Significance
Previous studies have attempted to unravel the complex phylogeographic patterns of the malaria parasites Plasmodium vivax and Plasmidium falciparum diversity and also to understand its evolutionary affinities. However, all these studies are constrained by the lack of evidence from the eradicated European strains that could be central to some dispersals. This study successfully retrieves massive genetic data from old slides treated with traditional staining techniques to be observed under the microscope, thus providing a new material source for the study of past pathogens that could place value in historical medical collections. We generated sequence data from the eradicated European malaria parasites and shed light on the genetic diversity patterns of P. vivax and P. falciparum.
Keywords: malaria, Europe, Plasmodium, paleogenetics, microscopy slides
Abstract
Phylogenetic analysis of Plasmodium parasites has indicated that their modern-day distribution is a result of a series of human-mediated dispersals involving transport between Africa, Europe, America, and Asia. A major outstanding question is the phylogenetic affinity of the malaria causing parasites Plasmodium vivax and falciparum in historic southern Europe—where it was endemic until the mid-20th century, after which it was eradicated across the region. Resolving the identity of these parasites will be critical for answering several hypotheses on the malaria dispersal. Recently, a set of slides with blood stains of malaria-affected people from the Ebro Delta (Spain), dated between 1942 and 1944, have been found in a local medical collection. We extracted DNA from three slides, two of them stained with Giemsa (on which Plasmodium parasites could still be seen under the microscope) and another one consisting of dried blood spots. We generated the data using Illumina sequencing after using several strategies aimed at increasing the Plasmodium DNA yield: depletion of the human genomic (g)DNA content through hybridization with human gDNA baits, and capture-enrichment using gDNA derived from P. falciparum. Plasmodium mitochondrial genome sequences were subsequently reconstructed from the resulting data. Phylogenetic analysis of the eradicated European P. vivax mtDNA genome indicates that the European isolate is closely related to the most common present-day American haplotype and likely entered the American continent post-Columbian contact. Furthermore, the European P. falciparum mtDNA indicates a link with current Indian strains that is in agreement with historical accounts.
The genus Plasmodium contains two of the most significant human pathogenic organisms. One, Plasmodium vivax, is the most widely distributed human malaria parasite outside of Africa, with a range that extends well into temperate zones (1), whereas the other, Plasmodium falciparum, is the predominant malaria parasite of humans in subsaharan Africa and the one that causes >90% malarial deaths (2).
Although an increasing number of studies have attempted to investigate the present-day distribution of Plasmodium parasites using DNA sourced from modern isolates of the pathogens, their origin and dispersal around the planet remain controversial. For example, the current global distribution of P. vivax includes Asia, the Middle East, South and Central America, and parts of Africa (3) and has likely resulted from complex dispersals involving intercontinental human population movements (4, 5). Although an African origin is likely (6), the current strains from this continent show little genetic polymorphism compared with the rest of the world (4), a fact that has been related to the emergence of Duffy-negative blood types—resistant to P. vivax—in human populations from this continent (7). Alternatively, it has been suggested that modern African parasites, at least in the Horn of Africa and Madagascar, could have been reintroduced via the import of East Asian/Indian P. vivax to the coastal areas of East Africa by early sea-going traders and that this could explain their lack of diversity (4). Intriguingly, the diversity of the American strains is comparable to those found in Oceania and Asia (8), raising questions regarding the origin of these strains. Some of the American strains are thought to have been brought there from Europe by Spanish sailors and colonists in the last few centuries (4, 9). An alternative possibility would imply a pre-Columbian entrance through the Bering Straits during the initial peopling of the Americas, although this seems unlikely due to the climatic conditions of this passage (9) and the long Beringian standstill that likely lasted up to several thousands of years (10). However, the existence of divergent mitochondrial lineages in the Americas seems to rule out the possibility of a single, recent introduction of P. vivax from Europe into this continent (8). At the nuclear level, the American isolates show geographic differentiation but also a common clustering regarding the Old World populations (5).
With regard to the evolutionary history of P. falciparum, genetic analyses of Laveriana parasite species found in African chimpanzees and gorillas, considered to be precursors of the human parasites, prove an African origin (11), but the time by which the parasite dispersed out of Africa remains controversial. Although some suggest that the pathogen followed the original Homo sapiens dispersals around 60,000 y ago (12), others support a recent expansion within the last 5,000–10,000 y associated to the advent of agriculture (2). The recent analysis of 44 Indian field isolates showed, however, a higher than expected diversity and suggested that the Indian strains are also part of the ancestral distribution range of P. falciparum (13). Historical accounts seem to indicate that malaria spread from India into the Mediterranean at the time of Classical Greece; therefore, the now extinct European P. falciparum could be genetically related to some of these Indian parasites.
The current controversies on the recent evolutionary history of P. vivax and P. falciparum partly relate to the lack of genetic evidence from the European parasite that was eradicated more than half century ago. In Spain, malaria had remained endemic until the early 20th century, in particular in Andalucia, Extremadura, and Ebro delta regions; it was declared oficially eradicated in 1964 (14). P. vivax and P. falciparum have been transmitted in southern Spain by two former malaria vectors: Anopheles atroparvus and Anopheles labranchiae. Both were present in the country but whereas An. atroparvus is still widely spread, An. labranchiae has not been found since the middle of the last century (15). An. atroparvus from different geographic origins are infected in laboratory with different world strains of P. vivax, whereas it is very difficult to do the same in the case of P. falciparum (16). Thus, the lack of genetic evidence of the former Plasmodium European parasites also hampers our understanding of their biology.
