Significance
Highly regulated and precise positioning of Ca2+ channels at the active zone (AZ) controls Ca2+ nanodomains at release sites. Their exact localization affects vesicular release probability (PVR) and is important for proper synaptic transmission during repetitive stimulation. We provide a detailed analysis of synaptic transmission combined with superresolution imaging of the AZ organization in mouse hippocampal synapses lacking Rab-interacting molecule-binding protein 2 (RIM-BP2). By dual- and triple-channel time-gated stimulated emission depletion (gSTED) microscopy, we directly show that RIM-BP2 fine-tunes voltage-gated Ca2+ channel 2.1 (CaV2.1) localization at the AZ. We reveal that RIM-BP2 likely regulates the Ca2+ nanodomain by positioning CaV2.1 channels close to synaptic vesicle release sites. Loss of RIM-BP2 reduces PVR and alters short-term plasticity.
Keywords: RIM-BP2, calcium channel coupling, release probability, short-term plasticity, active zone structure
Abstract
The tight spatial coupling of synaptic vesicles and voltage-gated Ca2+ channels (CaVs) ensures efficient action potential-triggered neurotransmitter release from presynaptic active zones (AZs). Rab-interacting molecule-binding proteins (RIM-BPs) interact with Ca2+ channels and via RIM with other components of the release machinery. Although human RIM-BPs have been implicated in autism spectrum disorders, little is known about the role of mammalian RIM-BPs in synaptic transmission. We investigated RIM-BP2–deficient murine hippocampal neurons in cultures and slices. Short-term facilitation is significantly enhanced in both model systems. Detailed analysis in culture revealed a reduction in initial release probability, which presumably underlies the increased short-term facilitation. Superresolution microscopy revealed an impairment in CaV2.1 clustering at AZs, which likely alters Ca2+ nanodomains at release sites and thereby affects release probability. Additional deletion of RIM-BP1 does not exacerbate the phenotype, indicating that RIM-BP2 is the dominating RIM-BP isoform at these synapses.
At the presynapse, coupling between action potentials (APs) and synaptic vesicle fusion is exquisitely precise, ensuring high temporal fidelity of neuron-to-neuron signaling in the nervous system. Two properties are thought to be responsible for this remarkable precision: a highly efficient release apparatus that transduces Ca2+ signals into vesicle fusion and a tightly organized active zone (AZ), where the release apparatus and voltage-gated Ca2+ channels (CaVs) are spatially coupled. Rab-interacting molecules (RIM) are thought to contribute to both properties, because loss of RIM impairs vesicle priming (1) and CaV localization at the AZ (2). RIM-binding proteins (RIM-BPs) directly interact with RIM (3), the pore-forming subunits of CaV1 and CaV2 channels (2, 4, 5), and Bassoon (5), and have therefore been suggested to play a role in presynaptic CaV localization. The Drosophila homolog of RIM-binding proteins (DRBP) is indeed crucial for neurotransmitter release at the AZ of neuromuscular junctions (NMJs) because loss of DRBP reduces CaV abundance and impairs the integrity of the AZ scaffold (6). DRBP-deficient flies show severe impairment of neurotransmitter release along with increased short-term facilitation (6, 7).
Recently, Acuna et al. (8) published a report on the combined loss of RIM-BP1 and RIM-BP2 in mouse synapses. The authors report that although RIM-BPs are not essential for synaptic transmission, AP-triggered neurotransmitter release is more variable and the sensitivity to the Ca2+ chelator EGTA is increased at the Calyx of Held, suggesting a larger coupling distance of CaV and the release machinery.
In the present study, we further investigated the consequences of constitutive deletion of RIM-BP2 on the structure and function of mouse hippocampal synapses. We show that loss of RIM-BP2 leads to a moderate reduction in initial release probability, which translates into profound changes in short-term plasticity (STP). This deficit can be overcome by increasing extracellular Ca2+. We established triple-channel time-gated stimulated emission depletion (gSTED) microscopy for RIM-BP2, Munc13-1, and Bassoon, as well as for CaV2.1, RIM, and the postsynaptic marker protein Homer1. Using this technique, we demonstrate that although synapse number and molecular architecture appear essentially intact, RIM-BP2 is necessary for proper coclustering of the P/Q-type CaV subunit CaV2.1 with the AZ protein Bassoon at hippocampal CA3-CA1 synapses. We hypothesize that the observed change in CaV localization causes a discrete alteration in the coupling of Ca2+ influx and exocytosis, and thereby modifies release probability and, consequently, STP. Additional deletion of RIM-BP1 did not strengthen the changes in short-term facilitation, supporting our hypothesis that RIM-BP2 is the major RIM-BP paralog at glutamatergic hippocampal synapses.
Results
RIM-BP2 Localization at the AZ of Hippocampal CA3-CA1 Synapses.
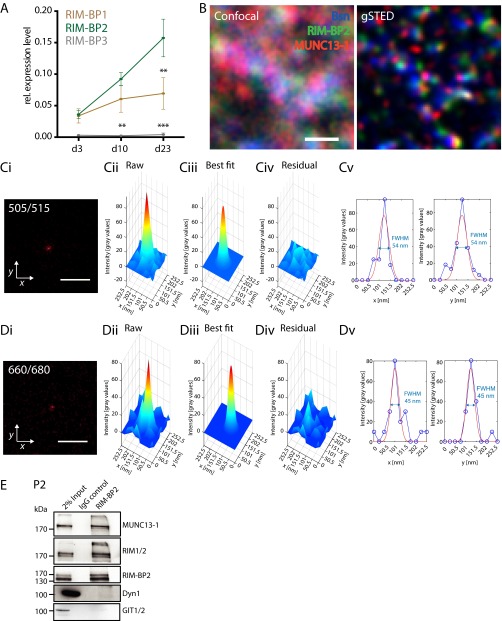
STED microscopy revealed that DRBP localizes close to the membrane near the AZ center of Drosophila NMJ synapses (6), but comparable studies on RIM-BPs at mammalian AZs with nanometer scale resolution are lacking. Quantitative real-time PCR suggested that RIM-BP2 is the predominant paralog in cultured hippocampal neurons (Fig. S1A). We analyzed the spatial relationship between RIM-BP2 and two AZ components, Bassoon and MUNC13-1, at CA3-CA1 hippocampal synapses of mouse brain cryosections. On the confocal level, RIM-BP2 colocalized with both AZ proteins (Fig. S1B). To dissect the AZ nanoscale architecture, we established triple-channel gSTED with a lateral resolution of ∼50 nm in all three channels (Fig. 1A and Fig. S1 B–D). Analysis of the mean distance between nearest neighbors (k-nearest neighbor analysis) revealed that RIM-BP2 is localized at a short distance to Bassoon and MUNC13-1, whereas MUNC13-1 is equidistant to RIM-BP2 and Bassoon (k = 1). In coimmunoprecipitations from P2 fractions of mouse brains, RIM-BP2 coprecipitated with RIM and Munc13-1, but not with the Arf GTPase-activating protein GIT, a binding partner of Piccolo (9) and of endocytotic proteins such as Dynamin1 (Fig. S1E) and Stonin 2 (10). Together, these results indicate that RIM-BP2 is part of the presynaptic AZ scaffold and forms a complex with the priming factors RIM and Munc13-1.
Fig. S1.
RIM-BP expression and interaction with RIM and Munc13-1. (A) Relative (rel.) expression levels of RIM-BP1–3 in cultured murine hippocampal neurons over time. The mRNA levels are normalized to Rpl4. Data are expressed as mean ± SEM, and tested against RIM-BP2 expression levels (pDIV23,RIM-BP1 = 0.001, pDIV10, RIM-BP3 = 0.001, pDIV23,RIM-BP3 < 0.001, Tukey’s multiple comparisons test; n = 4). *P < 0.05; **P < 0.01; ***P < 0.001. (B) Confocal (Left) and corresponding gSTED (Right) images of RIM-BP2 localization to Bassoon (Bsn) and MUNC13-1 at CA3-CA1 hippocampal synapses on a WT mouse brain cryosection. (Scale bar: 500 nm.) (C, i–v and D, i–v) Estimation of the effective lateral point spread function of the gSTED microscope. The gSTED images of a 40-nm fluorescent bead with excitation and emission maxima at 505 and 515 nm (C, i) or 660 and 680 (D, i), respectively, are shown. (Scale bar: 500 nm.) Three-dimensional surface plot of the local intensity profile (C, ii and D, ii) approximated with a 2D Gaussian fit (C, iii and D, iii). (C, iv and D, iv) Deviation of the 2D Gaussian fit from the intensity profile. (C, v and D, v) Intensity profile and 2D Gaussian fit values along the x axis for constant y (126.25 nm) (Left) and the y axis for constant x (126.25 nm) (Right). In both channels, the smallest bead had a FWHM of 54 nm (C, v) or 45 nm (D, v), indicating a lateral resolution of ∼50 nm. (E) Representative Western blot showing input, IgG control, and immunoprecipitation of RIM-BP2 probed with antibodies against different synaptic proteins. RIM-BP2 forms a complex with the AZ proteins RIM1/2 and MUNC13-1, but not with the endocytic protein Dynamin1 (Dyn1) or the cytomatrix protein GIT.
Fig. 1.
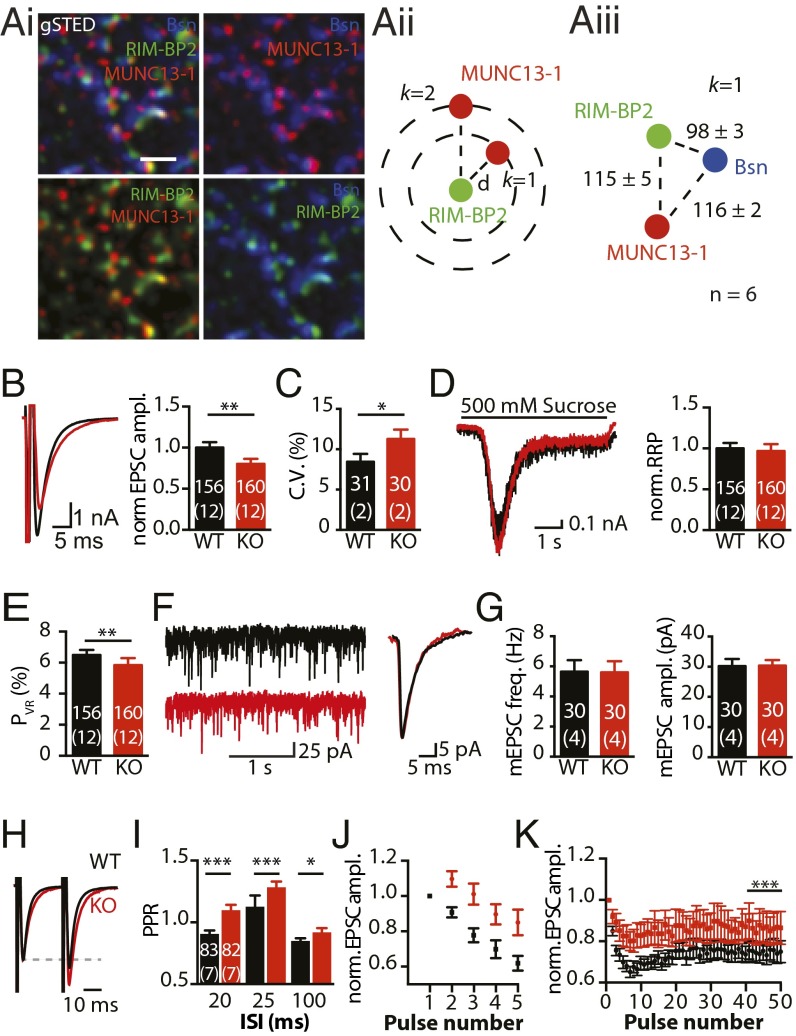
RIM-BP2 localization at the hippocampal AZ, and effect of RIM-BP2 deletion on synaptic transmission in autaptic hippocampal neurons. (A, i–iii) Spatial organization of RIM-BP2 in relation to Bassoon (Bsn) and MUNC13-1 at the AZ of CA3-CA1 synapses in WT mouse brain cryosections imaged by gSTED and analyzed using the mean k-nearest neighbor distance between clusters. (Scale bar: 500 nm.) (B) EPSCs evoked by 2-ms somatic depolarization in RIM-BP2 WT (black) and KO (red) autaptic neurons. Amplitudes (ampl.) were normalized (norm.) to WT mean of the same culture. (C) C.V.s of 24 EPSC amplitudes recorded during a period of 2 min. (D) Synaptic responses to application of hypertonic sucrose (500 mM) solution probing the RRP. (E) PVR of the same cells as in B and D. (F) Spontaneous release and averages of miniature EPSCs (mEPSCs) from the same cells. (G) mEPSC amplitudes and frequencies (freq.). (H) Normalized traces of two EPSCs at an ISI of 25 ms. (I) PPR calculated for the indicated ISIs. Normalized amplitudes of the same cells as in I in response to 5 APs triggered at 50 Hz (J) or 50 APs at 10 Hz (K). The last 10 EPSCs of the 10-Hz train are larger in KO neurons compared with WT. The numbers of neurons and independent cultures analyzed are shown within the bars. Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Generation of a RIM-BP2 Constitutive Knockout Mouse Line.
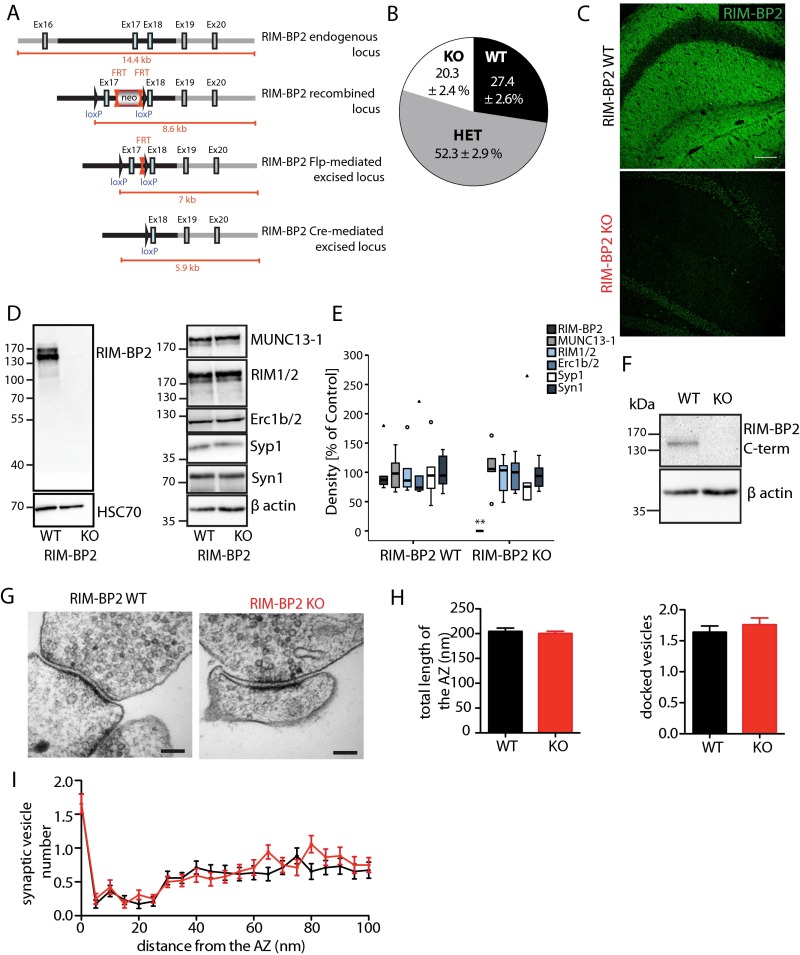
We generated knockout (KO) mice constitutively lacking RIM-BP2 by deleting exon 17 of the RIM-BP2 gene that encodes part of the second SH3 domain (Fig. S2A). RIM-BP2–deficient mice were born at Mendelian ratios (Fig. S2B) and survived into adulthood. Complete loss of RIM-BP2 protein expression in KO animals was confirmed by immunostaining of hippocampal cryosections (Fig. S2C) and immunoblot analysis of P2 fractions using two antibodies targeting different RIM-BP2 epitopes (Fig. S2 D and F). The expression of other presynaptic proteins such as RIM1/2, MUNC13-1, Erc1b/2, Synaptophysin1, and Synapsin1 was unaltered (Fig. S2 D and E). Transmission electron microscopy showed that the ultrastructure of the presynaptic AZ was not grossly altered by the absence of RIM-BP2 (Fig. S2 G–I).
