Significance
Because of its practical importance and scientific significance, heterosis (hybrid vigor) is an interesting topic for both breeders and biologists. However, although heterosis has been applied successfully to increase crop yields, the molecular mechanisms involved remain obscure. In this study, using an integrative approach, we found that multiple quantitative trait loci (QTLs) cumulatively drive yield heterosis in hybrid rice by regulating two grain-yield component traits in which the RH8 (rice heterosis 8) gene plays a major role. Our research highlights the importance of integrative methods to uncover the molecular mechanism of heterosis and thus pave a way toward revealing the molecular mechanisms in rice heterosis in detail.
Keywords: hybrid rice, heterosis, yield, QTL, RH8
Abstract
Hybrid rice is the dominant form of rice planted in China, and its use has extended worldwide since the 1970s. It offers great yield advantages and has contributed greatly to the world’s food security. However, the molecular mechanisms underlying heterosis have remained a mystery. In this study we integrated genetics and omics analyses to determine the candidate genes for yield heterosis in a model two-line rice hybrid system, Liang-you-pei 9 (LYP9) and its parents. Phenomics study revealed that the better parent heterosis (BPH) of yield in hybrid is not ascribed to BPH of all the yield components but is specific to the BPH of spikelet number per panicle (SPP) and paternal parent heterosis (PPH) of effective panicle number (EPN). Genetic analyses then identified multiple quantitative trait loci (QTLs) for these two components. Moreover, a number of differentially expressed genes and alleles in the hybrid were mapped by transcriptome profiling to the QTL regions as possible candidate genes. In parallel, a major QTL for yield heterosis, rice heterosis 8 (RH8), was found to be the DTH8/Ghd8/LHD1 gene. Based on the shared allelic heterozygosity of RH8 in many hybrid rice cultivars, a common mechanism for yield heterosis in the present commercial hybrid rice is proposed.
Hybrids often present phenotypes that surpass their parents in terms of growth and fertility, a phenomenon known as “hybrid vigor” or “heterosis,” which was first described by Charles Darwin in 1876 (1) and was rediscovered by George H. Shull 32 y later (2). Since then, because of its practical importance and scientific significance, heterosis has become a primary interest for both breeders and biologists. Beginning with the breeding of hybrid maize in the 1930s, and later continued by commercialization of hybrid rice in the 1970s, crop heterosis has been applied extensively to the agricultural production of several species, offering significant yield advantages over the respective traditional inbred lines worldwide (3).
However, despite the successful agronomic exploitation of yield heterosis in crop production, progress in uncovering the molecular mechanisms underlying crop heterosis has lagged, although three main competing but nonmutually exclusive hypotheses—dominance (4, 5), over-dominance (6, 7), and epistasis (8, 9)—have been proposed to explain heterosis at the genetic level. Recently, Birchler et al. (10) suggested that additive partial dominance and over-dominance might be different points on a continuum of dosage effects of alleles. Thus far, by quantitative trait locus (QTL) mapping and genome-wide association studies (GWAS), a number of Mendelian factors, some of which are dominant and others that function in an over-dominant or an epistatic manner, have been identified in hybrid maize, rice, and other crops (11–15), Nevertheless, only a limited number of these factors have been characterized systematically for their involvement in yield heterosis (16). On the other hand, high-throughput gene-expression profiling in heterotic cross combinations has been carried out in both maize and rice, and a large number of genes have been found to be differentially expressed in the hybrids and their parents (17); some of these genes were reported to display nonadditive expression, in support of over-dominance, but others were mainly additive, supporting the dominance hypothesis. In addition, allelic variation in gene expression in hybrids has been reported in maize and rice (18). However, very few of the data obtained from high-throughput transcriptome analysis have been integrated with phenotypic profiles or mapped to single genetic loci associated with yield heterosis. Thus, a causative link between heterotic phenotypes and the underlying molecular events has yet to be established in grain crops. Therefore describing heterosis in a predictable manner is a major challenge, mainly because the genetic and molecular parameters of heterosis are far from elucidated (19).
In the model plant Arabidopsis thaliana, vegetative growth heterosis has been well studied. Hybrid vigor occurs in both inter- and intraspecies combinations, to different degrees depending on developmental stages, tissues, and cross combinations and even with maternal effects in some crosses (17, 20). Arabidopsis hybrid vigor can be observed in such varying traits as photosynthetic efficiency, seedling viability, seed number, phosphate efficiency, biomass, freezing tolerance, seed size, flowering time, metabolite contents, and leaf area (21, 22). Of note, genes involved in flowering time and circadian clock control were found to be linked to heterosis by mediating physiological and metabolic pathways (21–25). Recent studies further confirm the role of epigenetic regulation of transcription in hybrid vigor in Arabidopsis (25–27). Although Arabidopsis is not a staple crop, these advanced studies gave deep insight into the molecular mechanism of heterosis and highlighted the importance of integrating molecular approaches with phenomics methods for the study of crop heterosis.
To understand the mechanisms underlying heterosis in rice, we have undertaken sequential research steps to identify the genes responsible for yield heterosis in Liang-you-pei 9 (LYP9), a model two-line superhybrid rice from the cross of Peiai64S (PA64S) × 93-11 (28–30) that was ranked as the most widely planted hybrid rice cultivar in China from 2002–2007. We first sequenced the genome of the paternal variety 93-11 (31) and then profiled and compared the transcriptomes of LYP9 and its two parents at various life stages (32). Subsequently, we released the whole-genome sequence of the other parental line, PA64S (33), and developed a recombinant inbred line (RIL) population consisting of 219 RILs derived from LYP9 and a backcross population (RILBC1) derived from crossing each RIL with the female parent PA64S (34). In this study, we integrated phenome analyses, QTL mapping by genome resequencing, and transcriptome profiling to identify genes that drive yield heterosis in LYP9s.
Results
The Grain Yield Heterosis in LYP9 Is the Result of Hybrid Vigor in Only Two Yield-Component Traits, Spikelet Number per Panicle and Effective Panicle Number.
A two-line rice hybrid has a maternal parent with photo-thermogenic male sterility and a paternal parent that possesses fertility restoration capacity. The paternal line itself is usually an excellent inbred variety as well. To be commercially advantageous, a two-line hybrid should outperform its paternal parent with respect to agronomic traits, especially the traits related to grain yield. Thus, heterotic traits in the two-line rice hybrid are those that exceed either the better parent value (BPV) or the midparent value (MPV) on the basis of the good performance of the paternal parent value (PPV). Accordingly, heterosis in a two-line rice hybrid is actually referred as “heterobeltiosis” [i.e., better parent heterosis (BPH)], “middle parent heterosis” (MPH), or “paternal parent heterosis” (PPH).