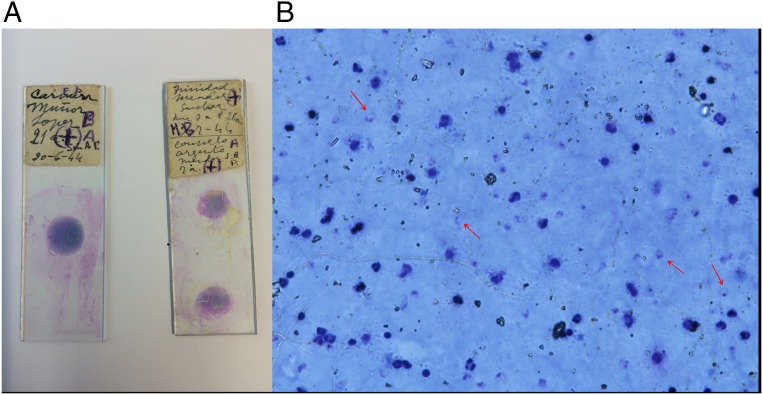
To clarify the phylogenetic position of the extinct European Plasmodium strains, we report here the retrieval of the European P. vivax and P. falciparum genetic data from the analysis of three recently discovered old slides with blood drops from malaria-infected people living at the Ebro Delta in Spain in the first half of the 20th century that corresponds to local epidemics (Fig. 1).
Fig. 1.
(A) Two of the Giemsa-stained slides analyzed in this study, labeled CM and CA (inferior stain). (B) Visualization of some Plasmodium parasites (arrows) in the CM slide under the microscope (400×).
Results
The three samples (CA, CM, and POS) contained Plasmodium and human sequences. The “depleted” libraries mapped to P. vivax nuclear genome in fold coverage values that range from no coverage in POS sample to 0.0889× (CM) and to P. falciparum from 0.0157× (POS) to 0.0182× (CA); the captured libraries mapped to P. vivax from 0.0013× (POS) to 0.0260 (CM) and to P. falciparum from 0.0092 (POS) to 0.4291 (CA) (Table 1 and Tables S1–S3). Therefore, samples CA and CM seem to be coinfected with both malaria parasites (the former having more P. falciparum and the latter more P. vivax), whereas sample POS carries minimal amounts of P. vivax. The differences (being CA the most efficient sample) do not seem to be a consequence of the original amount of DNA present in the slides, because the depleted libraries of the three samples mapped to the human genome—deriving from a similar amount of sequences previously generated—yielded similar mean nuclear depths: 0.2116× (CA), 0.1937× (CM), and 0.2290× (POS). Additionally, these figures indicate that the depletion strategy to remove human DNA from the slides was not totally efficient. The high ratio of coinfection could be attributable to the fact that the slides are a biased sample and likely were taken from patients that showed acute malaria symptoms.
Table 1.
Summary statistics of the DNA reads mapped to P. vivax and P. falciparum mtDNA
| Sample | Total reads | Mapped reads (P. vivax/P .falciparum) | Unique reads (P. vivax/P .falciparum) | MtDNA coverage (P. vivax/P .falciparum) |
| CA depletion | 41,669,177 | 573/1,325 | 228/569 | 2.56/6.66 |
| CM depletion | 36,781,114 | 965/600 | 268/229 | 3.06/2.66 |
| POS depletion | 47,712,546 | 279/426 | 129/229 | 2.79/2.84 |
| CA capture | 52,310,688 | 474,175/1,093,502 | 336/814 | 3.86/9.73 |
| CM capture | 32,441,831 | 576,598/509,355 | 264/250 | 3.00/2.86 |
| POS capture | 32,323,796 | 38,160/178,290 | 112/210 | 1.32/2.56 |
| Consensus mt DNA | — | — | 459/1,220 | 5.02/14.75 |
Data were generated with two experimental approaches on the three slides (CM, CA, and POS). Depletion refers to the mapped reads from the genomic libraries depleted by whole human genome capture; capture refers to the mapped reads after capturing the genomic libraries with P. falciparum baits.
Table S1.
P. vivax nuclear reads
| Sample | Total reads | Mapped reads | Unique reads | Nuclear coverage |
| CA depletion | 41,669,177 | 8,939 | 3,036 | 0.0000 |
| CM depletion | 36,781,114 | 40,047 | 35,072 | 0.0889 |
| POS depletion | 47,712,546 | 2,348 | 710 | 0.0000 |
| CA capture | 52,310,688 | 8,388 | 2,600 | 0.0053 |
| CM capture | 32,441,831 | 13,940 | 10,678 | 0.0259 |
| POS capture | 32,323,796 | 2,013 | 547 | 0.0010 |
Table S3.
Human nuclear reads
| Sample | Total reads | Mapped reads | Unique reads | Nuclear coverage |
| CA depletion | 41,669,177 | 12,667,791 | 12,033,290 | 0.2116 |
| CM depletion | 36,781,114 | 6,221,402 | 5,841,497 | 0.1937 |
| POS depletion | 47,712,546 | 4,309,690 | 4,111,376 | 0.2290 |
| CA capture | 52,310,688 | 2,468,011 | 2,321,087 | 0.2073 |
| CM capture | 32,441,831 | 1,734,974 | 1,611,254 | 0.1881 |
| POS capture | 32,323,796 | 219,493 | 203,647 | 0.0451 |
Table S2.