Fig. S2.
Generation and characterization of RIM-BP2 KO mice. (A) Schematic representation of the RIM-BP2 targeting strategy showing the endogenous and recombined locus, as well as the RIM-BP2 locus after Flp-mediated and Cre-mediated excision. The RIM-BP2 locus after Cre-mediated excision corresponds to the constitutive RIM-BP2 KO. (B) Mendelian distribution of RIM-BP2–deficient progeny [total number of litters analyzed = 34; total number of animals: WT, 77; heterozygous (HET), 151; KO, 57]. (C) RIM-BP2 expression in the CA1 area of the hippocampus of WT and KO mice. Note the loss of RIM-BP2 immunoreactivity in the KO (WT, n = 4; KO, n = 2). (Scale bar: 100 μm.) (D, Left) RIM-BP2 protein expression in WT (n = 6) and KO (n = 6) mouse crude P2 membrane preparations probed with an RIM-BP2–specific antibody recognizing amino acids 589–869 of rat RIM-BP2, upstream of the deletion site. (D, Right) Expression levels of the AZ proteins RIM1/2, MUNC13-1, and Erc1b/2 and synaptic vesicle proteins Synaptophysin1 (Syp1) and Synapsin1 (Syn1) in the WT and KO mouse are not affected. HSC70 and β-actin were used as loading controls. (E) Quantification of signals from the Western blot analysis in D showing complete loss of RIM-BP2 levels (n = 6; **P < 0.01, Mann–Whitney U test), but no significant alterations in the levels of other synaptic proteins analyzed (n = 6; P > 0.05, ERC1b/2/Syp1, Mann–Whitney U test; P > 0.05, MUNC13-1/RIMs/Syn1, unpaired Student t test). Data are expressed as median (25th–75th percentiles). Circles indicate outliers, and triangles indicate extremes. (F) RIM-BP2 protein expression in WT (n = 6) and KO (n = 6) mouse crude P2 membrane preparations probed with RIM-BP2 antibody recognizing the last 20 amino acids of mouse RIM-BP2 C terminus. β-Actin was used as a loading control. (G) Representative electron microscopic images of WT and RIM-BP2 KO synapses. (Scale bars: 100 nm.) (H) Bar graph of total length of the AZ (Left) and number of docked vesicles (Right). (I) Distribution of synaptic vesicles relative to the AZ. Data are expressed as mean ± SEM.
RIM-BP2 Deletion Moderately Decreases Vesicular Release Probability and Leads to Increased Short-Term Facilitation in Cultured Neurons.
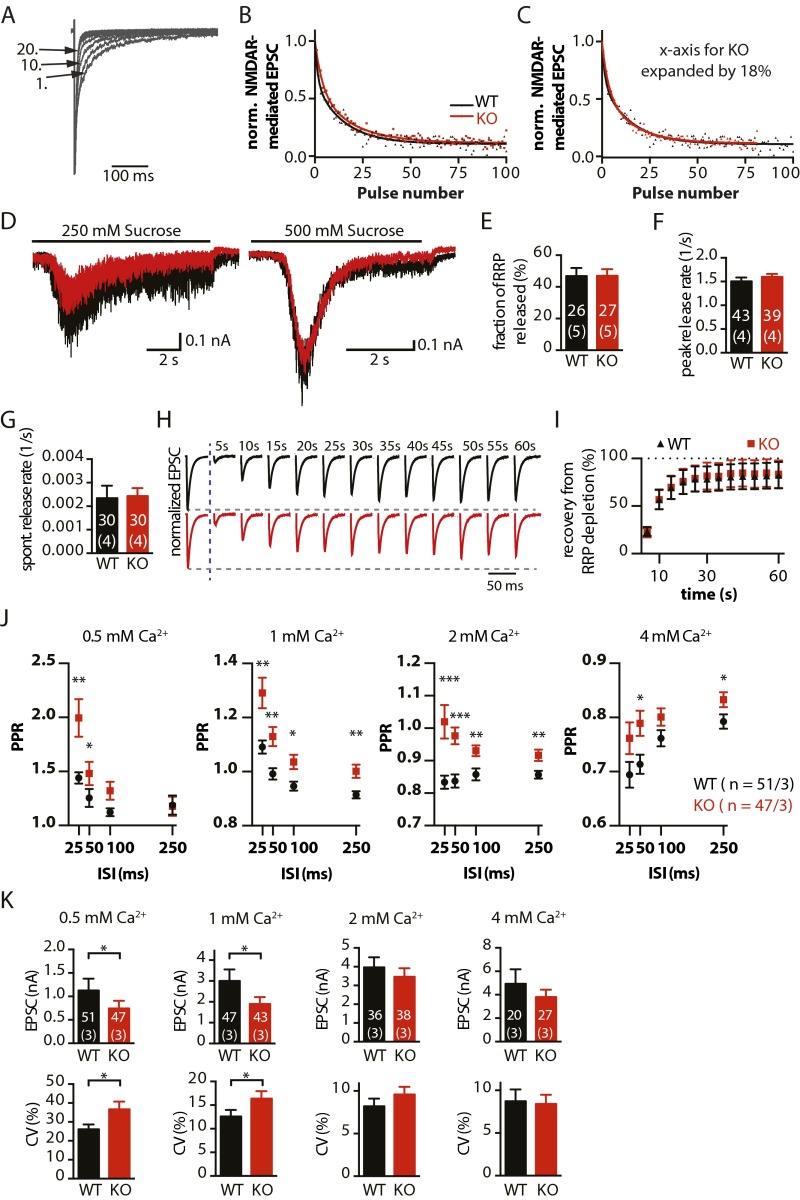
To investigate the role of RIM-BP2 in synaptic transmission, we analyzed basic synaptic properties and STP in autaptic hippocampal glutamatergic neurons from RIM-BP2 WT and KO mice. Evoked excitatory postsynaptic current (EPSC) amplitudes were decreased by 20% in RIM-BP2 KO neurons compared with WT (Fig. 1B). We further analyzed the coefficient of variation (C.V.) of EPSCs as a measure for the reliability of evoked release. Consistent with previous results from RIM-BP1/2 double KOs (DKOs) (8), we found an increased C.V. in RIM-BP2 KO neurons (Fig. 1C). The size of the readily releasable pool (RRP) (11) was not significantly altered (Fig. 1D). The probability of a synaptic vesicle being released by an AP [vesicular release probability (PVR)] was calculated as the ratio of the EPSC and the RRP charge. PVR was reduced by 10% in RIM-BP2 KO neurons (Fig. 1E). Additionally, we assessed release probability (PR) by monitoring the progressive block of the NMDA receptor-mediated component of the EPSC by the noncompetitive NMDA receptor blocker (5S,10R)-(+)-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine maleate (MK-801). The block rate, which is proportional to PR (12), was indeed decreased by ∼18% in RIM-BP2 KO neurons (Fig. S3 A–C). Spontaneous miniature release, in contrast, was unaltered (Fig. 1 F and G).
Fig. S3.
PR, molecular priming and RRP replenishment in RIM-BP2 KO neurons. (A–C) Progressive block of the NMDA receptor-mediated component of the EPSC by MK-801. (A) Example traces normalized to the AMPA component of the EPSC. The first, 10th, and 20th EPSCs in the presence of MK-801 are labeled. (B) Normalized NMDA receptor-mediated EPSCs during application of 5 μM MK-801 and double exponential fits. The rate of MK-801 block is reduced in RIM-BP2 KO neurons. (C) Same data as in B, but the x axis was expanded by 18% for RIM-BP2 KO. The alignment of the manipulated curves indicates a reduction in PR in RIM-BP2 KO neurons by ∼18%. (D) Representative traces of synaptic responses induced by 250 mM or 500 mM sucrose solution in autaptic neurons (WT, black; KO, red). (E) Fraction of RRP released by 250 mM sucrose application. (F) Peak release rates in 500 mM sucrose. (G) Spontaneous (spont.) release rates, calculated as the ratio of mEPSC frequency and number of vesicles in the RRP. (H) Representative traces of EPSCs before and after RRP depletion by sucrose application (blue vertical dashed line indicates time point of sucrose application). Responses are normalized to the average EPSC before RRP depletion. Stimulation artifacts were blanked for better visibility. (I) Summary graph of average recovery of EPSC amplitudes after depletion of the RRP by hypertonic sucrose application (WT, n = 35; KO, n = 39). (J) PPRs for different ISIs and in different [Ca2+]exts. (K) EPSC amplitudes and C.V.s of EPSC amplitudes in different [Ca2+]exts. The numbers of neurons analyzed, and independent experiments are shown within the bars. Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
We next investigated how STP is affected by loss of RIM-BP2. Autaptic RIM-BP2 KO neurons showed a robust increase in paired-pulse ratio (PPR) when stimulated with pairs of APs at different interstimulus intervals (ISIs) compared with WT neurons (Fig. 1 H and I). During short bursts of five APs at 50 Hz (Fig. 1J), EPSCs showed initial facilitation, followed by moderate depression in RIM-BP2 KO neurons but significant depression in WT neurons. Similarly, RIM-BP2 KO neurons exhibited significantly reduced depression of EPSC amplitudes during 10-Hz trains compared with WT neurons (Fig. 1K).
RIM-BP2 Deletion Alters STP in Acute Hippocampal Slices.
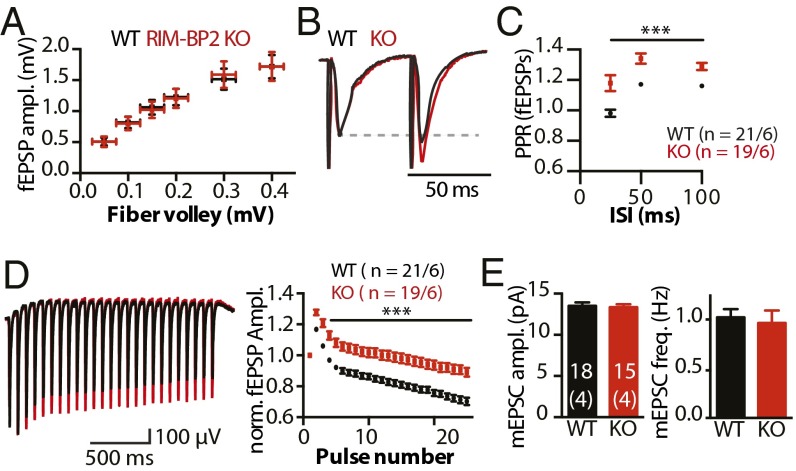
To verify the results independently in autaptic culture, we analyzed synaptic transmission in the CA1 area of acute hippocampal slices. The input/output function relating field recordings of excitatory postsynaptic potentials (fEPSPs) and fiber volley amplitudes were unchanged (Fig. 2A), suggesting that the loss of RIM-BP2 does not cause major alterations in basal synaptic transmission.
Fig. 2.
RIM-BP2 deletion alters STP in acute hippocampal slices. (A) Input/output curves relating the amplitude of the presynaptic fiber volley to the fEPSP amplitude in the stratum radiatum of area CA1 of acute hippocampal slices at different stimulus intensities (0.05 mV–0.2 mV: WT, n = 21; KO, n = 19; 0.3 mV: WT, n = 18; KO, n = 14; 0.4 mV: WT, n = 16; KO, n = 13). (B) Normalized fEPSPs in response to a paired pulse with 50-ms ISI. (C) PPRs of fEPSPs at different ISIs. (D) fEPSPs elicited by a 14-Hz stimulation train, with stimulation artifacts blanked for better visibility. (E) mEPSC amplitudes and frequencies recorded from CA1 pyramidal neurons. The numbers of slices and independent animals are indicated. Data are expressed as mean ± SEM. ***P < 0.001.
However, the PPR of fEPSPs was significantly elevated for all ISIs (Fig. 2 B and C), corroborating the cell culture results. Train stimulations with 25 pulses at 14 Hz caused greater initial facilitation and less depression of fEPSPs in RIM-BP2 KO compared with WT mice (Fig. 2D). As in autaptic cultures, spontaneous miniature release was unaltered in hippocampal slices (Fig. 2E). Together, the data from acute slices and autaptic culture demonstrate enhanced short-term facilitation in the absence of RIM-BP2.
Additional RIM-BP1 Deletion Does Not Exacerbate the RIM-BP2 KO Phenotype.
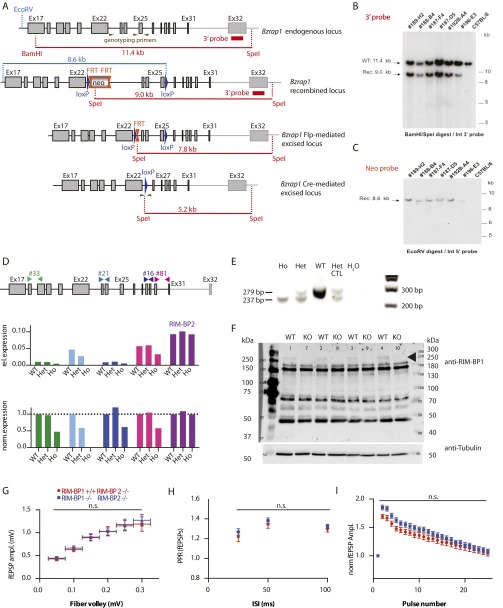
We crossed RIM-BP2 KO mice with constitutive RIM-BP1 KO mice (Fig. S4 A–F) to generate RIM-BP1/2 DKO mice. In area CA1 of acute hippocampal slices, input/output functions of RIM-BP2 KO and RIM-BP1/2 DKO were indistinguishable (Fig. S4G). We then compared PPRs (Fig. S4H) and STP (Fig. S4I) in RIM-BP2 single-KO and RIM-BP1/2 DKO slices. Additional deletion of RIM-BP1 did not exacerbate the increased facilitation observed in the single RIM-BP2 KO. Thus, our data support the hypothesis that RIM-BP2 is the major RIM-BP paralog in murine hippocampal neurons.
Fig. S4.