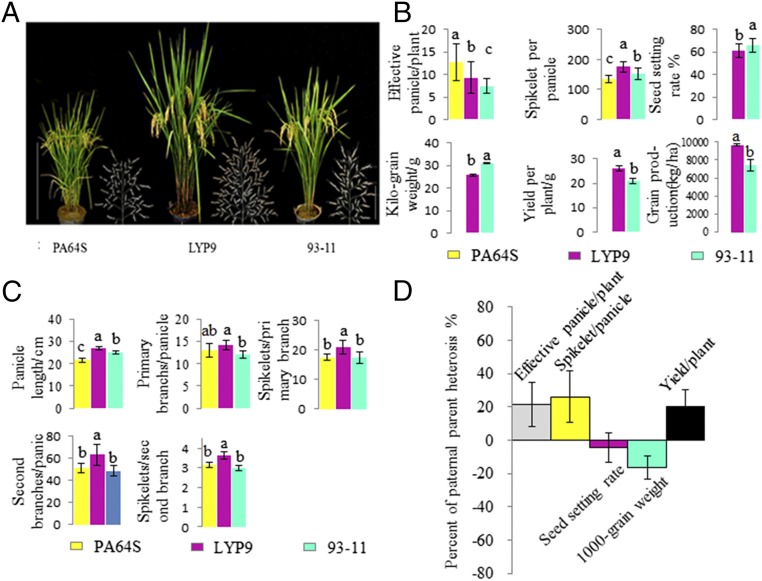
Normally, the grain yield of hybrid LYP9 in the field is about 9.5–11.0 metric tons/hectare (MTH), corresponding to an increase of 10.6–25.2% over the model three-line hybrid variety Shanyou 63 (SY63) (30, 35). However, the degree to which LYP9 outperforms its parents in yield-related traits had not been well defined before the current study. To determine the causal traits for yield heterosis, we carried out a phenomics investigation focusing on the performance of the yield and yield-component traits in LYP9 compared with its two parents during five consecutive years under standard agricultural conditions in Changsha, China (28°12′N, 112°58′E, long-day conditions) and in Sanya, China (18°15′N, 109°30′E, short-day conditions). The yield traits were evaluated in terms of field yield (FY) and yield per plant (YPP), and the yield-component traits included grains per panicle (GPP) [with its two components, spikelet numbers per panicle (SPP) and seed set rate (SRT)], effective panicle number (EPN), and 1,000-grain weight (KGW). For the maternal parent PA64S, no yield traits other than EPN and SPP were recorded because it cannot produce filled grains during the hybrid growing season. The data are collectively presented in Dataset S1, and, as an example, the results obtained in 2013 in Changsha, a typical growing region for two-line hybrid rice cultivars in China, are shown in Fig. 1. Under our field conditions, the FY of LYP9 could reach 9.8 MTH, a 13.8–37.9% increase over its male parent 93-11 (Fig. 1 A and B); the YPP for LYP9 was 31.3 g, 25.3% higher than for 93-11 (Fig. 1B). Thus, LYP9 shows obvious BPH and PPH for both field yield and grain yield per plant. However, when all the component traits of YPP were compared in the hybrid and the two parents, we found that no obvious BPH in the yield components except for GPP. Furthermore, when SPP and SRT, the two component traits of GPP, were investigated, a complicated scenario appeared. The SPP in LYP9 was 175.1, 23.8 more than in 93-11 and 41.1 more than in PA64S on average, representing an increase of 15.7% over the PPV and/or BPV (Fig. 1B). Moreover, the constituents of the SPP, including primary branch number, secondary branch number, and spikelet number in each branch, were all significantly higher in the hybrid than in either parent, with a heterosis level of 4.5, 1.6, 7.7, and 6.4%, respectively, over the BPV of PA64S and 13.3, 43.8, 18.7, and 5.9% over the PPV of 93-11 (Fig. 1C). However, the SRT of LYP9 fell below 65.9%, 14.1% lower than the PPV of 80.0%. Cumulatively, LYP9 developed 12.5% more filled grains per panicle than did 93-11, giving a PPH of 10.4% (Fig. 1B). Also, LYP9 had an EPN of 8.2 on average, lower than the 10.6 EPN of in PA64S but higher than the EPN of 7.30 in 93-11, which was 8.1% lower than the MPV but 12.6% higher than the PPV (Fig. 1B). Moreover, the KGW of LYP9 was 26.4 g, 5.2 g less than that of 93-11, representing a 16.5% reduction compared with the PPV (Fig. 1B). In addition, the agronomic traits that indirectly impact rice yield, such as plant height (PH), tiller number, biomass, heading date (HD), photosynthesis capacity in leaf, and vascular number in stem, were also analyzed dynamically at four agronomically important stages: in 20-d-old seedlings, at tillering, at heading, and at maturity. No significant differences were detected between LYP9 and 93-11 except for PH, tiller number, and consequently biomass in vegetative growth (SI Appendix, Fig. S1).
Fig. 1.
Phenotypes of the rice hybrid LYP9 and its parental inbred lines. (A) Whole-plant and panicle morphology of the two-line rice hybrid LYP9 and its parental lines Peiai64S (PA64S) and 93-11. The images were taken when the plants had reached maturity. (Scale bars: left, 50 cm, plant height; right, 10 cm, panicle length.) (B) Phenomics investigation into the yield performance and yield-component traits in LYP9 and its two parents. (C) Phenomics investigation into the performance of panicle-related traits in LYP9 and its two parents. B and C show examples of results obtained in 2013 under normal agricultural conditions in Changsha, China (28°12′N, 112°58′E, long-day conditions). Different letters above the bars represent significant differences (P < 0.05) by t test. (D) The percent of yield and yield-component PPH.
When grain yield and its component traits in the hybrid and its parents were compared year by year, the heterotic expressions varied significantly at different geographical locations (Dataset S1); however all heterotic traits other than the SRT could be stably detected across all 5 y (although at varied levels) in both Changsha and Sanya. The SRT for either LYP9 or 93-11 changed significantly with year and geographic location, so that the SRT of the hybrid could be higher, lower, or equal to that of 93-11, and in general no reliable PPH was observed for the SRT. When these observations are considered together, the yield heterosis of LYP9 is not reflected in all yield-related traits but only in two specific yield-related components: the outperformance of the SPP over the BPV and of the EPN over the PPV in the hybrid, which in most cases must offset the negative effects of the grain weight (GW) and the reduced SRT in the hybrid (Fig. 1D).