P. falciparum nuclear reads
| Sample | Total reads | Mapped reads | Unique reads | Nuclear coverage |
| CA depletion | 41,669,177 | 90,132 | 80,570 | 0.0182 |
| CM depletion | 36,781,114 | 9,516 | 4,784 | 0.0130 |
| POS depletion | 47,712,546 | 6,653 | 4,965 | 0.0157 |
| CA capture | 52,310,688 | 24,196,904 | 132,642 | 0.4290 |
| CM capture | 32,441,831 | 78,769 | 3,559 | 0.0100 |
| POS capture | 32,323,796 | 489,464 | 2,792 | 0.0100 |
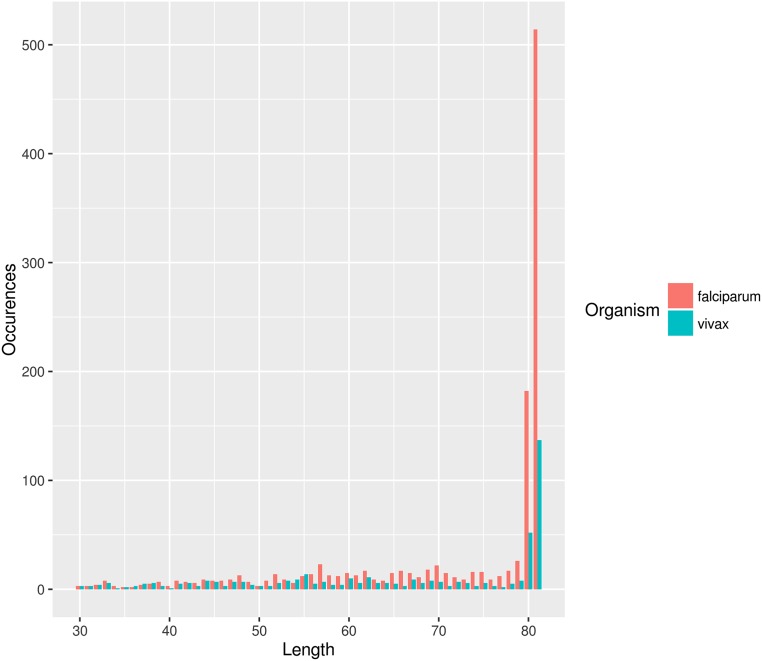
Both Plasmodium and human sequences display a weak but discernible signal of ancient DNA nucleotide misincorporation patterns (C to T substitutions at the 5′ ends and G to A at the 3′ ends) (17), reaching a deamination rate of 2.5% at the read extremes. This observation is in agreement with values found in museum specimens of similar age (18), which, along with the fragmentation pattern, supports the authenticity of the analyzed DNA sequences (Figs. S1 and S2).
Fig. S1.
Damage pattern at the end of the DNA reads retrieved from the slides, consistent with those being ancient. Frequencies of C to T (red) and G to A (blue) misincorporations of the 5′ end (left) and 3′ end (right) are shown. From top to bottom: P. falciparum mtDNA reads; P. vivax mtDNA reads; and human reads.
Fig. S2.
Fragment length distribution of the P. vivax and P. falciparum reads.
Each of the three samples exhibited different, common human European mtDNA haplogroups (CA: J2a1a1; CM: H1e1a6; and POS: H2a2b), which indicates that each blood source had a unique individual origin (human mtDNA coverage ranged from 6× at CM to 24.7× at CA). Some background contamination from two people—one of them a researcher that handled the slides—could be detected in figures of 6.81% in CM, 10.86% in CA, and 23.83% in POS. The fact that the predominant haplotypes are different among slides supports that no major DNA mixing occurred among patients' samples, although it cannot be totally discarded.
Although it was impossible to separate the nuclear genomic sequences from both parasites due to the low number of Plasmodium nuDNA reads, the relatively higher depth of coverage in the mtDNA genomes allowed us to map the reads either to P. falciparum or to P. vivax sequences (the former ranged from 2.8× in POS to 9.9× in CA and the latter from 1.6× in POS to 3.9× in CA).
In total, 459 mtDNA reads passed the quality filters for P. vivax, yielding a 5.02× depth of coverage and 67% of the P. vivax mtDNA genome covered with at least two reads (total length available of 4,045 nucleotides). We detected in our genome a European-specific nucleotide change that cannot be attributed to the typical C to T or G to A ancient DNA postmortem damage (17): a T to C substitution at nucleotide position 1033. This mutation is present among reads from two of the slides analyzed (CM and POS) but is absent in four reads on this position for sample CA. Although it is impossible to generate confident haplotypes for each of the slides due to the low coverage, this could indicate that CA carries in fact the haplotype without this substitution.
The P. falciparum mtDNA sequence obtained reached 14× coverage and represented the full length of the mitochondrial genome, with 5,827 nucleotides. We found three mutations present in the three slides that are also shared with present-day Indian individuals: 276 (G > A), 725 (C > T), and 2763 (C > T). No haplotype differences among the three individuals were detected.
