of PER2 on circadian gene expression as follows:
-
i)
Nr1d1 (repressed by PER2): This gene is predominantly regulated by an E-box. Binding of CRY1 (or CRY2) to the CLOCK–BMAL1–E-box complex represses transcription. When PER2 is abundant enough, it removes the CRY1–CLOCK–BMAL1 complex from the promoter as part of the dual repression mechanism of CRY and PER (15).
-
ii)
Bmal1 (de-repressed by PER2): This gene is predominantly regulated by NR1D1/NR1D2 nuclear receptors; CRY1/CRY2 de-repress its transcription. We show that de-repression is through the effects of CRY1/2 and PER1/2 on NR1D1/NR1D2 expression. When CRY and PER are at high enough concentrations, they inhibit NR1D1/NR1D2 expression through the E-box and thus down-regulate the repressors of Bmal1 causing up-regulation of Bmal1 transcription. Knockout of Nr1d1/Nr1d2 eliminates the effect of CRYs and PERs on Bmal1 transcription, which is mediated by NR1D1/NR1D2.
-
iii)
Cry1 (de-repressed by PER2): This gene has several transcriptional elements, three of which have been studied previously—D-box (day element), E-box (morning element), and RRE (evening element)—which are the targets of transcription factors DBP, CLOCK–BMAL1, and NR1D1/NR1D2, respectively (14). Up-regulation (de-repression) of the Cry1 promoter by PER2 seen in our experiments is not consistent with the accepted role of PER2 as a repressor. Kondratov et al. (19) and Liu et al. (12) observed related, unexpected results, namely the up-regulation of Cry1 in Bmal1 knockout cells. Kondratov et al. (19) suggested that with some genes, such as Cry1, there is a high “basal” level of transcription that is not substantially increased by CLOCK–BMAL1 binding to the promoter, and binding of CRY1 to the CLOCK–BMAL1 complex inhibits both CLOCK–BMAL1 activated and basal transcription. In this vein, our findings are consistent with the existence of an additional transcriptional activator (factor T) that binds to a regulatory element (T element) in the Cry1 promoter (12) and is inhibited by the CRY1–CLOCK–BMAL1 complex. In this scenario, removal of CRY1-CLOCK-BMAL1 from the Cry1 promoter by PER2 restores transcription via factor T.
Regarding the elevated Cry1 level in the Bmal1 knockout cells, Liu et al. (12) hypothesized that the absence of BMAL1 led to less NR1D1/NR1D2, which normally binds to the RRE element and represses transcription of Cry1. In fact, in their experiments, overexpression of NR1D1 in Bmal1 knockout cells did inhibit Cry1 expression. Thus, their interpretation of BMAL1 indirectly inhibiting Cry1 expression through NR1D1/NR1D2 appears appropriate to their defined experimental system, and it is reasonable to expect that NR1D1/NR1D2 influence Cry1 expression in vivo. However, our cell-based (Fig. 3) and reporter assay (Fig. 7) data show that PER2-mediated expression of Cry1 is independent of NR1D1 (our reporter lacked the NR1D1 binding site). These seemingly conflicting results are reconciled and incorporated in the revised clock model shown in Fig. 8, which also shows levels of NR1D1, CRY1, and PER2 proteins in mouse liver at various circadian times. At ZT10, NR1D1 is high and it likely represses Cry1 by binding to the RRE element, whereas at ZT18 NR1D1 is low, and this likely contributes to initiate Cry1 expression. Also, at ZT18, PER2 is high and PER2 at this time likely helps initiate Cry1 transcription by removing the inhibitory CRY1–CLOCK–BMAL1 complex. Thus, in this model, PER2 has dual roles in regulation of Cry1. First, through displacement of CRY–CLOCK–BMAL1 from the Nr1d1/Nr1d2 promoters, it represses NR1D1/NR1D2 and thus indirectly activates Cry1 expression. Second, through displacement of CRY–CLOCK–BMAL1 from the Cry1 promoter, it directly de-represses Cry1 expression.