The Yield-Related Heterotic Traits of LYP9 Are Shared by Other Commercial Hybrid Rice Cultivars.
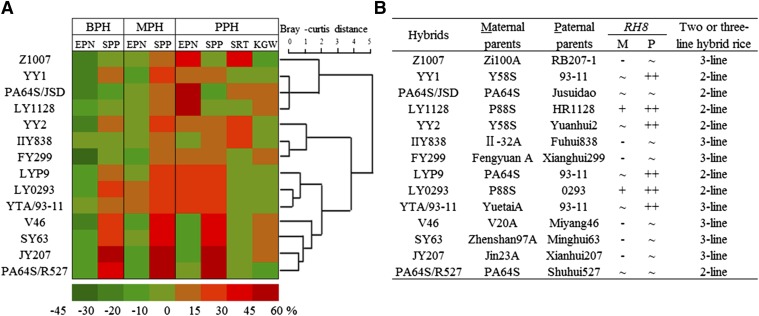
The yield traits and yield-related heterosis were surveyed further in 13 other representative commercial hybrid rice cultivars, including the most frequently planted superhybrids and the three-line hybrid rice SY63, to see whether our observations in LYP9 are common features of hybrid rice combinations (Fig. 2). We performed cluster analyses of the heterotic performances of the hybrids for each of the yield-related traits (Fig. 2A and Dataset S1). Significant yield outperformance of the hybrids over the PPV was observed in all combinations, with the level of the PPH ranging from 8.6–43.6%; in all instances the SPP was a major causal trait for yield heterosis. In all the combinations hybrids exhibited MPH for the SPP; nine of these combinations presented BPH, and all but three showed PPH. We found that the EPN also was a determinant yield component for yield heterosis in most combinations. Similar to our observations in LYP9, no hybrids developed more tillers than the BPV, and all but two developed fewer tillers than the MPV. However, PPH for the EPN was still detected in 9 of 13 combinations. In contrast, no consensus PPH was observed for KGW. Similarly, we found that the SRT was higher than, lower than, or similar to the PPV, depending on the combinations. Thus, our observations suggest that the cumulative outperformance of PPH in the SPP (ascribed to either BPH or MPH) and of the PPH in tiller number (ascribed to MPH) are conserved features in these commercial hybrid rice cultivars, whereas the effects of SRT and GW are case dependent.
Fig. 2.
Heterosis performance of LYP9 and 13 other commercial hybrids and RH8 genotype analysis. (A) Cluster analysis of LYP9 and 13 other rice hybrids based on yield-related traits. Cluster analysis was performed with the Vegan package in R using Bray–Curtis distance (https://cran.r-project.org/). Abbreviations are as defined in the text. (B) Genotypes of RH8 in 14 hybrid combinations. +, weak allele; ++, strong allele; - and ∼, null allele caused by either frameshift mutation or deletion of 1116 bp.
Construction of a High-Density SNP Marker Linkage Map and Detection of Yield-Related QTLs in the LYP9-Derived RIL Population.
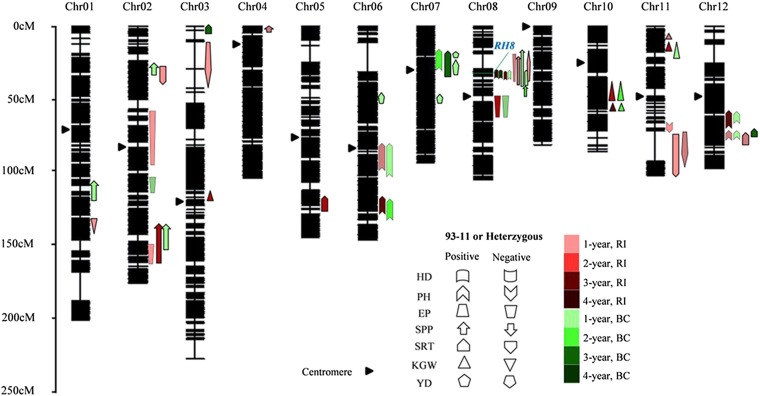
To identify the candidate genes for yield-component traits of the parents of LYP9, we used an LYP9-derived RIL population for QTL mapping. All the phenotypic values of the yield-component traits except for the HD obeyed a normal distribution, indicative of quantitative inheritance and suitable for QTL mapping (SI Appendix, Fig. S2). The genomes of 219 RIL lines were sequenced to an average approximate depth of 1.8-fold on an Illumina HiSEq 2500 instrument. Based on the sequence variations in 93-11 and PA64S and the variant sequences among the RILs, 780,717 loci were randomly selected for linkage analysis. The loci that cosegregated with one another were anchored into the same blocks, called “bins”; a total of 2,972 bins were used to construct the molecular linkage map using Highmaps software (SI Appendix, Fig. S3). The phenotypic datasets for the RILs collected for 4 y in Changsha were first used for QTL analysis. A total of 27 QTLs for all traits were mapped independently on rice chromosomes 1, 2, 4, 6, 8, 10, 11, and 12 (Fig. 3 and Dataset S2), 16 of which had been reported previously, with nine detected in another LYP9-derived RIL population (36) and six detected in a set of chromosome segment substitution lines carrying PA64S genomic segments in the 93-11 genetic background (37). Three of these QTLs on chromosomes 2, 4, and 8 were identified for SPP. The positive alleles of these QTLs were all from 93-11, and each explained less than 10% of the variance. Two QTLs were detected for EPN on chromosomes 2 and 8, with positive alleles originating from PA64S; these QTLs explained 7.0–12.8% of the phenotypic variance. We detected four QTLs for SRT and four for GW with minor effects. In addition, one QTL was mapped for HD, and six were mapped for PH. With regard to the QTL positions, two QTL clusters are highlighted: qSPP2/qEP2 and qHD8/qPH8/qSPP8/qYD8 are on chromosomes 2 and 8, respectively (Fig. 3), suggesting that the two groups of loci may be controlled either by one gene with pleiotropy or by a group of closely linked genes.
Fig. 3.
QTL analysis of heterosis in LYP9 based on the high-density bin maps derived from the RIL and RILBC populations. At left is the scale for the genetic length of each chromosome. The upward direction indicates that the 93-11 parental allele for each locus increases the phenotype in the RIL population or that the heterozygous genotype increases the phenotypes in the backcrossed (BC) population. The downward direction indicates that the 93-11 allele for each locus decreases the phenotype in the RIL population or that the heterozygous genotype decreases the phenotypes in the BC population. The red shapes indicate QTLs detected in the RIL population; green shapes represent QTLs in the BC population. The intensity of the colors indicates QTLs detected for 1 to 4 y.