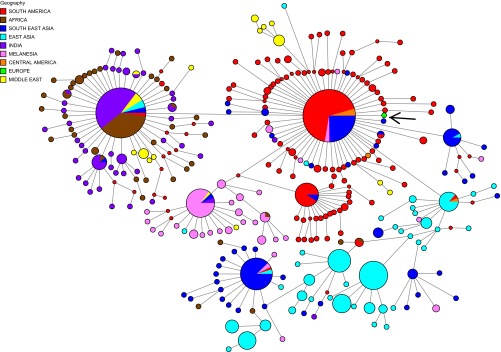
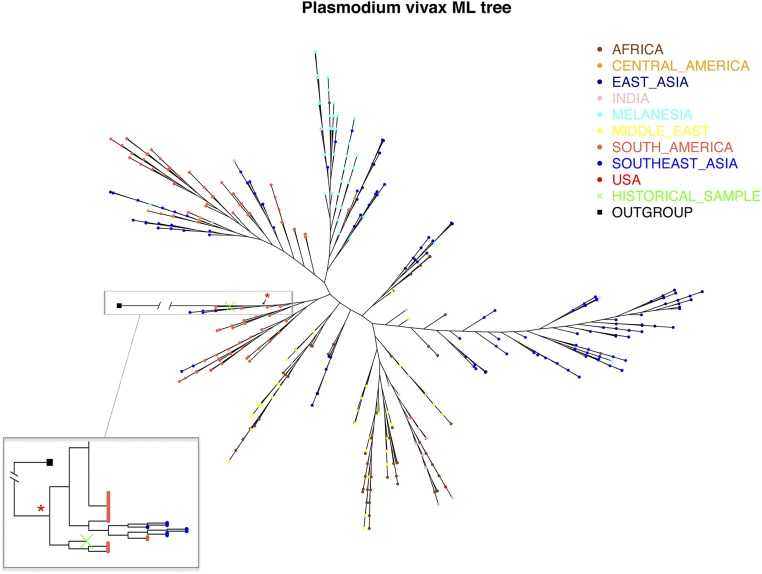
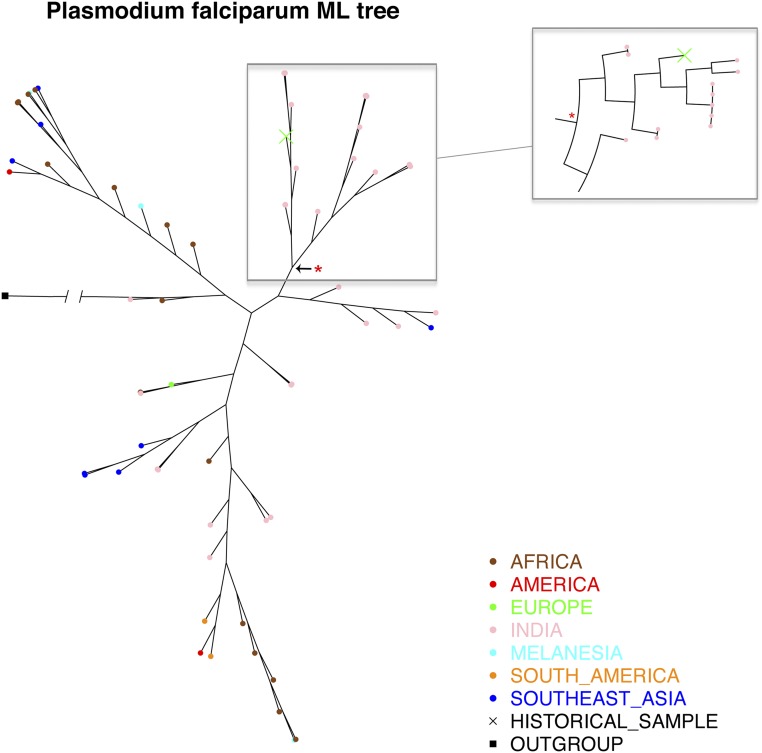
In the phylogenetic analysis, we found that the extinct European P. vivax mtDNA clusters close to the haplotype most commonly found nowadays in the Americas: also described in some Southeast Asians from Vietnam, Bangladesh, and Thailand and also in one Melanesian sample (Fig. 2). In the maximum-likelihood tree, the historical sample appears relatively early in terms of branching events (this is the closest sample to the root) and clusters with a group of Brazilian strains (SI Materials and Methods) with a 0.84 bootstrap support (Fig. S3).
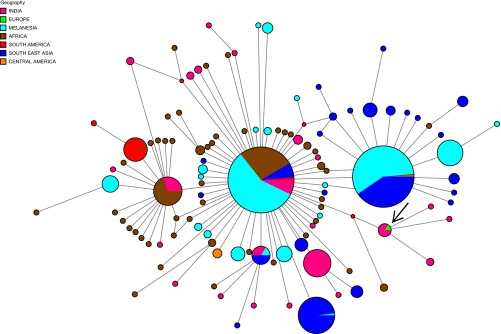
Fig. 2.
Haplotype network constructed using NETWORK 5,0.0, incorporating 790 P. vivax mitochondrial DNA genomes previously published and the one retrieved in this study (arrow-marked European isolate). Size circles equal to the frequency of a given haplotype; lines separating haplotypes represent mutational steps. Each color shows the geographic origin of the samples.
Fig. S3.
Maximum likelihood tree generated with a geographical diverse dataset of P. vivax mtDNA genomes (the green cross, marked with an arrow, corresponds to the European isolate). Red start represents a clade with 0.84 bootstrap support.
The European P. falciparum clusters in a node with nine Indian samples (SI Materials and Methods), described as H-41 in ref. 13 (Fig. 3), supported by a 0.80 bootstrap value in the maximum-likelihood tree (Fig. S4).