We note that our findings contradict a recent report (20) that claimed that PER2 up-regulates Cry1 transcription by removing CRY1 from the CRY1–CLOCK–BMAL1 complex at the Cry1 E-box and at the E-boxes of other genes regulated by CLOCK–BMAL1. Here, we confirm our earlier finding (15) that in fact PER2 nuclear entry causes the removal of CLOCK–BMAL1 from the Cry1 promoter in a CRY-dependent manner while at the same time up-regulating Cry1 transcription.
Because the D-box in the Cry1 promoter has been shown to play a role in Cry1 regulation, we considered transcription factors (DBP, TEF, and HLF) that bind to the D-box as potential Cry1 transcriptional activators when PER2 removes the CRY–CLOCK–BMAL1 complex from the Cry1 promoter. We tested this possibility by using reporter gene assays with a 1.5 kbp fragment carrying the Cry1 promoter including the D-box and E-box elements. As expected, in this system, CRY1 repressed and PER2 together with CRY1 de-repressed transcription of Cry1. MutE and double mutant E-box/D-box constructs, with greatly reduced CLOCK–BMAL binding, demonstrated reduced transcription stimulation following PER2 induction. Thus, DBP cannot be factor T whose activity is revealed by dissociation of the CRY1–CLOCK–BMAL1 complex. Additional studies are needed to identify the postulated T element and T factor that acts in a tonic manner (12) to regulate Cry1 transcription and whose effect is rendered circadian in the context of the entire circadian ensemble.
Materials and Methods
Cells and Antibodies.
Cell lines (Table S1) were maintained in DMEM supplemented with 10% (vol/vol) FBS in 5% (vol/vol) CO2 at 37 °C. Blasticidin (5 μg/mL) was present during cell culture and absent during experiments. To induce PER2-ER* nuclear entry, 4-hydoxytamoxifen was added to the medium to 1 μM when cells were ∼90% confluent.
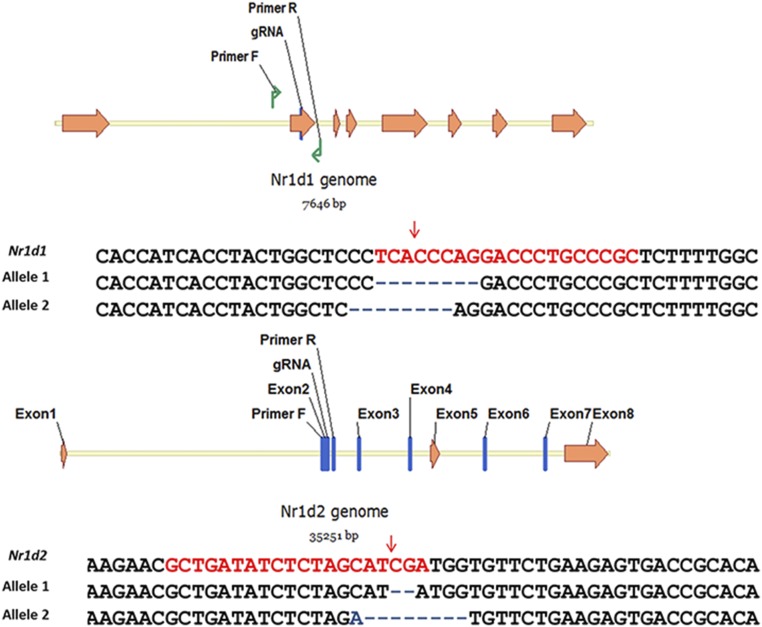
The Bmal1−/−, Per1/2−/−, and Cry1/2−/−;Per1/2−/− mouse embryonic fibroblasts have been described (15, 32). The Per1/2−/−;Nr1d1/2−/- mouse embryonic fibroblast cell line was made by CRISPR technology using LentiCRISPRv2 (35) obtained from Addgene to mutate the Nr1d1/2 alleles in Per1/2−/− fibroblasts. Single colonies lacking NR1D1/2 proteins were isolated and screened by Western blot, and mutational inactivation was confirmed by genomic DNA sequencing. Mutated sequences in the isolated clone (PN-P2ER) are shown in Fig. S3. Lack of NR1D1/2 protein and movement of PER2-ER* to the nucleus are shown in Fig. S4.
Fig. S3.
