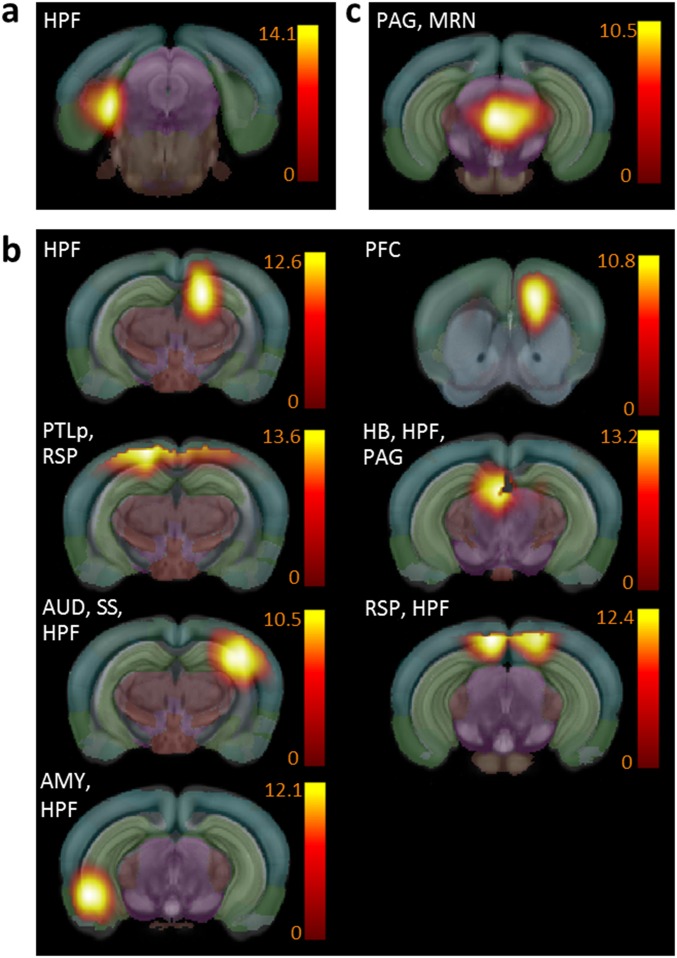
Significance
Mice manipulated by targeted deletion of a specific brain gene show diverse pathological phenotypes, apparent, for example, in behavioral experiments. To explain observed findings, connectome genetics attempts to uncover how brain functional connectivity is affected by genetics. However the causal impact of a single gene on whole-brain networks is still unclear. Here the sole targeted deletion of the mu opioid receptor gene (Oprm1), the main target for morphine, induced widespread remodeling of brain functional connectome in mice. The strongest perturbations occurred within the so-called reward/aversion-circuitry, predominantly influencing the negative affect centers. We present a hypothesis-free analysis of combined structural and functional connectivity data obtained via MRI of the living mouse brain, and identify a specific Oprm1 gene-to-network signature.
Keywords: mouse brain connectivity, resting-state functional MRI, diffusion tensor imaging, mu opioid receptor, reward/aversion network
Abstract
Connectome genetics seeks to uncover how genetic factors shape brain functional connectivity; however, the causal impact of a single gene’s activity on whole-brain networks remains unknown. We tested whether the sole targeted deletion of the mu opioid receptor gene (Oprm1) alters the brain connectome in living mice. Hypothesis-free analysis of combined resting-state fMRI diffusion tractography showed pronounced modifications of functional connectivity with only minor changes in structural pathways. Fine-grained resting-state fMRI mapping, graph theory, and intergroup comparison revealed Oprm1-specific hubs and captured a unique Oprm1 gene-to-network signature. Strongest perturbations occurred in connectional patterns of pain/aversion-related nodes, including the mu receptor-enriched habenula node. Our data demonstrate that the main receptor for morphine predominantly shapes the so-called reward/aversion circuitry, with major influence on negative affect centers.
Neuronal connectivity is at the foundation of brain function (1) and the concept that brain connectivity patterns are dynamically shaped by experience, pathology, and genetics has gained increasing importance. In humans, MRI has opened the era of connectome/imaging genetics to elucidate how genetic factors affect brain organization and connectivity in healthy individuals and disease, and to correlate genotype to phenotype (2). However, the causal impact of a single gene on overall functional connectivity (FC) remains largely unknown, and animal research is best suited to this goal. Here we tested whether combined functional/structural MRI in live animals (3–8) coupled to open-ended postprocessing analysis would reveal connectivity alterations upon targeted inactivation of a single gene. The mu opioid receptor (MOR) mediates the remarkably potent analgesic and addictive properties of opiates, like morphine (9), and belongs to the endogenous opioid system that controls sensory, emotional, and cognitive processes. This receptor is broadly distributed throughout the nervous system (10). It is a key component to facilitate reward (11) and relieves the negative experience of pain (12–14). In this report we show that targeted deletion of the MOR gene (Oprm1) significantly alters the brain connectome in living mice and predominantly reshapes the so-called reward/aversion network involved in pain, depression, and suicide (15).
Results and Discussion
Fine-Grained Mapping of the Mouse Brain Functional Connectome.
In a first step, we established fine-grained mapping of the mouse brain functional connectome (MBFC) in control and Oprm1−/− living mice. Using data-driven spatial independent component analysis (100-ICASSO) (4) of combined blood oxygenation level–dependent (BOLD) resting-state functional MRI (rsfMRI) datasets (Materials and Methods, Data Analysis), we identified 87 functional components, the patterns of which covered neuroanatomical regions defined by automatic coregistration on the Allen Mouse Brain Atlas (AMBA; mouse.brain-map.org/static/atlas) (Fig. S1). We tested the reproducibility of the group ICASSO [a tool for reliability investigation of independent component analysis (ICA) estimates] patterns in each animal and in each experimental group separately via back-reconstruction (SI Materials and Methods, Statistical and Algorithmic Reliability of Group ICA Results and Fig. S2). These examples illustrate low intragroup variability of the ICA patterns and extremely high similarity between group patterns, supporting our further approach of using the 87 group ICA functional clusters (ICASSO components) as nodes in the generation of brain FC matrices of both Oprm1−/− and a control (Ctrl) group of animals (Materials and Methods, Data Analysis and SI Materials and Methods). These matrices, including both, correlations (positive) and anticorrelations (negative) between brain nodal activities (Fig. S3), were further used to examine whether global topological properties and organizational principles of the MBFC (4, 16) are modified in Oprm1−/− mice using graph theory (17). We probed small-world network hallmarks (SI Materials and Methods, Assessment of Global Topological Features of the MBFC in Ctrl and Oprm1−/− Mice) and found similar features (Fig. S3) for both genotypes: a short average path length between all node pairs with high local clustering. We also tested modular properties (17) of the MBFC, a key feature of mammalian brain networks (18), and found partitioning into four stable functional modules (SI Materials and Methods, Assessment of Global Topological Features of the MBFC in Ctrl and Oprm1−/− Mice) in both animal groups, indicating again that general organization principles of the MBFC are preserved in Oprm1−/− mice.
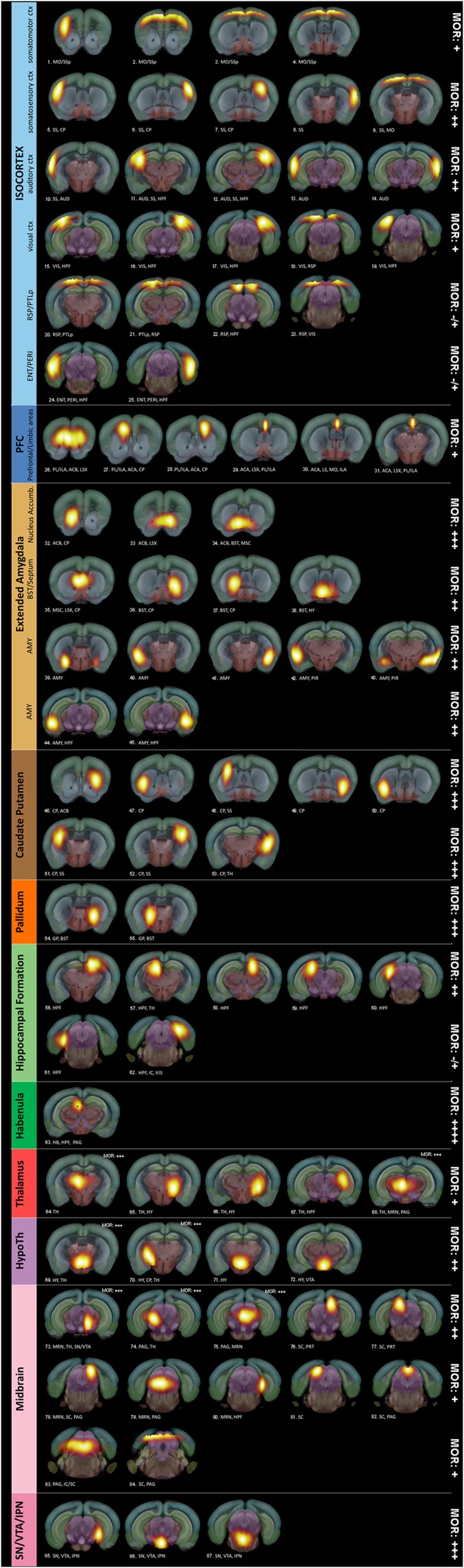
Fig. S1.