Mapping of QTLs for Yield-Related Heterotic Traits in the RILBC1 Population.
We then used the RILBC1 population (34) to detect QTLs for yield-component heterosis that are associated with the effects of the heterozygous genotype of 93-11 on the PA64S genetic background. Twenty-five loci for the respective phenotypes were detected on chromosomes 1, 2, 3, 6, 7, 8, 10, 11, and 12 in the 4-y results (Fig. 3 and Dataset S2). Twelve of these loci overlapped with QTLs detected in the RIL population, and 13 were detected independently in the RILBC1 population only, presenting a mode of dominance or superdominance in the heterozygote. A set of five QTLs for SPP were detected: three superdominant QTLs were mapped to chromosomes 1, 2, and 8, one partially dominant QTL was mapped to chromosome 2, and another displaying complete dominance was mapped to chromosome 8. The percent of variance explained by these loci ranged from 6.3–13.7%. These five loci function together to drive the BPH of LYP9 for the SPP. For the EPN, we detected a completely dominant QTL on chromosome 2 and a partially dominant locus on chromosome 8, both of which were from PA64S; each explained less than 8% of the variance. This result is consistent with our observation of PPH in LYP9 for the EPN. In addition, four loci for the SRT and three for KGW were also detected, explaining 6.2–17.2% of the phenotypic variance. QTLs for HD and PH in the hybrids were also analyzed. Only one QTL with partial dominance for HD on chromosome 8, named qhHD8, was identified in the RILBC1 population. As a major QTL, qhHD8 explained more than 40% of the variance and had a genetic effect of 5.7 d on average. For PH, six QTLs with incomplete dominance, over-dominance, or superdominance were detected. A major QTL was detected on chromosome 8 that displayed over-dominance and explained 28.4% of the variance, thus explaining why LYP9 had BPH for plant height (SI Appendix, Fig. S1).
Notably, only one QTL cluster, qhHD8/qhPH8/qhSPP8/qhEP8, was found in the RILBC1 population, on chromosome 8. This cluster overlaps with the QTL cluster qHD8/qPH8/qSPP8 that was detected in the RIL population (Fig. 3). Because we later identified one gene with pleiotropic function in this cluster (see below), we tentatively named this QTL cluster rice heterosis 8 (RH8). It can explain >40% of the variance for HD in each population, and it overlaps with QTLs for SPP and PH. Thus, we regard this QTL cluster as the major effective locus contributing to yield heterosis in LYP9.
Transcription Profiling Reveals Significant Differences in Gene Expression Between LYP9 and Its Parents.
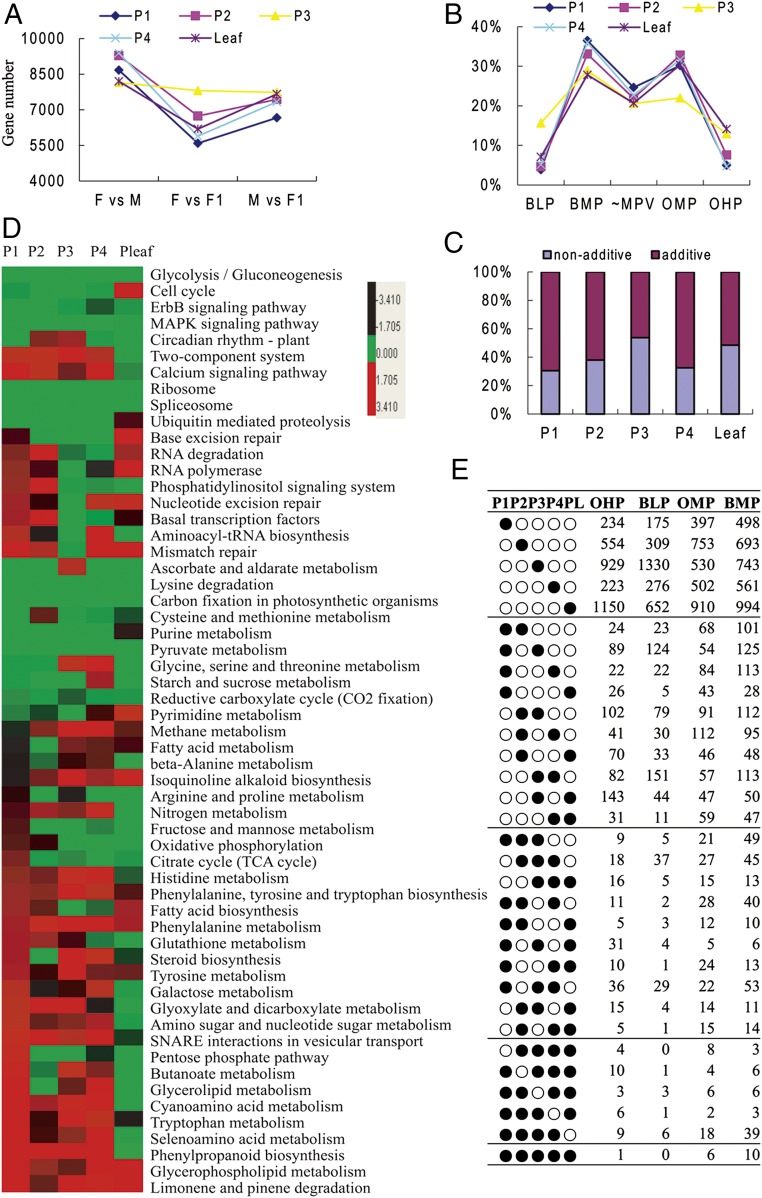
To explore further the possible mechanisms and genes involved in the yield heterosis performance of LYP9, we profiled the transcriptomes of young inflorescence buds in the hybrid and its parents at the four successive early developmental stages at which the spikelet numbers were determined (SI Appendix, Fig. S4) (38) and in the 6-wk-old leaf blades at the key stage for the rice reproductive growth transition (39) by genome-wide transcriptional profiling using the SOLiD next-generation sequencing system. In total, 21 different poly-A–enriched mRNA-sequencing libraries, including three biological replicates for the inflorescence buds (3–4 mm in length), were constructed and sequenced (SI Appendix, Fig. S5 and Dataset S3). We identified 15,843 differentially expressed genes (DEGs) (P < 0.05) out of 17,993 genes expressed in the panicle and 10,821 DEGs out of 15,145 genes expressed in the leaf between any two cultivars (Fig. 4A and SI Appendix, Table S1). Validating the RNA-sequencing data for dozens of genes by quantitative RT-PCR supported the reliability of our transcriptome data (SI Appendix, Fig. S6). Genes showing differential expression between the hybrid and its parental lines were classified into five major expression patterns based on gene-expression level: over higher parent (OHP), below lower parent (BLP), over midparent (OMP), below midparent (BMP), and similar to midparents (MPV), with the BMP and OMP patterns being the highest, ranging from 27–35% for each stage, and the OHP and BLP patterns being the lowest, ranging from 3.8–14% (Fig. 4B and SI Appendix, Table S2). We further identified a stack of nonadditive expressed genes (3,224–6,266) that differ significantly from the MPV (Fig. 4C). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for the nonadditively expressed genes in each stage revealed that they are involved in a variety of biological pathways (Fig. 4D). Most of the enriched pathways are for translation, transcription, circadian rhythm, carbon fixation, and starch and sucrose metabolism. Consistent with the reports that the circadian rhythm regulation and flowering time pathways are involved in Arabidopsis vegetative heterosis (23), some genes involved in circadian rhythm regulation and flowering time pathways were expressed at higher levels in the hybrid than in either parental line at one or more developmental stages (Fig. 4E and Dataset S4). Although we harvested the samples for transcriptome profiling only in the morning (SI Appendix, SI Materials and Methods), and thus the evening-expressed circadian genes were missing in our current data, 16 known flowering-related genes, 34 circadian-associated factors, and 11 panicle-branching regulators were found to be nonadditively expressed in one or all stages of panicle and leaf development (Fig. 4E and Dataset S4). This list includes genes related to circadian rhythm and flowering time, such as OsPHYA, B, and C, and OsPRR73/37/95/59, OsCCA1, OsLHY, OsGI, and genes related to panicle branching and panicle development such as FZP, Cga1, APO2, LAX1, TWA1, and DEP1/2/3.
