Significance
Understanding the functions of cellular transcripts based on their sequence is challenging, in particular for noncoding RNAs, which tend to lack easily recognizable motifs. A more functionally relevant criterion is the association of RNAs with cognate RNA-binding proteins. Here, we describe the gradient profiling by sequencing (Grad-seq) approach to draft global RNA landscapes through partitioning all cellular transcripts into diverse coding and noncoding groups based on their shared RNA–protein interactions. Grad-seq has enabled us to define a large class of structured small RNAs that commonly associate with the conserved RNA-binding protein ProQ and appears to constitute a new branch of posttranscriptional control in bacteria. The generic nature of the Grad-seq approach will help to rapidly describe functional RNA landscapes in numerous understudied microbes.
Keywords: small RNA, noncoding RNA, RNA–protein interaction, ProQ, Hfq
Abstract
The functional annotation of transcriptomes and identification of noncoding RNA (ncRNA) classes has been greatly facilitated by the advent of next-generation RNA sequencing which, by reading the nucleotide order of transcripts, theoretically allows the rapid profiling of all transcripts in a cell. However, primary sequence per se is a poor predictor of function, as ncRNAs dramatically vary in length and structure and often lack identifiable motifs. Therefore, to visualize an informative RNA landscape of organisms with potentially new RNA biology that are emerging from microbiome and environmental studies requires the use of more functionally relevant criteria. One such criterion is the association of RNAs with functionally important cognate RNA-binding proteins. Here we analyze the full ensemble of cellular RNAs using gradient profiling by sequencing (Grad-seq) in the bacterial pathogen Salmonella enterica, partitioning its coding and noncoding transcripts based on their network of RNA–protein interactions. In addition to capturing established RNA classes based on their biochemical profiles, the Grad-seq approach enabled the discovery of an overlooked large collective of structured small RNAs that form stable complexes with the conserved protein ProQ. We show that ProQ is an abundant RNA-binding protein with a wide range of ligands and a global influence on Salmonella gene expression. Given its generic ability to chart a functional RNA landscape irrespective of transcript length and sequence diversity, Grad-seq promises to define functional RNA classes and major RNA-binding proteins in both model species and genetically intractable organisms.
The genomes of many studied organisms are pervasively transcribed, and a significant proportion of this RNA output is accounted for by noncoding RNA (ncRNA) (1, 2). This collective term encompasses different types of transcripts such as ribosomal, transfer, and other housekeeping RNAs [e.g., signal recognition particle (SRP) and RNase P RNA components, tmRNA, or CRISPR RNAs], and also many regulatory RNA species. The latter group is particularly vast and heterogeneous, and several new classes, such as siRNAs, micro RNAs (miRNAs), PIWI-interacting RNAs (piRNAs), long noncoding RNAs, and various bacterial regulatory small RNAs (sRNAs), have emerged as important modulators of gene expression (3, 4).
The discovery of new RNA classes has been greatly facilitated by next-generation sequencing, which, by reading transcript sequences, theoretically allows the rapid profiling of all RNA molecules in a cell (5, 6). However, the sequence per se has limited predictive power when used to identify classes of functionally related ncRNAs, as these often lack easily identifiable motifs such as the translation signals of protein-coding transcripts. This has been particularly evident for the bacterial sRNAs, which dramatically vary in length and structure within and among bacteria (4, 7). Therefore, an informative description of the ncRNA landscape in any organism necessitates the use of more direct and functionally relevant criteria.
One powerful diagnostic trait for ncRNA classes is their complex formation with particular RNA-binding proteins (RBPs), which, for example, has been used to define Argonaute-associated miRNAs or piRNAs (8, 9). In bacteria, the largest class of posttranscriptional regulators is represented by the sRNAs that associate with the Hfq protein (10, 11). This RNA chaperone both stabilizes bound sRNAs and helps them regulate their mRNA targets via imperfect base pairing (12–15). Together, Hfq and its associated sRNAs impact the expression >20% of all genes in enteric model bacteria such as Escherichia coli and Salmonella enterica (16, 17). A second widespread class of bacterial sRNAs interact with proteins of the CsrA/RsmA family via GGA motifs. In E. coli, the CsrB/C and McaS sRNAs sequester the translational repressor CsrA, which in turn regulates hundreds of mRNAs (18, 19).
Importantly, even the well-characterized model bacteria E. coli and Salmonella contain many additional sRNAs that lack the motifs recognized by Hfq and CsrA (20–22), whereas other prokaryotes possess functional ncRNAs but no Hfq homolog (7, 23, 24). This strongly suggests that additional global RBPs and associated classes of sRNAs with roles in posttranscriptional regulation exist, but have escaped identification by conventional genetic and biochemical approaches. Here, we have combined a classic biochemical technique with high-throughput analysis to reveal the complete functional RNA landscape of a bacterial cell. Our global partitioning of cellular transcripts based on their biochemical behavior has resulted in the discovery of a domain of posttranscriptional control by ProQ, a widespread RBP with a previously unknown large suite of cellular targets, which include many highly structured regulatory sRNAs.
Results and Discussion
Global Partitioning of Cellular RNAs by Grad-Seq.