Open-ended high-dimensional spatial ICA reveals anatomically well-defined brain components or nodes. Spatial ICA using ICASSO (Gift-Group ICA of fMRI Toolbox v.1.3i, www.nitrc.org/projects/gift/) was performed on 28 combined control and Oprm1−/− datasets. From the 100-ICASSO results, 13 artifactual components related to CSF; vascular- or movement-related pseudoactivations were excluded from analysis. The analysis revealed 87 meaningful components or nodes, displayed as spatial color-coded z-maps (threshold 3.0) onto the AMBA, and arranged according to their affiliation to broader brain areas: Isocortex (MO, SS, AUD, VIS, RSP and PTLp areas, ENT and PIR), PFC (prefrontal and limbic areas), extended amygdala (ACB, BST, AMY), caudate putamen, pallidum, HPF, HB, TH, HY, MB, and VTA/IPN. MOR expression levels were estimated for each broad brain area, according to Erbs et al. (10), and marked on the right side of each image row.
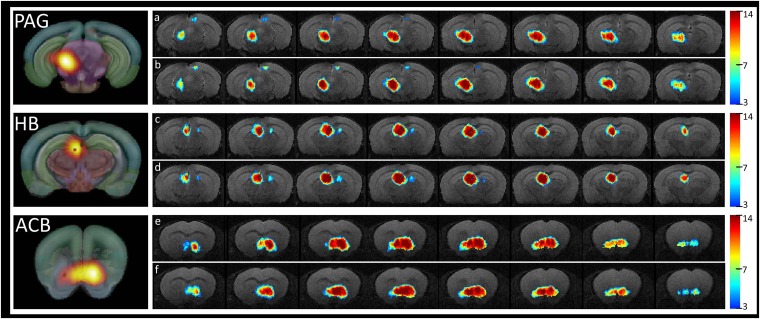
Fig. S2.
Incidence maps showing the spatial distribution and reproducibility of the PAG (A and B), HB (C and D), and ACB (E and F) functional clusters revealed via ICASSO, shown for each animal group separately (A, C, E correspond to the Ctrl group and B, D, F corresponding to the Oprm1−/− group). The color codes the number of animals in which the spatial map overlay the indicated anatomical area. We only show the patterns replicated more than three times. On the left side, the ICASSO group spatial components corresponding to PAG, HB, and ACB are provided.
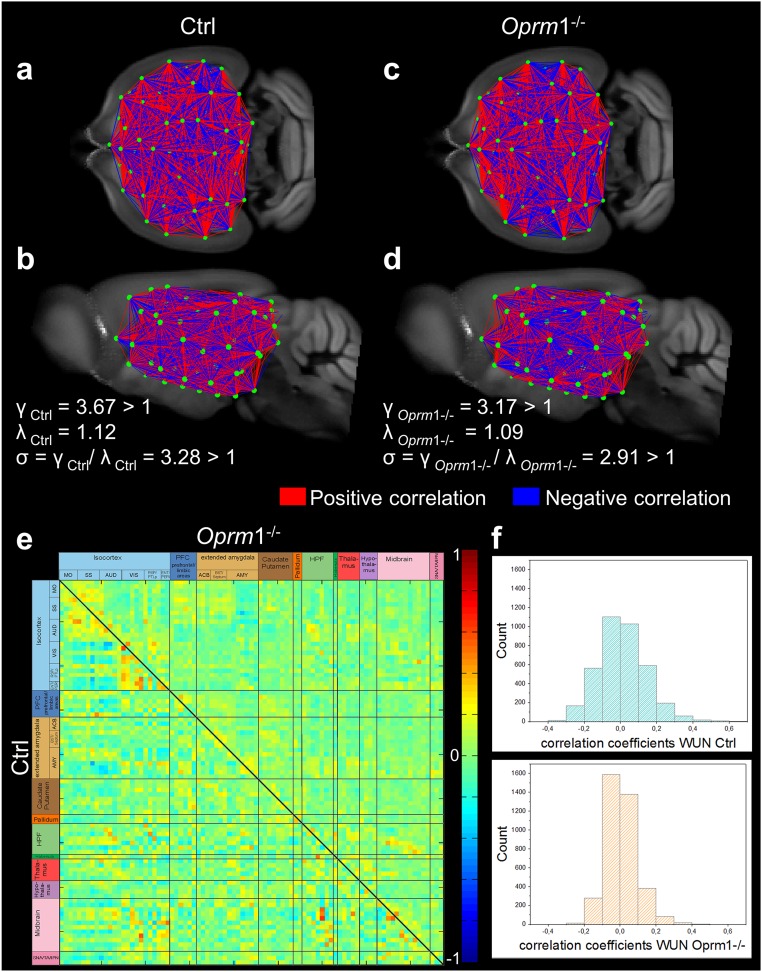
Fig. S3.
Fine-grained 3D-mapping of the mouse brain resting-state functional networks in control (Ctrl, n = 14) and MOR knockout (Oprm1−/−, n = 14) living mice reveals distinct connectivity matrices. (A–D) Whole-brain connectivity matrix in Ctrl (A, coronal; B, sagittal views) and Oprm1−/− (C, coronal; D, sagittal views) mice. Key elements of these networks are nodes (green dots) and connections (red and blue lines) between the nodes. Network nodes were identified via blind-source spatial separation of the low-frequency fluctuations (<0.01Hz) of the BOLD signal, using ICA. The connections between pairs of nodes were defined as direct statistical correlations of BOLD signal fluctuations overtime in the connected areas. Nodes linked in red (positive correlations) showed coherent BOLD signal fluctuations, whereas the blue lines (negative correlations) marked anticorrelated relationships in spontaneous signal fluctuations. The genetic deletion remodels the FC matrix in live Oprm1−/− mice, which show distinct clustering pattern of correlated and anticorrelated connections in both cortical and subcortical areas. Note that despite widespread changes in the brain FC matrix, the network of Oprm1−/− mice displayed small-world features (D) similar to the control group (B). In both genotypes, the ratio γ = C/Crand was greater than 1, the ratio λ = L/Lrand was close to 1, and the scalar measure of small worldness σ = γ/λ was ≥ 1 (see SI Materials and Methods and ref. 58), indicating together that optimal configurations for global information transfer and local processing remains in mutant mice. (E) Strength of the internode connectivity (correlation coefficients) is presented in the PC matrices of Ctrl (lower triangular matrix) and Oprm1−/− groups, rearranged according to broader brain associated with anatomical areas (as also presented in Fig. 1A). (F) Histogram showing the distribution of internode FC correlation coefficients. The width of this distribution is remarkably smaller in Oprm1−/− compared with the Ctrl group.
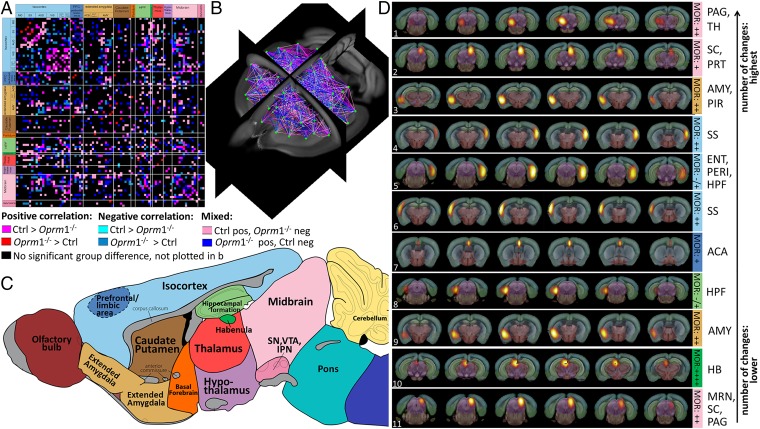
However, this global analysis revealed that the recruitment of brain regions as network hubs (4, 19), defined as functional nodes showing above-mean normalized connectivity strength and diversity (SI Materials and Methods, Assessment of Global Topological Features of the MBFC in Ctrl and Oprm1−/− Mice), was significantly modified in Oprm1−/− mice. In the positive correlation analysis (Fig. S4), several components lost their hub status in Oprm1−/− mice, suggesting decreased relay function in brain structures involved in positive affect and motivational processes [nucleus accumbens (ACB), prefrontal cortex (PFC)], as well as negative sensory and emotional experiences [midbrain reticular nucleus (MRN), periaqueductal gray (PAG), habenula (HB), somatosensory areas (SS)]. Concurrently, other nodes appeared as functional hubs in Oprm1−/− mice only [caudoputamen (CP), bed nuclei of stria terminalis (BST), hippocampal formation (HPF) and peri-HPF cortex, thalamus (TH), superior colliculus (SC)/PAG, MRN/SC/PAG], which, without exception, covered areas integrated into the so-called core aversion-related network (20, 21). In addition, connectivity of PAG, which is a major opioid-sensitive pain-modulatory structure in both rodents (14, 22) and humans (23) and is engaged in aversive learning (24), appeared entirely remodeled in mutant mice (Fig. S4 B and C). Finally, the application of stronger exclusion criteria (combined positive and negative correlations) (Fig. S5) designated the ventro-medial rostral MRN/PAG as the sole remaining Oprm1-dependent functional hub. Together, these substantial hub alterations suggest facilitated communication across pain/aversion-processing centers and perhaps less-efficient integration of reward-related information.