Fig. 4.
Expression patterns of DEGs. (A) The number of DEGs between any two cultivars in each sequenced sample. F, paternal line 93-11 (father); M, maternal line PA64S (mother); F1, F1 hybrid LYP9. P1, P2, P3, and P4 indicate young inflorescences collected in the morning from panicles with lengths of <1 mm (P1), 1–2 mm (P2), 2–3 mm (P3), and 3–4 mm (P4). (B) The five major expression patterns of DEGs in the F1 hybrids and their parental lines, based on reads per kilobase of transcript per million reads mapped (RPKM) values. (C) The percentage distribution of nonadditive and additive DEGs at each developmental stage. (D) Enriched KEGG pathways for nonadditive DEGs. (E) The number of nonadditive DEGs for four major expression patterns (OHP, BLP, OMP, BMP) in those sequenced developmental periods showed as filled circles.
In addition, based on the genomic sequence SNPs discovered by resequencing the PA64S and 93-11 genomes and the expressed SNPs (eSNPs) found by surveying the transcriptome sequences (see details in Materials and Methods), we constructed a heterozygosity map of LYP9 (SI Appendix, Fig. S7 A and B) to identify allelic DEGs in the hybrid. Of the total 3,767–5,770 DEGs with eSNPs (Materials and Methods), 0.26–0.45% of the DEGs showed monoallelic expression (MAE) i.e., expressing only one maternal or paternal allele in a parent-dependent fashion, 11.63–13.65% of the DEGs showed preferential allelic expression (PAE), i.e., more than a twofold difference between the two expressed alleles, and 85.91–88.11% of the DEGs showed biallelic expression (BAE).
Several Differentially Expressed Genes Are Located in the Yield-Related QTL Regions.
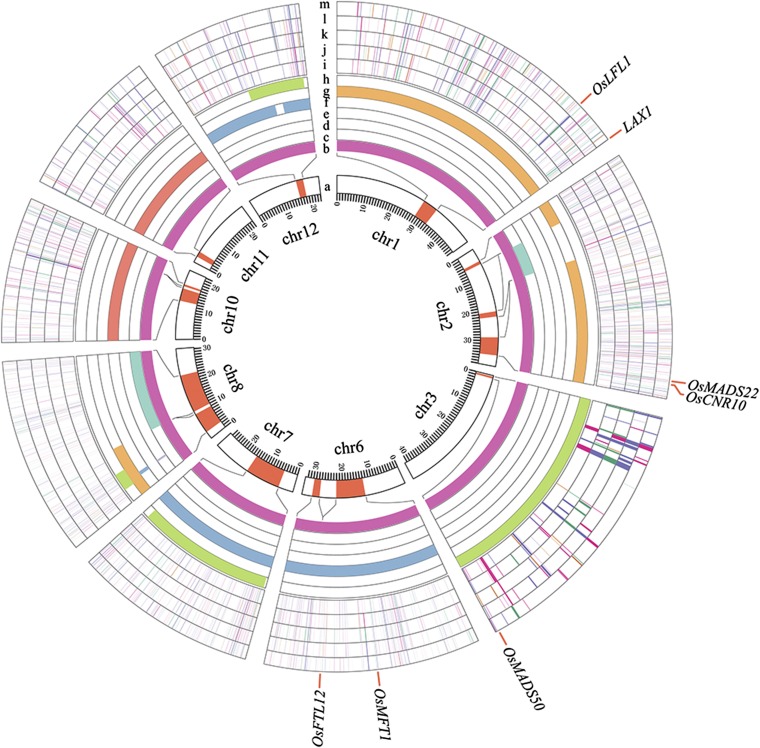
To integrate the data from transcriptome profiling and QTL mapping, we mapped the DEGs onto the QTL regions (Fig. 5). In the five QTL regions for SPP, 673 genes were differentially expressed in LYP9 and its parents at the young inflorescence bud stages (Dataset S5). Of these, 17 overlapped with the QTL peak signals (SI Appendix, Table S3). Interestingly, we found one known flowering-related gene, OsMADS22 (LOC_Os02g52340, spikelet meristem indeterminacy) (40), and one known circadian rhythm-related gene, OsLFL1 (LOC_Os01g51610), which are known to be involved in regulating flowering time and panicle development (41, 42), located in the overlapping regions of qhSPP2.2 and qhSPP1 and expressed in the BMP and OHP modes, respectively. In addition, there are 236 DEGs in the QTL loci for EPN, 4 in the QTL loci for HD, 349 in the QTL loci for SRT, 199 in the QTL loci for KGW, and 521 in the QTL loci for PH (Fig. 5). The DEGs located in the identified heterosis-related QTL regions, especially those in the QTL peak regions, could provide clues about the genes responsible for yield-related trait heterosis in the hybrid.