STP of fEPSPs in the stratum radiatum of RIM-BP1/2 DKO mice. (A) Schematic representation of the Bzrap1 (RIM-BP1) targeting strategy showing the endogenous and recombined locus, as well as the Bzrap1 locus after Flp-mediated and Cre-mediated excision. The Bzrap1 locus after Cre-mediated excision corresponds to the constitutive Bzrap1 KO lacking exons 23–25, which leads to a frameshift and a premature stop codon in exon 27. If the resulting RNA were translated, the resulting truncated 174-kDa protein would lack the second and third SH3 domains, and therefore would likely not be functional. (B) Southern blot analysis for 3′ homologous recombination (Rec) in ES cells performed by genOway. The genomic DNA of the tested ES cell clones was compared with WT DNA (C57BL/6). The digested DNA samples were blotted on nylon membrane and hybridized with an external 3′ probe (LA-E-A probe) hybridizing downstream of the targeting vector homology sequence (genOway). (C) Southern blot analysis for 5′ homologous recombination in ES cells performed by genOway. The genomic DNA of tested ES cell clones was compared with WT DNA (C57BL/6). The digested DNA samples were blotted on nylon membrane and hybridized with the Neo probe detecting the EcoRV fragment to screen for 5′ homologous recombination events (genOway). (D) Schematic representation of different real-time PCR (RT-PCR) assays for RIM-BP1 and RT-PCR results of cDNA obtained from RIM-BP1 WT, heterozygous (Het), and homozygous (Ho) KO mouse brain. Expression relative to Rpl4 was normalized (norm.) to WT. The RT-PCR assay 21 confirms that exons 23–25 are deleted in the RIM-BP1 KO. Assays spanning exons before and after the deleted region show a 50% reduction of mRNA levels in the KO. The remaining signals are likely due to the presence of frameshift mRNA that has not been completely degraded. This mRNA would not yield a functional protein. (E) Genotyping results obtained using the three primers indicated in A and genomic DNA from tails of WT, Het, and RIM-BP2 Ho KO littermates. The WT band is larger than the KO band, as indicated. In Het animals, both bands are present. (F) Western blot of P2 fractions of RIM-BP1 WT (n = 5) and KO (n = 5) mice using the commercially available N-terminal antibody against RIM-BP1 (Synaptic Systems). In RIM-BP1 KO samples, the upper band (arrow) is missing. This band likely corresponds to the RIM-BP1 isoforms with a molecular mass of ∼200 kDa. However, the RIM-BP1 antibody recognizes several bands at molecular mass that do not correspond to any of the isoforms described so far, indicating unspecific binding. (G) Input/output curves relating the fEPSP amplitude to the amplitude of the presynaptic fiber volley in stratum radiatum of the CA1 region in acute hippocampal slices (0.05 mV: WT, n = 20; KO, n = 21; 0.1 mV: WT, n = 26; KO, n = 26; 0.15 mV: WT, n = 24; KO, n = 25; 0.2 mV: WT, n = 19; KO, n = 26; 0.25 mV: WT, n = 11; KO, n = 18; 0.3 mV: WT, n = 9; KO, n = 19). (H) Summary graph of PPRs of fEPSPs in the stratum radiatum in response to paired stimulation with indicated ISIs (nWT = 28, nKO = 34). (I) Summary graph of fEPSPs in response to a 14-Hz stimulation train (nWT = 10, nKO = 17). Data are expressed as mean ± SEM. n.s., not significant.
RIM-BP2 Deletion Does Not Affect Vesicle Priming or Replenishment.
RIM and Munc13 are known to mediate priming of synaptic vesicles (1, 13). Thus, could the increased short-term facilitation be caused by RIM-BP2 modulating priming through its interactions with the RIM/Munc13-1 complex? We can exclude this possibility for several reasons. First, the unaltered spontaneous release (Figs. 1G and 2E and Fig. S3G) argues against a priming defect in RIM-BP2 KO synapses (14). Second, the transmitter release induced by subsaturating hypertonic stimulation (250 mM sucrose) compared with release induced by a saturating stimulus (500 mM sucrose) was identical in autaptic RIM-BP2 WT and KO neurons, indicating no difference in vesicle fusogenicity (14) (Fig. S3 D and E). Third, due to a higher energy barrier for fusion, sucrose-induced release kinetics would be slower in the case of a priming deficit (15). However, peak release rates were unchanged (Fig. S3F). Additionally, we tested recovery from pool depletion but did not detect any difference between WT and RIM-BP2 KO neurons (Fig. S3 H and I), suggesting that RIM-BP2 deletion does not affect RRP replenishment.
RIM-BP2 Deletion Alters Ca2+ Sensitivity of Release.
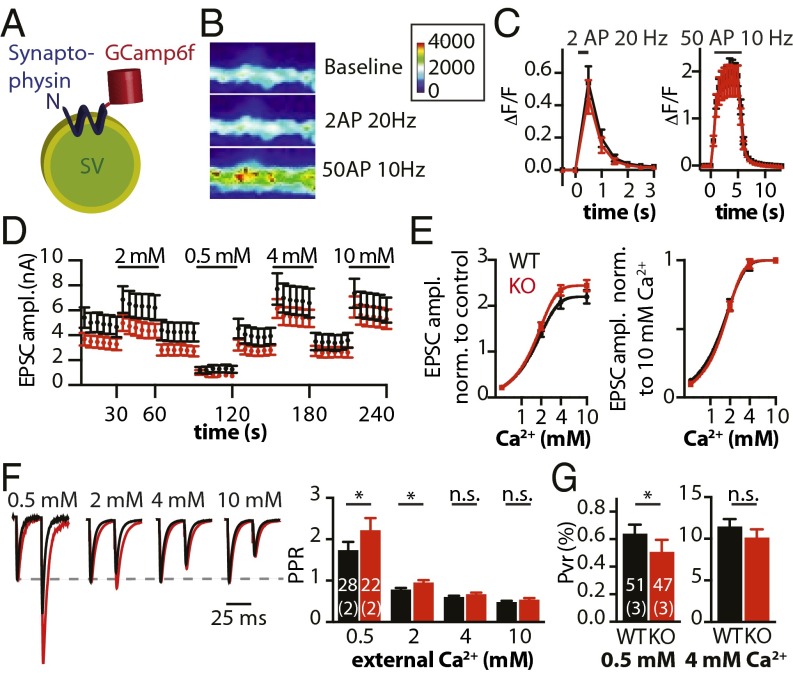
Work at Drosophila NMJ synapses demonstrated that deletion of DRBP results in defective CaV localization, reduced Ca2+ influx, impaired synaptic transmission, and increased short-term facilitation (6). We examined presynaptic Ca2+ influx in cultured hippocampal neurons using the fast Ca2+ sensor GCamp6f coupled to synaptophysin (SynGCamp6f) that specifically localizes the sensor to the presynapse (Fig. 3A). We did not detect significant differences in global Ca2+ signals in response to two (at 20 Hz) or 50 (at 10 Hz) APs (Fig. 3 B and C). Furthermore, we analyzed the dependence of transmitter release on varying external Ca2+ concentrations ([Ca2+]exts) in RIM-BP2 WT and KO autaptic neurons in detail (Fig. 3 D and E). The relative sensitivity of EPSC amplitudes to varying [Ca2+]exts (0.5–10 mM Ca2+/1 mM Mg2+) was determined by intermittent measurements of EPSC amplitudes from control solution (2 mM Ca2+/4 mM Mg2+; Fig. 3D). Normalizing to control EPSCs, we found that the relative potentiation of release is increased in RIM-BP2 KO at 4 mM and 10 mM [Ca2+]exts (Fig. 3E, Left). To fit a Hill function, we normalized the same dataset to saturating 10 mM [Ca2+]ext, resulting in almost identical Ca2+ dose–response curves for WT and KO neurons (Fig. 3E, Right), suggesting no change in the Ca2+ cooperativity n for release and similar numbers of CaVs at the AZ of both genotypes. These findings, however, do not exclude changes in residual Ca2+ at local micro- or nanodomains in the RIM-BP2 KO.
Fig. 3.
RIM-BP2 deletion alters Ca2+ sensitivity of release. (A) Schematic representation of the genetically encoded Ca2+ indicator synGCamp6f at the membrane of a synaptic vesicle. Heat-colored images of a WT dendrite (B) and quantification of synGCamp6f fluorescence change (ΔF/F) during stimulation with two APs at 20 Hz and 50 APs at 10 Hz (C). (D) Mean EPSCs of WT (black) and RIM-BP2 KO (red) autaptic neurons in different [Ca2+]exts. The [Ca2+]exts of test solutions are indicated (all 1 mM Mg2+). A control solution (2 mM Ca2+/4 mM Mg2+) was applied in between. (E) EPSC amplitudes at different [Ca2+]exts normalized to alternating control responses (Left) and to 10 mM [Ca2+]ext (Right). Hill functions were fitted to the data. (F) Normalized EPSCs in response to a paired stimulus and mean PPRs at indicated [Ca2+]exts. (G) PVR of autaptic hippocampal neurons in 0.5 mM and 4 mM [Ca2+]exts. The numbers of neurons and independent cultures analyzed are shown within the bars. Data are expressed as mean ± SEM. *P < 0.05. n.s., not significant.
We next determined PPRs at different [Ca2+]exts (Fig. 3F). In 0.5–2 mM [Ca2+]ext, PPRs (25-ms ISI) were significantly increased in RIM-BP2 KO neurons, whereas PPRs were indistinguishable in 4–10 mM [Ca2+]ext. In a second set of experiments, we analyzed PPRs at different ISIs (25–250 ms) and found similar results for all chosen ISIs (Fig. S3J). PPRs were significantly increased in lower [Ca2+]exts (≤2 mM) and also for ISIs up to 250 ms with the exception of 0.5 mM [Ca2+]ext, where PPRs at 100 ms and 250 ms were identical in WT and RIM-BP2 KO. Overall, the differences in PPRs became smaller with increasing [Ca2+]ext. At 4 mM [Ca2+]ext, only the PPR values recorded at 50-ms and 250-ms ISIs were significantly increased in RIM-BP2 KO neurons.
We also measured PVR at low and high [Ca2+]exts and found that in 0.5 mM [Ca2+]ext, PVR was reduced by 21% in RIM-BP2 KO neurons (Fig. 3G), whereas the difference in 4 mM [Ca2+]ext was not significant. Similarly, RIM-BP2 KO had significantly reduced EPSC amplitudes, accompanied by an increased C.V. of the EPSCs in low, but not high, [Ca2+]ext (Fig. S3K).
Altogether, these data show that loss of RIM-BP2 leads to an impairment of Ca2+-secretion coupling that can be overcome by elevating [Ca2+]ext.
RIM-BP2 KO Alters Presynaptic CaV2.1 Channel Localization.
If priming, global presynaptic Ca2+ influx, and Ca2+ cooperativity are unaltered in the RIM-BP2 KO neurons, could altered CaV positioning explain the changes in release probability and PPR?
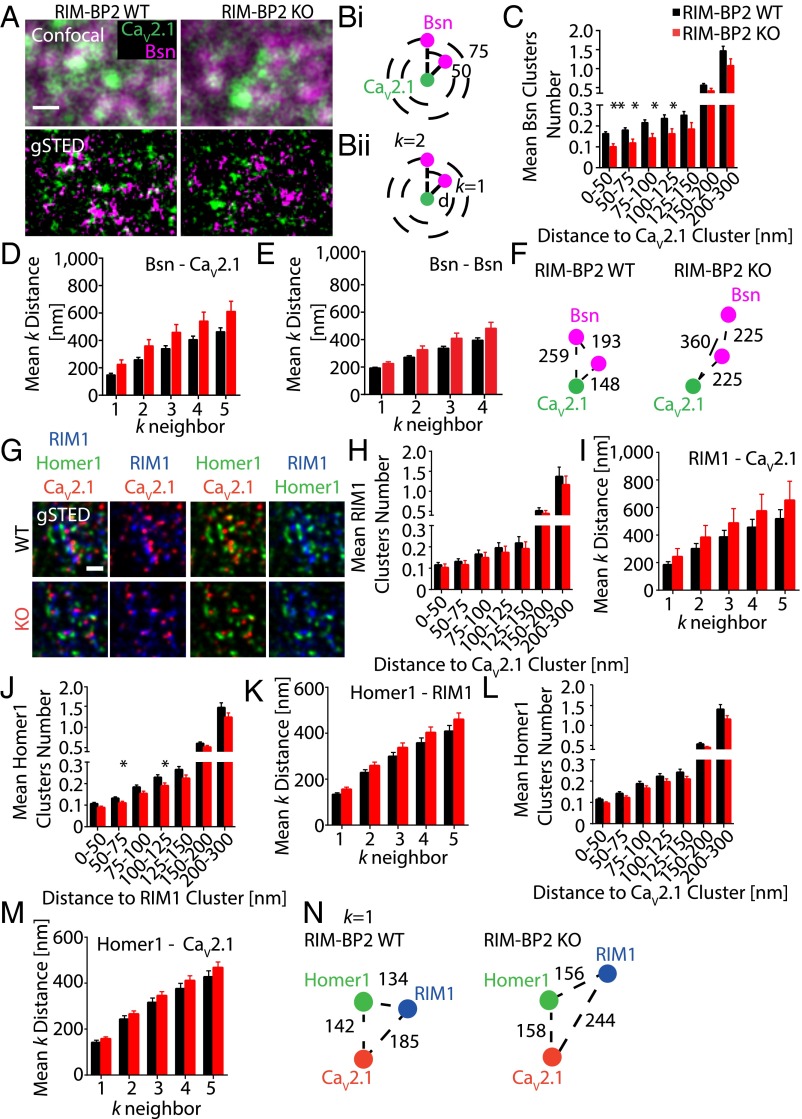
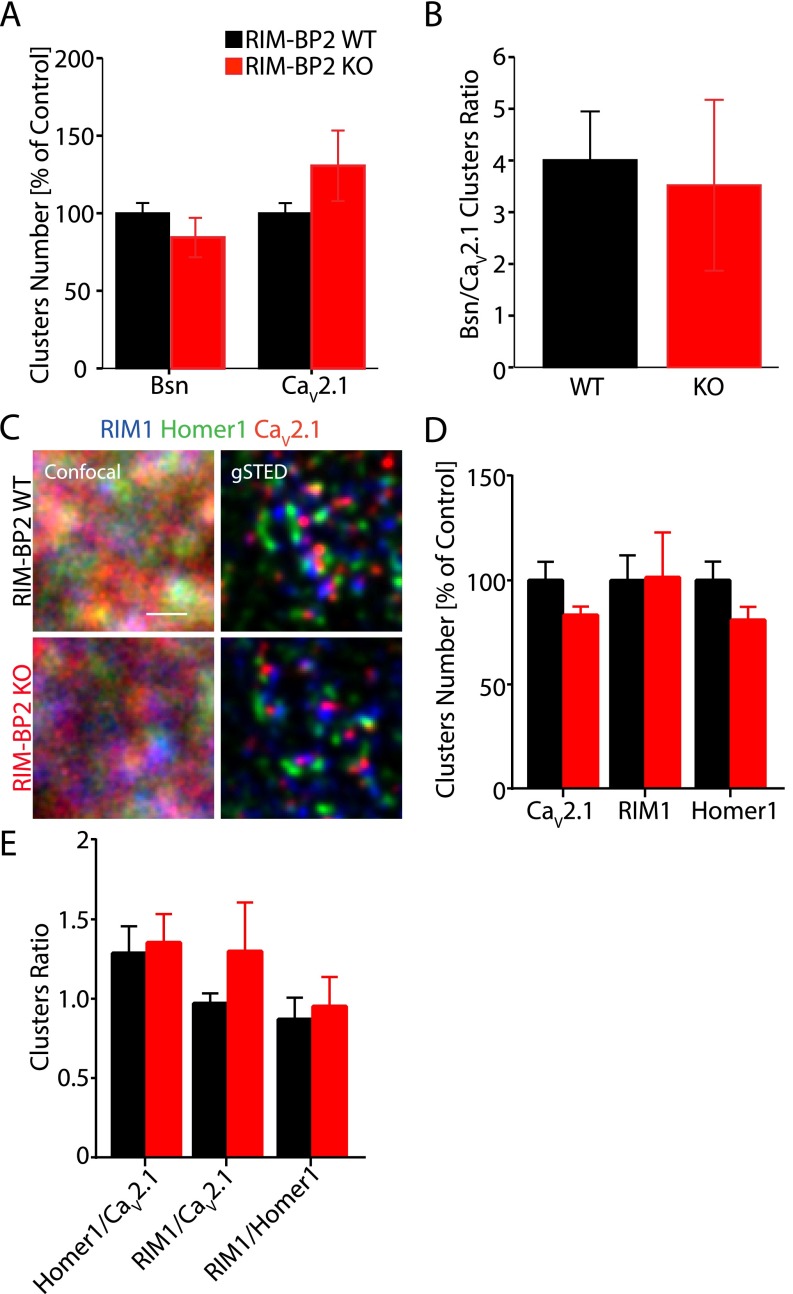
To address this question, we turned to superresolution microscopy and tested if deletion of RIM-BP2 alters the subsynaptic positioning of the P/Q-type CaV subunit CaV2.1, because interference with RIM-BP2 and Bassoon interaction affects their synaptic localization (5). We first measured the synaptic distribution of CaV2.1s in relation to the AZ protein Bassoon in the stratum radiatum of the hippocampal area CA1 of WT and RIM-BP2 KO mice by dual-channel gSTED (Fig. 4 A–F). We found that loss of RIM-BP2 did not significantly affect either the total number of CaV2.1 and Bassoon clusters or their ratio (Fig. S5 A and B), in agreement with the unaltered total Ca2+ influx observed by Ca2+ imaging (Fig. 3 A–C). Notably, however, the average number of Bassoon clusters at short distance intervals from CaV2.1 clusters was significantly reduced by more than 30% (Fig. 4 B, i and C). This finding indicates that RIM-BP2 deletion alters CaV2.1 localization at short distances from the AZ. This effect is not due to an overall change in total cluster number. Supporting these results, at RIM BP2 KO synapses, we observed a 50% increase in the mean k distance of Bassoon clusters surrounding a given CaV2.1 cluster, but the P value reached was only 0.068 (k = 1; Fig. 4 B, ii and D). The mean k distance between individual Bassoon clusters did not significantly change in the absence of RIM-BP2, suggesting that although the distance between single AZs is unaltered in RIM-BP2 KO mice, CaV2.1s are localized more distal from the AZ (Fig. 4 E and F).