Fig. S4.
Mouse brain functional hubs defined as nodes displaying both above-mean normalized connection strength and diversity, and identified from positive correlation analysis, differ between Ctrl and Oprm1−/− mice. (A) Functional hubs found in both Ctrl (normalized strength > 0.24; diversity > 0.36) and Oprm1−/− (normalized strength > 0.17; diversity > 0.34) mice (see SI Materials and Methods). (B) Hubs detected only in the Ctrl group (normalized strength > 0.24; diversity > 0.36 in Ctrl; but normalized strength < 0.17 and diversity < 0.34 in the Oprm1−/− group). These hubs have lost their importance as connectivity relays in Oprm1−/− mice. Among the hubs ACB, HB, and PAG, are known for their high expression of MORs in the normal brain. (C) Hubs detected only in the Oprm1−/− group (normalized strength < 0.24 and diversity < 0.36 in Ctrl; but normalized strength > 0.17 and diversity > 0.34 in the Oprm1−/− group). These hubs have gained importance in mutant mice connectional matrix and cover well-known pain/aversion processing areas, including the anterior cingulate cortex, somatosensory cortex, BNST, and HPF.
Fig. S5.
Mouse brain functional hubs identified from combined positive and negative correlation analysis. (A) Functional hubs found in both Ctrl (positive correlations: normalized strength > 0.24 and diversity > 0.36; and negative correlations: normalized strength > |−0.19| and diversity > 0.47) and Oprm1−/− (positive correlations: normalized strength > 0.17 and diversity > 0.34; and negative correlations: normalized strength > |−0.11| and diversity > 0.39) mice (see SI Materials and Methods as well). (B) Hubs detected only in the Ctrl group include HB (selection criteria: Ctrl group: positive correlations, normalized strength > 0.24 and diversity > 0.36; and negative correlations, normalized strength > |−0.19| and diversity > 0.47; whereas for the Oprm1−/− group the following criteria was not fulfilled: positive correlations, normalized strength > 0.17 and diversity > 0.34; and negative correlations, normalized strength > |−0.11| and diversity > 0.39). (C) Ventro-medial rostral PAG/MRN remains the only hub detected in Oprm1−/− mice (for the Ctrl group the following criteria was not fulfilled: positive correlations, normalized strength > 0.24 and diversity > 0.36; and negative correlations, normalized strength > |−0.19| and diversity > 0.47; whereas for the Oprm1−/− group positive correlations, normalized strength > 0.17 and diversity > 0.34; and negative correlations, normalized strength > |−0.11| and diversity > 0.39).
Quantitative Intergroup Comparison of Ctrl and Oprm1−/− Functional Connectomes Reveals an Oprm1−/−-Specific Fingerprint.
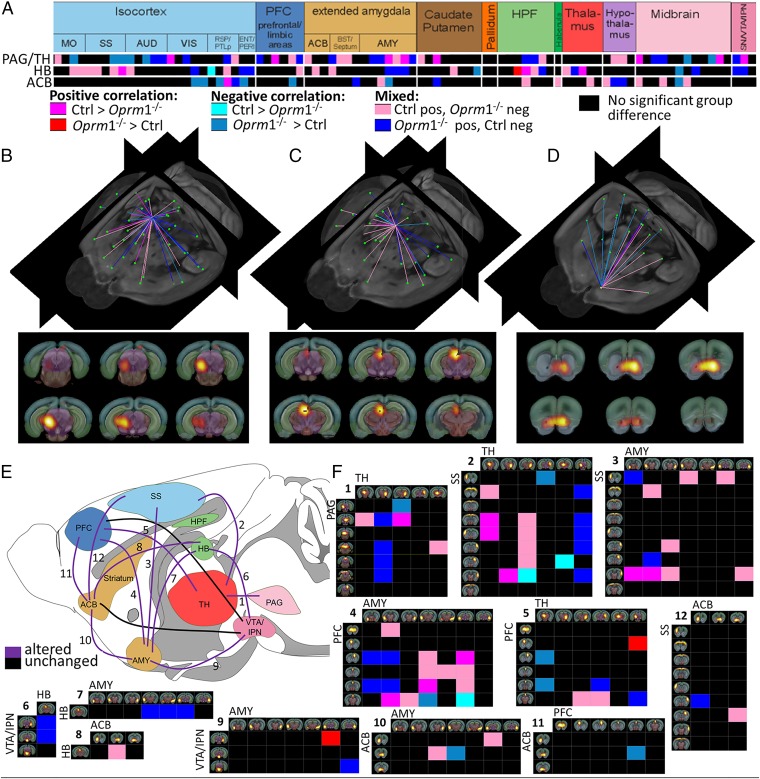
In a second step, we quantified remodeling of the Oprm1−/− functional connectome using a direct statistical intergroup comparison of Ctrl and Oprm1−/− MBFC matrices (Materials and Methods, Direct Intergroup (Ctrl. vs. Oprm1−/−) Statistical Analysis of MBFC and Fig. 1). We detected significant and widespread alterations of internodes connectivity (Fig. 1) [P < 0.05, false-discovery rate (FDR) -corrected]. The 2D-matrix representation (Fig. 1A) captured the causal effect of targeted Oprm1 gene disruption at the level of whole-brain networks, and the extent of Oprm1-dependent connectional activity appeared surprisingly broad. To establish characteristic features of this Oprm1 FC signature, we ranked nodes on the basis of highest number of statistically significant differences in connectivity across the two genotypes (Materials and Methods, Direct Intergroup (Ctrl. vs. Oprm1−/−) Statistical Analysis of MBFC and Fig. 1D). There was a clear dominance of connectivity changes for pain/aversion-related nodes [PAG, hippocampal region (HIP), amygdala (AMY), SS, anterior cingulate areas (ACA), MRN, HB], with the first top 10 of this hierarchy being core players of the aversion-related network (20, 21). The intergroup comparative evaluation therefore leads to conclusions similar to the hub analysis (i.e., predominant reshaping of networks known to process information with negative valence).
Fig. 1.
Quantitative mapping of functional network alterations in Oprm1−/− mice reveals a MOR-dependent activity signature in live animals. (A–C) Direct intergroup (Ctrl vs. Oprm1−/−) statistical comparison of connectivity matrices (P < 0.05, FDR-corrected) is shown as a 2D-matrix (A) or a 3D view (B). Functional nodes were grouped and color-coded as assigned in the sagittal brain view from C. The Oprm1 genetic inactivation induced widespread modifications of internode connectivity. (D) Nodes with the highest number of statistically significant connectivity changes are ranked. Their functional pattern is overlaid on the Allen Brain Atlas, for precise anatomical identification. The top-10 nodes correspond to brain areas associated with pain/aversion processing or double players involved in both pain and reward (PAG/TH, SC/PRT, bilateral AMY, bilateral SS, and MRN/SC/PAG, ACA, HPF, HB). Information on MOR density (10) is included [from low “−/+” barely detectable in the entorhinal area (ENT)/perihinal area (PERI) cortex and HPF to “++++” highest expression in HB].
Specifically, the ventro-lateral PAG (Fig. 1D, rank 1) showed the highest number of changes (Fig. 1D, Top, and Movie S1). In addition, the hippocampus, involved in early memory formation and responsive to pain in humans (25); the AMY, regulating affective dimensions of pain (26) (Fig. 1D; HPF ranks 5 and 8 and AMY ranks 3 and 9); and cortical connectivity, involved in aversion processing at high-order level (27) (Fig. 1D; SS ranks 4 and 6; ACA ranks 7) all showed strong FC perturbations. HB, covering the habenular complex that conveys negative reward-related information (28), was further ranked among nodes with highest connectivity changes (Fig. 1D, rank 10, and Movie S2). Of note, accumbens-related components were not among the top 10, although one ACB component showed above-threshold FC alterations (rank 37) (Movie S3). Coincident with the loss of hub function for the ACB/PFC node (Fig. S4), our data indicate detectable but only modest remodeling within this well-established brain substrate for reward processing (11, 29).
Genetic Inactivation of the MOR Reshapes the Reward/Aversion Runctional Circuitry.