Fig. 5.
Distribution of mapped heterosis QTLs and DEGs in QTL regions. a and b: Mapped QTLs for related phenotypes shown in red blocks (a) and in purple bars (b) at amplified modes. c–h: QTL blocks for EPN (c, cyan), HD (d, yellow), KGW (e, red), PH (f, blue), SPP (g, orange), and SRT (h, greeb) phenotypes. i–m: DEGs located in the QTL regions in panicles with lengths of 0–1 mm (i, P1), 1–2 mm (j, P2), 2–3 mm (k, P3), and 3–4 mm (l, P4) and leaves (m). The names of some known flowering genes are indicated on the outside of the circle.
We next investigated the allelic DEGs in the QTL regions of the hybrid. A total of 868 DEGs with eSNPs were found to be located in the yield-related QTL regions, 649 of which showed MAE or PAE patterns, suggesting their possible relationships to yield-related component heterosis. For example, a gene putatively regulating cell number, OsCNR10 (LOC_Os02g52550) (43), located in the qEP2.3-, qSPP2.2-, and qhSPP2.2-overlapping regions, was expressed nonadditively in the LYP9 hybrid, with the expression of the paternal allele 2.3 times that of the maternal allele. Similarly, for LOC_Os06g29800, a member of the pentatricopeptide repeat (PPR) family located in the qhPH6.1 region, the paternal allele is expressed preferentially in LYP9 at about twofold the frequency of the maternal allele. For LOC_Os10g33620, a ubiquitin protein located in the qKGW10 regions, the maternal allele is dominantly expressed in LYP9 (SI Appendix, Fig. S7C). These genes that show preferential allelic expression in QTL regions might play roles in yield-related trait heterosis.
The Heterotic Locus for RH8 Is the Known Gene DTH8/Ghd8/LHD1.
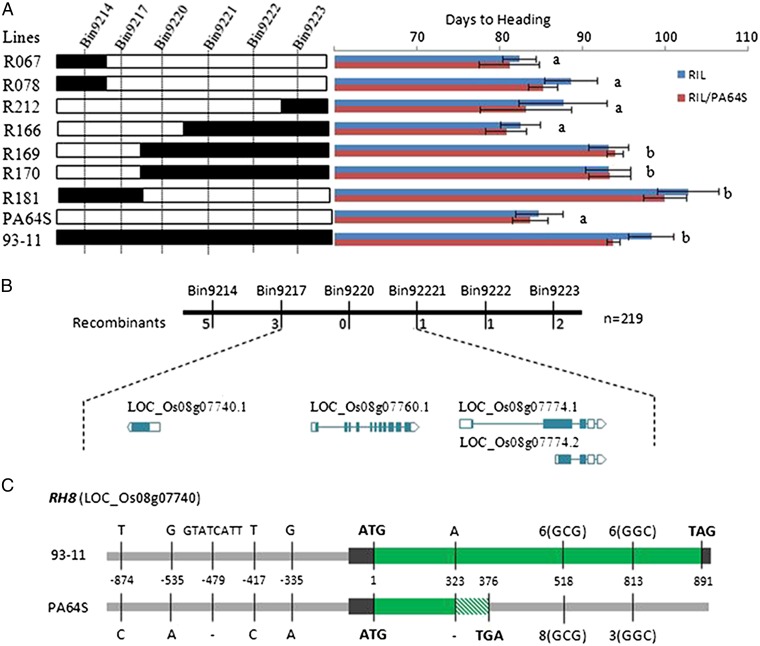
The RH8 QTL cluster on chromosome 8 explained >40% of the variance for HD, 28.7% for PH, and 6.3% for SPP in the RILBC1 population and also showed strong effects in the RIL population. To identify further the exact gene(s) responsible for these QTLs, HD was chosen as the first trait for fine mapping, and it was mapped to a 31.8-kb interval in bin 9,220 (Fig. 6A; also see SI Appendix, SI Materials and Methods). There are three annotated genes, LOC_Os08g07740 (DTH8/Ghd8/LHD1), LOC_Os08g07760 (BRI1-associated receptor kinase), and LOC_Os08g07774 (disease resistance protein RPM1) in this bin (Fig. 6 B and C). Notably, one of these genes, DTH8/Ghd8/LHD1, has been shown previously to be a pleiotropic major QTL responsible for HD, PH, and grain yield (44–46) and also for tiller number in a genetic background-dependent manner (44). Moreover, this gene has been confirmed to be qSN8 and qHD8, which are responsible for HD and SPP, respectively, in a previously reported population consisting of 132 LYP9-derived RILs (36). In that study, the 93-11 allele was shown to be able to complement the PA64S allele in HD, PH, and grain yield simultaneously. Of note, in the RILBC1 population the DTH8/Ghd8/LHD193-11 lines had significantly higher SPP (∼18.2%) and were later maturing (∼14.6%) and taller (∼14.8%) than the DTH8/Ghd8/LHD1PA64S lines (SI Appendix, Fig. S8). Thus, we can conclude that DTH8/Ghd8/LHD1 is the responsible gene in the RH8 QTL cluster.
Fig. 6.
Fine mapping and sequence comparison of RH8 in LYP9 and its parental lines. (A) Fine mapping the RH8 locus using the heading date as the first trait. The letters “a” and “b” at the right of bars represent significant differences (P < 0.05) as determined by Student’s t test. RH8 was mapped to a 31.8-kb interval in bin 9,220. (B) Three annotated genes located within the interval (rice.plantbiology.msu.edu/). (C) Sequence comparison of RH8/DTH8 (LOC_Os08g07740) in 93-11 and PA64S. Four SNPs (T–873→C–873, G–537→A–537, T–418→C–418, and G–333→A–333), an 8-bp indel (TATCATTG) in the promotor region and two deletions (+322 A+323 and +813GGCGGCGGC+822) and one insertion (+519GGC+522) in the coding region were detected. The 1-bp deletion (+322A+323) is predicted to result in a frameshift mutation that would lead to a premature termination codon in PA64S.
We then sequenced the promoter region and the coding region of RH8 in 93-11 and PA64S and confirmed the presence of a previously reported deletion of 8 bp in the promoter region of the PA64S allele (36). In addition, we found four SNPs (T–873→C–873, G–537→A–537, T–418→C–418, and G–333→A–333) in the promotor region and two deletions (+322A+323, +813GGCGGCGGC+822) and one insertion (+519GGC+522) in the coding region (Fig. 6C). Notably, the 1-bp deletion (+322A+323) results in a frameshift mutation that leads to a premature termination codon (Fig. 6C). Thus the RH8PA64S allele is nonfunctional. As a result, the genetic effect of RH8 in LYP9 is caused mainly by allelic heterozygosity of one functional allele and one nonfunctional allele in the hybrid.