Fig. 4.
RIM-BP2 loss results in defective CaV2.1 clustering at the AZ. (A) CaV2.1 and Bsn clusters imaged in situ at CA3-CA1 hippocampal synapses of RIM-BP2 WT and KO mice by dual-channel gSTED (Bottom) compared with confocal acquisition (Top). (B) Schematic representation of two kinds of cluster analysis. (B, i) Bsn clusters within indicated sampling distances (e.g., 50 nm, 75 nm) to a given CaV2.1 cluster were quantified and averaged on thousands of CaV2.1 clusters per image. (B, ii) Mean distance between the k-nearest neighbor Bsn cluster and CaV2.1 cluster (k = 1, k = 2, k = 3, k = 4, k = 5). (C) Bsn cluster numbers at short distances from CaV2.1 clusters (WT, n = 5; KO, n = 6). (D–F) Mean k distance between Bsn and CaV2.1 clusters (D) and between neighboring Bsn clusters (E) in RIM-BP2 KO mice. (G) Triple-channel gSTED images of CaV2.1, RIM1, and Homer1 at CA3-CA1 hippocampal synapses (WT, n = 9; KO, n = 9). (H) RIM1 clusters found in proximity to CaV2.1 channels. (I) Mean k distance of RIM1 clusters to CaV2.1 clusters. Homer1-RIM1 clustering (J) and Homer1-RIM1 mean k distance (K). Homer1 clusters close to CaV2.1 channels (L), and mean k distances between Homer1 clusters and CaV2.1 (M) are shown. (N) CaV2.1 spatial organization relative to RIM1 and Homer1 at excitatory hippocampal synapses. (Scale bars: A and G, 500 nm.) Distances between clusters are represented in nanometers. Values are expressed as mean ± SEM. *P < 0.05; **P < 0.01.
Fig. S5.
RIM-BP2 does not significantly alter the number of Cav2.1 clusters. Cluster analysis performed on gSTED images of CA3-CA1 hippocampal synapses in situ shows that RIM-BP2 deletion does not significantly alter either the number of Bsn and Cav2.1 clusters (A) or their ratio (B) (WT, n = 5; KO, n = 6). (C) Cav2.1, RIM1, and Homer1 distribution at CA3-CA1 hippocampal synapses imaged by conventional confocal (Left) and gSTED (Right) microscopy in brain cryosections of RIM-BP2 WT (Upper) and KO (Lower) mice. (Scale bar: 500 nm.) (D) Quantification of the number of clusters imaged by gSTED (WT, n = 9; KO, n = 9). Also, this independent experiment showed that the number of Cav2.1 clusters does not significantly change in RIM-BP2 KO mice. (E) Similarly, RIM-BP2 deletion does not significantly alter either the number of Homer1 and RIM1 clusters or their ratio to Cav2.1 channels. However, as shown in D and E, we observed a higher variability in the total number of RIM1 clusters, and therefore in the ratio of RIM1/Cav2.1 clusters at RIM-BP2 KO synapses.
To map CaV2.1 localization precisely relative to the AZ protein RIM1 and the postsynaptic marker Homer1, we established triple-channel gSTED (Fig. 4 G–N). The total number of CaV2.1, RIM1, and Homer1 clusters did not significantly differ between WT and RIM-BP2 KO (Fig. S5 C–E). However, RIM-BP2 KO mice showed higher variability in RIM1 cluster number as expressed in a highly variable RIM1/CaV2.1 ratio (Fig. S5 D and E). This variability did not affect net RIM1 clustering relative to CaV2.1s, although we also observed a trend here toward increased mean k distance of RIM1 surrounding CaV2.1 clusters (Fig. 4 H and I). We also found a significantly reduced mean number of Homer1 clusters surrounding a given RIM1 cluster at 50–75 nm and 100–125 nm in RIM-BP2 KO (Fig. 4J), suggesting an effect of RIM-BP2 on RIM1 clustering. However, the mean k distance of neighboring Homer1 toward RIM1 clusters did not significantly change (Fig. 4K). These data indicate that RIM-BP2 might exert a minor effect on the exact RIM1 spatial distribution at the AZ. The mean number of Homer1 clusters relative to CaV2.1 was not altered in RIM-BP2 KO (Fig. 4L). At RIM-BP2 WT synapses, the closest neighboring Homer1 cluster was found at 142 ± 10 nm from CaV2.1 clusters (Fig. 4 M and N). In RIM-BP2 KO, this distance increased by 11% with a statistical P value at the edge of significance (P = 0.05), demonstrating that RIM-BP2 deletion does not grossly alter CaV2.1 positioning versus the postsynaptic density.
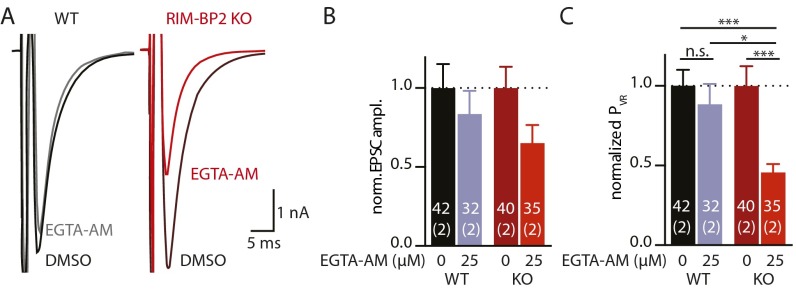
Our structural analysis shows that loss of RIM-BP2 modifies the molecular architecture of the AZ by changing the relative distribution of CaV2.1s versus the AZ scaffold protein Bassoon. Increased distance should result in a larger functional coupling distance between the Ca2+ channel and Ca2+ sensor, and therefore a stronger effect of the slow Ca2+ chelator EGTA (16). Thus, we tested whether application of the membrane-permeable EGTA acetoxymethyl ester (EGTA-AM) would have differential effects on EPSC amplitudes and PVR of WT and RIM-BP2 KO autaptic neurons (Fig. S6 A–C). Indeed, preincubation with 25 μM EGTA-AM decreased EPSC amplitudes more in RIM-BP2 KO than in WT (Fig. S6A). Although this effect was not statistically significant, EGTA-AM did reduce PVR by more than 50% in RIM-BP2 KO without altering WT PVR (Fig. S6B). Thus, together with our structural data, the EGTA-AM effect on PVR in RIM-BP2 KO neurons supports the idea of a larger distance between Ca2+ channels and Ca2+ sensors at release sites in the absence of RIM-BP2. Our analysis provides evidence that RIM-BP2 contributes to proper positioning of CaV2.1s at AZs of hippocampal glutamatergic synapses that allows precise Ca2+-secretion coupling.
Fig. S6.
EGTA-AM reduces PVR significantly more in RIM-BP2 KO than in WT. (A) Representative traces of average EPSCs of WT and RIM-BP2 KO neurons preincubated with 25 μM EGTA-AM or DMSO control. (B) Normalized EPSC amplitudes of neurons preincubated with 25 μM EGTA-AM or DMSO control. EPSCs are reduced in EGTA-AM–treated neurons, but the effect is not significant. (C) PVR of the same neurons as in A, normalized to control. EGTA-AM has a significantly stronger effect on the PVR of RIM-BP2 KO neurons than on WT, supporting larger coupling distances. Data are expressed as mean ± SEM. *P < 0.05; ***P < 0.001.
Discussion
Presynaptic neurotransmitter release is a highly orchestrated process ensuring high-fidelity neuronal communication. The RIM/RIM-BP complex has been implicated in enhancing the efficiency of the fusion machinery and the positioning of synaptic vesicles in close proximity to CaVs to optimize Ca2+-secretion coupling (2, 5, 6). Recently, Acuna et al. (8) showed that deletion of RIM-BP1 and RIM-BP2 in the murine Calyx of Held impairs the reliability of evoked neurotransmitter release, presumably due to an uncoupling of CaVs from release sites. At the Drosophila NMJ, the RIM-BP ortholog DRBP, together with Bruchpilot, is an AZ core component, and elimination of DRBP causes severe structural deficits in AZ organization, associated with strongly impaired basal transmission and STP (6, 7).
Our analysis of the RIM-BP2 KO in glutamatergic hippocampal neurons revealed a moderate disruption of the AZ architecture in comparison to the Drosophila phenotype, resulting in a mild decrease in release efficiency but a pronounced alteration in STP. Thus, rather subtle modifications in the fine positioning of the CaVs within the AZ are sufficient to promote changes in release probability and induce robust STP alterations.
The altered PVR, PPR, and STP of RIM-BP2–deficient neurons are most plausibly explained by changes in the nanoscale organization of protein architectures within the AZs (i.e., the mislocalization of CaV2.1s in proximity to Bassoon). We interpret these results as an indication of increased coupling distances between Ca2+ channels and release sites, also based on our EGTA-AM experiments. These data are also consistent with previously published results (5, 8). A larger distance between CaV2.1s and release sites would result in an altered local Ca2+ profile “seen” by the Ca2+ sensor for release, which determines vesicle fusion and strongly depends on the coupling distance (17). RIM-BPs have been identified in a number of studies as candidate genes for autism spectrum disorder (ASD) (18–21). Although neurons might be able to compensate for the 10% decrease in PVR by homeostatic mechanisms, it is likely that the robust changes in STP might ultimately severely alter the computational properties and function of the affected neuronal networks, and thereby perturb synaptic information processing (22).
It should be noted that a very similar phenotype with mildly reduced PVR and robustly increased short-term facilitation is also evident in Ras-related protein Rab3-deficient neurons (23). Schlüter et al. (23) speculate that Rab3 “superprimes” a subset of vesicles, specifically increasing their release probability. One possible mechanism is that Rab3 directs vesicles to release sites closer to Ca2+ channels, where they would have an intrinsically higher release probability. This scenario might explain the similarities in both phenotypes. Whereas Rab3 would control coupling of synaptic vesicles and Ca2+ channels from the vesicle side, RIM-BP2 might act from the Ca2+ channel side in a subset of AZs. Indeed, in our analysis, we see only a relatively small fraction of AZs having a CaV2.1 within 125 nm. At this subset of AZs, proper CaV2.1 localization depends on RIM-BP2.
We established triple-channel gSTED to determine precise distances between clusters of specific synaptic components at CA3-CA1 synapses in situ. At other excitatory synapses, previous studies using direct stochastical optical reconstruction microscopy (dSTORM) reported a Bassoon-Homer1 average axial distance of 154 nm (24). Accordingly, we find that at WT synapses, Homer1 is located at 142 nm and 134 nm from CaV2.1 and RIM1, respectively, indicating that gSTED can be used reliably to map protein cluster localization, and thus synaptic substructures at mammalian synapses in situ. However, with both superresolution techniques, true cluster distances are obviously influenced by using indirect immunolabeling, because the size of the primary/secondary IgG sandwich (∼20 nm) and the position of the epitopes recognized by the antibodies likely influence the exact measured distances (epitopes targeted by each antibody are listed in Table S1). Still, comparing the values between mutant and WT constellation should be meaningful.
Table S1.
Primary antibodies for immunohistochemistry and immunoblot
| Antigen | Species | Dilution | Source |
| Bsn (N-terminal) | ms | 1:1,200 (IHC) | Abcam |
| Cav2.1 (rat amino acids 1,921–2,212) | Rb | 1:500 (IHC) | Synaptic Systems |
| Dynamin1 | ms | 1:1,000 (WB) | Kind gift of P. De Camilli (Boyer Center for Molecular Medicine, New Haven, CT) |
| ERC1b/2 | Rb | 1:500 (WB) | Synaptic Systems |
| GIT1/2 (p95PKL) | ms | 1:1,000 (WB) | BD Biosciences |
| Homer1 (human amino acids 1–186) | GP | 1:200 (IHC) | Synaptic Systems |
| HSC70 | ms | 1:2,000 | Thermo Fisher Scientific |
| MUNC13-1 (rat amino acids 3–317) | Rb | 1:150 (IHC) | Synaptic Systems |
| MUNC13-1 | ms | 1:1,000 (WB) | Synaptic Systems |
| RIM1 (rat amino acids 602–723) | ms | 1:200 (IHC) | BD Pharmingen |
| RIM1/2 | Rb | 1:2,000 (WB) | Synaptic Systems |
| RIM-BP1 (mouse amino acids 1–335) | Rb | 1:500 (WB) | Synaptic Systems |
| RIM-BP2 (last 20 aa of RIM-BP2 C terminus) | Rb | 1:1,000 (IP, WB) | Self-raised |
| RIM-BP2 (rat amino acids 589–869) | GP | 1:600/1:1,000 (IHC) | Kind gift of A. Fejtova and Eckart Gundelfinger, Leibniz Institute for Neurobiology, Magdeburg, Germany |
| 1:10,000 (WB) | |||
| Synapsin1 | ms | 1:1,000 (WB) | Synaptic Systems |
| Synaptophysin1 | ms | 1:10,000 (WB) | Synaptic Systems |
| β-Actin | ms | 1:2,000 (WB) | Sigma |
| β-Tubulin | ms | 1:2,000 (WB) | Sigma |
GP, guinea pig; IHC, immunohistochemistry; ms, mouse; Rb, rabbit; WB, Western blot.