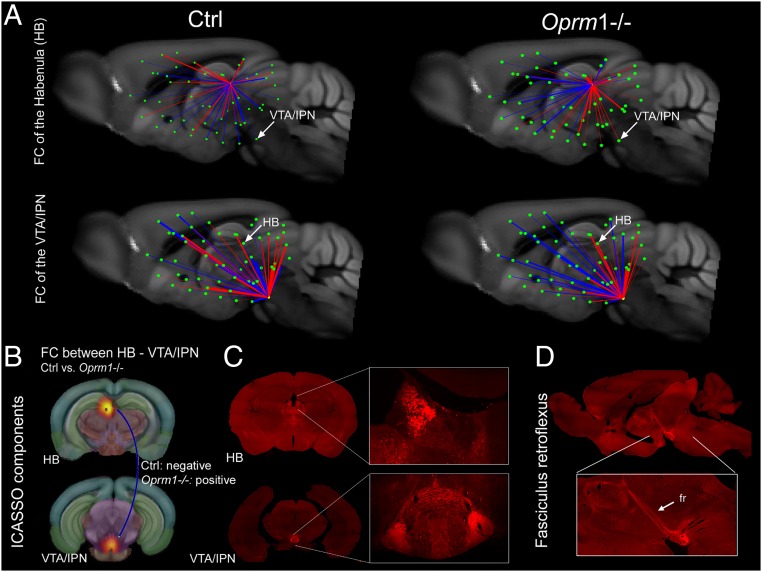
There is rising evidence that aversive and appetitive states interact to optimize adaptive behavioral choices and the existence of a reward/aversion circuitry (RAC) that would act as a unitary salience network has been proposed (30, 31). Because our statistical analysis reveals that the top-10 nodes all belong to the RAC (Fig. 1D), we isolated the Oprm1 signature for this particular network. Fig. 2 (see also Movies S1–S3) shows the major impact of Oprm1 gene activity on core components of the RAC in living mice and illustrates the notion that the Oprm1 fingerprint covers circuits encoding negative (PAG, HB, SS) rather than positive (ACB) dimensions of affective processing. We also extracted connectional patterns of the HB and ventral tegmental area/interpeduncular nucleus (VTA/IPN) nodes (Fig. 3 A and B), which represent key RAC circuitry components, expressing the highest density of MORs in the brain (Fig. 3 C and D). The FC organization was remarkably altered for these two nodes. In particular, highly mixed rostro-caudal correlated/anticorrelated connections in control mice opposed prominent spatial segregation of correlated (mainly caudal) and anticorrelated (mainly rostral) connections in mutants (Fig. 3A). Thus, major changes of connectivity strength for the two nodes demonstrate concerted perturbation of the entire dorsal diencephalic conduction pathway (32) in Oprm1−/− mice.
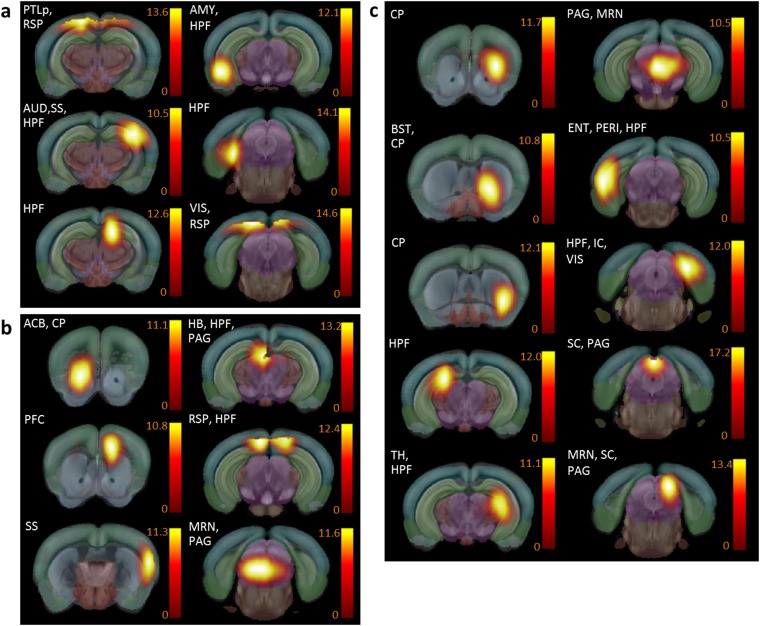
Fig. 2.
MOR deletion predominantly reshapes the RAC, with a major impact on aversion-related components. (A) Detailed view into the matrix of significant connectivity alterations, corresponding to the three main nodes of RAC: PAG/TH (rank 1, see Fig 1D), HB (rank 10, see Fig 1D), and ACB (rank 37). Predominant alterations within reward/aversion pathways correlate with major behavioral modifications reported in mutant mice for pain, emotional, and reward-related behaviors (Dataset S1). (B–D) Three-dimensional display of significantly altered connections of functional nodes from A. The anatomical assignment corresponds to PAG/TH (B and Movie S1), pain/aversion component; HB (C and Movie S2), involved in both reward and aversion processing; and ACB (D and Movie S3), dominant role in reward processing. The three areas are also sites of high MOR density in the normal mouse brain. (E and F) Unified view of connectivity changes in the RAC circuity of live Oprm1−/− brain. Key players of this circuitry are identified as follows: PFC, ACB, AMY, VTA/IPN, TH/PAG, HB, and SS (E). All modified connections are numbered and corresponding detailed connectivity patterns are provided in F. Functional pathways between two regions were considered altered when at least two functional nodes assigned to the respective anatomical areas change their direct connectional pattern.
Fig. 3.
Comparative 3D mapping of FC in Oprm1−/− and control mice for the MOR-enriched HB-VTA/IPN pathway. (A) FC mapping of HB and VTA/IPN nodes in control (Left, sagittal views) and Oprm1−/− brains (Right, sagittal views), extracted from the whole-brain FC matrices (Fig. S3) shows strong spatial segregation of anticorrelated (blue) and correlated (red) connections along the rostro-caudal brain axis in mutant animals. Highly mixed rostro-caudal correlated/anticorrelated connections are seen in control mice. The impact of the MOR deletion on internode connectivity strength is also represented (bar thickness). (B) The selected nodes are representative components of ICASSO analysis, anatomically assigned to HB and VTA/IPN (Upper). Statistical analysis (extracted from Fig. 1A) shows significant modification of FC between the two nodes, with negative correlation in the Ctrl and positive correlation in the Oprm1−/− group, respectively (see blue line). (C and D) MOR expression in HB and VTA/IPN, and along the fasciculus retroflexus (fr), with subcellular resolution (32). These brain areas are particularly rich in MOR expression, as shown in coronal (C) and sagittal (D) sections from MOR-mCherry knockin mice, with images acquired on slide scanner. (Magnification: Inset, 20×.) Reprinted with permission from ref. 32. In these mice the MOR protein, fused to a red-fluorescent protein, is directly visible in mouse tissues. Arrows point to MOR at the level of medial HB and IPN. Views correspond to both sagittal (A) and coronal (B) representations from the rsfMRI.
Rich Remodeling of Oprm1−/− Functional Connectome Is Accompanied by Only Subtle Modifications of Structural Scaffolding Measured via Diffusion Tractography.
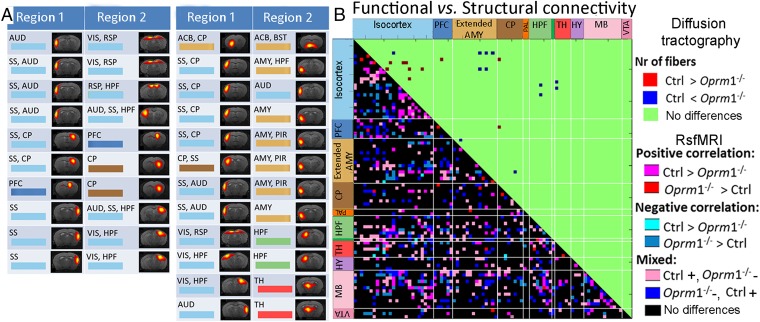
Finally, we tested whether remodeling upon Oprm1 gene knockout was paralleled by modifications of the brain microstructure. We performed high-resolution fiber mapping of the structural connectivity (Movie S4) in the same animals (Materials and Methods, Mouse Brain Tractography-Based Structural Network Analysis). We used high angular-resolution diffusion imaging (HARDI) and global fiber tracking (3, 33). We found only subtle modifications of structural scaffolding (Fig. 4), contrasting the rich remodeling of FC and consistent with the neuromodulatory nature of the single missing gene (13, 34, 35).
Fig. 4.
Limited alteration of tractography-based structural compared with FC in Oprm1−/− mice. (A) Modifications of internode structural connectivity: changes were assessed based on the number of fibers directly connecting functional nodes of the brain connectivity matrix (between region 1 and region 2). (Left) Significant change in fiber numbers Ctrl > Oprm1−/−. (Right) Significant change in fiber numbers Oprm1−/− > Ctrl. (B) Direct comparison of significant functional and structural connectivity showed widespread FC modifications in mutant mice, whereas structural adaptations were limited. The few alterations of structural connectivity determined from tractography included SS, AMY, and ACB, as well as SS–AMY connections. Diffusion tractography also showed remodeling within ACB (A, first row right) in MOR depleted brains but no modification for midbrain centers (i.e., PAG).
SI Materials and Methods
Animal Preparation, Anesthesia, and Physiological Parameters.