Target sequences (highlighted in red) for Nr1d1 and Nr1d2 knockout are both located in Exon2 of the genes. Sequences of Nr1d1 and Nr1d2 near the target site from Per1/2−/−;Nr1d1/2−/−;PER2-ER* cells indicate frame-shifted mutation of both genes on both alleles (Nr1d1: –8 nt, –8 nt; Nr1d2: –2 nt, –8 nt). Locations of primers used to amplify genomic DNA for sequencing and location of gRNA target sequence are shown. Red arrows indicate the expected cutting sites on DNA.
Fig. S4.
Western blot data show that there is no detectable NR1D1 or NR1D2 protein in Per1/2−/−;Nr1d1/2−/−;PER2-ER* cells (Left). Nuclear PER2-ER* level is induced in these cells after 4-OHT treatment (Right).
DNA containing mPER2-ER* was subcloned from pBABE-puro-mPER2-ER* into plasmid pWZL-blast to create pWZL-blast-mPER2-ER*. Cells stably expressing P-P2ER, PN-P2ER, and B-P2ER were made by retrovirus infection using pWZL-blast-mPER2-ER* (15).
Anti-mCRY1 (IgM-type monoclonal) antibodies were described previously (18). Anti-CLOCK (Bethyl Laboratories), anti-BMAL1 (Bethyl Laboratories), anti-CRY2 (Bethyl Laboratories), anti-PER2 (Alpha Diagnostic International), anti–Rev-Erbαα (NR1D1) (Cell Signaling Technology), anti–Rev-Erbβ (NR1D2) (Santa Cruz Biotechnology), anti-MEK2 (BD Biosciences), and anti-Lamin A/C (EMD Millipore) antibodies were obtained from commercial sources.
ChIP and mRNA Real-Time PCR.
ChIP and mRNA real-time PCR were performed as previous described with minimal modifications (15). Protein G Dynabeads (Thermo Fisher Scientific) were used for purification of immune complexes, and eluted DNA was purified using phenol-chloroform extraction and ethanol precipitation. Real-time PCR assays were performed using an ABI 7500 system (Applied Biosystems) using primers shown in Tables S3 and S4.
Table S3.
Primer sets used for RT-quantitative PCR
| Gene name | Forward primer | Reverse primer |
| mGapdh | AACTTTGGCATTGTGGAAGG | ACACATTGGGGGTAGGAACA |
| mNr1d1 | CCTGACTCAAGGTTGTCCCACAT | CATGGCCACTTGTAGACTTCCTG |
| mBmal1 | TCATTGATGCCAAGACTGGA | CAAATAGCCTGTGCTGTGGA |
| mCry1 | TCCCCTCCCCTTTCTCTTTA | TGAGTCATGATGGCGTCAAT |
| mCry2 | CACTGGTTCCGCAAAGGACTAC | GCTTTCTTAAGCTTGTGTCCAGA |
| mDec1 | TTGGGTCACTTGGAAAAAGC | CAGGTCCCGAGTGTTCTCAT |
| mNr1d2 | AGCAGTGCCGCTTTAAGAAG | TCCTGAGCTGGTAGTGCTGA |
Table S4.
Primer sets used for ChIP-quantitative PCR
| Name | Sequence | Genome position, mm10 |
| mNr1d1_–19 | GCTGCTGGAAAAGTGTGTCA | chr11(–): 98775377–98775396 |
| mNr1d1_+215 | ATGGAGAAATGAGGCACCAG | chr11(+): 98775162–98775181 |
| mBmal_+24 | CGGATTGGTCGGAAAGTAGG | chr7(+): 113207489–113207508 |
| mBmal_+197 | AGCCATGCCGACACTCAC | chr7(–): 113207645–113207662 |
| mCry1_–67 | GGCACCTCACGTTTCTGAAG | chr10(+): 85185121–85185140 |
| mCry1_–273 | TATGCCTCCTCCCCCTTCT | chr10(-): 85185309–85185327 |
Real-time PCR assays measured expression of RNA following addition of 4-OHT. For each gene, in each cell line, the values for RNA expressed after 4-OHT addition were normalized to give the percent expression relative to the level expressed at zero time. It should be noted that at zero time, the RNA levels of some clock RNAs varied between cell lines, compared with expression of control RNA (GAPDH). Thus, in Fig. 3A, Right, although all cell lines showed a similar level of Cry1 RNA expression at zero time, in Bmal1−/− cells, Cry1 RNA was expressed constitutively at a high level.