Here, we show that at hippocampal CA3-CA1 AZs, RIM-BP2 is located close to Bassoon and Munc13-1 in a complex in which each nearest neighbor is rather equidistant (∼100 nm). According to ultrastructural studies, cortical pyramidal neuron synapses usually have a single AZ with a highly variable area of about 0.04 μm2 (25, 26). Our cluster analysis at CA3-CA1 synapses maps two adjacent Bassoon clusters at less than 200 nm. We therefore assume that the first (k = 1) nearest neighbor of our analysis might indicate a neighboring cluster within a single AZ. To address this point more precisely, 3D reconstruction of AZ components imaged at subdiffraction axial resolution will be necessary. Our data also show that the localization of CaV2.1 in close apposition to the PSD marker Homer1 is relatively stable even in the absence of RIM-BP2. Although the overall RIM1 expression level was not affected in crude synaptosomal membranes, we observed increased variability in RIM1 total cluster number in RIM-BP2–deficient synapses. This increased variability may reflect altered nanoscale distribution of RIM1 localization within AZs and toward the postsynapse, which is supported by a trend toward a larger RIM1-to-CaV2.1 mean k distance and slightly, but significantly, altered Homer1 clustering relative to RIM1 in RIM-BP2 KO.
We performed a detailed analysis of RIM-BP2 loss of function mostly in autaptic neurons, whereas our structural analysis was done in situ to provide information on the organization of the AZ within the hippocampus. Nevertheless, our PPR and STP experiments in slices demonstrate that RIM-BP2 KO results in a similar functional defect in both preparations. On the other hand, our EGTA-AM experiments provide evidence that RIM-BP2 is necessary for proper Ca2+ channel localization at the AZ also in vitro.
Besides coupling of CaVs to release sites, other functions have been suggested for RIM-BPs. At the Drosophila NMJ, DRBP is required for homeostatic modulation of presynaptic Ca2+ influx and the size of the RRP, as well for as recovery from pool depletion (7). In contrast, at the murine Calyx of Held, RRP size and the kinetics of priming into the RRP are not RIM-BP–dependent (8). However, the release of the RRP was significantly decelerated in RIM-BP1/2 DKO. In our autaptic culture system, we observed no differences in RRP size, fusogenicity, or peak release rates, and we did not detect changes in the recovery from pool depletion. These results suggest that loss of RIM-BPs manifests distinctively at different specialized synapse types.
Why are the effects of RIM-BP2 deletion on presynaptic structure and function of hippocampal synapses rather subtle compared with the severe phenotype at Drosophila NMJ AZs? Differences in the exact structure of the AZ scaffolds and the level of genetic redundancy might well be involved here. In fact, different isoforms of AZ proteins likely contribute to shaping specific functions that differ from synapse to synapse. For both hippocampal and NMJ synapses, however, the RIM/RIM-BP complex is crucial for precise CaV localization at the AZ and expression of STP and/or long-term plasticity. The comparison of KO phenotypes suggests that in Drosophila, DRBP plays a more pivotal role (6, 27), whereas judged from single gene KOs at mammalian synapses, RIM is functionally most important (1, 2, 8, 28–30). Still, the importance of RIM-BPs for STP is conserved between flies and mammals (6, 8, 31), consistent with the highly conserved molecular interactions between RIM-BPs, CaVs, and RIM (2, 4, 6).
Further investigation is required to understand fully the molecular role of RIM-BPs in different synapse types, which might also depend on the type of presynaptic CaV present. Studying potential behavioral deficits in RIM-BP1 and RIM-BP2 KO mice and the function of RIM-BPs in neuronal circuits implicated in ASD (32) will likely advance our understanding of how the disruption of RIM-BP function might lead to behavioral and cognitive deficits.
Methods
KO Mouse Generation.
RIM-BP2 and RIM-BP1 targeting vector construction and KO mouse generation by standard homologous recombination were performed by genOway. All animal experiments were approved by the animal welfare committee of Charité Universitätsmedizin Berlin and the Landesamt für Gesundheit und Soziales Berlin.
Cell Culture and Electrophysiological Recordings.
Primary neuronal cultures were prepared as described by Arancillo et al. (14). Whole-cell patch-clamp recordings in autaptic neurons were performed at days in vitro 13–21 as described (14).
Slice Preparation and Electrophysiological Recordings.
Acute hippocampal slices were prepared as described by Stempel et al. (33). The 300-μm-thick horizontal slices were maintained for 30 min at 35 °C in sucrose-artificial cerebrospinal fluid (ACSF) and subsequently stored in ACSF at room temperature. Experiments were started after 30 min and no longer than 6 h after preparation. A detailed description of electrophysiological experiments is provided in SI Methods.
Immunohistochemical Analysis and gSTED Imaging.
Following immunostaining, sagittal cryosections (10 μm) of RIM-BP2 WT and KO brains were imaged by gSTED with a Leica SP8 gSTED microscope (Leica Microsystems) equipped with two depletion lasers (592 nm and 775 nm). Cluster analysis on deconvolved images was performed with Amira (Visualization Sciences Group) and a MATLAB (The MathWorks, Inc.) custom-written script.
SynGCamp6f Imaging.
SynGCamp6f was generated analogous to synGCamp2 (34) by fusing GCamp6f (35) to the C terminus of the synaptic vesicle protein synaptophysin. Imaging was done as previously described (34).
SI Methods contains figures, a detailed description of the methods used, antibodies (Table S1), raw values, and statistical analysis (Tables S2–S4).
Table S2.
Summary of all electrophysiological parameters of RIM-BP2 KO
| RIM-BP2 WT | RIM-BP2 KO | Statistic | |||||||
| Parameter | Mean | SEM | n | Mean | SEM | n | P | Test | |
| Normalized EPSC amplitudes | 1.00 | 0.07 | 156 | 0.80 | 0.06 | 160 | 0.0055 | Mann–Whitney U test | |
| C.V. of EPSC, % | 8.46 | 0.98 | 31 | 11.29 | 1.16 | 30 | 0.0351 | Mann–Whitney U test | |
| Normalized RRP size | 1.00 | 0.07 | 156 | 0.97 | 0.08 | 160 | 0.2315 | Mann–Whitney U test | |
| PVR, % | 6.51 | 0.32 | 156 | 5.85 | 0.46 | 160 | 0.0041 | Mann–Whitney U test | |
| mEPSC frequency: culture, Hz | 5.65 | 0.76 | 30 | 5.61 | 0.73 | 30 | 0.9715 | Unpaired Student t test | |
| mEPSC amplitude: culture, pA | 30.24 | 2.37 | 30 | 30.37 | 1.82 | 30 | 0.786 | Mann–Whitney U test | |
| PPR, 20-ms ISI: culture | 0.91 | 0.03 | 83 | 1.10 | 0.05 | 82 | 0.0001 | Mann–Whitney U test | |
| PPR, 25-ms ISI: culture | 1.13 | 0.09 | 83 | 1.29 | 0.05 | 82 | <0.0001 | Mann–Whitney U test | |
| PPR, 100-ms ISI: culture | 0.85 | 0.02 | 83 | 0.92 | 0.03 | 82 | 0.0366 | Mann–Whitney U test | |
| Train of 10 Hz: mean of last 10 pulses, nA | 0.749 | 0.003 | 83 | 0.866 | 0.005 | 82 | <0.0001 | Mann–Whitney U test | |
| fEPSPs, mV | FV: 0.05 mV | 0.51 | 0.06 | 21 | 0.51 | 0.08 | 19 | 0.8285 | Mann–Whitney U test |
| FV: 0.1 mV | 0.82 | 0.09 | 21 | 0.80 | 0.11 | 19 | 0.9137 | ||
| FV: 0.15 mV | 1.06 | 0.12 | 21 | 1.02 | 0.13 | 19 | 0.7685 | ||
| FV:0.2 mV | 1.23 | 0.13 | 21 | 1.21 | 0.15 | 19 | 0.7046 | ||
| FV: 0.3 mV | 1.52 | 0.17 | 18 | 1.59 | 0.21 | 14 | 0.8942 | ||
| FV: 0.4 mV | 1.72 | 0.19 | 16 | 1.72 | 0.23 | 13 | 0.9959 | ||
| Train of 14 Hz: slices fEPSPs, mV | Pulse 1 | 1.00 | 0.00 | 17 | 1.00 | 0.00 | 18 | >0.05 | Repeated measurement ANOVA |
| Pulse 2 | 1.16 | 0.01456 | 1.27 | 0.02 | <0.05 | ||||
| Pulse 3 | 1.05 | 0.01644 | 1.21 | 0.03 | <0.001 | ||||
| Pulse 4 | 0.96 | 0.01833 | 1.13 | 0.03 | <0.001 | ||||
| Pulse 5 | 0.91 | 0.01774 | 1.08 | 0.03 | <0.001 | ||||
| Pulse 6 | 0.89 | 0.01940 | 1.06 | 0.03 | <0.001 | ||||
| Pulse 7 | 0.88 | 0.01811 | 1.05 | 0.03 | <0.001 | ||||
| Pulse 8 | 0.87 | 0.02 | 1.04 | 0.03 | <0.001 | ||||
| Pulse 9 | 0.86 | 0.02 | 1.02 | 0.03 | <0.001 | ||||
| Pulse 10 | 0.85 | 0.02 | 1.02 | 0.03 | <0.001 | ||||
| Pulse 11 | 0.84 | 0.02 | 1.02 | 0.03 | <0.001 | ||||
| Pulse 12 | 0.83 | 0.02 | 1.00 | 0.03 | <0.001 | ||||
| Pulse 13 | 0.82 | 0.02 | 1.00 | 0.03 | <0.001 | ||||
| Pulse 14 | 0.81 | 0.02 | 0.99 | 0.03 | <0.001 | ||||
| Pulse 15 | 0.80 | 0.02 | 0.98 | 0.03 | <0.001 | ||||
| Pulse 16 | 0.79 | 0.02 | 0.98 | 0.03 | <0.001 | ||||
| Pulse 17 | 0.77 | 0.02 | 0.96 | 0.03 | <0.001 | ||||
| Pulse 18 | 0.76 | 0.02 | 0.96 | 0.03 | <0.001 | ||||
| Pulse 19 | 0.76 | 0.02 | 0.95 | 0.03 | <0.001 | ||||
| Pulse 20 | 0.74 | 0.02 | 0.94 | 0.03 | <0.001 | ||||
| Pulse 21 | 0.73 | 0.02 | 0.92 | 0.03 | <0.001 | ||||
| Pulse 22 | 0.72 | 0.02 | 0.92 | 0.03 | <0.001 | ||||
| Pulse 23 | 0.71 | 0.02 | 0.90 | 0.03 | <0.001 | ||||
| Pulse 24 | 0.71 | 0.02 | 0.90 | 0.03 | <0.001 | ||||
| Pulse 25 | 0.70 | 0.02 | 0.88 | 0.04 | <0.001 | ||||
| Fraction of RRP released, % | 46.88 | 5.02 | 26 | 47.06 | 4.20 | 27 | 0.8496 | Mann–Whitney U test | |
| Peal release rate, 1/s | 1.51 | 0.08 | 44 | 1.50 | 0.06 | 43 | 0.0841 | Mann–Whitney U test | |
| Spontaneous release rate, 1/s | 0.002 | 0.0005 | 30 | 0.002 | 0.0003 | 30 | 0.2554 | Mann–Whitney U test | |
| PPR (fEPSP): slice | 25-ms ISI | 0.98 | 0.03 | 21 | 1.18 | 0.05 | 19 | 0.0003 | Mann–Whitney U test |
| 50-ms ISI | 1.17 | 0.02 | 21 | 1.34 | 0.03 | 19 | <0.0001 | Mann–Whitney U test | |
| 100-ms ISI | 1.16 | 0.01 | 21 | 1.29 | 0.02 | 19 | <0.0001 | Mann–Whitney U test | |
| mEPSC frequency: slice, Hz | 1.00 | 0.13 | 15 | 1.06 | 0.09 | 18 | 0.71 | Unpaired Student t test | |
| mEPSC amplitude: slice, pA | 13.36 | 0.35 | 15 | 13.51 | 0.44 | 18 | 0.79 | Unpaired Student t test | |
| Hill curve | logEC50 | 0.25 | 0.05 | 22 | 0.25 | 0.03 | 28 | 0.1158 | Extra sum-of squares F test |
| Hill slope | 3.37 | 1.95 | 22 | 3.74 | 1.97 | 28 | |||
| PPR (Fig. 3) | 0.5 mM Ca2+ | 1.74 | 0.20 | 22 | 2.22 | 0.29 | 28 | 0.0215 | Mann–Whitney U test |
| 2 mM Ca2+ | 0.79 | 0.04 | 22 | 0.95 | 0.06 | 28 | 0.0152 | Unpaired Student t test | |
| 4 mM Ca2+ | 0.61 | 0.04 | 22 | 0.67 | 0.04 | 28 | 0.4946 | Unpaired Student t test | |
| 10 mM Ca2+ | 0.49 | 0.03 | 22 | 0.54 | 0.04 | 28 | 0.2875 | Unpaired Student t test | |
| PVR, % (Fig. 3) | 0.5 mM Ca2+ | 0.64 | 0.07 | 51 | 0.51 | 0.09 | 47 | 0.0281 | Mann–Whitney U test |
| 4 mM Ca2+ | 11.49 | 0.87 | 51 | 10.15 | 1.00 | 47 | 0.1303 | Mann–Whitney U test | |
| PPR, 0.5 mM Ca2+ (Fig. S3) | 25-ms ISI | 1.44 | 0.05 | 51 | 2.00 | 0.18 | 47 | 0.0251 | Mann–Whitney U test |
| 50-ms ISI | 1.26 | 0.08 | 1.48 | 0.11 | 0.0409 | Mann–Whitney U test | |||
| 100-ms ISI | 1.12 | 0.04 | 1.32 | 0.08 | 0.1798 | Mann–Whitney U test | |||
| 250-ms ISI | 1.19 | 0.09 | 1.18 | 0.09 | 0.4442 | Mann–Whitney U test | |||
| PPR, 1 mM Ca2+ (Fig. S3) | 25-ms ISI | 1.09 | 0.02 | 1.29 | 0.06 | 0.0196 | Mann–Whitney U test | ||
| 50-ms ISI | 0.99 | 0.02 | 1.13 | 0.03 | 0.0013 | Unpaired Student t test | |||
| 100-ms ISI | 0.95 | 0.02 | 1.04 | 0.03 | 0.0164 | Mann–Whitney U test | |||
| 250-ms ISI | 0.91 | 0.01 | 1.00 | 0.02 | 0.0131 | Mann–Whitney U test | |||
| PPR, 2 mM Ca2+ (Fig. S3) | 25-ms ISI | 0.83 | 0.02 | 1.02 | 0.05 | 0.0003 | Mann–Whitney U test | ||
| 50-ms ISI | 0.84 | 0.02 | 0.98 | 0.03 | <0.0001 | Mann–Whitney U test | |||
| 100-ms ISI | 0.86 | 0.02 | 0.93 | 0.02 | 0.0049 | Unpaired Student t test | |||
| 250-ms ISI | 0.86 | 0.01 | 0.92 | 0.02 | 0.0085 | Mann–Whitney U test | |||
| PPR, 4 mM Ca2+ (Fig. S3) | 25-ms ISI | 0.69 | 0.02 | 0.76 | 0.03 | 0.084 | Mann–Whitney U test | ||
| 50-ms ISI | 0.71 | 0.02 | 0.79 | 0.02 | 0.0108 | Unpaired Student t test | |||
| 100-ms ISI | 0.76 | 0.02 | 0.80 | 0.02 | 0.0813 | Unpaired Student t test | |||
| 250-ms ISI | 0.79 | 0.01 | 0.83 | 0.01 | 0.0364 | Unpaired Student t test | |||
| EPSC, nA (Fig. S3) | 0.5 mM Ca2+ | 1.13 | 0.25 | 51 | 0..75 | 0.16 | 47 | 0.0494 | Mann–Whitney U test |
| 1 mM Ca2+ | 3.01 | 0.54 | 47 | 1.90 | 0.33 | 43 | 0.0332 | Mann–Whitney U test | |
| 2 mM Ca2+ | 3.98 | 0.52 | 36 | 3.47 | 0.46 | 38 | 0.3461 | Mann–Whitney U test | |
| 4 mM Ca2+ | 4.92 | 1.22 | 20 | 3.82 | 0.63 | 27 | 0.6621 | Mann–Whitney U test | |
| C.V. of EPSC, % (Fig. S3) | 0.5 mM Ca2+ | 26.24 | 2.52 | 51 | 36.7 | 4.09 | 47 | 0.0494 | Mann–Whitney U test |
| 1 mM Ca2+ | 12.6 | 1.40 | 47 | 16.36 | 1.62 | 43 | 0.0382 | Mann–Whitney U test | |
| 2 mM Ca2+ | 8.22 | 0.88 | 36 | 9.61 | 0.88 | 38 | 0.1757 | Mann–Whitney U test | |
| 4 mM Ca2+ | 8.73 | 1.40 | 20 | 8.42 | 1.09 | 27 | 0.9745 | Mann–Whitney U test | |
| MK-801 | Kfast | 0.68 | 0.30 | 12 | 0.25 | 0.09 | 24 | 0.0197 | Extra sum-of squares F test |
| Kslow | 0.07 | 0.01 | 0.05 | 0.01 | |||||
| PercentFast | 39.87 | 7.85 | 47.33 | 14.87 | |||||
| EPSC: EGTA-AM, nA | DMSO | 1.00 | 0.15 | 42 | 1.00 | 0.14 | 40 | >0.2 | Kruskal–Wallis test with Dunn’s multiple comparison |
| 25 μM EGTA-AM | 0.83 | 0.15 | 32 | 0.65 | 0.11 | 35 | |||
| PVR, %: EGTA-AM, nA | DMSO | 1.00* | 0.10 | 42 | 1.00† | 0.12 | 40 | 0.0187‡ | Kruskal–Wallis test with Dunn’s multiple comparison |
| 25 μM EGTA-AM | 0.88‡ | 0.13 | 32 | 0.46*,†,‡ | 0.05 | 35 | 0.0005† | ||
| 0.0001* | |||||||||
Comparison of PVR between WT DMSO and KO 25 μM EGTA-AM.