Brain MRI was performed on two groups of 12-wk-old male mice: the group of Oprm1−/− mice [n = 14, created by MOR gene disruption via homologous recombination (9)] and the wild-type control mice (n = 14). The animals were briefly anesthetized with isoflurane during imaging preparation (stereotaxic fixation on the mouse head, attachment of physiological monitoring devices). To avoid the inhibitory effects of the isoflurane on BOLD signal, the anesthesia was further switched to MD (Domitor, Pfizer). Moderate MD sedation was initially induced by a subcutaneous bolus injection (0.3 mg MD per kilogram body weight in 100 µL 0.9% NaCl-solution). Fifteen minutes later, the animals received a continuous subcutaneous infusion of MD through a MRI compatible catheter (0.6 mg per kilogram body weight in 200 µL per hour) subcutaneously inserted at the mouse shoulder level. Several previous studies in rodents indicated that anesthesia using MD alone (45–47) or combined with a low dose of isoflurane (5) currently represents the best trade-off for minimal impact on the rsfMRI patterns. However, the timing of the acquisitions is important because Nasrallah et al. (47) observed an impact of MD on the thalamocortical connectivity after long sedation times (longer than 2 h). All of our acquisitions were performed within 45 min after MD bolus injection to avoid such effects. Throughout the imaging sessions, body temperature, respiration rate, blood oxygen saturation, and heart rate were monitored.
Mouse Brain MRI Data Acquisition.
Imaging was performed with a 7T small-bore animal scanner (Biospec 70/20, Bruker) and a mouse head-adapted cryocoil (MRI CryoProbe, Bruker). Before data acquisition, a whole-brain shimming protocol was applied, including adjustment of basic frequency, reference pulse gain, and field homogeneity.
rsfMRI data were collected using single-shot Gradient Echo EPI (TE/TR = 10 ms/1,700 ms). The whole mouse brain (excluding the cerebellum) was covered using 12 axial slices (0.7-mm slice thickness), with a field-of-view (FOV) of 1.92 × 1.2 cm2 and an acquisition matrix of 128 × 80, which resulted in a planar spatial resolution of 150 × 150 μm2. Next, 200 volumes were recorded with slice acquisition in interlaced fashion for each run. The rsfMRI scans were performed at 30 min after MD bolus injection.
After the rsfMRI acquisition, MD infusion was stopped and replaced by anesthesia via isoflurane (∼2.5 vol%) for further scanning performed on respiration triggering. First, high-resolution morphological images were acquired using Turbo RARE T2 (TE/TR = 50 ms/6,514 ms), two averages at RARE factor of 4. The whole brain, including the cerebellum, was covered using 48 slices at spatial resolution of 51 × 51 × 300 μm3 with a FOV of 1.3 × 1.0 cm2.
Second, HARDI was performed using four-shot DTI–EPI sequence with 15 axial slices at a resolution of 94 × 94 × 500 μm3 covering the equivalent partition of the brain as for the rsfMRI scan (TE/TR= 27 ms/3,750 ms); Δ = 10 ms, diffusion gradient duration (δ) = 5 ms, bfactor of 0 and 1,000 s/mm2, 30 diffusion gradient directions.
Data Preprocessing.
For direct comparison of functional and structural mouse brain information, an initial processing step was implemented in MATLAB (The MathWorks) and used in combination with the fMRI tool of Statistical Parametric Mapping (SPM8, www.fil.ion.ucl.ac.uk/spm/) and its SPMmouse toolbox (www.spmmouse.org). We refined the SPMmouse brain template by including additional high-resolution mouse brain images and we used this template for spatial normalization and alignment of rsfMRI mouse brain images, morphological T2-weighted images, and the A0 images obtained from HARDI (HARDI acquisition with a diffusion weighting factor bfactor = 0 mm2/s, no diffusion gradient applied) and the parametric maps derived from diffusion tractography [fiber density (FD) maps] (see also Mouse Brain Structural Network Analysis, below).
The preprocessing pipeline included the following steps: (i) Registration of the morphological T2-weighted contrast based images to the A0 images performed for each mouse individually. (ii) The registered morphological T2-weighted images of each mouse (size: 256 × 196 × 48) were further aligned to the mouse brain template, including information on standard locations of gray matter, white matter, and cerebrospinal fluid, which were then segmented from the T2-weighted image. This alignment involved a linear registration (12-parameter affine transformation, accounting for major differences in head shape and position between subjects), as well as a nonlinear registration (warping, accounting for small-scale anatomical differences). (iii) The individual fourth degree B-Spline deformation information gained from the alignment and segmentation of the T2-weighted image was further applied to the rsfMRI contrast data and diffusion-based parametric maps (FD maps obtained after global tractography of each mouse brain) to bring them into a common group space with the same resolution and physical voxel size as well as a congruent orientation (final resolution: 76 × 86 × 51 voxels at 0.15 × 0.15 × 0.15 mm3). (iv) Smoothing of normalized rsfMRI data were performed with a Gaussian kernel of FWHM 0.4 × 0.4 × 1 mm3. (v) A brain mask was created, guided by white and gray matter segmentation in group space, and applied to all datasets before further processing steps.
Statistical and Algorithmic Reliability of Group ICA Results.
ICASSO (49) was used to assess pattern stability via bootstrapping and randomizing initial conditions for different numbers of independent components. Thereby, the “quality index” Iq (values ranging from 0 to 1) was used as a quantitative measure of robustness of the identified components evaluating compactness and isolation of each cluster (4). We tested the consistency of the results when progressively achieving a high spatial definition (in accordance to fine anatomical details) of the functional clustering patterns with 20-, 40-, 60-, 80-, 100-, 120-, and 140-ICASSO, respectively. ICASSO group analysis resulted in a quality index Iq > 0.8 in 92%, 86%, and 82% of the components for 60-, 80-, and 100-ICASSO respectively. Of the 100 components, 55% had Iq > 0.9, 94% of the components showed Iq > 0.65, and all components exceeded an Iq of 0.56, demonstrating a high level of stability in the obtained network pattern. However, using the percentage of components revealing Iq > 0.6 as a stability criterion, we observed a clear degradation of the IC estimates for 120- and 140-ICASSO, justifying our choice of 100-ICASSO analysis. Selecting the number of components is a critical step when using ICA. Underestimation may result in mixing various components (51, 52), whereas overestimation can result in splitting reliable networks (53, 54), decreasing the stability of IC estimates (55).
To further test the reproducibility of the Group ICA patterns in each experimental group, we exploited information and results generated with GIFT tools (Group ICA of fMRI Toolbox, v1.3i, www.nitrc.org/projects/gift/) via back reconstruction.
Indeed, the patterns of functional elementary clusters resulting from 100-ICASSO (presented in Fig. S1) represent group components. In the display, the color coding represents the dependence of the time course in each voxel compared with the mean time course of the respective component, in arbitrary units. However, from these aggregate components and the original data, GIFT toolbox computes spatial back-reconstructed individual subject components using a spatial-temporal regression approach (details are given in the GIFT toolbox manual: mialab.mrn.org/software/gift/documentation.html).
We used the back-reconstructed individual spatial maps to create “incidence maps” (Fig. S2) for each IC. The incidence maps are illustrating the spatial distribution and the reproducibility of the IC pattern over each animal group (the Oprm1−/− and Ctrl groups). The color-coded incidence of a voxel reflects in how many of the animals it was found to belong.
Relevant examples are the incidence maps illustrating the patterns of PAG, HB, and ACB functional clusters. These examples are included in Fig. S2, and illustrate low intragroup variability of the ICA patterns and extremely high similarity between group patterns. These results substantiate our further approach of using the Group ICA functional clusters as “nodes” in the generation of brain FC matrices of both Oprm1−/− and Ctrl group of animals.
Partial Pearson Correlation.
The PC coefficients (Pearson) between each pair of IC were calculated using the time courses of the other 86 components as controlling variables, investigating the extent of temporal correlation between the spatial IC. With this approach we estimated the direct statistical association by controlling correlation mediated by other components (56). The data (probed separately for both groups via an Anderson–Darling test) meets the assumption of normal distribution. This procedure generated a WUM for each group, containing statistically relevant/significant correlation values. Based on WUM, the normalized strength of connectivity (separately calculated for positive and negative correlations) for each IC and for each group of mice was further determined as the total weight of relevant connections of a node divided to the number of these connections. The mean normalized strength for positive correlations was equal to 0.24 in Ctrl mice and 0.17 in Oprm1−/− animals. The mean normalized strength for negative correlations was equal to −0.19 in Ctrl mice and −0.11 in Oprm1−/− animals.
The PC matrices (MBFC) were arranged in association to broad anatomical areas as follows: isocortex [somato-motor areas (MO), SS, auditory (AUD), visual (VIS), retrosplenial (RSP) and posterior parietal association (PTLp) areas, ENT and piriform areas (PIR), PFC (prefrontal and limbic areas), extended amygdala (ACB, BST, AMY), caudate putamen, pallidum, HPF, HB, TH, hypothalamus (HY), midbrain (MB), and VTA/IPN. For 3D visualization of the MBFC, a Matlab-based toolbox was developed (see next section).
Visualization of Results.
For 3D visualization of the MBFC, a Matlab-based toolbox was developed. The information contained into the WUMs was mapped on the mouse brain template and the AMBA. Each node of the matrix, corresponding to the 87 identified ICs, was represented by a dot positioned at its center of mass. The coloring distinguishes between positive (red) and negative (blue) correlations, as presented in Fig. S3 A–D. Additional visualization of the strength of correlation is exemplified in Fig. 3A, where the values were normalized with respect to the highest absolute value to enhance differentiation. The thicker the line, the higher (more positive/more negative depending on the sign) the correlation coefficient.