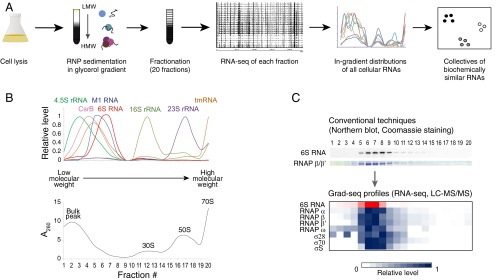
To describe the RNP landscape in the model enteric bacterium S. enterica serovar Typhimurium (henceforth Salmonella), we applied a high-throughput biochemical profiling approach (Grad-seq) (Fig. 1A). Grad-seq relies on the sedimentation of cellular RNAs and proteins in a glycerol gradient, which sorts complexes by size and shape and offers a means to assess their involvement in various macromolecular assemblies (25, 26). Following this biochemical partitioning step, we analyzed the RNA content of each of the 20 gradient fractions by Illumina cDNA sequencing and visualized the sedimentation profiles of 3,969 individual Salmonella transcripts (SI Appendix, Figs. S1 and S2 and Dataset S1). These profiles readily matched the expected distributions of housekeeping RNAs in glycerol gradients, including the 16S and 23S rRNAs, which cosediment with the 30S and 50S ribosomal subunits, respectively, tmRNA and the SRP and RNase P RNA components (Fig. 1B). Transfer RNAs primarily exist in small RNPs (average s020,w ∼5S). Importantly, the tRNA sedimentation profiles (SI Appendix, Fig. S1) correspond to those of their main protein partners, namely aminoacyl-tRNA synthetases, tRNA modification enzymes and elongation factor Tu, based on liquid chromatography-tandem MS (LC-MS/MS) detection of 1,326 proteins in total from the same gradient fractions (SI Appendix, Fig. S3 and Dataset S2).
Fig. 1.
Grad-seq visualizes the Salmonella RNA interactome. (A) Grad-seq experimental strategy. (B) RNA-seq–based in-gradient distributions of housekeeping RNAs (all profiles are standardized to the range from 0 to 1). M1 and 4.5S RNAs are the RNA subunits of RNase P and SRP, respectively. CsrB is a CsrA-sequestering ncRNA. The UV profile of the corresponding gradient showing the bulk peak of low molecular weight complexes and the positions of ribosomal subunits is provided below. (C) The 6S RNA (in complex with the RNA polymerase holoenzyme) visualized with conventional techniques (Top, cropped from SI Appendix, Fig. S3A) and by Grad-seq (heat map below, all profiles are standardized to the range from 0 to 1).
Likewise, 6S RNA associates with RNA polymerase holoenzyme (RNAP) to modulate the activity of the transcription machinery (27). We obtained almost congruent profiles of 6S RNA reads and RNAP-derived peptides (Spearman’s 0.62 < r < 0.91, P < 0.0034), which matched in-gradient profiles determined by standard techniques (Fig. 1C): both indicated the formation of a ∼16S complex, corresponding to a previously reported particle of 500–600 kDa (27).
Regulatory ncRNAs were more heterogeneously distributed. For instance, Hfq-associated sRNAs (11) sedimented broadly in fractions 3–7 (average s020,w ∼11S, or ∼350 kDa) with additional peaks in the 30S or 70S ribosome fractions (SI Appendix, Fig. S1). Representatives of other functional RNA classes (attenuators, antisense RNAs, Hfq-independent sRNAs) displayed even more disparate distributions, suggesting that they are associated with several distinct RBPs. As expected, mRNAs abundantly populate both the 30S and 70S ribosome fractions (SI Appendix, Fig. S1). The mRNA reads in lower molecular weight fractions likely represent untranslated mRNAs in complex with regulatory proteins or stable decay intermediates. Of note, the profile of the dual-function tmRNA, which rescues stalled ribosomes (28), combined profile features of both coding and noncoding RNAs with pronounced peaks in both low molecular weight and 70S fractions (Fig. 1B and SI Appendix, Fig. S1A).
Topology of a Bacterial RNA Interactome.