Comparison of PVR between KO DMSO and KO 25 μM EGTA-AM.
Comparison of PVR between WT 25 μM EGTA-AM and KO 25 μM EGTA-AM.
Table S4.
Summary of gSTED-based cluster analysis
| RIM-BP2 WT | RIM-BP2 KO | Statistic | |||||||
| Parameter | Distance, nm/k neighbor | Mean | SEM | n | Mean | SEM | n | P | Test |
| Bsn cluster number,* % of control | — | 100 | 6.65 | 5 | 84.42 | 12.70 | 6 | 0.311 | Unpaired Student t test |
| CaV2.1 cluster number,* % of control | — | 100 | 6.59 | 5 | 130.62 | 22.73 | 6 | 0.245 | Unpaired Student t test |
| Bsn/CaV2.1 ratio* | — | 4.01 | 0.94 | 5 | 3.52 | 1.65 | 6 | 0.814 | Unpaired Student t test |
| Mean Bsn cluster number at tested distance to a given CaV2.1 cluster* | 0–50 | 0.164 | 0.009 | 5 | 0.101 | 0.014 | 6 | 0.005 | Unpaired Student t test |
| 50–75 | 0.181 | 0.011 | 0.119 | 0.018 | 0.022 | ||||
| 75–100 | 0.216 | 0.013 | 0.143 | 0.020 | 0.017 | ||||
| 100–125 | 0.236 | 0.017 | 0.163 | 0.023 | 0.037 | ||||
| 125–150 | 0.251 | 0.018 | 0.185 | 0.031 | 0.112 | ||||
| 150–200 | 0.562 | 0.044 | 0.411 | 0.068 | 0.110 | ||||
| 200–300 | 1.466 | 0.121 | 1.078 | 0.178 | 0.119 | ||||
| Mean k distance (Bsn-CaV2.1),* nm | k = 1 | 148 | 12 | 5 | 225 | 33 | 6 | 0.068 | Unpaired Student t test |
| k = 2 | 259 | 18 | 360 | 46 | 0.082 | ||||
| k = 3 | 339 | 22 | 459 | 57 | 0.096 | ||||
| k = 4 | 406 | 26 | 540 | 66 | 0.114 | ||||
| k = 5 | 463 | 29 | 611 | 74 | 0.119 | ||||
| Mean k distance (Bsn- Bsn),* nm | k = 1 | 193 | 6 | 5 | 225 | 14 | 6 | 0.074 | Unpaired Student t test |
| k = 2 | 271 | 11 | 326 | 27 | 0.112 | ||||
| k = 3 | 337 | 14 | 410 | 37 | 0.123 | ||||
| k = 4 | 395 | 18 | 482 | 44 | 0.127 | ||||
| CaV2.1 cluster number, % of control | — | 100 | 8.88 | 9 | 83.35 | 4.11 | 9 | 0.116 | Unpaired Student t test |
| RIM1 cluster number, % of control | — | 100 | 12.04 | 9 | 101.46 | 21.64 | 9 | 0.954 | Unpaired Student t test |
| Homer1 cluster number, % of control | — | 100 | 9.01 | 9 | 80.98 | 6.37 | 9 | 0.104 | Unpaired Student t test |
| RIM1/CaV2.1 ratio | — | 0.971 | 0.064 | 9 | 1.300 | 0.308 | 9 | 0.325 | Unpaired Student t test |
| RIM1/Homer1 ratio | — | 0.871 | 0.136 | 9 | 0.953 | 0.184 | 9 | 0.725 | Unpaired Student t test |
| Homer1/ CaV2.1 ratio | — | 1.288 | 0.171 | 9 | 1.355 | 0.180 | 9 | 0.790 | Unpaired Student t test |
| Mean RIM1 cluster number at tested distance to a given CaV2.1 cluster | 0–50 | 0.115 | 0.011 | 9 | 0.103 | 0.018 | 9 | 0.559 | Unpaired Student t test |
| 50–75 | 0.131 | 0.014 | 0.116 | 0.020 | 0.551 | ||||
| 75–100 | 0.166 | 0.019 | 0.150 | 0.025 | 0.625 | ||||
| 100–125 | 0.195 | 0.025 | 0.174 | 0.030 | 0.581 | ||||
| 125–150 | 0.218 | 0.031 | 0.192 | 0.032 | 0.566 | ||||
| 150–200 | 0.510 | 0.078 | 0.441 | 0.079 | 0.545 | ||||
| 200–300 | 1.371 | 0.237 | 1.161 | 0.220 | 0.526 | ||||
| Mean k distance (RIM1-CaV2.1), nm | k = 1 | 185 | 23 | 9 | 244 | 59 | 9 | 0.730 | Mann–Whitney U test |
| k = 2 | 301 | 37 | 385 | 85 | 0.796 | ||||
| k = 3 | 386 | 49 | 488 | 104 | 0.730 | ||||
| k = 4 | 456 | 58 | 575 | 121 | 0.730 | ||||
| k = 5 | 518 | 67 | 653 | 137 | 0.730 | ||||
| Mean Homer1 cluster number at tested distance to a given RIM1 cluster | 0–50 | 0.107 | 0.006 | 9 | 0.089 | 0.007 | 9 | 0.060 | Unpaired Student t test |
| 50–75 | 0.132 | 0.007 | 0.111 | 0.007 | 0.045 | ||||
| 75–100 | 0.184 | 0.010 | 0.154 | 0.011 | 0.056 | Mann–Whitney U test† | |||
| 100–125 | 0.229 | 0.012 | 0.191 | 0.013 | 0.043 | ||||
| 125–150 | 0.265 | 0.015 | 0.225 | 0.015 | 0.077 | ||||
| 150–200 | 0.603 | 0.037 | 0.515 | 0.040 | 0.122 | ||||
| 200–300 | 1.491 | 0.120 | 1.252 | 0.107 | 0.222† | ||||
| Mean k distance (Homer1-RIM1), nm | k = 1 | 134 | 7 | 9 | 156 | 9 | 9 | 0.064 | Unpaired Student t test |
| k = 2 | 229 | 12 | 259 | 15 | 0.113† | ||||
| k = 3 | 299 | 17 | 338 | 20 | 0.161† | Mann–Whitney U test† | |||
| k = 4 | 358 | 22 | 403 | 24 | 0.113† | ||||
| k = 5 | 409 | 25 | 461 | 28 | 0.113† | ||||
| Mean Homer1 cluster number at tested distance to a given CaV2.1 cluster | 0–50 | 0.114 | 0.007 | 9 | 0.096 | 0.007 | 9 | 0.077 | Unpaired Student t test |
| 50–75 | 0.142 | 0.009 | 0.123 | 0.008 | 0.136 | ||||
| 75–100 | 0.188 | 0.011 | 0.167 | 0.011 | 0.193 | Mann–Whitney U test† | |||
| 100–125 | 0.223 | 0.013 | 0.197 | 0.013 | 0.136† | ||||
| 125–150 | 0.241 | 0.015 | 0.210 | 0.013 | 0.127 | ||||
| 150–200 | 0.534 | 0.038 | 0.448 | 0.030 | 0.094 | ||||
| 200–300 | 1.406 | 0.118 | 1.156 | 0.092 | 0.050† | ||||
| Mean k distance (Homer1-CaV2.1), nm | k = 1 | 142 | 10 | 9 | 158 | 8 | 9 | 0.050† | Unpaired Student t test |
| k = 2 | 243 | 15 | 266 | 13 | 0.161† | ||||
| k = 3 | 316 | 19 | 346 | 17 | 0.265 | Mann–Whitney U test† | |||
| k = 4 | 376 | 23 | 412 | 21 | 0.270 | ||||
| k = 5 | 428 | 26 | 468 | 24 | 0.267 | ||||
Double immunolabeling Bassoon/CaV2.1 in hippocampal cryosections.
Mann–Whitney U test, exact significance [2 * (one-tailed)].
Table S3.
Summary of all electrophysiological parameters of RIM-BP1/2 DKO
| RIM-BP2 KO | RIM-BP1/2 DKO | Statistic | |||||||
| Parameter | Mean | SEM | n | Mean | SEM | n | P | Test | |
| fEPSPs, mV | FV: 0.05 mV | 0.43 | 0.04 | 20 | 0.45 | 0.04 | 21 | 0.5212 | Mann–Whitney U test |
| FV: 0.1 mV | 0.63 | 0.05 | 26 | 0.67 | 0.5 | 26 | 0.6148 | ||
| FV: 0.15 mV | 0.88 | 0.07 | 24 | 0.90 | 0.08 | 25 | 0.9671 | ||
| FV: 0.2 mV | 1.03 | 0.10 | 19 | 1.03 | 0.08 | 26 | 1.0000 | ||
| FV: 0.25 mV | 1.15 | 0.11 | 11 | 1.18 | 0.12 | 18 | 0.6488 | ||
| FV: 0.3 mV | 1.19 | 0.16 | 9 | 1.27 | 0.13 | 19 | 0.9608 | ||
| PPR, fEPSP | 25-ms ISI | 1.22 | 0.05 | 28 | 1.27 | 0.03 | 34 | 0.1026 | Mann–Whitney U test |
| 50-ms ISI | 1.35 | 0.04 | 1.38 | 0.03 | 0.129 | Mann–Whitney U test | |||
| 100-ms ISI | 1.30 | 0.03 | 1.32 | 0.02 | 0.2690 | Mann–Whitney U test | |||
| Train of 14 Hz (fEPSPs) | Pulse 1 | 1.00 | 0.00 | 10 | 1.00 | 0.00 | 17 | >0.9999 | Repeated measurement ANOVA |
| Pulse 2 | 1.69 | 0.04 | 1.85 | 0.04 | 0.4137 | ||||
| Pulse 3 | 1.66 | 0.05 | 1.83 | 0.04 | 0.3003 | ||||
| Pulse 4 | 1.57 | 0.04 | 1.70 | 0.05 | 0.6672 | ||||
| Pulse 5 | 1.48 | 0.04 | 1.63 | 0.05 | 0.4986 | ||||
| Pulse 6 | 1.45 | 0.05 | 1.57 | 0.04 | 0.8907 | ||||
| Pulse 7 | 1.42 | 0.05 | 1.55 | 0.05 | 0.8047 | ||||
| Pulse 8 | 1.41 | 0.04 | 1.52 | 0.05 | 0.9056 | ||||
| Pulse 9 | 1.37 | 0.05 | 1.50 | 0.04 | 0.8488 | ||||
| Pulse 10 | 1.36 | 0.05 | 1.47 | 0.04 | 0.9551 | ||||
| Pulse 11 | 1.34 | 0.05 | 1.46 | 0.04 | 0.8304 | ||||
| Pulse 12 | 1.31 | 0.05 | 1.43 | 0.04 | 0.8703 | ||||
| Pulse 13 | 1.30 | 0.05 | 1.39 | 0.04 | 0.9966 | ||||
| Pulse 14 | 1.27 | 0.05 | 1.36 | 0.05 | 0.9944 | ||||
| Pulse 15 | 1.24 | 0.05 | 1.34 | 0.04 | 0.9862 | ||||
| Pulse 16 | 1.23 | 0.05 | 1.31 | 0.04 | 0.9995 | ||||
| Pulse 17 | 1.21 | 0.05 | 1.28 | 0.04 | 0.9999 | ||||
| Pulse 18 | 1.18 | 0.05 | 1.23 | 0.04 | >0.9999 | ||||
| Pulse 19 | 1.17 | 0.05 | 1.23 | 0.05 | >0.9999 | ||||
| Pulse 20 | 1.15 | 0.05 | 1.19 | 0.05 | >0.9999 | ||||
| Pulse 21 | 1.13 | 0.06 | 1.18 | 0.04 | >0.9999 | ||||
| Pulse 22 | 1.11 | 0.06 | 1.14 | 0.04 | >0.9999 | ||||
| Pulse 23 | 1.10 | 0.06 | 1.11 | 0.04 | >0.9999 | ||||
| Pulse 24 | 1.08 | 0.06 | 1.10 | 0.05 | >0.9999 | ||||
| Pulse 25 | 1.06 | 0.06 | 1.07 | 0.04 | >0.9999 | ||||
SI Methods
KO Mouse Generation.
RIM-BP2 and RIM-BP1 KO targeting strategies were designed together with genOway. Targeting vector construction and KO mouse generation by homologous recombination were performed at the genOway facility (Figs. S2A and S4A). Briefly, long- and short-homology regions were cloned from C57bl6 mouse genomic DNA. The targeting vectors contained two loxP sites flanking exon 17 (RIM-BP2) and exons 23–25 (RIM-BP1), respectively, and a neomycin resistance gene flanked by FRT sites for positive selection (Figs. S2A and S4A). Additionally, the vectors contained diphtheria toxin A as a negative selection marker. The linearized targeting constructs were transfected into ES cells, and positive clones were selected starting 48 h afterward. After PCR and Southern blot verification of 5′ and 3′ homologous recombination (Fig. S4 B and C), ES cells were injected into recipient blastocysts and implanted into pseudopregnant C57BL/6 females. The F1 chimeric males were crossed to a C57BL/6 Cre deleter line to excise the loxP flanked sequences, resulting in generation of the constitutive RIM-BP1 and RIM-BP2 KO mouse lines (Figs. S2A and S4A). For both single-KO lines, heterozygous animals were backcrossed to C57BL/6 mice and progeny were expanded to establish colonies.