Assessment of Global Topological Features of the MBFC in Ctrl and Oprm1−/− Mice.
Small-worldness properties.
The clustering coefficient (C) and the minimum path length (L) represent two key metrics of small-worldness introduced by Watts and Strogatz (57). To evaluate the topological properties of the Ctrl and Oprm1−/− mouse brain connectivity, these parameters were compared with the mean clustering coefficient (Crand) and path length (Lrand) estimated in a random network with the same number of nodes, edges, and degree distribution as our current mouse brain network (WUM).
For a small-world network, the ratio γ = C/Crand is defined to be greater than 1 and the ratio λ = L/Lrand has to be ∼1. The scalar measure of small worldness σ = γ/λ was further calculated and compared with 1, for each experimental group, to determine if the Ctrl and Oprm1−/− networks have the characteristic property of greater-than-random clustering and near-random path length (σ ≥ 1) or not (σ < 1) (58). The results are given in Fig. S3 B and D.
Modularity (Q).
Based on WUM, the community structures (modules) of the Ctrl and Oprm1−/− mouse brains were identified by graph theory and spectral partitioning method (59).
The optimal community structure is a subdivision of the network into nonoverlapping groups of nodes in a way that maximizes the number of within-group connections, and minimizes the number of between-group connections. The modularity is therefore a statistic that quantifies the degree to which the network may be subdivided into such clearly delineated groups. Comparable Q-values for the Ctrl (Q = 0.33) and Oprm1−/− (Q = 0.36) could be obtained, suggesting a prominent modular structure of intrinsic connectional brain architecture for each group. A distribution of Q values and respective partitions were computed. If the number of modules was stable over all runs, the association of each component to a particular module and the respective stability of this configuration were assessed. We found that both, the Ctrl and Oprm1−/− functional networks were subdivided in four very stable modules.
Finally, from WUMs and modular partitioning results, the “diversity” of each IC (separately evaluated for positive and negative correlations in each experimental group) was calculated, quantifying the extent of its intermodular connectivity (60). The mean diversity value for positive correlations was equal to 0.36 in Ctrl mice and 0.34 for Oprm1−/− animals. The mean diversity value for negative correlations was equal to 0.47 in Ctrl mice and 0.39 for Oprm1−/− animals.
Hub regions.
The nodes revealing a high level of connectivity within and even across modules are considered as being of main importance for the proper functioning of the brain. Nodes with simultaneously above mean normalized strength (39 for Ctrl and 44 for Oprm1−/−) and diversity (44 for Ctrl and 46 for Oprm1−/−) were thus defined as mouse brain network hubs. In a first step we identified the network hubs for each group based on the positive correlations only as described in our previous paper (4) (see results in Fig. S4) and we next included in our evaluation both positive and negative correlations (Fig. S5). In Figs. S4 and S5 we grouped the hubs into three categories: hubs common for both groups (i), hubs unique to Oprm1−/− (ii), and hubs missing in Oprm1−/− mice (iii).
Mouse Brain Structural Network Analysis.
Global mouse brain fiber tractography.
HARDI data were acquired for all animals of our study and fiber tracking was performed via a global fiber tracking algorithm developed in our group (33), optimized and validated for in vivo mouse brain tractography (3). Diffusion data postprocessing, was performed using the FiberTool package developed in-house (https://www.uniklinik-freiburg.de/mr-en/research-groups/postprocessing/globaltractography.html?L=).
The method used in our study is reconstructing all fiber bundles simultaneously for the whole brain without initially defining seed or target regions. The approach is therefore offering resistance to the local imaging artifacts, avoiding the cumulative errors generally arising when sequentially integrating local fiber directions from predefined seed-points. This method allows for the reconstruction of a larger FOV when an ambiguous area has to be resolved. The reconstructed fibers are built with small line segments (particles) described by a spatial position and orientation. These segments are the basic building blocks, bonding together and forming longer chains that represent the individual fibers. Their orientation and number are adjusted simultaneously to best match the measured data. We denote the measured data by D(x, n) = S(x, n)/S0(x), where S(x, n) is the diffusion-weighted HARDI signal at position x with gradient direction n with a fixed b-value (1,000 s/mm2); S0(x) denotes the measurement without diffusion gradient. The connections between segments are formed based on a probabilistic approach.
The reconstructions contain several hundred of these segments per voxel. Their behavior is governed by certain parameters, classified in two categories: (i) geometrical parameters and (ii) parameters concerning the iteration process. The influence of these parameters on the results was discussed by Reisert et al. (33). For the global reconstruction of the mouse brain fibers, we previously optimized these parameters (3) based on the comparison of the in vivo global fiber tracking results with histological tract tracings in the same animals.
Geometrical parameters.
Each fiber is constructed from small segments that have a width (σ), a length (l), and a weight (w). Width and weight influence the numbers of fibers, whereas length influences the curvatures of the fibers. Weight acts more like a threshold: for example, for low weights (like in our settings) the gray matter fibers could be generated because high weights only appear in strongly anisotropic areas. The width is actually the diameter of the cylindrical segments. During the optimization for tracking the mouse brain nerve fibers we found that values of σ = 0.078 mm, l = 0.234 mm, and w = 0.041 produce a high number of segments per voxel, optimized with respect to the computation time and the complexity of the reconstruction. The trackable area was defined by a mouse brain mask generated after thresholding on the A0 image. During the tracking process the number of particles, the number of connections and fibers, and the time per iteration could be checked. A particle vs. connections plot informs about the connection degree during the iteration process. The connection degree is a number between 0 and 1; low values are indicating small, short, and curly fibers; high values are indicating long fibers. Typically, the process starts with low values below 0.5 and ends with values of about 0.8. A fiber length distribution plot gives information about the number of fibers with a specific length, where the number of fibers is given in logarithmic scale.
Iteration parameters.
Because geometrical parameters determine the quality and characteristics of the reconstruction, the iteration parameters have to be adapted to these needs: complex models (e.g., mouse brain fiber architecture) need more iterations than simple sparse reconstructions. The iteration parameters are the start/stop “temperature” of the optimizer (for our reconstructions the start temperature was 0.1 and the stop temperature 0.001), the number of update steps (30 steps for our reconstructions), and the number of iterations (3 × 108 in our study). The number of update steps determines how often the iteration process is interrupted for cooling down the temperature.
We applied global tractography for each individual mouse brain included in the study. We used these tractography results of each individual brain and the spatial patterns of the 87 independent components obtained from rsfMRI data to generate group-specific “structural connectivity matrices.” More precisely, the “nodes” of the structural matrices were represented by the spatial patterns of the IC, whereas the connectivity measure was represented by the information on the number of fibers interconnecting each pairs of nodes.
Generation of group-specific mouse brain structural connectivity matrices.
To achieve straightforward comparison between functional and structural mouse brain connectomes, we included functional nodes in the structural brain matrix and used the number of interconnecting fibers as the main parameter.
First, the spatial patterns of the retained 87 components generated via a group ICASSO approach were transferred into individual mouse space via inverse fourth-degree B-Spline deformation for each animal. Z score values were thresholded at 3.0, similarly to all displays of IC components (Fig. S1) to create component masks. These masks—registered on the tractography data of each animal—were used for extraction of normalized number of fibers interconnecting pairs of ICs for each mouse. Two group-specific (Ctrl vs. Oprm1−/−) adjacency matrices, containing information on number of fibers connecting pairs of the 87 network nodes, were further generated. Direct statistical intergroup comparison (two-sided two-sample t test, P < 0.05) was further performed and significant group differences were plotted in a structural GCM (Fig. 4B, Upper). This matrix highlighted internodes structural modifications.
We further examined whether correlation may exist between structural and functional alterations in brain areas with marked structural connectivity modifications (Fig. 4B, Upper). Direct area-based correlation analysis between numbers of significant structural and FC changes showed weak, although significant correlation (Pearson correlation coefficient R = 0.26).
We also used the individual fiber-tracking results to generate high-resolution fiber maps (as described in ref. 3) that also contain the fiber-orientation information coded by color (resolution of 12 × 12 × 50 µm3) (Movie S4).
Supplementary to structural connectivity analysis based on numbers of fibers interconnecting pairs of nodes, we also investigated axial and radial diffusivity, fractional anisotropy, and trace modifications along tracts interconnecting the resting-state functional nodes. Intergroup comparison didn’t reveal statistically significant group differences when using these parameters (two-sided two-sample t test, P < 0.05), highlighting once again reduced structural connectivity modifications in the Oprm1−/− mice.
One has to keep in mind that diffusion tractography still represents an indirect measure of structural connections in the brain, relying on the principles of water diffusion in the tissue. Many factors could influence the outcome of the tractography results, including acquisitions parameters and the selected tractography approach. One limitation in our study is the acquisition of raw diffusion data with nonisotropic resolution (94 × 94 × 500 μm3). Nonisotropic resolutions make the tractography process more difficult. However, acquiring isotropic diffusion data using 3D diffusion-based sequences would have considerably increased the acquisition time, making the in vivo experiments challenging and prone to accumulative movement artifacts.