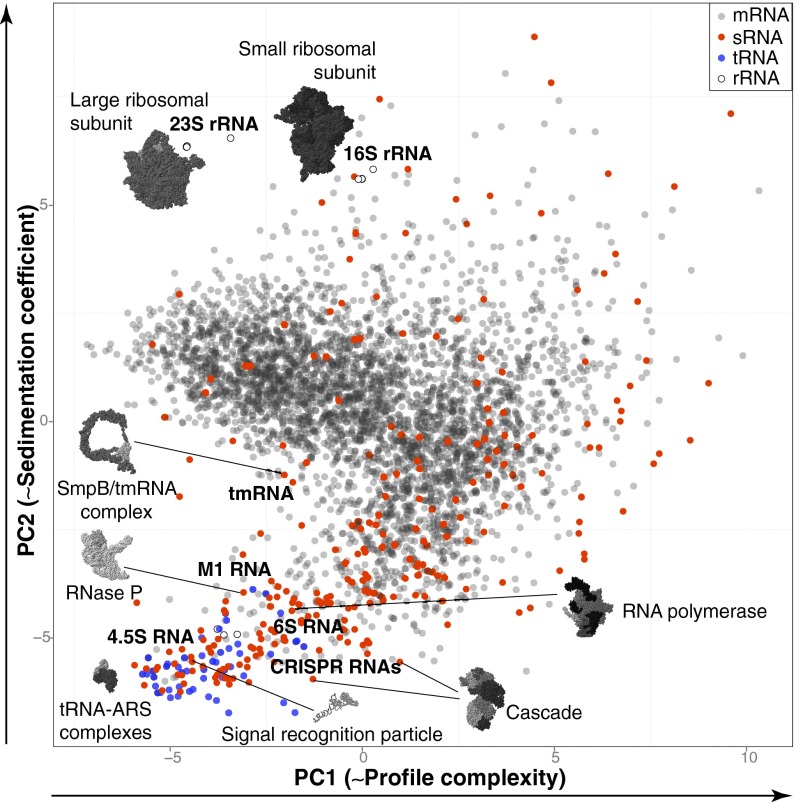
Applying principal component analysis (PCA) to the 3,969 RNA profiles obtained (Fig. 2 and SI Appendix, Dataset S1), we observed that transcripts with similar biochemical behavior cosegregated, based on the first two principal components that correlate with the profile complexity, i.e., the number of individual peaks in a profile (PC1), and the sedimentation coefficient of complexes (PC2). This bacterial “RNA universe” exhibits two major branches: the upper protein-coding branch is dominated by mRNAs whose behavior is determined by ribosomal components, whereas the noncoding branch toward the bottom is enriched in ncRNAs that are associated with a variety of low molecular weight complexes. The ncRNAs found in the upper branch of the map mostly consisted of mRNA-derived sRNAs (overlapping and often processed from UTRs or coding regions of messengers) (16, 29) or misidentified short mRNAs, which explains their association with ribosomal components. Overall, this simple and logical structure reveals the fundamental dichotomy of coding and noncoding RNA functions within the cell, reflected at the level of RNA–protein interactions.
Fig. 2.
Topology of the Salmonella RNA interactome revealed by PCA of Grad-seq profiles from 3,969 Salmonella transcripts. The number of distinct RNPs formed by an RNA increases from Left to Right (PC1, ∼41% of variance), whereas sedimentation coefficients of major RNPs increase from Bottom to Top (PC2, ∼26% of variance). Select examples of corresponding RNPs are provided.
A Cluster of Noncoding RNAs That Interact with Protein ProQ.
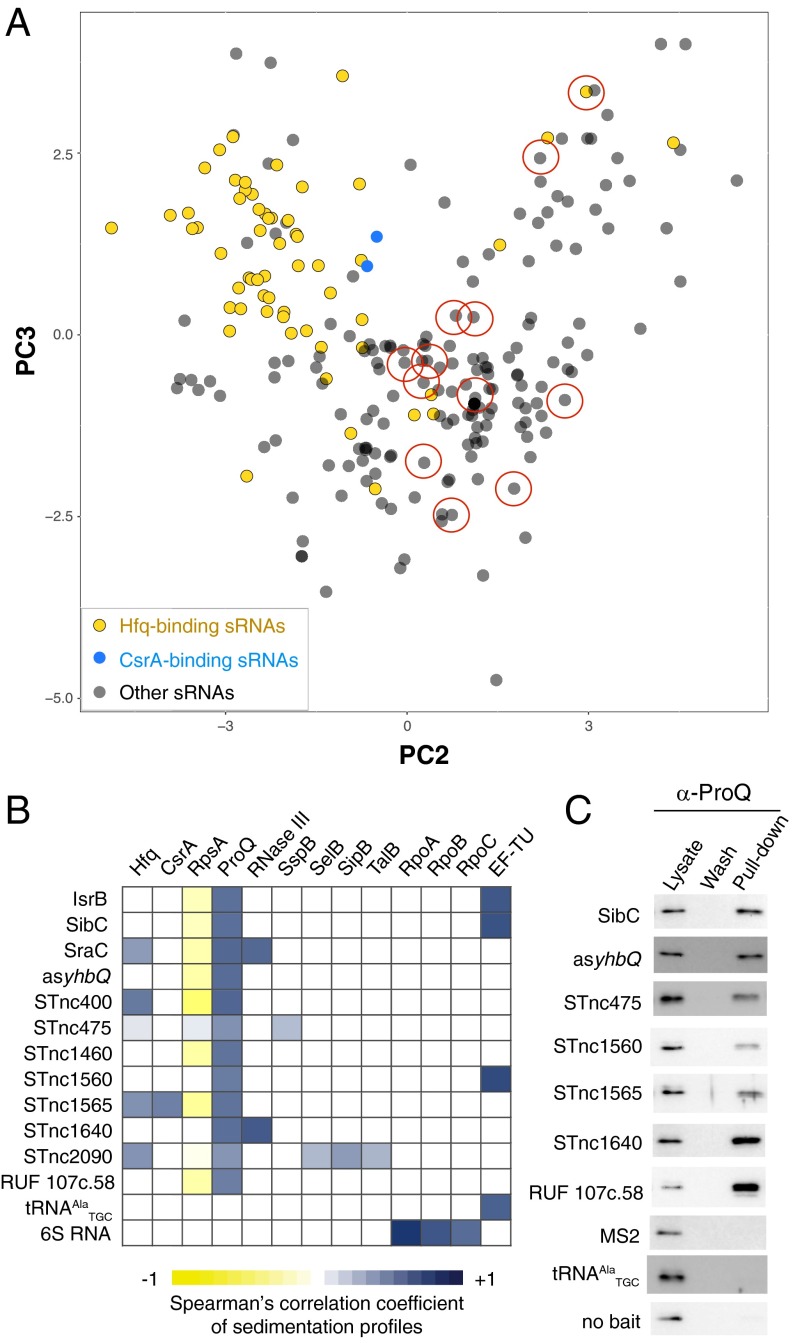
Focusing the PCA on sRNAs revealed several distinct transcript clusters (Fig. 3A). Most well-characterized Hfq-dependent sRNAs (11) form a dense group on this map. Remarkably, even Hfq-associated sRNAs that differ fourfold in length, such as ArcZ (57 nt) and GcvB (206 nt) clustered together (SI Appendix, Fig. S4), indicating that biochemical properties rather than sequence determined their in-gradient distributions. Likewise, the CsrA-binding sRNAs (19) CsrB and CsrC (360 and 240 nt, respectively) almost overlapped on the map (Fig. 3A). Intriguingly, there are many annotated Salmonella sRNAs, including attenuators, cis-antisense RNAs and uncharacterized species, that populated the map outside the Hfq- or CsrA-related clusters (Fig. 3A and SI Appendix, Fig. S4). This raised the possibility that some of these ncRNAs interact with an unknown global RBP.