Heterozygous RIM-BP2 mice were bred to obtain homozygous, heterozygous, and WT littermates for all subsequent experiments. Mice from both strains were crossed to obtain RIM-BP1/2 DKO animals. Genotyping PCR on genomic DNA obtained from tail biopsies was performed before neuronal cultures were made, as well as before and after slice experiments (Fig. S4E).
The following primers were used for genotyping: RBP1 WT Safw: ttatcactgctgcctgccacacgcc, RBP1 LA2 forward (fw): agcatcgagacaaccagtctctgc, RBP1 LA2 reverse (rev): acatctgaacctctcaggatgggac, RIM-BP2 KO fw: gagtccagtatctgcatgagggagtctg, RIM-BP2 WT fw: gaacataggacaggcagtacacttacttcacctag, and RIM-BP2 WT/KO rev: caacctctcacatacatcacctggatcg.
Quantitative Real-Time PCR.
RNA was isolated from neuronal cultures at days in vitro (DIV) 3, 10, and 23, DNase I digested to eliminate genomic DNA and transcribed into cDNA. Quantitative real-time PCR was carried out using the LightCycler 480 Probes Master Kit and the LightCycler 480 (Roche Diagnostics). The following primers and Universal ProbeLibrary (UPL) probes were used: RIM-BP1: fw: gacctatggtggctgcattt, rev: acatcccctgctctgaagg, UPL probe 16 [efficiency (eff.) 2.1]; RIM-BP2: fw: gtggcggagggtctcatag, rev: ctcgcccggtgtattcat, UPL probe 58 (eff.: 1.9); RIM-BP3: fw: ccctagccaccgtaagca, rev: ggactgttgtcagtttcaggtg, UPL probe 1 (eff.: 2.0); and Rpl4: fw: gatgagctgtatggcacttgg, rev: cttgtgcatgggcaggtt, UPL probe 38 (eff.: 1.9).
For RIM-BP1 KO real-time PCR, mRNA was isolated from whole brain of WT, heterozygous, and homozygous littermates; DNase I digested; and transcribed into cDNA. The following primers and UPL probes were used (also Fig. S4D): assay 16: fw: gacctatggtggctgcattt, rev: acatcccctgctctgaagg, UPL probe 16; assay 33: fw: aggctatgccatctatgctga, rev: tgcgtacagtcacctcatgg, UPL probe 33; assay 21: fw: aggtctggtaccactgagctg, rev: catcagggttcggtgacatt, UPL probe 21; and assay 81: fw: gcctggttccatccaactt, rev: ctggactcttctcctcctcgt, UPL probe 81.
Immunoprecipitation from P2 Fraction.
WT brains (4-wk-old mice) were homogenized in 10 mL of precooled homogenization buffer [0.32 M sucrose, 4 mM Hepes (pH 7.4)] containing 1 mM PMSF and protease inhibitor mixture (Roche Diagnostics). All steps were performed at 4 °C. Homogenate was centrifuged at 800 × g for 10 min. The supernatant S1 was centrifuged at 10,000 × g for 15 min. The resulting pellet was suspended in 10 mL of homogenization buffer and spun again at 10,000 × g for 15 min. The washed P2 fraction was suspended in 1.8 mL of immunoprecipitation (IP) buffer [25 mM Hepes (pH 8.3), 150 mM NaCl, 1 mM MgCl2, 10% (vol/vol) glycerol] containing 0.4% deoxycolate sodium salt and 1% Triton X-100. The P2 fraction was solubilized for 1 h and centrifuged at 17,000 × g for 30 min. The supernatant was transferred to the antibody-coupled beads (Protein A beads; Biorad) and incubated overnight. A self-raised rabbit anti–RIM-BP2 antibody (sera generated against peptide covering the last 20 amino acids of mouse RBP2 C terminus) was used, as well as random rabbit IgGs (Sigma–Aldrich) as controls. The input was collected after centrifugation at 1,000 × g for 3 min. The beads were extensively washed in washing buffer [25 mM Hepes (pH 7.4), 150 mM NaCl, 10% glycerol, 0.5% Triton X-100]. Elution of the bound complex was performed by addition of 100 μL of 2× Laemmli sample buffer, boiling the samples at 95 °C for 5 min. For immunoblotting, samples were run on 8% (wt/vol) SDS-polyacrylamide gels. For the P2 input and IP, samples were loaded at 2% and 25%, respectively (discussed below).
Immunohistochemistry.
Brains freshly removed from 4- to 6-wk-old RIM-BP2 WT and KO mice were immediately shock-frozen on dry ice. Sagittal brain sections (10 μm) were cut with a cryostat (Leica) and collected on SuperFrost Plus slides (Menzel GmbH). For immunostaining, cryosections of RIM-BP2 WT and KO were processed simultaneously. Sections were fixed with 4% (wt/vol) paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB), pH 7.4, for 10 min at room temperature (RT). Blocking was performed with 10% (vol/vol) normal goat serum (NGS) and 0.3% Triton X-100 in 0.1 M PB for 3 h at RT. Sections were then incubated with goat anti-mouse IgG Fab fragments (1:10; Jackson Immunoresearch) in 0.1 M PB for 1 h at RT. Primary antibodies diluted in 5% (vol/vol) NGS and 0.3% Triton X-100 in 0.1 M PB were applied overnight at 4 °C. Secondary antibodies were incubated for 2 h at RT. For gSTED imaging, we used the following secondary antibodies: goat anti-guinea pig Alexa 488 (1:200), goat anti-guinea pig Alexa 594 (1:200 or 1:400 for confocal microscopy), goat anti-mouse Alexa 488 (1:200), goat anti-mouse Alexa 532 (1:200), goat anti-mouse ATTO647N (1:100; Active Motif), goat anti-rabbit Alexa 488 (1:200), goat anti-rabbit Alexa 594 (1:200), and goat anti-rabbit ATTO647N (1:100). All secondary antibodies conjugated to Alexa dyes were purchased from Invitrogen. Afterward, sections were washed, postfixed with 3% (wt/vol) PFA and 0.05% glutaraldehyde in 0.1 M PB, and mounted with Mowiol (pH 8) or ProLong Gold (gSTED imaging; Life Technologies) on high-precision glass coverslips. Conventional confocal images of RIM-BP2 immunofluorescence in the hippocampal CA1 area were acquired with a Leica SP5 laser confocal microscope equipped with a 20× 0.7-N.A. oil immersion objective. Primary antibodies used are described in Table S1.
Immunoblotting.
For KO mouse analysis, the crude membrane fractions were isolated from WT and KO brain samples to investigate expression of RIM-BP2 and different synaptic proteins (Fig. S2 D–F) or RIM-BP1 (Fig. S4F). Brain samples from 4- to 6-wk-old mice were snap-frozen with liquid nitrogen and stored at −80 °C. Samples were homogenized in 10 vol of homogenization buffer, pH 7.4 (0.32 M sucrose, 10 mM Hepes, 2 mM EDTA), precooled at 4 °C containing protease inhibitors. Next, the homogenates were centrifuged at 1,000 × g for 15 min to remove the pelleted nuclear fraction (P1). The supernatant (S1) was then taken and centrifuged at 17,000 × g for 30 min. The obtained crude membrane pellet (P2) was resuspended in 200–300 μL of cold Hepes lysis buffer (50 mM Hepes, 2 mM EDTA). Protein concentrations were determined by Bradford assay. Samples were separated on 8% and 12% (wt/vol) SDS-polyacrylamide gels (30 μg of protein per lane), transferred to a nitrocellulose membrane (Amersham Protran; GE Healthcare), and probed with different primary antibodies followed by incubation with secondary antibodies conjugated to horseradish peroxidase (Jackson Immunoresearch). Bound antibodies were visualized using an enhanced chemiluminescence detection system (PerkinElmer Immuno Labs) and imaged with a Biorad ChemiDoc XRS+ imager (Biorad). Protein expression levels were quantified with Image laboratory software (Biorad). All primary antibodies used are described in Table S1.
Electron Microscopy.
Hippocampal neurons were plated on sapphire disks and frozen at RT using an HPM 100 (Leica) at DIV 14 as described previously (36). Freeze substitution was performed in AFS2 (Leica). After staining with 1% uranyl acetate, samples were infiltrated and embedded into Epon and cured for 48 h at 60 °C. Serial 40-nm sections were cut using a microtome (Leica) and collected on formvar-coated single-slot grids (Science Services GmbH). Before imaging, sections were contrasted with 2.5% (wt/vol) uranyl acetate. Random profiles were imaged using a Zeiss 900 electron microscope with a digital Proscan 1K Slow-Scan CCD-Camera (Fa. Proscan elektronische Systeme GmbH) and analyzed with a custom-written ImageJ (NIH) and MATLAB (The MathWorks, Inc.) routine.
Electrophysiological Recordings.
Whole-cell patch-clamp recordings in autaptic neurons were performed using a Multiclamp 700B amplifier (Molecular Devices) at DIV 13–21. The patch pipette solution contained 136 mM KCl, 17.8 mM Hepes, 1 mM EGTA, 0.6 mM MgCl2, 4 mM ATP-Mg, 0.3 mM GTP-Na, 12 mM phosphocreatine, and 50 units/mL phosphocreatine kinase (300 mOsm, pH 7.4). The recording chamber was constantly perfused with extracellular solution containing 140 mM NaCl, 2.4 mM KCl, 10 mM Hepes, 2 mM CaCl2, 4 mM MgCl2, and 10 mM glucose (pH adjusted to 7.3 with NaOH, 300 mOsm). Solutions were applied using a fast-flow system. Data were filtered at 3 kHz, digitized at 10 kHz, and recorded with pClamp 10 (Molecular Devices). Data were analyzed with Axograph X (AxoGraph Scientific) and Prism 6 (GraphPad Software).
In autaptic cell culture, EPSCs were evoked by a brief 2-ms somatic depolarization to 0 mV from a holding potential of −70 mV. PPRs were calculated as the ratio from the second and first EPSC amplitudes. Hypertonic 500 mM sucrose solution was applied for 5 s to assess the size of the RRP of neurotransmitter vesicles (11). The RRP was assessed by integrating the transient current component evoked by application of hypertonic extracellular solution. Spontaneous release events were detected using a template-based algorithm in Axograph X (37). The spontaneous release rate is the fraction of the RRP released per second by spontaneous release and was calculated by dividing the miniature EPSC (mEPSC) frequency by the number of vesicles within the RRP. Synaptic vesicle fusogenicity was measured by applying 250 mM sucrose solution onto the neuron for 10 s and analyzed as described previously (38). Briefly, to obtain the fraction of RRP released at 250 mM sucrose solution, the charge transfer of the transient synaptic current was measured and divided by the RRP size obtained by application of 500 mM sucrose (5 s) from the same neuron. The peak release rate was calculated by normalizing and integrating the response to 500 mM sucrose. The maximal slope was then quantified as a measure for peak release rate (15).
For the (5S,10R)-(+)-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine maleate (MK-801) assay, EPSCs were recorded at 0.33 Hz from autaptic hippocampal neurons. EPSCs were recorded in extracellular solution containing 140 mM NaCl, 2.4 mM KCl, 10 mM Hepes, 1 mM CaCl2, 0 mM MgCl2, 10 mM glucose, and 10 μM glycine (pH adjusted to 7.3 with NaOH, 300 mOsm) until reaching a stable baseline. Progressive block rate was measured in the presence of 5 μM MK-801. The AMPA component was used to normalize for the NMDA block-independent changes in synaptic transmission
For calcium sensitivity assays (Fig. 3 F and G and Fig. S3 J and K), EPSCs from autaptic hippocampal neurons were measured in extracellular solution containing 0.5 mM, 2 mM, 4 mM, or 10 mM CaCl2 and 1 mM MgCl2. To control for rundown and cell-to-cell variability, test responses were normalized to average EPSCs in standard external solution (2 mM CaCl2, 4 mM MgCl2) that were recorded between treatments. Normalized responses were then normalized to the response in 10 mM CaCl2. The normalized values were fitted into a standard Hill equation [y = 1 + 10^((logEC50 − x) * h), where h is the Hill slope] to plot dose–response curves. For analysis of C.V. of EPSCs, only cells with an overall stable response were included.
For the functional coupling distance assay (Fig. S6), autaptic neurons were preincubated in 25 μM EGTA-AM or DMSO control for 15 min, washed three times in extracellular solution, and left for 10–15 min in conditioned Neurobasal A medium (Gibco) to allow for complete cleavage of AM esters. Neurons were recorded within ∼30 min thereafter.
For slice preparation, mice were anesthetized and decapitated. The brain was removed and chilled in ice-cold sucrose-artificial cerebrospinal fluid (sACSF) containing 87 mM NaCl, 26 mM NaHCO3, 10 mM glucose, 50 mM sucrose, 2.5 mM KCl, 1.25 mM NaH2PO4, 0.5 mM CaCl2, and 3 mM MgCl2 saturated with 95% (vol/vol) O2/5% (vol/vol) CO2, pH 7.4. Horizontal slices were cut at 300-μm thickness on a microslicer (VT1200S; Leica), maintained for 30 min at 35 °C in sACSF and subsequently stored in ACSF containing 119 mM NaCl, 26 mM NaHCO3, 10 mM glucose, 2.5 mM KCl, 1 mM NaH2PO4, 2.5 mM CaCl2, and 1.3 mM MgCl2 saturated with 95% (vol/vol) O2/5% (vol/vol) CO2, pH 7.4, at RT. Experiments were started after 30 min and no longer than 6 h after the preparation.
For recordings of fPSPs in the stratum radiatum of area CA1, slices were placed in a recording chamber continuously superfused with ACSF (oxygenated with 95% (vol/vol) O2/5% (vol/vol) CO2) at RT. Compound polysynaptic potentials were evoked by electrical stimulation of the Schaffer collaterals (0.05-ms duration) via a bipolar insulated stimulation electrode. Extracellular recordings to record from a large number of parallel fiber bundles were performed with a low-resistance patch-pipette filled with recording ACSF placed in the stratum radiatum. Amplitudes of evoked synaptic potentials were measured from averaged (n = 5–10) sweeps. Recordings were performed with a MultiClamp 700B amplifier. Signals were filtered at 2 kHz and digitized (BNC-2090; National Instruments Germany GmbH) at 5 kHz. IGOR Pro software was used for signal acquisition (WaveMetrics, Inc.). Data were analyzed with MATLAB or the Igor plug-in NeuroMatic (neuromatic.thinkrandom.com) software. Statistical analysis was performed with Prism 6.
For whole-cell recordings of spontaneous release in acute slices, intracellular electrodes were pulled from borosilicate glass (1.2-mm OD) and filled with KMeSO3-based intracellular solutions containing 130 mM KMSO3, 10 mM Hepes, 10 mM KCl, 4 mM NaCl, 4 mM MgATP, 0.5 mM NaGTP, and 5 mM phosphocreatine-Na (pH 7.35). Electrode resistance was 2–5 MΩ. Series resistance (Rs) and input resistance were constantly monitored by applying a 4-mV hyperpolarizing voltage step for 50 ms. Experiments were discarded if changes in the Rs were >15%. The mEPSCs in slices were isolated at −60 mV in the presence of 1 μM tetrodotoxin (TTX), 1 μM GABAzine, 50 μM d(−)-2-amino-5-phosphonopentanoic acid (D-APV), and 100 μM cyclothiazide.
SynGCamp6f Imaging.