Conclusions
In sum, unbiased analysis of MBFC in live Oprm1−/− mice reveals an Oprm1-specific FC signature, with strongest impact on the RAC connectome. Pain and pleasure are essential to shape learning and decision-making. The well-known dual analgesic/rewarding effects of morphine and the behavioral phenotypes of Oprm1−/− mutant mice showing increased pain perception (36) and reduced drug (37) or social (38, 39) reward, posit MOR as a central player for these fundamental processes. Indeed, two decades of Oprm1−/− mouse studies have unambiguously established the pivotal role of MOR in both pain and pleasure (Dataset S1 and references therein), recognized as intermingled processes at circuit level (40) and for pathology (41).
In our analysis, the major influence of Oprm1 inactivation on aversion/pain-related, rather than reward connectivity, may reflect a stronger inhibitory MOR tone or developmental influence on negative affect centers, at least under resting-state conditions. From an evolutionary perspective, pain represents a key signal for survival, and successful coping with a pain stimulus is essential to gain a selective advantage (42). Despite the antique notion that pain and pleasure form a continuum, it is only recently that the rewarding value of pain relief has been recognized (40, 41). The key implication of MOR activity in dampening physical, emotional, and social pain, evidenced in human PET imaging studies (see ref. 43 and references therein), and our own FC analysis of live Oprm1-deficient mice, together suggest that pain relief may be a primary MOR function.
Importantly, our data unequivocally reveal pronounced causal effects of a single gene on whole-brain FC in live animals, with subtle modifications of the tractography-based structural connectome. This report is among the very first studies (44) that open the way to targeted connectome genetics (2) in basic research and, to the best of our knowledge, this is the first hypothesis-free analysis of combined rsfMRI/diffusion tractography data in the mouse, leading to the identification of a specific gene-to-network signature.
Materials and Methods
Ethics.
All experiments were performed in accordance with the German and French laws and guidelines regarding ethics on animal experimentation (ethics allowance 35_9185.81/G-13/15).
Animal Preparation, Anesthesia, and Physiological Parameters.
Animal preparation, anesthesia, and physiological parameters during imaging are described in the first part of SI Materials and Methods. The rsfMRI data were acquired under continuous Medetomidine (MD, an α-2 adrenergic agonist) sedation through a MRI compatible catheter (initial intraperitoneal injection of 0.3 mg MD per kilogram body weight in 100 µL 0.9% NaCl-solution followed by subcutaneous infusion of 0.6 mg per kilogram body weight in 200 µL/h). MD was selected among other anesthetics based on previous reports suggesting minimal impact on FC (5, 45–47).
Mouse Brain MRI Data Acquisition.
Mouse brain MRI data acquisition (see also SI Materials and Methods) was performed with a 7T animal scanner (Biospec 70/20) and a mouse head-adapted cryocoil (both from Bruker). rsfMRI data were collected (30 min after MD bolus injection) using single-shot Gradient Echo Echo Planar Imaging (EPI) [12 axial slices, 200 volumes, image resolution 150 × 150 × 700 μm3, echo time (TE)/repetition time (TR) = 10 ms/1,700 ms]. High-resolution morphological imaging was done using Turbo RARE T2 (51 × 51 × 300 μm3, TE/TR = 50 ms/6,514 ms). HARDI was performed using a four-shot Diffusion Tensor Imaging–EPI (DTI–EPI) sequence (15 axial slices, resolution of 94 × 94 × 500 μm3, TE/TR = 27 ms/3,750 ms); Δ = 10 ms, diffusion gradient duration (δ) = 5 ms, bfactor = 1,000 s/mm2, 30 diffusion gradient directions.
Data Analysis.
The data preprocessing pipeline is described in SI Materials and Methods.
rsfMRI data analysis.
Identification of elementary functional clusters as nodes of the MBFC matrix was performed via high-dimensional ICA (100 components). Spatial group ICA (48) via the MATLAB based toolbox GIFT (Group ICA of fMRI Toolbox, v1.3i, www.nitrc.org/projects/gift/) was carried out on all of the mouse brain rsfMRI data (Oprm1−/− and Ctrl mice) using the Infomax algorithm. ICASSO (49) was used to assess pattern stability for the identified components (SI Materials and Methods, Statistical and Algorithmic Reliability of Group ICA Results). The mean resulting patterns were displayed as spatial color-coded z-maps onto T2 weighted images and on coregistered AMBA (50) (see, for example, Figs. 1–3, Figs. S1, S4, and S5, and Movies S1–S3). Coregistration with AMBA allowed for automatic identification of anatomic brain areas covered by IC patterns. From the 100-ICASSO results, 13 artifactual components were excluded from analysis. The meaningful 87 functional clusters were further used as nodes (Fig. S1) in the generation of the MBFC matrix, via partial correlation (PC).
PC analysis (SI Materials and Methods, Partial Pearson Correlation) was performed for each experimental group (Oprm1−/− and Ctrl) separately. The time courses associated with each relevant independent component (IC, node) obtained from 100-ICASSO were used in PC analysis using an in-house developed MATLAB tool (4). The PC coefficients (Pearson) between each pair of IC were calculated and used to create a 87 × 87 adjacency PC matrix for each animal, as well as two average matrices, representative for each experimental group (Oprm1−/− and Ctrl) (Fig. S3E; see also and histogram display of correlation coefficients in Fig. S3F). Each element of the matrix represented the strength of direct connectivity between two components (nodes). The PC matrices were then normalized using Fisher's z transformation. The significance of positive and negative correlations between pairs of components was further assessed via a two-sided one-sample t test, for P < 0.05 (4). This procedure generated a weighted undirected matrix (WUM) for each group, containing statistically relevant/significant correlation values. For 3D visualization of the MBFC, a Matlab-based toolbox was developed (SI Materials and Methods, Visualization of Results).
Assessment of global topological features of the MBFC in Ctrl and Oprm1−/− mice is described in SI Materials and Methods.
Direct intergroup (Ctrl vs. Oprm1−/−) statistical analysis of MBFC.
The analysis of the FC remodeling of the Oprm1−/− mouse brain was done via direct statistical comparison between the PC matrices (unthresholded z matrices) generated for each experimental group. We tested the hypothesis that there are no differences in connectivity between the two groups via a two-sided two-sample t test (similar variation within each group). The hypothesis was rejected at a significance level of 0.05, under FDR control for multiple comparisons.
A group comparison matrix (GCM) was generated (Fig. 2A) that color-coded the statistically significant intergroup differences of connectivity. Each node was associated to a broader brain area, based on the anatomical overlapping assigned via coregistration of the ICA results on the AMBA. The GCM was arranged to cluster the connectivity changes in association to anatomical areas (Fig. 1 A and C and Fig. S1). Three-dimensional visualization of the changed connections was also generated (Fig. 1B). The color-code associated with the GCM was maintained for the 3D displays. Only nodes showing changes in their FC are plotted. The GCM was further used to count the significantly changed connections for each node (IC) and we further ranked nodes on the basis of highest number of such statistically significant differences in connectivity across the two genotypes (Fig. 1D).
Mouse brain tractography-based structural network analysis.
Mouse brain tractography-based structural network analysis are detailed in SI Materials and Methods, Mouse Brain Structural Network Analysis.
Supplementary Material
Acknowledgments
We thank Robin Simpson for his help in creating Movie S1. This work was supported by the French Academy of Sciences and the NIH (National Institute on Alcohol Abuse and Alcoholism Grant 16658); the Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale; Strasbourg University; and grants from the Brain Links Brain Tools cluster of excellence from Freiburg (MouseNet) and European Research Area Network (ERANET-Neuron), AF12-NEUR0008-01-WM2NA.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601640113/-/DCSupplemental.