Fig. 3.
Salmonella sRNA interactome and identification of ProQ as a recurrent sRNA binder. (A) Grad-seq PCA plot of 238 Salmonella sRNAs. PC2 and PC3 are selected to visualize sRNA clusters with finer detail (Materials and Methods provides further information). Only Hfq- and CsrA-associated sRNAs are highlighted (SI Appendix, Fig. S4 shows a complete functional sRNA assignment). sRNAs selected for MS2 aptamer tagging and pull down are circled. (B and C) Pull down of the selected MS2 aptamer-tagged sRNAs from Salmonella lysates and identification of their binding partners. (B) Heat map showing proteins specifically copurified with each MS2 aptamer-tagged sRNA (most ribosomal proteins are omitted for clarity) and Spearman’s correlation coefficients of their sedimentation profiles with the Grad-seq profiles of the bait sRNAs. ProQ is a particularly frequent partner of the selected sRNAs and their sedimentation profiles match, suggesting a stable interaction. (C) ProQ is enriched in the MS2 aptamer-tagged sRNA pull downs, compared with control baits or no bait, as visualized with specific antibodies.
To identify the associated RBP(s) we tagged 12 sRNAs from outside the Hfq and CsrA clusters with the MS2 aptamer (30) and pulled down interacting proteins from Salmonella cell lysates (Fig. 3A and SI Appendix, Fig. S5). In each case, we detected several candidate protein binding partners. Comparison of sedimentation profiles from our reference proteomics dataset (SI Appendix, Fig. S3) with those of the bait sRNAs identified the ProQ protein as the most common and highly correlated partner (Spearman’s 0.48 < r < 0.7, P < 0.033, Fig. 3B). Western blot analysis confirmed the enrichment of ProQ in sRNA pull-down samples compared with control RNA pull downs (Fig. 3C).
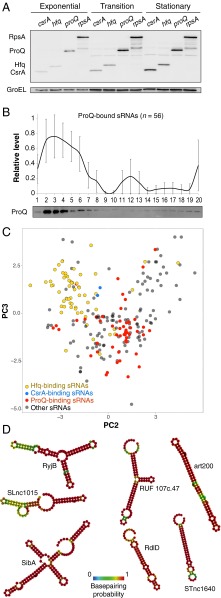
ProQ is a FinO-like osmoregulatory protein required for optimal expression of proline channel ProP (31, 32). However, ProQ homologs can be predicted in the chromosomes, plasmids, and bacteriophages of α-, β- and γ-proteobacteria (SI Appendix, Fig. S6), many of which lack a proP gene. Similarly, the high abundance and constitutive expression of ProQ (33, 34) are also inconsistent with a specialized function: in a semiquantitative Western blot analysis, ProQ levels compare with those of the highly expressed general RBPs, CsrA, Hfq, or ribosomal protein S1 (Fig. 4A). Moreover, we find that deletion of proQ affects the levels of hundreds of transcripts with no known function in osmoprotection (SI Appendix, Fig. S10A). Together, these observations suggest a much broader cellular role for ProQ than fine-tuning ProP expression.
Fig. 4.
ProQ is a conserved abundant RNA-binding hub associated with a distinct class of highly structured sRNAs. (A) ProQ is an abundant, constitutively expressed protein. Equal amounts of cells with chromosomally FLAG-tagged csrA, hfq, proQ, or rpsA genes were analyzed by Western blotting with anti-FLAG antibodies in three growth phases. (B) Averaged Grad-seq distribution of ProQ-bound sRNAs. As on SI Appendix, Fig. S1C, all individual profiles of ProQ-associated sRNAs were cumulated and presented as an average along the gradient ± SD. Corresponding Western blot probed for ProQ is shown below. Only the 56 ProQ-binding sRNAs that are sufficiently covered in the Grad-seq dataset are shown. (C) Grad-seq PCA plot of Salmonella sRNAs showing segregation of Hfq-, CsrA- and ProQ-binding transcripts (Fig. 3A and SI Appendix, Fig. S9 A and B for a complete functional sRNA assignment). (D) Minimum free energy secondary structures of representative highly enriched ProQ ligands.