Fig. 3.
Haplotype network constructed using NETWORK 5.0.0 incorporating 698 P. falciparum mitochondrial DNA genomes previously published and the one retrieved in this study (arrow-marked European isolate).
Fig. S4.
Maximum likelihood tree generated with a geographical diverse dataset of P. falciparum mtDNA genomes (the green cross, marked with an arrow, corresponds to the European isolate). Red start represents a clade with 0.80 bootstrap support.
SI Materials and Methods
DNA Extraction.
Following lysis, the sample was extracted with the following protocol: the supernatant was added to 15 mL binding buffer containing 5 M guanidine hydrochloride, 40% (vol/vol) isopropanol, 0.05% Tween-20, and 90 mM sodium acetate. A binding apparatus was constructed by fitting an extension reservoir removed from a Zymo-Spin V column (Zymo Research) into a MinElute silica spin column (Qiagen) and placing them into a 50-mL Falcon tube. The 25- to 30-mL solution containing binding buffer and the extraction supernatant was poured into the extension reservoir and centrifuged for 5 min at 269 × g. The MinElute columns were subsequently placed into 2-mL Eppendorf tubes, washed with PE buffer (Qiagen), and dried by centrifugation at maximum speed. The DNA was finally eluted with 25 μL Tris-EDTA-Triton X-100 buffer, incubated for 2 min, and collected by centrifugation for 1 min at maximum speed.
Library Preparation.
Given the fragmented nature of aDNA, the initial nebulization step was skipped. The first module of the protocol, corresponding to the end-repair reaction, was performed in 50-μL reactions using 30 μL aDNA extract, 5 μL NEBNext 10× end-repair buffer, 2.5 μL end-repair enzyme mix, and 12.5 μL molecular grade water. The reactions were incubated for 20 min at 12 °C and 15 min at 37 °C, purified using MinElute spin columns (Qiagen) using 10× PB buffer, and eluted in 30 μL buffer EB after a 10-min incubation at 37 °C. The second module, the adapter ligation step, was performed in 50-μL reactions using 30 μL end-repaired DNA, 5 μL Illumina-specific adapters from with a final concentration of 0.25 μM, 10 μL 5× Quick ligation buffer, and 5 μL Quick T4 Ligase. The reactions were incubated for 15 min at 20 °C and purified using QiaQuick spin columns (Qiagen) and 5× PB before being eluted in 30 μL buffer EB after a 10-min incubation at 37 °C. The third module, the adaptor fill-in step, was performed in a 50-μL reaction with 30 μL adapter-ligated DNA, 5 μL adapter fill-in reaction buffer, 3 μL Bst polymerase, and 12 μL molecular grade water. The reaction was incubated for 20 min at 37 °C, followed by 20 min at 80 °C to inactivate the Bst polymerase.
Library Amplification.
A 20-μL reaction was prepared using 2 μL 1:10 dilution of library aDNA template, 0.1 U/µL AmpliTaq Gold Polymerase (Applied Biosystems), 1× AmpliTaq Gold Buffer, 2.5 mM MgCl2, 0.8 mg/mL BSA, 0.25 mM dNTPs, 1 μL 1× SYBR Green/Rox dye mix (Invitrogen), and 0.2 μM forward and reverse indexing primer. Thermocycling conditions were 3 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 60 °C, and 60 s at 72 °C.
Based on qPCR results, samples were amplified for 23 cycles. Two 100-μL reactions were prepared: one for capture-depletion experiments and another one for Plasmodium capture. We used 15 μL aDNA library as template, 0.1 U/µL AmpliTaq Gold polymerase, 1× AmpliTaq Gold Buffer, 2.5 mM MgCl2, 0.8 mg/mL BSA, 0.25 mM dNTPs, and 0.2 μM forward and reverse indexing primer for each reaction. Thermocycling conditions were 3 min at 95 °C, followed by 23 cycles of 30 s at 95 °C, 30 s at 60 °C, and 60 s at 72 °C, and a final 5-min elongation step at 72 °C. The amplified libraries were then purified following the manufacturer’s instructions using the QiaQuick PCR Purification Kit (Qiagen) and eluted in 30 µL buffer EB after a 10-min incubation at 37 °C.
Bioinformatic and Phylogenetic Analysis.
We mapped the reads to P. vivax, P. falciparum, and human genome using BWA and the following options: aln option, 0.01 edit distance, no seed, a penalization of 2 for gap open, and no read trimming; duplicated reads were removed using Picard tools.
For the P. vivax phylogenetic analysis, we used the following modern samples: JN788737–JN788776, JQ240329–JQ240429, AY791517–AY791692, AY598035–AY598140, AB550270–AB550280, KC330370–KC330678, and KF668361–KF668429. The different P. vivax haplotypes were assigned to geographic locations: East Asia, Southeast Asia, South and Central America, Africa, India, Melanesia, and Middle East: the latter including specimens from Turkey and Iran. The European isolate clusters with a group of Brazilian strains, with the following accession numbers: AY598093–AY598084, AY598085, AY598084, and AY598093.
For the P. falciparum phylogenetic analysis, we included the following modern strains for comparison: AY282924–AY282927, AY282929, AY282931–AY282982, AY282984–AY283006, AY283008–AY283019, AJ276844–AJ276847, KT119847–KT119883, and AB570434–AB570951. In this analysis, the haplotypes were geographically distributed among Central and South America, Africa, Southeast Asia, India, and Melanesia. The Indian haplotypes that cluster with the European isolate were as follows: KJ569479, KJ569484, KJ569488, KJ569480, KJ418723, KT119881, and KT119847.