References
- 1.Van Essen DC. Cartography and connectomes. Neuron. 2013;80(3):775–790. doi: 10.1016/j.neuron.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson PM, Ge T, Glahn DC, Jahanshad N, Nichols TE. Genetics of the connectome. Neuroimage. 2013;80:475–488. doi: 10.1016/j.neuroimage.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harsan LA, et al. Mapping remodeling of thalamocortical projections in the living reeler mouse brain by diffusion tractography. Proc Natl Acad Sci USA. 2013;110(19):E1797–E1806. doi: 10.1073/pnas.1218330110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mechling AE, et al. Fine-grained mapping of mouse brain functional connectivity with resting-state fMRI. Neuroimage. 2014;96:203–215. doi: 10.1016/j.neuroimage.2014.03.078. [DOI] [PubMed] [Google Scholar]
- 5.Grandjean J, Schroeter A, Batata I, Rudin M. Optimization of anesthesia protocol for resting-state fMRI in mice based on differential effects of anesthetics on functional connectivity patterns. Neuroimage. 2014;102(Pt 2):838–847. doi: 10.1016/j.neuroimage.2014.08.043. [DOI] [PubMed] [Google Scholar]
- 6.Gozzi A, Schwarz AJ. Large-scale functional connectivity networks in the rodent brain. Neuroimage. 2016;127:496–509. doi: 10.1016/j.neuroimage.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Stafford JM, et al. Large-scale topology and the default mode network in the mouse connectome. Proc Natl Acad Sci USA. 2014;111(52):18745–18750. doi: 10.1073/pnas.1404346111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zerbi V, et al. Resting-state functional connectivity changes in aging apoE4 and apoE-KO mice. J Neurosci. 2014;34(42):13963–13975. doi: 10.1523/JNEUROSCI.0684-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthes HW, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383(6603):819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 10.Erbs E, et al. A mu-delta opioid receptor brain atlas reveals neuronal co-occurrence in subcortical networks. Brain Struct Funct. 2015;220(2):677–702. doi: 10.1007/s00429-014-0717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Merrer J, Becker JA, Befort K, Kieffer BL. Reward processing by the opioid system in the brain. Physiol Rev. 2009;89(4):1379–1412. doi: 10.1152/physrev.00005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leknes S, Tracey I. A common neurobiology for pain and pleasure. Nat Rev Neurosci. 2008;9(4):314–320. doi: 10.1038/nrn2333. [DOI] [PubMed] [Google Scholar]
- 13.Navratilova E, Porreca F. Reward and motivation in pain and pain relief. Nat Neurosci. 2014;17(10):1304–1312. doi: 10.1038/nn.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields H. State-dependent opioid control of pain. Nat Rev Neurosci. 2004;5(7):565–575. doi: 10.1038/nrn1431. [DOI] [PubMed] [Google Scholar]
- 15.Elman I, Borsook D, Volkow ND. Pain and suicidality: Insights from reward and addiction neuroscience. Prog Neurobiol. 2013;109:1–27. doi: 10.1016/j.pneurobio.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Z, Li T, King J, Zhang N. Mapping thalamocortical networks in rat brain using resting-state functional connectivity. Neuroimage. 2013;83:237–244. doi: 10.1016/j.neuroimage.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullmore E, Sporns O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10(3):186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 18.Fornito A, Zalesky A, Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16(3):159–172. doi: 10.1038/nrn3901. [DOI] [PubMed] [Google Scholar]
- 19.Liska A, Galbusera A, Schwarz AJ, Gozzi A. Functional connectivity hubs of the mouse brain. Neuroimage. 2015;115:281–291. doi: 10.1016/j.neuroimage.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 20.Hayes DJ, Northoff G. Identifying a network of brain regions involved in aversion-related processing: A cross-species translational investigation. Front Integr Nuerosci. 2011;5:49. doi: 10.3389/fnint.2011.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes DJ, Northoff G. Common brain activations for painful and non-painful aversive stimuli. BMC Neurosci. 2012;13:60. doi: 10.1186/1471-2202-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linnman C, Moulton EA, Barmettler G, Becerra L, Borsook D. Neuroimaging of the periaqueductal gray: State of the field. Neuroimage. 2012;60(1):505–522. doi: 10.1016/j.neuroimage.2011.11.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wager TD, Scott DJ, Zubieta JK. Placebo effects on human mu-opioid activity during pain. Proc Natl Acad Sci USA. 2007;104(26):11056–11061. doi: 10.1073/pnas.0702413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy M, et al. Representation of aversive prediction errors in the human periaqueductal gray. Nat Neurosci. 2014;17(11):1607–1612. doi: 10.1038/nn.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forkmann K, et al. Pain-specific modulation of hippocampal activity and functional connectivity during visual encoding. J Neurosci. 2013;33(6):2571–2581. doi: 10.1523/JNEUROSCI.2994-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neugebauer V. Amygdala pain mechanisms. Handbook Exp Pharmacol. 2015;227:261–284. doi: 10.1007/978-3-662-46450-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bushnell MC, Ceko M, Low LA. Cognitive and emotional control of pain and its disruption in chronic pain. Nat Rev Neurosci. 2013;14(7):502–511. doi: 10.1038/nrn3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hikosaka O. The habenula: from stress evasion to value-based decision-making. Nat Rev Neurosci. 2010;11(7):503–513. doi: 10.1038/nrn2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14(9):609–625. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borsook D, et al. Reward-aversion circuitry in analgesia and pain: Implications for psychiatric disorders. Eur J Pain. 2007;11(1):7–20. doi: 10.1016/j.ejpain.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Lammel S, et al. Input-specific control of reward and aversion in the ventral tegmental area. Nature. 2012;491(7423):212–217. doi: 10.1038/nature11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardon O, et al. Expression of mu opioid receptor in dorsal diencephalic conduction system: New insights for the medial habenula. Neuroscience. 2014;277:595–609. doi: 10.1016/j.neuroscience.2014.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reisert M, et al. Global fiber reconstruction becomes practical. Neuroimage. 2011;54(2):955–962. doi: 10.1016/j.neuroimage.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 34.Loseth GE, Ellingsen DM, Leknes S. State-dependent μ-opioid modulation of social motivation. Front Behav Neurosci. 2014;8:430. doi: 10.3389/fnbeh.2014.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navratilova E, et al. Endogenous opioid activity in the anterior cingulate cortex is required for relief of pain. J Neurosci. 2015;35(18):7264–7271. doi: 10.1523/JNEUROSCI.3862-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kieffer BL, Gavériaux-Ruff C. Exploring the opioid system by gene knockout. Prog Neurobiol. 2002;66(5):285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 37.Contet C, Kieffer BL, Befort K. Mu opioid receptor: A gateway to drug addiction. Curr Opin Neurobiol. 2004;14(3):370–378. doi: 10.1016/j.conb.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Moles A, Kieffer BL, D'Amato FR. Deficit in attachment behavior in mice lacking the mu-opioid receptor gene. Science. 2004;304(5679):1983–1986. doi: 10.1126/science.1095943. [DOI] [PubMed] [Google Scholar]
- 39.Becker JA, et al. Autistic-like syndrome in mu opioid receptor null mice is relieved by facilitated mGluR4 activity. Neuropsychopharmacology. 2014;39(9):2049–2060. doi: 10.1038/npp.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navratilova E, Atcherley CW, Porreca F. Brain circuits encoding reward from pain relief. Trends Neurosci. 2015;38(11):741–750. doi: 10.1016/j.tins.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elman I, Borsook D. Common brain mechanisms of chronic pain and addiction. Neuron. 2016;89(1):11–36. doi: 10.1016/j.neuron.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 42.Fields HL. Neuroscience. More pain; less gain. Science. 2014;345(6196):513–514. doi: 10.1126/science.1258477. [DOI] [PubMed] [Google Scholar]
- 43.Hsu DT, et al. It still hurts: Altered endogenous opioid activity in the brain during social rejection and acceptance in major depressive disorder. Mol Psychiatry. 2015;20(2):193–200. doi: 10.1038/mp.2014.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhan Y, et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci. 2014;17(3):400–406. doi: 10.1038/nn.3641. [DOI] [PubMed] [Google Scholar]
- 45.Weber R, Ramos-Cabrer P, Wiedermann D, van Camp N, Hoehn M. A fully noninvasive and robust experimental protocol for longitudinal fMRI studies in the rat. Neuroimage. 2006;29(4):1303–1310. doi: 10.1016/j.neuroimage.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 46.Pawela CP, et al. A protocol for use of medetomidine anesthesia in rats for extended studies using task-induced BOLD contrast and resting-state functional connectivity. Neuroimage. 2009;46(4):1137–1147. doi: 10.1016/j.neuroimage.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nasrallah FA, Tay HC, Chuang KH. Detection of functional connectivity in the resting mouse brain. Neuroimage. 2014;86:417–424. doi: 10.1016/j.neuroimage.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 48.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14(3):140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Himberg J, Hyvärinen A, Esposito F. Validating the independent components of neuroimaging time series via clustering and visualization. Neuroimage. 2004;22(3):1214–1222. doi: 10.1016/j.neuroimage.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 50.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 51.van de Ven VG, Formisano E, Prvulovic D, Roeder CH, Linden DE. Functional connectivity as revealed by spatial independent component analysis of fMRI measurements during rest. Hum Brain Mapp. 2004;22(3):165–178. doi: 10.1002/hbm.20022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Margulies DS, et al. Resting developments: A review of fMRI post-processing methodologies for spontaneous brain activity. MAGMA. 2010;23(5–6):289–307. doi: 10.1007/s10334-010-0228-5. [DOI] [PubMed] [Google Scholar]
- 53.Esposito F, et al. Real-time independent component analysis of fMRI time-series. Neuroimage. 2003;20(4):2209–2224. doi: 10.1016/j.neuroimage.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 54.Moritz CH, Carew JD, McMillan AB, Meyerand ME. Independent component analysis applied to self-paced functional MR imaging paradigms. Neuroimage. 2005;25(1):181–192. doi: 10.1016/j.neuroimage.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 55.Li YO, Adali T, Calhoun VD. Estimating the number of independent components for functional magnetic resonance imaging data. Hum Brain Mapp. 2007;28(11):1251–1266. doi: 10.1002/hbm.20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith SM, et al. Network modelling methods for FMRI. Neuroimage. 2011;54(2):875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- 57.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393(6684):440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 58.Humphries MD, Gurney K, Prescott TJ. The brainstem reticular formation is a small-world, not scale-free, network. Proc Biol Sci. 2006;273(1585):503–511. doi: 10.1098/rspb.2005.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103(23):8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage. 2010;52(3):1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.