ProQ-Associated sRNAs Form a Distinct Group of Riboregulators.
To obtain a better understanding of the target suite of ProQ in Salmonella, we used RNA immunoprecipitation coupled with deep sequencing. This analysis suggested that ProQ associates with >400 cellular transcripts from diverse cellular pathways (SI Appendix, Figs. S7 and S8 and Dataset S3), with small RNAs being significantly overrepresented (98/422, or ∼23.2%, compared with the annotated 547/5,205 or ∼10.5%, P < 0.0001, Fisher’s exact test). Regarding these noncoding targets (SI Appendix, Table S1), ProQ preferentially bound to Hfq-independent sRNAs (P < 0.0007, Mann–Whitney test, P < 0.003, Kolmogorov–Smirnov test; only two ProQ-bound sRNAs, SraC and STnc520, are known to be Hfq dependent). This suggests the existence of a distinct class of noncoding transcripts, which comprised ∼18% of all currently known Salmonella sRNAs, most (>80%) of which are currently uncharacterized. The few ProQ-enriched sRNAs of known function included attenuators (SraF), trans-acting base-pairing sRNAs (SraL) (35), sRNA-sequestering sponges (STnc2180) (36), and type I antitoxins (Sib, Rdl, IstR) (24, 37). Diverse types of antisense RNAs (antitoxins, chromosomal, phage and transposon associated), which typically operate in a Hfq-independent manner (24), are particularly overrepresented among ProQ ligands (53/98, or 54%, compared with the annotated 216/547, or ∼39%, P = 0.008, Fisher’s exact test). As expected, the group of sRNAs used as bait in MS2 aptamer pull-down assays (Fig. 3) showed significantly higher median enrichment in ProQ coimmunoprecipitation compared with all sRNAs (P < 0.003, Mann–Whitney test).
ProQ-bound sRNAs formed relatively small RNPs (average s020,w ∼7S, corresponding to 100–150 kDa, Fig. 4B), and clearly segregated from Hfq- and CsrA-bound transcripts within the sRNA PCA plot (Fig. 4C and SI Appendix, Fig. S9 A and B). This separation illustrates the discriminatory power of Grad-seq in identifying collectives of biochemically similar RNAs. EMSAs with purified Salmonella ProQ confirmed the high affinity and specific binding of several enriched sRNAs in vitro, supporting their involvement in stable ProQ-containing RNP particles (SI Appendix, Fig. S9 C and D). Top-enriched ProQ sRNA ligands, such as SibA and STnc2090, formed complexes with apparent dissociation constants in the low nanomolar range, similar to the affinities typically observed in interactions of Hfq and CsrA with cognate RNAs (7, 18).
Of note, most ProQ-associated sRNAs form extensively base-paired structures, in some cases resembling eukaryotic miRNA precursors (Fig. 4D). A significant positive relationship was observed between the predicted folding energy of Salmonella sRNAs and ProQ binding (SI Appendix, Fig. S9E), suggesting that, similar to the RNA chaperone FinO (32, 38), interactions between ProQ and RNAs are at least partially structure driven. These binding preferences contrast with the typical modes of Hfq (12) or CsrA (39) binding, which use single-stranded regions. Indeed, Hfq-dependent sRNAs, whose only recurrent structured region is the intrinsic terminator stem loop (12), appear to be significantly less folded than ProQ-associated ones (SI Appendix, Fig. S9F). Thus, ProQ-interacting sRNAs form a structurally distinct class of ncRNAs, and ProQ may fill the niche of a global RBP that associates specifically with highly structured transcripts in bacteria.
Effect of ProQ on Associated RNAs.