Discussion
Previous phylogeographic analyses on mtDNA from P. vivax have commented on the anomalous central position of the American haplotypes, placed intermediate between Melanesian and Southeast Asian clusters (19). In fact the most common American haplotype—in which the European isolate sequenced here differs by only one substitution—was identified by outgroup probability analysis as the most likely ancestral type (4), despite the presumed African origin of the parasite. This finding led the previous authors to argue that the most parsimonious explanation being that American strains (or at least some of them) derive from the now extinct European P. vivax, which would itself be related to an ancient “African” stock related to modern strains found in India and Western Asia. Furthermore, they argued that the surprising lack of variation previously observed among a large proportion of contemporary African samples—at least those from the Horn of African and Madagascar—would indicate that they are relatively young strains (1, 4) that might derive from Indian or West Asian stocks introduced in parts of the Africa continent in historical times (1, 4, 5), whereas the original African diversity would have been obscured by the rise of Duffy antigen null allele.
In addition, there is substantial geographic differentiation among P. vivax mitochondrial lineages, not only between continents, but also between neighboring countries such as those in South America itself (8, 19). The American data are regarded as consistent with two alternative but not mutually exclusive hypotheses: an ancient, pre-Columbian introduction with the original Amerindian settlers and a recent introduction from European colonizers. There is no solid evidence for an introduction with the original Amerindian settlers. None of the American lineages seems to share a common ancestry with those present in East Asian countries such as China and Korea that would be close to the potential entrance route from Asia into the American continent through Beringia (5, 20). In addition, it is doubtful if the climate in Beringia at the time of people crossing from North Asia into North America would have been sufficiently warm to allow transmission.
Our results provide strong support for the hypothesis that the dominant present-day South American haplotypes were indeed carried from Europe by post-Columbian travelers and that these haplotypes are in fact the closest modern representatives of the ancestral Eurasian P. vivax stock. The limitation on the genetic data available from the putative source population implies that we cannot know at present if all American strains derive from the European continent. It will be necessary to examine further P. vivax specimens form archival medical collections from previously malaria endemic areas of Europe to better understand the possible sources of post-Columbian introduction of P. vivax into the Americas.
According to historical accounts, intermittent fever conditions with the hallmarks of severe malaria—thus likely associated to P. falciparum—spread to India about 3,000 y ago and reached Greece maybe 500 y later. By the beginning of the Christian Era, it was present around all of the Mediterranean shores (2, 21). Therefore, the phylogeographic position of the European P. falciparum isolate indicates a potential link with Indian strains that is in agreement with these historical accounts. In contrast to the case of P. vivax, the American strains likely derive from Africa and took place during the slave trade (22). This finding is in agreement with the position of the European isolate in the worldwide haplotype network, which is clearly unrelated to the American haplotypes (Fig. 3).
Recent studies on the nuclear genome of both parasites have uncovered not only regional diversity patterns, especially in the case of P.vivax, but also the signals of recent adaptive responses to antimalarial drugs such as sulfadoxine, pyrimethamine, and other antifolate drugs (5, 20). The European Plasmodium samples have to predate these selective sweeps and thus could be used to test the observed genetic signals in modern isolates. The same approach could be used to determine the molecular resistance mechanism underlying the fact that present-day P.vivax is increasingly resistant to the first-line treatment, which is based on chloroquine (23). We could also obtain a thorough estimate of the evolutionary rate of the parasite with extra historical data from dated slides.
We are currently searching for additional malaria slides among antique medical collections along Europe to obtain further information on the genetic diversity of the European Plasmodium strains. With the methodology developed in the present research, we hope we could be able to retrieve also a substantial fraction of the nuclear genome of these parasites.
Materials and Methods
Sample Selection.
Ildefonso Canicio, who was in charge of the antimalaria center inaugurated in 1925 by the Generalitat de Catalunya at Sant Jaume d’Enveja (Ebro Delta), worked with malaria-infected patients from the rice fields for several decades and eventually he caught the disease himself. The slides selected for this study were among those collected by his daughter and his family after his death in 1961 and are dated between the years 1942 and 1944; the preserved patient records indicate these were local people with no travel histories. We selected two slides (thereafter, labeled CM and CA for an abbreviation of the patient's names) stained with Giemsa, on which some Plasmodium parasites could still be seen under the microscope (Fig. 1), and one additional slide of anonymous dried blood (labeled POS) for analysis from the original medical collection from Dr. Canicio. We designed an ad hoc extraction method based on previous experiences with ancient museum samples. The extraction procedure was done in a dedicated ancient DNA laboratory at the Institute of Evolutionary Biology in Barcelona where no previous malaria work has ever been conducted.
DNA Extraction.
The slides were immersed (except for one end, to preserve the handwritten labels) in 10–15 mL lysis solution and left in Falcon 50-mL tubes at 37 °C overnight. The lysis buffer was composed of 0.5% SDS, 0.25 mg/mL proteinase K, 10 mM Tris, and 0.5 M EDTA, pH 8.0. The supernatant was subsequently concentrated with a silica column-based method following a protocol that was originally designed to retrieve very short and highly degraded DNA fragments from very ancient specimens (24) (SI Materials and Methods).
Library Preparation and Amplification.