Library Generation and Next Generation Sequencing.
ChIP-seq.
DNA libraries of input or BMAL1-ChIP from Per1/2−/−;Nr1d1/2−/−;PER2-ER* cells with or without 4-OHT treatment (4 h) were made using ThruPLEX DNA-seq Kit (RUBICON GENOMICS). Libraries were sequenced using Illumina HiSEq. 2000 (1 × 50). Two independent experiments were performed. The number of reads obtained from each sample was around 15–20 million.
RNA-seq.
Total RNA was extracted from PN-P2ER cells with or without 4-OHT treatment (4 h) using TRIzol RNA extraction (Thermo Fisher Scientific). After phase separation, RNA was purified using the PureLink RNA Mini Kit (Thermo Fisher Scientific). Libraries were made using TruSeq stranded mRNA preparation kit (Illumina) and sequenced using Illumina HiSEq. 2000 (1 × 50 × 2 lanes). The number of reads obtained from each sample was around 80∼90 million.
Next Generation Sequencing Data Analysis.
The sequences were trimmed to remove adaptor sequences using BBDuk tool (bbmap/35.82) (36) using parameters “ktrim = r k = 23 hdist = 1 minlen = 50.”
ChIP-Seq analysis.
Sequencing reads from each experiment were mapped to the mouse genome (mm10) using Bowtie2/2.2.8 (37) using default parameters. Low-quality reads were filtered using Samtools/1.3 (38) with option “-q 10.” Enrichment regions (peaks) relative to the matching input control for each experiment were determined using Callpeaks function in MACS/2015–04-20 (39) with options “-f BAM -g mm -B -q 0.01.” Given the peak regions called in each experiment, we next determined the common peak regions using Intersect function in Bedtools/2.25.0 (40). From two experiments, 4,740 common peak regions were determined in the condition without 4-OHT treatment, and 483 regions were determined in the condition with 4-OHT treatment. Because the regions determined in the condition with 4-OHT treatment were included in the regions determined in the condition without 4-OHT, we used these 4,740 regions for further quantitative analysis. To verify the specificity of our data, we performed motif analysis of these regions by MEME-ChIP (41) and the result showed high enrichment of the E-box sequence (CACGTG). To compare with random regions from the genome, we generated random regions with the same length distribution using the ShuffleBed function in Bedtools/2.25.0.
To quantify the relative binding strength of BMAL1, we first tabulated the number of unique mapping reads per region using BamtoBed and Intersect functions (with option “-c”) in Bedtools/2.25.0. Then, the numbers were normalized to number of reads per 10 million reads. We then determined the relative peak binding strength, defined as (number of reads in ChIP + c)/(number of reads in Input + c) in these regions. The constant “c” was used to smooth fluctuations in fold change due to very small denominator values, shrinking the fold change toward 1 for small counts. We chose c = 5 in this case as a conservative measure.
RNA-Seq.
Sequencing reads from each experiment were mapped to the mouse genome (mm10) using bbmap/35.82 (36) with the option “maxindel = 100000 intronlen = 10 ambig = random qin = 33.” Mapped reads were quantified using featureCounts (42) with option “-T 4 -t exon.” Gene expression analysis was performed using Deseq2 (43). To analyze CLOCK–BMAL1 controlled genes, the transcription start sites of annotated genes were compared with the BMAL1 binding sites (extended to 10 kb) identified in our ChIP-seq using Intersect functions in Bedtools/2.25.0. Data of annotated genes with a transcription start site within 5 kb from a BMAL1 binding site were selected from the Deseq2 analysis and were shown by Volcano blot.
Reporter Gene Assay and Biotin-DNA Pull Down.