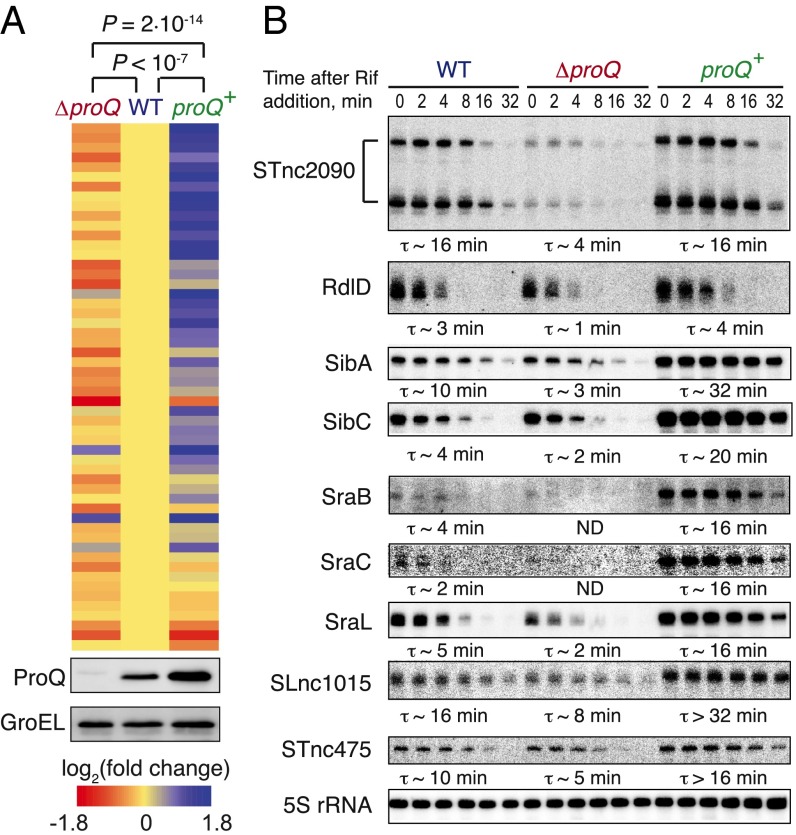
Association of an RNA with a cellular protein may not necessarily impact its function. However, we found that ProQ affected the abundance of many of its sRNA ligands (Fig. 5), as their cellular levels decreased upon proQ deletion (ΔproQ); conversely, overexpressing the protein (proQ+) increased the levels of most ProQ-associated sRNAs. Furthermore, using the drug rifampicin to arrest bacterial transcription, we observed that in the absence of ProQ the half-lives of several selected ProQ-bound sRNAs were reduced, whereas increased levels of ProQ generally resulted in sRNA overstabilization (Fig. 5B). Therefore, ProQ likely protects some of its ligands from degradation and thus may extend the time window for their cellular activity, similar to the FinO protein with its sole known RNA substrate, FinP (40).
Fig. 5.
ProQ acts as a stability factor for most of its sRNA ligands. (A) ProQ positively affects the steady-state levels of most of its sRNA ligands. The heat map shows the changes in the abundance of ProQ-associated noncoding RNAs (n = 54) upon proQ deletion (ΔproQ) or complementation (proQ+). Significance of the differences is evaluated by Wilcoxon matched-pairs signed-ranks test. (Lower) Corresponding levels of ProQ in these strains, as revealed by Western blotting with a ProQ-specific antiserum. Only those sRNAs that have been sufficiently covered in the transcriptome dataset are shown. (B) ProQ stabilizes its associated sRNAs in vivo. Samples from WT, ΔproQ, and complemented proQ+ strains were collected in the stationary phase after transcription arrest with rifampicin and analyzed by Northern blotting. Approximate half-lives for major detected species are shown below the blots. ND, not determined (<1 min). A representative of three independent experiments is shown.
As expected from the large suite of ProQ-bound RNAs, proQ deletion dramatically affects gene expression in Salmonella, changing the levels of >800 transcripts (∼16% of genome, SI Appendix, Fig. S10A and Dataset S4). The pathways significantly overrepresented in the group of differentially regulated genes include energy production, amino acid metabolism, and translation (SI Appendix, Fig. S10B), indicating that ProQ pervasively impacts bacterial physiology. ProQ seemed to exert both direct and indirect effects on its ligands as ∼36% of ProQ-associated RNAs showed significant changes in expression levels (of which ∼28% are up-regulated and ∼72% are down-regulated upon proQ deletion, P < 0.0001, Fisher’s exact test). Of note, mRNA targets of the few ProQ-dependent sRNAs whose functions are known, such as cis-acting Sib antitoxin RNAs (37), were derepressed in response to proQ deletion and overrepressed in the proQ+ strain (SI Appendix, Fig. S10C and Dataset S4), suggesting that some of the observed gene expression changes are sRNA mediated. Altogether, these data reveal the existence of a large ProQ-dependent regulon and position ProQ as the third global posttranscriptional gene expression modulator in bacteria besides Hfq and CsrA.
Conclusions
The Grad-seq approach provides an experimental and analytical framework to rapidly visualize the major RNA collectives of an organism of interest, which will be particularly useful in drafting initial functional RNA landscapes for many understudied microbes with unexplored RNA biology (41, 42). With rapidly improving sequencing technologies and decreasing cost, Grad-seq will be able to partition major RNA classes susceptible to therapeutic intervention in medically important microbial communities such as the gut microbiota, the vast majority of whose species remain unculturable.
Grad-seq has conceptual parallels with other methods that have been used to describe the RNP landscape of a cell and efficiently combines their individual strengths. For instance, ribosome profiling (43) employs a similar approach to partition and sequence cellular transcripts but focuses exclusively on those associated with translating ribosomes, whereas Grad-seq also describes smaller RNPs associated with noncoding RNA functions. Similarly, RNPomics (26) aims to enrich and sequence functionally relevant cellular RNA species by separating them from unbound RNA by glycerol gradient centrifugation. However, the resulting RNPs are subsequently studied in bulk, without fractionation, decreasing the resolution of the biochemical information obtained on transcripts. RNA interactome capture (44, 45) is an extremely powerful approach that aims to globally characterize cellular RBPs by deep mass-spectrometric analysis of proteins cross-linked to cellular RNA, but it also operates in bulk, without distinguishing individual RNPs.