Double-stranded libraries were built using the NEBNext DNA Sample Prep Master Mix Set 2 (E6070; New England Biolabs) and Illumina specific adapters from ref. 24, following the manufacturer’s instructions with minor modifications to the protocol (SI Materials and Methods). Using quantitative PCR (qPCR), we determined the optimal number of cycles required to amplify the samples to get the right amount of DNA necessary for capture experiments (100–500 ng) (SI Materials and Methods).
Capture Depletion.
As tiny amounts of parasite DNA are expected to be present in the slides, along with abundant human DNA from the hosts, we used several strategies to increase the Plasmodium DNA yield: first, we depleted the human content by performing whole genome capture with human baits and shotgun-sequenced what would normally be considered the waste product, and second, we carried out a capture enrichment approach using whole genome bait synthesized from P. falciparum genomic (g)DNA.
The capture-depletion experiment using whole-genome human bait was conducted using the MYbait Human Whole Genome Capture Kit (MYcroarray) following the manufacturer’s instructions according to manual version 3.01 (from www.mycroarray.com/pdf/MYbaits-manual-v3.pdf).
Because we were interested in the nonhuman fraction of the samples, after hybridizing the ancient (a)DNA libraries with the human bait for 24 h, we let it bind to streptavidin magnetic beads for 30 min at 65 °C. Afterward, we collected the supernatant (fraction that did not bind to the beads) and cleaned it up using QiaQuick PCR Purification Kit (Qiagen) following the manufacturer’s instructions. Samples were eluted in 30 µL elution buffer (EB) buffer after a 10-min incubation at 37 °C.
P. falciparum Capture.
P. falciparum baits were prepared according to a previously published protocol (25). Briefly, genomic DNA obtained from the P. falciparum African strain 3D7 in vitro culture (MRA-102G, MR4; ATCC) was sheared and built into libraries containing a custom T7 adapter. Then, Plasmodium T7 libraries were used to prepare biotinylated RNA baits by in vitro transcription. Using these homemade probes, we carried out a second capture experiment following the MyBaits manual version 3.01.
Amplification of Capture-Depleted Products.
After estimating the optimal number of cycles with qPCR, capture-depletion products were amplified for 5 cycles and P. falciparum capture products for 22 cycles using 2× KAPA HotStart ReadyMix and reamplification primers IS5 and IS6 (26). Samples were quantified on an Agilent 2100 Bioanalyzer (Agilent Technologies) and pooled in equimolar amounts. The pool was sequenced on one lane of an Illumina HiSEq 4500 run in 80 SR mode. Library blank controls were included and showed no evidence of contamination with exogenous P. vivax DNA.
Bioinformatic Analysis.
We used AdapterRemoval (27) to remove adapter sequences from the 3′ end of the reads and to remove stretches of consecutive bases with zero, one, or two quality scores. Before mapping, we discarded sequences shorter than 30 bp. All reads were mapped against P. vivax, P. falciparum, and human [National Center for Biotechnology Information (NCBI) 37, hg19] reference genomes using Burrows–Wheeler Aligner (BWA) (28) (SI Materials and Methods).
To reconstruct the P. vivax mtDNA genome, we first removed P. falciparum reads from the P. vivax mtDNA alignment files using a homemade script. Briefly, for each sequence we recorded the edit distance to both the P. vivax and P. falciparum mtDNA genomes and retained reads with equal or lower edit distance to the P. vivax genome. The resulting alignment files were visually inspected with IGV software (29). Next, to increase resolution, we merged the mtDNA sequences from the three samples and generated a consensus P. vivax mtDNA genome using SAMtools and bcftools, restricting to positions with more than two reads and accepting all substitutions that were present in more reads than alternative nucleotides. The P. falciparum mtDNA genome was reconstructed using the same methodology.
Phylogenetic Analysis.
The obtained consensus sequence and 790 additional P. vivax sequences from different continental regions (SI Materials and Methods) were aligned using MUSCLE 3.8 with default parameters (30). The alignments were used to construct a haplotype network using the NETWORK 5.0.0 software (Fluxus Technology). A median joining tree was calculated (31), and the network was postprocessed with maximum parsimony (MP) calculation (32).
The obtained P. falciparum sequence and additional 698 mtDNA sequences (SI Materials and Methods) from different continents (11, 13, 33, 34) were used to reconstruct a haplotype using the same methodology previously described with P. vivax.
Maximum-likelihood trees for both P. falciparum and P. vivax were constructed in PhyML 3.0 (35) under the GTR+I+G model with a BioNJ starting tree, the best of either nearest neighbor interchange (NNI) or subtree pruning and regraphing (SPR) tree improvement and aBayes branch support. Public mitogenomes of Plasmodium knowlesi (AY722797.1) and Plasmodium reichenowi (AJ251941.1) were used as outgroup for the P. vivax and the P. falciparum phylogenies, respectively. The best estimated trees were imported in R and plotted using the ape package (36).
Acknowledgments
We thank the descendants of Dr. Canicio and especially Mr. Miquel Oliveras for offering old malaria slides for the realization of this study. Sequencing was performed at the Danish National High-Throughput DNA-Sequencing Centre, University of Copenhagen. This research was supported from European Regional Development Fund and Ministry of Economy and Competitiveness of Spain Grant BFU2015-64699-P (to C.L.-F.) and a European Research Council Consolidator Grant 681396 - Extinction Genomics (to M.T.P.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: P. vivax and P. falciparum mtDNA sequences are available through the Sequence Read Archive (SRA) (accession no. SRP081144).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611017113/-/DCSupplemental.