The mouse Cry1 promoter (1.5 kb from –1208 to +328) was amplified by PCR from genomic DNA of PN-P2ER* cells using the primer sequences provided in a previous publication (14). The amplified DNA fragment was cloned into pGL4.16 plasmid (Promega) using NheI and XhoI. Two E-boxes identified in this region (AACGTG, CACGTG) were mutated to AAGCTG and CAGCTG (mutE). D-boxes were mutated using the published sequence (14) and plasmid mutE as template to generate the mCry1 promoter with mutDE. Mutagenesis was performed by Q5 mutagenesis kit (NEB). Reporter gene assay was performed by transfection of pGL4.16 (250 ng), pBind (50 ng), pcDNA3-mCry1 (50 ng), pcDNA3-mPER2 (100, 200, 400, and 800 ng), and pcDNA4-myc-his (to final 1,150 ng) into NIH 3T3 cells in 24-well plates using Lipofetamine 3000 (Thermo Fisher Scientific). After 24 h, reporter gene expression was analyzed using the Dual-Luciferase Reporter Assay System (Promega).
For biotin–DNA pull-down analysis, the 1.5 kbp NheI–XhoI WT and mutant Cry1 promoter fragments were first purified after NheI and EcoRV (next to XhoI) digestion of the reporter plasmids. Biotin-11-dUTP was incorporated into the DNA fragments by fill-in of the 5′-overhangs of the NheI-digested sites using exo− Klenow (NEB). DNA was purified by microSpin G50 column (GE Healthcare Life Sciences) and phenol-chloroform/ethanol precipitation to remove residual Biotin-dUTP. Binding of CLOCK–BMAL1 was performed in 30 μL reactions with 600 ng of poly-dI:C, 30 μg of NIH 3T3 nuclear extract, 0.33 M urea, 0.33% Nonidet P-40, 100 mM NaCl, and 300 fmol biotin–DNA. After 30 min incubation on ice, 20 μL of each reaction was mixed with 5 μL Dynabeads M280 streptavidin (Thermo Fisher Scientific) and incubated for another 30 min with rotation. After washing with TE buffer, BMAL1 was eluted with SDS sample buffer and detected by Western blot.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612917113/-/DCSupplemental.
References
- 1.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24(2):90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardin PE, Panda S. Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol. 2013;23(5):724–731. doi: 10.1016/j.conb.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 4.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 5.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96(21):12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shearman LP, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288(5468):1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 7.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 8.Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105(5):683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 9.Zheng B, et al. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400(6740):169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 10.Bugge A, et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26(7):657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu AC, et al. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4(2):e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 14.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144(2):268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Ye R, et al. Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev. 2014;28(18):1989–1998. doi: 10.1101/gad.249417.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421(6919):177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 17.Xu H, et al. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat Struct Mol Biol. 2015;22(6):476–484. doi: 10.1038/nsmb.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen KD, et al. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341(6153):1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. 2006;20(3):530–532. doi: 10.1096/fj.05-5321fje. [DOI] [PubMed] [Google Scholar]
- 20.Akashi M, et al. A positive role for PERIOD in mammalian circadian gene expression. Cell Reports. 2014;7(4):1056–1064. doi: 10.1016/j.celrep.2014.03.072. [DOI] [PubMed] [Google Scholar]
- 21.Chappuis S, et al. Role of the circadian clock gene Per2 in adaptation to cold temperature. Mol Metab. 2013;2(3):184–193. doi: 10.1016/j.molmet.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampp G, et al. Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr Biol. 2008;18(9):678–683. doi: 10.1016/j.cub.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430(6998):467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 24.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 25.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24(4):345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vielhaber E, Eide E, Rivers A, Gao ZH, Virshup DM. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol Cell Biol. 2000;20(13):4888–4899. doi: 10.1128/mcb.20.13.4888-4899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J Biol Chem. 2002;277(19):17248–17254. doi: 10.1074/jbc.M111466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129(5):1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoo SH, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152(5):1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem. 2011;286(29):25891–25902. doi: 10.1074/jbc.M111.254680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nangle SN, et al. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. eLife. 2014;3:e03674. doi: 10.7554/eLife.03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107(7):855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 35.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bushnell B. 2014 BBMap. Available at https://sourceforge.net/projects/bbmap/
- 37.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma W, Noble WS, Bailey TL. Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat Protoc. 2014;9(6):1428–1450. doi: 10.1038/nprot.2014.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]