A key advantage of Grad-seq is the biochemical grouping of cellular transcripts into RNA collectives of likely similar function, to subsequently identify common protein partners. Although currently a two-step technique, improving the resolution of Grad-seq through finer fractionation, more comprehensive protein detection, and the decreasing costs for RNA-seq may soon permit a more direct approach to match RNA–protein profiles, enabling the analysis of complex eukaryotic systems.
The power of Grad-seq is here illustrated by unveiling ProQ as a global RNA-binding hub in addition to Hfq and CsrA, which is remarkable in light of the extensive previous work on E. coli and Salmonella as the current workhorses of bacterial RNA research (7). Our discovery of a large class of ProQ-associated sRNAs opens the door for further studies of this potential domain of posttranscriptional control, which will likely reveal molecular mechanisms and physiological roles of RNA in bacteria. Indeed, a recent study in Legionella identified a ProQ-like protein as a matchmaker of sRNA–mRNA interactions in the regulation of bacterial competence (46).
Materials and Methods
Bacteria and Media.
All bacterial strains and growth conditions are described in SI Appendix, Tables S2 and S3.
Grad-Seq.
Salmonella cells grown to OD600 = 2 were lysed in 20 mM Tris⋅HCl, pH 7.5, 150 mM KCl, 1 mM MgCl2, 1 mM DTT, 1 mM PMSF, 0.2% Triton X-100, 20 units/mL DNase I (Thermo Fisher Scientific), 200 units/mL SUPERase-IN (Life Technologies) with 0.1-mm glass beads (BioSpec Products) on a Retsch MM400 machine. Cleared lysates were centrifuged through linear 10–40% (wt/vol) glycerol gradients in the same buffer formed in Beckman SW40Ti tubes at 100,000 × g for 17 h at 4 °C and fractionated in 20 equal fractions. Fractions were deproteinized with 1% SDS and 1 volume of hot phenol by shaking at 55 °C for 5 min, centrifuged, and RNA was precipitated with isopropanol.
For Grad-seq, two biological replicates have been analyzed. Eighty-five picograms per microliter of the spike-in RNA [5′ P-CUCGUCCGACGUCACCUAGA (IBM GmbH)] had been added to each fraction prior to the library preparation. RNA-seq libraries were prepared by Vertis Biotechnologie and sequenced on an Illumina HiSeq 2000 platform. The reads were mapped to the Salmonella Typhimurium SL1344 genome with the use of the READemption pipeline version 0.3.4 (47). Fractionwise gene-specific RNA sequencing (RNA-seq) read counts were normalized by corresponding spike-in counts and any remaining uniformly distorting biases were manually removed. All profiles with ≥30 reads in at least one fraction were power transformed to improve linearity and standardized to the range from 0 to 1. To derive averaged distributions for RNA classes, profiles of individual RNAs were summed up and averaged fractionwise. PCA was performed in R and visualized in R and Python. For sRNAs, PC1 mostly reflects the influence of mRNA-derived and overlapping sRNAs (following, like mRNAs, strong ribosomal components) and is uninformative for our purpose of revealing groups of ncRNAs. We opted for PC2 and PC3 analysis instead, which allowed a better resolution and clustering of typical sRNAs. Both PC1 vs. PC2 and PC2 vs. PC3 plots are available on SI Appendix, Fig. S4. RNA-seq data are available in Gene Expression Omnibus (accession no. GSE62988). The workflow implemented as Shell script and the analysis-specific tools are deposited at Zenodo at DOI:10.5281/zenodo.35176. All molecular weight estimates provided in this work were made assuming a most frequently encountered moderately elongated shape of particles (25).
Other Methods.
Affinity chromatography, RNA coimmunoprecipitation, sRNA turnover, and all statistical analyses are described in detail in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Gorski for editing the manuscript; D. Sheidy, M. Springer, and A. Carpousis for antisera; C. Wang for EMSA and stability samples; R. Rieder, M. Raabe, and S. Grund for the assistance with affinity chromatography and MS; M. Hecker for proteomics infrastructure; T. Achmedov for technical assistance; and members of the J.V. laboratory for comments on the manuscript. The project was supported by funds from the Bavarian BioSysNet Program, the Max Planck Society, and Bundesministerium für Bildung und Forschung Grants Next-Generation Transcriptomics of Bacterial Infections and eBio RNAsys. E.H. is supported by the Wenner-Gren Foundations.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE62988).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609981113/-/DCSupplemental.