The Allelic Heterozygosity of RH8 is a Significant Feature in Commercial Two-Line Hybrid Rice Cultivars.
We surveyed the allelic variation of RH8 in other commercial hybrid cultivars in which the heterosis for SPP and EPN were ubiquitously expressed (Fig. 2A). As shown in Fig. 2B, RH8 was heterozygous in five of the two-line hybrids, each with a paternal functional allele identical to RH893-11 and a maternal nonfunctional or weakly functional allele identical to RH8Nipponbare (47). In the other eight hybrids, which included two two-line and six three-line combinations, both parents contributed nonfunctional alleles. It seems that the LYP9-like heterozygosity of RH8 is a notable feature shared by most two-line rice hybrids.
We next analyzed the allelic combinations of RH8 in a set of 361 commercial hybrid rice populations comprising 125 two-line hybrids and 236 three-line hybrids (SI Appendix, Table S4). RH8 heterozygosity, consisting of a strong allele and a nonfunctional allele, was found in 89 hybrids, of which 51 were two-line hybrids and 38 were three-line hybrids, accounting for 40.8% and 16.1% of the two- and three-line hybrids, respectively. Thus we can postulate that heterozygosity at the RH8 locus could be a significant feature, but not the only one, in commercial hybrid rice cultivars and particularly in the two-line hybrids.
Discussion
Hybrid vigor resulting from crossing rice varieties was first described in 1926 by Jones (48) in the United States, and the first commercial hybrid rice variety was bred in 1973 by Yuan (49) in China. Currently, hybrid rice dominates rice production in China and also has taken root worldwide. A large number of elite commercial combinations have been developed, with yield increases ≥20% compared with their inbred counterparts (50). The most popular hybrid combinations are SY63 and LYP9, both of which have been adopted as model systems for studying the molecular mechanism of heterosis for three-line hybrids and two-line hybrids, respectively (51–53). Despite the great achievements made in rice breeding programs, our understanding of the molecular mechanism of rice heterosis is still in its infancy. Here we integrated phenomics analysis with genome resequencing, transcriptional profiling, and QTL mapping to identify the genes (QTLs) responsible for yield-related trait components.
In the current study, yield heterosis in LYP9 and the other investigated rice hybrids turned out to be a complicated quantitative- and component trait-specific phenotype. The expression of yield-related heterosis in rice is the accumulative output of many phenotypic components (Fig. 1C). Moreover, most of the yield trait components are environmentally sensitive (Dataset S1). Here we clearly demonstrated that the yield heterosis of LYP9 over its paternal parent is a complex trait contributed mainly by the outperformance of two yield components, SPP and EPN, over the BPV the PPV, respectively, but compromised in most cases by the negative effects of two other component traits, GW and SRT (Fig. 1D). We then explored the possible genes underlying the two yield heterosis-related component traits by an integrative genetic approach, because neither transcriptome profiling based on expressed sequence differences nor genome mapping based on genomic sequence variations alone can identify all heterosis-related genes. Finally, with all the data from phenome, genome, transcriptome, and genetic analyses, we were able to identify a number of candidate genes for the traits related to yield heterosis. Thus, by using systems genetics, our study has paved the way toward a greater understanding of the molecular mechanisms of hybrid vigor in rice and other grain crops.
By QTL mapping in the LYP9-derived RIL and RILBC1 populations, we identified a stack of QTLs that are responsible for the yield or yield-related heterosis (Fig. 3 and Dataset S3); some of these had already been detected in other LYP9-derived populations (36, 37). Five QTLs were detected for the BPH heterosis of SPP with the positive alleles from either parent, whereas for EPN two QTLs were detected with the positive alleles only from PA64S, suggesting that both parents contribute to the yield heterosis and do so mainly in a dominant way. DHT8/Ghd8/LHD1, as a member of the group of circadian genes controlling flowering time, was confirmed here as the major heterosis-related QTL, RH8, for SPP and also for HD and PH because of its significant phenotypic effects in the two mapping populations (Fig. 6). Other detected QTLs also showed minor effects on SPP or EPN and were difficult to fine map. Nevertheless, our transcriptome data revealed some of the nonadditive DEGs (Fig. 4 and SI Appendix, Table S2) and DEGs with allelic expression variations (Fig. 4 and Dataset S4) enriched in the QTL regions that might be candidates for yield heterosis genes. For example, it is tempting to test the hypothesis that a gene controlling flowering time, such as OsLFL1, which is located in the region of qhSPP1 and has a known function in determining spikelet number, is involved in yield heterosis (42).
Our results suggest that hybrid rice cultivars share some common yield-related heterotic component traits to some extent. As described above, although hybrid advantages may differ in various traits at different developmental stages, depending on the parental combinations, MPH for SPP and PPH for EPN were observed in most of the hybrid combinations investigated (Fig. 2B). At the gene level, we found that the heterotic gene RH8/DTH8/Ghd8/LHD1 is one of the loci contributing to yield heterosis in hybrid rice cultivars. (Fig. 2). Of note, the heterozygote of RH8 was detected initially in five of seven two-line hybrid combinations that showed a yield heterotic phenotype pattern similar to that of LYP9 and then was detected in 51 of 125 two-hybrid commercial hybrid combinations that we later investigated (SI Appendix, Table S4). In parallel with our observation, DTH8 also was found to be one of a few genes with the largest effects on grain yield, which showed the highest correlation with panicle number and grain number in a GWAS survey of 1,495 hybrid rice combinations (15). The DTH8/Ghd8/LHD1 gene is mainly responsible for HD but also for grain yield in a genetic background-dependent manner. Previous studies have shown that the DTH8/Ghd8/LHD93-11 allele could increase grain number in the two male sterile lines, Zhengshan (a maternal parent for three-line hybrid rice) and PA64S, possibly by up-regulating MOC1 (44), but not in the inbred cultivar Teqing (36, 44–46). Thus, RH8 actually may be nonfunctional in some of the rice hybrids, and the RH8 heterozygosity would not be present in these hybrids.