References
- 1.Miao M, et al. Plasmodium vivax populations revisited: Mitochondrial genomes of temperate strains in Asia suggest ancient population expansion. BMC Evol Biol. 2012;12:22. doi: 10.1186/1471-2148-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter R, Mendis KN. Evolutionary and historical aspects of the burden of malaria. Clin Microbiol Rev. 2002;15(4):564–594. doi: 10.1128/CMR.15.4.564-594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price RN, et al. Vivax malaria: Meglected and not benign. Am J Trop Med Hyg. 2007;77(6) Suppl:79–87. [PMC free article] [PubMed] [Google Scholar]
- 4.Culleton R, et al. The origins of African Plasmodium vivax: Insights from mitochondrial genome sequencing. PLoS One. 2011;6(12):e29137. doi: 10.1371/journal.pone.0029137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hupalo DN, et al. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet. 2016;48(8):953–958. doi: 10.1038/ng.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, et al. African origin of the malaria parasite Plasmodium vivax. Nat Commun. 2014;5:3346. doi: 10.1038/ncomms4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamblin MT, Di Rienzo A. Detection of the signature of natural selection in humans: Evidence from the Duffy blood group locus. Am J Hum Genet. 2000;66(5):1669–1679. doi: 10.1086/302879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor JE, et al. The evolutionary history of Plasmodium vivax as inferred from mitochondrial genomes: Parasite genetic diversity in the Americas. Mol Biol Evol. 2013;30(9):2050–2064. doi: 10.1093/molbev/mst104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter R. Speculations on the origins of Plasmodium vivax malaria. Trends Parasitol. 2003;19(5):214–219. doi: 10.1016/s1471-4922(03)00070-9. [DOI] [PubMed] [Google Scholar]
- 10.Llamas B, et al. Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas. Sci Adv. 2016;2(4):e1501385. doi: 10.1126/sciadv.1501385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joy DA, et al. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300(5617):318–321. doi: 10.1126/science.1081449. [DOI] [PubMed] [Google Scholar]
- 12.Tanabe K, et al. Plasmodium falciparum accompanied the human expansion out of Africa. Curr Biol. 2010;20(14):1283–1289. doi: 10.1016/j.cub.2010.05.053. [DOI] [PubMed] [Google Scholar]
- 13.Tyagi S, Pande V, Das A. New insights into the evolutionary history of Plasmodium falciparum from mitochondrial genome sequence analyses of Indian isolates. Mol Ecol. 2014;23(12):2975–2987. doi: 10.1111/mec.12800. [DOI] [PubMed] [Google Scholar]
- 14.Pletsch D. Informe sobre una misión efectuada en España en septiembre-noviembre de 1963 destinada a la certificación de la erradicación del paludismo. Rev Sanid Hig Publica. 1965;39(7/9):309–367. [PubMed] [Google Scholar]
- 15.Blazquez J, De Zulueta J. The disappearance of Anopheles labranchiae from Spain. Parassitologia. 1980;22(1-2):161–163. [PubMed] [Google Scholar]
- 16.Daskova NG, Rasnicyn SP. Review of data on susceptibility of mosquitoes in the USSR to imported strains of malaria parasites. Bull World Health Organ. 1982;60(6):893–897. [PMC free article] [PubMed] [Google Scholar]
- 17.Briggs AW, et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci USA. 2007;104(37):14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS One. 2012;7(3):e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forero-Rodriguez J, Garzon-Ospina D, Patarroyo MA. Low genetic diversity and functional constraint in loci encoding Plasmodium vivax P12 and P38 proteins in the Colombian population. Malar J. 2014;13:58. doi: 10.1186/1475-2875-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson RD, et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet. 2016;48(8):959–964. doi: 10.1038/ng.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Zulueta J. Malaria and Mediterranean history. Parassitologia. 1973;15(1):1–15. [PubMed] [Google Scholar]
- 22.Yalcindag E, et al. Multiple independent introductions of Plasmodium falciparum in South America. Proc Natl Acad Sci USA. 2012;109(2):511–516. doi: 10.1073/pnas.1119058109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price RN, et al. Global extent of chloroquine-resistant Plasmodium vivax: A systematic review and meta-analysis. Lancet Infect Dis. 2014;14(10):982–991. doi: 10.1016/S1473-3099(14)70855-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dabney J, Meyer M, Pääbo S. Ancient DNA damage. Cold Spring Harb Perspect Biol. 2013;5(7):1–7. doi: 10.1101/cshperspect.a012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.March S, et al. A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax. Cell Host Microbe. 2013;14(1):104–115. doi: 10.1016/j.chom.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc 6:pdb.prot5448.
- 27.Lindgreen S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res Notes. 2012;5(1):337. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 32.Polzin T, Daneshmand SV. On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett. 2003;31(1):12–20. [Google Scholar]
- 33.Conway DJ, et al. Origin of Plasmodium falciparum malaria is traced by mitochondrial DNA. Mol Biochem Parasitol. 2000;111(1):163–171. doi: 10.1016/s0166-6851(00)00313-3. [DOI] [PubMed] [Google Scholar]
- 34.Tanabe K, et al. Plasmodium falciparum mitochondrial genetic diversity exhibits isolation-by-distance patterns supporting a sub-Saharan African origin. Mitochondrion. 2013;13(6):630–636. doi: 10.1016/j.mito.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 36.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]