SynGCamp6f was generated analogous to synGCamp2 (35) by fusing GCamp6f (36) to the C terminus of the synaptic vesicle protein synaptophysin. The construct was cloned into lentiviral shuttle vectors under control of the synapsin promoter. Lentiviral particles were produced as previously described (37) by the viral core facility of the Charité Berlin. Mass cultured hippocampal neurons were infected on DIV 1 and imaged on DIV 14–21 at a 2-Hz sampling rate with 100 ms of exposure time as previously described (38). A 490-nm LED system (pE2; CoolLED) was used. Three micromolar 2,3-Dihydroxy-6-nitro-7-sulfamoyl-benzo[f]chinoxalin-2,3-dion (NBQX) and 30 μM bicuculline were added to the external solution to block spontaneous activity.
gSTED Microscopy.
STED imaging with time-gated detection was performed using a commercial Leica SP8 TCS STED microscope (Leica Microsystems). Briefly, the system includes an inverted DMi8 CS microscope equipped with a pulsed white light laser (WLL; ∼80-ps pulse width, 80-MHz repetition rate; NKT Photonics) for flexible excitation wavelengths and two STED lasers for depletion (continuous wave at 592 nm, pulsed at 775 nm). The pulsed 775-nm STED laser was triggered by the WLL. Within each independent experiment, samples from RIM-BP2 KO and WT mice were acquired with equal settings. In dual-channel experiments Alexa 488 and Alexa 532 were excited using a pulsed WLL at 488 nm and 545 nm, respectively. Depletion occurred at 592 nm. For Alexa 488 and Alexa 594, time gating ranged from 0.8–1.2 ns to 6 ns and from 0.8–2.3 ns to 6 ns, respectively.
Triple-channel STED imaging was performed exciting Alexa 488, Alexa 594, and ATTO647N at 488 nm, 598 nm, and 646 nm, respectively. Alexa 488 was depleted at 592 nm, whereas the 775-nm STED laser was used to deplete both Alexa 594 and ATTO647N. Time-gated detection was set from 0.3–6 ns for both dyes.
Fluorescence signals were detected sequentially by hybrid detectors at appropriate spectral regions separated from the STED laser by corresponding dichroic filters. Single optical slices were acquired with an HC PL APO CS2 100×/1.40-N.A. oil objective (Leica Microsystems), a scanning format of 1,024 × 1,024, eight-bit sampling, and 4.5 zoom, yielding a pixel dimension of 25.25 nm and 25.25 nm in the x and y dimensions, respectively. To minimize thermal drift, the microscope was housed in a heatable incubation chamber (LIS Life Imaging Services).
Raw dual-channel gSTED images were deconvolved using the built-in algorithm of the Leica LAS-AF software (signal intensity, regulation parameter of 0.05). The point spread function was generated with a 2D Lorentz function having the full-width half-maximum (FWHM) set to 60 nm. Raw data obtained from triple-channel gSTED imaging were deconvolved with Huygens Professional software (Scientific Volume Imaging) using a theoretical point spread function automatically computed based on pulsed- or continuous-wave STED optimized function and the specific microscope parameters. Default deconvolution settings were applied. To measure the effective lateral point spread function of the SP8 TCS STED microscope, we used 40-nm fluorescent beads (Life Technologies; excitation and emission maxima at 505/515 nm or 660/680 nm). Fluorescent beads were diluted 1:80,000 in poly-l-lysine (Sigma) and mounted on coated slides using ProLong Gold (Life Technologies) and high-precision glass coverslips. gSTED imaging was performed at appropriate wavelengths with similar imaging parameters as used for brain cryosections.
Cluster Distance Analysis.
For cluster distance analysis, deconvolved images were thresholded and segmented by watershed transform with Amira software (Visualization Sciences Group) to identify individual clusters and to obtain their x and y coordinates. Within the same independent experiment, the same threshold and segmentation parameters were used. According to the lateral resolution achieved, clusters with a size smaller than 0.0036 μm2 (two-channel gSTED) or 0.0025 μm2 (three-channel gSTED) were not considered for analysis.
The average number of clusters at specific distances and the k-nearest neighbor distance were analyzed with a MATLAB custom-written script. In the first step, the script determined the Euclidean distance between all possible cluster pairs in two channels in a matrix. The number of clusters in channel 1 found within 50-nm, 75-nm, 100-nm, 125-nm, 150-nm, 200-nm, and 300-nm distances from each single cluster of channel 2 was calculated and averaged for all particles found in channel 2. Thousands of clusters per single image were automatically analyzed. To identify precisely at which specific distance changes in clustering may occur, the mean number of channel 1 clusters found at sampling distances from channel 2 was expressed in distance intervals (0–50 nm, 50–75 nm, 75–100 nm, 100–125 nm, 0.125–200 nm, and 200–300 nm). The k-nearest neighbor distance analysis was similarly based on the matrix containing the distances between all particles in both channels: The distances of all particles in channel 1 to the ones in channel 2 were sorted in ascending order to find the k-nearest neighbor. The k value was set to 1, 2, 3, 4, and 5. The k distance values were then averaged on the number of clusters in channel 2. Data from independent experiments were pooled, and five to nine mice per genotype were analyzed.
Estimation of the Effective gSTED Point Spread Function by Gaussian Fit.
To estimate the lateral resolution of the gSTED microscope, fluorescent beads with uniform diameters well below the diffraction limit (∼40 nm) were imaged at appropriate wavelengths. We then approximated local intensity profiles with a 2D Gaussian function:
where is a parameter vector. In its elements, represents a baseline pixel intensity value, corresponds to the peak of the Gaussian, and represent its x and y positions, and determines its width, which we assume to be equal in both dimensions. To quantify the deviation to the fluorescence profile of the image, a cost value was calculated:
where is the intensity value of the experimentally obtained image at positions and and and are the maximal pixel positions (i.e., the image size). The optimal solution of was found by minimizing the cost value using a genetic optimization algorithm, implemented in the function “ga” of MATLAB R2016a with a population size of 500 and lower bounds of zero for all parameter values and upper bounds for and of and . There was no upper bound for the other parameters. The FWHM amplitude of the Gaussian was calculated to quantify the lateral resolution from the following relationship:
Absolute distances were calculated from pixel distances by multiplying with a pixel size of 25.25 nm. For display purposes, the 2D Gaussian was calculated at higher sampling in the plots of Fig. S2 C and D.
Statistics.
For all electrophysiological datasets, Prism6 was used for statistical analysis. The D’Agostino–Pearson omnibus test was used to check for normal distribution of data. For WT vs. KO comparison, an unpaired t test with Welch’s correction was used for normally distributed data and the Mann–Whitney U test was used for not normally distributed data.
For the 10-Hz trains in autaptic cultures, the last 10 EPSCs of each cell were averaged and the averages of WT and KO cells were tested using the Mann–Whitney U test.
For 14-Hz trains in slices, normalized data were tested using repeated measures ANOVA.
For gSTED, statistical analysis was done with SPSS Statistics software (IBM). Data normality was tested with the Kolmogorov–Smirnov test. Pairwise comparisons were analyzed with an unpaired Student t test or Mann–Whitney U test according to normality, unless otherwise stated. Values are expressed as mean ± SEM, and n indicates the number of animal tested.
For Western blot analysis, statistical analysis was performed with SPSS Statistics software, data were tested for normality with the Kolmogorov–Smirnov test, and significances between WT and KO RIM-BP2 animals were analyzed by the Mann–Whitney U test or unpaired Student t test according to normality. Values are expressed as median (25th–75th percentiles), and n indicates the number of animals tested.
For quantitative real-time PCR analysis, data from four independent experiments were pooled. Relative expression levels were tested in Prism 6 using repeated measurement two-way ANOVA with Tukey’s multiple comparisons test.
Acknowledgments
We thank Annegret Felies, Berit Soehl-Kielczynski, Sabine Lenz, Katja Poetschke, Carola Schweynoch, Rike Dannenberg, and Bettina Brokowski for technical support; and Lauren Mamer for discussions. We thank Burkhard Wiesner and Martin Lehmann (FMP cellular imaging facility) for the use of the Leica gSTED microscope and for the fluorescent beads; Andreas Grasskamp for the point spread function estimation by Gaussian fit; Anna Fejtova and Eckart Gundelfinger for the RIM-BP2 antibody; and Pietro De Camilli for the Dynamin1 antibody. This work was supported by the Deutsche Forschungsgemeinschaft (Collaborative Research Grant SFB 958 [to V.H. (A1, A7), D.S. (A5), S.S. (A3, A6), C.R. (A5)] and Emmy-Noether Program (to A.M.W.), Studienstiftung des deutschen Volkes PhD Fellowship (to M.K.G.), and Excellence Cluster Neurocure Exc257 (to B.J.E., V.H., D.S., S.S., and C.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605256113/-/DCSupplemental.
References
- 1.Deng L, Kaeser PS, Xu W, Südhof TC. RIM proteins activate vesicle priming by reversing autoinhibitory homodimerization of Munc13. Neuron. 2011;69(2):317–331. doi: 10.1016/j.neuron.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaeser PS, et al. RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell. 2011;144(2):282–295. doi: 10.1016/j.cell.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Sugita S, Sudhof TC. The RIM/NIM family of neuronal C2 domain proteins. Interactions with Rab3 and a new class of Src homology 3 domain proteins. J Biol Chem. 2000;275(26):20033–20044. doi: 10.1074/jbc.M909008199. [DOI] [PubMed] [Google Scholar]
- 4.Hibino H, et al. RIM binding proteins (RBPs) couple Rab3-interacting molecules (RIMs) to voltage-gated Ca(2+) channels. Neuron. 2002;34(3):411–423. doi: 10.1016/s0896-6273(02)00667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davydova D, et al. Bassoon specifically controls presynaptic P/Q-type Ca(2+) channels via RIM-binding protein. Neuron. 2014;82(1):181–194. doi: 10.1016/j.neuron.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Liu KSY, et al. RIM-binding protein, a central part of the active zone, is essential for neurotransmitter release. Science. 2011;334(6062):1565–1569. doi: 10.1126/science.1212991. [DOI] [PubMed] [Google Scholar]
- 7.Müller M, Genç Ö, Davis GW. RIM-binding protein links synaptic homeostasis to the stabilization and replenishment of high release probability vesicles. Neuron. 2015;85(5):1056–1069. doi: 10.1016/j.neuron.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acuna C, Liu X, Gonzalez A, Südhof TC. RIM-BPs mediate tight coupling of action potentials to Ca(2+)-triggered neurotransmitter release. Neuron. 2015;87(6):1234–1247. doi: 10.1016/j.neuron.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 9.Kim S, et al. The GIT family of proteins forms multimers and associates with the presynaptic cytomatrix protein Piccolo. J Biol Chem. 2003;278(8):6291–6300. doi: 10.1074/jbc.M212287200. [DOI] [PubMed] [Google Scholar]
- 10.Podufall J, et al. A presynaptic role for the cytomatrix protein GIT in synaptic vesicle recycling. Cell Reports. 2014;7(5):1417–1425. doi: 10.1016/j.celrep.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 11.Rosenmund C, Stevens CF. Definition of the readily releasable pool of vesicles at hippocampal synapses. Neuron. 1996;16(6):1197–1207. doi: 10.1016/s0896-6273(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 12.Rosenmund C, Clements JD, Westbrook GL. Nonuniform probability of glutamate release at a hippocampal synapse. Science. 1993;262(5134):754–757. doi: 10.1126/science.7901909. [DOI] [PubMed] [Google Scholar]
- 13.Varoqueaux F, et al. Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc Natl Acad Sci USA. 2002;99(13):9037–9042. doi: 10.1073/pnas.122623799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arancillo M, et al. Titration of Syntaxin1 in mammalian synapses reveals multiple roles in vesicle docking, priming, and release probability. J Neurosci. 2013;33(42):16698–16714. doi: 10.1523/JNEUROSCI.0187-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basu J, Betz A, Brose N, Rosenmund C. Munc13-1 C1 domain activation lowers the energy barrier for synaptic vesicle fusion. J Neurosci. 2007;27(5):1200–1210. doi: 10.1523/JNEUROSCI.4908-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggermann E, Bucurenciu I, Goswami SP, Jonas P. Nanodomain coupling between Ca²⁺ channels and sensors of exocytosis at fast mammalian synapses. Nat Rev Neurosci. 2011;13(1):7–21. doi: 10.1038/nrn3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadkarni S, Bartol TM, Stevens CF, Sejnowski TJ, Levine H. Short-term plasticity constrains spatial organization of a hippocampal presynaptic terminal. Proc Natl Acad Sci USA. 2012;109(36):14657–14662. doi: 10.1073/pnas.1211971109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucan M, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5(6):e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussman JP, et al. A noise-reduction GWAS analysis implicates altered regulation of neurite outgrowth and guidance in autism. Mol Autism. 2011;2(1):1. doi: 10.1186/2040-2392-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corominas R, et al. Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat Commun. 2014;5:3650. doi: 10.1038/ncomms4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klyachko VA, Stevens CF. Excitatory and feed-forward inhibitory hippocampal synapses work synergistically as an adaptive filter of natural spike trains. PLoS Biol. 2006;4(7):e207. doi: 10.1371/journal.pbio.0040207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlüter OM, Basu J, Südhof TC, Rosenmund C. Rab3 superprimes synaptic vesicles for release: Implications for short-term synaptic plasticity. J Neurosci. 2006;26(4):1239–1246. doi: 10.1523/JNEUROSCI.3553-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68(5):843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schikorski T, Stevens CF. Quantitative ultrastructural analysis of hippocampal excitatory synapses. J Neurosci. 1997;17(15):5858–5867. doi: 10.1523/JNEUROSCI.17-15-05858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holderith N, et al. Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat Neurosci. 2012;15(7):988–997. doi: 10.1038/nn.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graf ER, et al. RIM promotes calcium channel accumulation at active zones of the Drosophila neuromuscular junction. J Neurosci. 2012;32(47):16586–16596. doi: 10.1523/JNEUROSCI.0965-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoch S, et al. RIM1α forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415(6869):321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- 29.Castillo PE, Schoch S, Schmitz F, Südhof TC, Malenka RC. RIM1α is required for presynaptic long-term potentiation. Nature. 2002;415(6869):327–330. doi: 10.1038/415327a. [DOI] [PubMed] [Google Scholar]
- 30.Kintscher M, Wozny C, Johenning FW, Schmitz D, Breustedt J. Role of RIM1α in short- and long-term synaptic plasticity at cerebellar parallel fibres. Nat Commun. 2013;4:2392. doi: 10.1038/ncomms3392. [DOI] [PubMed] [Google Scholar]
- 31.Müller M, Liu KSY, Sigrist SJ, Davis GW. RIM controls homeostatic plasticity through modulation of the readily-releasable vesicle pool. J Neurosci. 2012;32(47):16574–16585. doi: 10.1523/JNEUROSCI.0981-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courchesne E, et al. Mapping early brain development in autism. Neuron. 2007;56(2):399–413. doi: 10.1016/j.neuron.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 33.Stempel AV, et al. Cannabinoid type 2 receptors mediate a cell type- specific plasticity in the hippocampus. Neuron. 2016;90(4):795–809. doi: 10.1016/j.neuron.2016.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman MA, Ackermann F, Trimbuch T, Rosenmund C. Vesicular glutamate transporter expression level affects synaptic vesicle release probability at hippocampal synapses in culture. J Neurosci. 2014;34(35):11781–11791. doi: 10.1523/JNEUROSCI.1444-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen T-W, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe S, et al. Clathrin regenerates synaptic vesicles from endosomes. Nature. 2014;515(7526):228–233. doi: 10.1038/nature13846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clements JD, Bekkers JM. Detection of spontaneous synaptic events with an optimally scaled template. Biophys J. 1997;73(1):220–229. doi: 10.1016/S0006-3495(97)78062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue M, et al. Binding of the complexin N terminus to the SNARE complex potentiates synaptic-vesicle fusogenicity. Nat Struct Mol Biol. 2010;17(5):568–575. doi: 10.1038/nsmb.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]