References
- 1.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Altman S. Twenty years. RNA. 2015;21(4):513–514. doi: 10.1261/rna.049924.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patil VS, Zhou R, Rana TM. Gene regulation by non-coding RNAs. Crit Rev Biochem Mol Biol. 2014;49(1):16–32. doi: 10.3109/10409238.2013.844092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: Expanding frontiers. Mol Cell. 2011;43(6):880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barquist L, Vogel J. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu Rev Genet. 2015;49:367–394. doi: 10.1146/annurev-genet-112414-054804. [DOI] [PubMed] [Google Scholar]
- 6.Sorek R, Cossart P. Prokaryotic transcriptomics: A new view on regulation, physiology and pathogenicity. Nat Rev Genet. 2010;11(1):9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 7.Wagner EG, Romby P. Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv Genet. 2015;90:133–208. doi: 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Meister G. Argonaute proteins: Functional insights and emerging roles. Nat Rev Genet. 2013;14(7):447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 9.Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: Its biogenesis and functions. Annu Rev Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- 10.De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288(12):7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9(8):578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18(5):1062–1074. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schu DJ, Zhang A, Gottesman S, Storz G. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 2015;34(20):2557–2573. doi: 10.15252/embj.201591569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Updegrove TB, Shabalina SA, Storz G. How do base-pairing small RNAs evolve? FEMS Microbiol Rev. 2015;39(3):379–391. doi: 10.1093/femsre/fuv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutherford ST, Valastyan JS, Taillefumier T, Wingreen NS, Bassler BL. Comprehensive analysis reveals how single nucleotides contribute to noncoding RNA function in bacterial quorum sensing. Proc Natl Acad Sci USA. 2015;112(44):E6038–E6047. doi: 10.1073/pnas.1518958112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31(20):4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell. 2014;55(2):199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jørgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev. 2013;27(10):1132–1145. doi: 10.1101/gad.214734.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romeo T, Vakulskas CA, Babitzke P. Post-transcriptional regulation on a global scale: Form and function of Csr/Rsm systems. Environ Microbiol. 2013;15(2):313–324. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Göpel Y, Papenfort K, Reichenbach B, Vogel J, Görke B. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev. 2013;27(5):552–564. doi: 10.1101/gad.210112.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westermann AJ, et al. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature. 2016;529(7587):496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 22.Holmqvist E, et al. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016;35(9):991–1011. doi: 10.15252/embj.201593360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464(7286):250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 24.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev. 2011;75(2):286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rederstorff M, et al. RNPomics: Defining the ncRNA transcriptome by cDNA library generation from ribonucleo-protein particles. Nucleic Acids Res. 2010;38(10):e113. doi: 10.1093/nar/gkq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101(6):613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 28.Giudice E, Macé K, Gillet R. Trans-translation exposed: Understanding the structures and functions of tmRNA-SmpB. Front Microbiol. 2014;5:113. doi: 10.3389/fmicb.2014.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyakoshi M, Chao Y, Vogel J. Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr Opin Microbiol. 2015;24:132–139. doi: 10.1016/j.mib.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Said N, et al. In vivo expression and purification of aptamer-tagged small RNA regulators. Nucleic Acids Res. 2009;37(20):e133. doi: 10.1093/nar/gkp719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunte HJ, Crane RA, Culham DE, Richmond D, Wood JM. Protein ProQ influences osmotic activation of compatible solute transporter ProP in Escherichia coli K-12. J Bacteriol. 1999;181(5):1537–1543. doi: 10.1128/jb.181.5.1537-1543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaulk SG, et al. ProQ is an RNA chaperone that controls ProP levels in Escherichia coli. Biochemistry. 2011;50(15):3095–3106. doi: 10.1021/bi101683a. [DOI] [PubMed] [Google Scholar]
- 33.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157(3):624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheidy DT, Zielke RA. Analysis and expansion of the role of the Escherichia coli protein ProQ. PLoS One. 2013;8(10):e79656. doi: 10.1371/journal.pone.0079656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva IJ, Ortega AD, Viegas SC, García-Del Portillo F, Arraiano CM. An RpoS-dependent sRNA regulates the expression of a chaperone involved in protein folding. RNA. 2013;19(9):1253–1265. doi: 10.1261/rna.039537.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lalaouna D, et al. A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol Cell. 2015;58(3):393–405. doi: 10.1016/j.molcel.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Han K, Kim KS, Bak G, Park H, Lee Y. Recognition and discrimination of target mRNAs by Sib RNAs, a cis-encoded sRNA family. Nucleic Acids Res. 2010;38(17):5851–5866. doi: 10.1093/nar/gkq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Biesen T, Frost LS. The FinO protein of IncF plasmids binds FinP antisense RNA and its target, traJ mRNA, and promotes duplex formation. Mol Microbiol. 1994;14(3):427–436. doi: 10.1111/j.1365-2958.1994.tb02177.x. [DOI] [PubMed] [Google Scholar]
- 39.Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11(10):1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jerome LJ, van Biesen T, Frost LS. Degradation of FinP antisense RNA from F-like plasmids: The RNA-binding protein, FinO, protects FinP from ribonuclease E. J Mol Biol. 1999;285(4):1457–1473. doi: 10.1006/jmbi.1998.2404. [DOI] [PubMed] [Google Scholar]
- 41.Brown CT, et al. Unusual biology across a group comprising more than 15% of domain bacteria. Nature. 2015;523(7559):208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 42.Weinberg Z, Perreault J, Meyer MM, Breaker RR. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature. 2009;462(7273):656–659. doi: 10.1038/nature08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 45.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46(5):674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 46.Attaiech L, et al. Silencing of natural transformation by an RNA chaperone and a multitarget small RNA. Proc Natl Acad Sci USA. 2016;113(31):8813–8818. doi: 10.1073/pnas.1601626113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Förstner KU, Vogel J, Sharma CM. READemption-a tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics. 2014;30(23):3421–3423. doi: 10.1093/bioinformatics/btu533. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.