Interestingly, Ghd7, another flowering-time gene functioning parallel to DTH8, has been found to contribute significantly to high yield heterosis in the model three-line hybrid SY63 (54). In our phenotypically characterized hybrid combinations, we also found heterozygosity consisting of a strong allele and a weak or nonfunctional allele at the Ghd7 locus (SI Appendix, Table S5). It has been reported that Ghd7, Ghd8, and Hd1 all belong to the class of flowering-time genes and define yield potential and eco-geographical adaptation in cultivated rice varieties (47). Thus, it can be postulated that RH8, Ghd7, and perhaps other flowering-time genes may independently or cooperatively underlie yield heterosis in most, if not all, hybrid rice cultivars and make them suitable for cultivation mainly in the subtropical region where hybrid rice originated. Thus, our study may have revealed a common genetic mechanism for the present commercial hybrid rice.
Our study also shed some light on future directions in hybrid rice breeding. The hybrid rice has reached a yield ceiling, and ways to improve it further are needed. Our phenotypic data showed that in every instance the GW, and in some environments the SRT, reduced the yields in LYP9 and in most other hybrid rice varieties tested. Developing hybrids with a stably increased SRT is highly desirable (55). Because the SRT is environmentally sensitive, the identification of environmentally insensitive QTLs and their integration into hybrids to mitigate the unstable status of expression of SRT will be greatly beneficial. In contrast, KGW performance in hybrid rice is very stable, and the related QTLs function mainly in an additive manner, as shown in our study and by others (15, 36). Thus, improved seed yield will benefit from pyramiding more QTLs from both parents, especially those that do not have a negative effect on other traits (56–58). Genetic analysis revealed that most QTLs detected in this study showed incomplete dominance and dominance effects, implying that dominance plays a leading role in driving yield heterosis and that it is possible to generate higher-yield hybrid rice by pyramiding more superior alleles. So far, most of the paternal parents already are excellent, with inbred varieties reaching a yield plateau, whereas the yield-related performance of maternal parents has been less focused. Thus, developing maternal parent lines with superior QTLs will complement the excellent paternal parent breeding for further improvement of hybrid rice. Yuan (59) has proposed an ideal of “new type morphology,” the main concept of which is to increase PH and subsequently plant biomass to raise the rice yield ceiling further and to break through the yield limitation imposed by harvest index. In this study, the heterosis of PH and biomass was observed in the vegetative growth in LYP9 and other combinations, although it is not very significant. Notably, both RH8/DTH8/Ghd8/LHD1 and Ghd7 are photoperiod-sensitive genes in rice responsible for PH and grain yield (44). Because the heterosis of PH and biomass underlies the yield heterosis indirectly, the new type morphology of hybrid rice also may be realized by manipulating some other photoperiod genes, as has been demonstrated in the model plant Arabidopsis (23).
Materials and Methods
Materials and methods are described in SI Appendix, Materials and Methods. The original sequencing datasets have been deposited in the Genome Sequence Archive of Beijing Institute of Genomics, Chinese Academy of Sciences (gsa.big.ac.cn) under accession no PRJCA000131. Phenotyping data are available in Dataset S1.
Supplementary Material
Acknowledgments
We thank Dr. Zhikang Li and Dr. Pedro Rocha for their critical reading of and helpful comments on the manuscript. This work was supported by State Key Laboratory of Plant Genomics, China, Grant 2015B0129-03 (to L.Z.); National Natural Science Foundation of China Grants 31123007 (to L.Z.), 31270426 (to C.C.), and 30900831 and 31271372 (to S.S.); National Key Research and Development Program of China Grant 2016YFD0100904 (to X. C. and D.L.); Beijing Nova Program Grant Z121105002512060 (to S.S.); National Key Basic Research and Development (973) Program of China Grant 2006CB101700 (to Y.X.); Doctor of Science Research Foundation of the Ministry of Education Grant 20060533064 (to Y.X.); and National Key Technology Support Program Grant 2012BAC01B07 (to Y.X.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence datasets reported in this paper have been deposited in the Genome Sequence Archive of Beijing Institute of Genomics, Chinese Academy of Sciences, gsa.big.ac.cn (accession no. PRJCA000131).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610115113/-/DCSupplemental.
References
- 1.Darwin C. The Effects of Cross and Self Fertilisation in the Vegetable Kingdom. Murray; London: 1876. [Google Scholar]
- 2.Shull GH. The composition of a field of maize. Journal of Heredity. 1908;4(1):296–301. [Google Scholar]
- 3.Xu Y. Developing marker-assisted selection strategies for breeding hybrid rice. Plant Breed Rev. 2003;23:73–174. [Google Scholar]
- 4.Bruce AB. The Mendelian theory of heredity and the augmentation of vigor. Science. 1910;32(827):627–628. doi: 10.1126/science.32.827.627-a. [DOI] [PubMed] [Google Scholar]
- 5.Jones DF. Dominance of linked factors as a means of accounting for heterosis. Genetics. 1917;2(5):466–479. doi: 10.1093/genetics/2.5.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shull GH. The genotypes of maize. Am Nat. 1911;45(4):234–252. [Google Scholar]
- 7.East EM. Heterosis. Genetics. 1936;21(4):375–397. doi: 10.1093/genetics/21.4.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minvielle F. Dominance is not necessary for heterosis - a 2-locus model. Genet Res. 1987;49(3):245–247. [Google Scholar]
- 9.Schnell FW, Cockerham CC. Multiplicative vs. arbitrary gene action in heterosis. Genetics. 1992;131(2):461–469. doi: 10.1093/genetics/131.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birchler JA, Johnson AF, Veitia RA. Kinetics genetics: Incorporating the concept of genomic balance into an understanding of quantitative traits. Plant Sci. 2016;245(2):128–134. doi: 10.1016/j.plantsci.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Xiao J, Li J, Yuan L, Tanksley SD. Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics. 1995;140(2):745–754. doi: 10.1093/genetics/140.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li ZK, et al. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics. 2001;158(4):1737–1753. doi: 10.1093/genetics/158.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semel Y, et al. Overdominant quantitative trait loci for yield and fitness in tomato. Proc Natl Acad Sci USA. 2006;103(35):12981–12986. doi: 10.1073/pnas.0604635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riedelsheimer C, et al. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet. 2012;44(2):217–220. doi: 10.1038/ng.1033. [DOI] [PubMed] [Google Scholar]
- 15.Huang X, et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat Commun. 2015;6:6258. doi: 10.1038/ncomms7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet. 2010;42(5):459–463. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- 17.Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010;15(2):57–71. doi: 10.1016/j.tplants.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song G, et al. Global RNA sequencing reveals that genotype-dependent allele-specific expression contributes to differential expression in rice F1 hybrids. BMC Plant Biol. 2013;13:221. doi: 10.1186/1471-2229-13-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birchler JA. Heterosis: The genetic basis of hybrid vigour. Nat Plants. 2015;1(3):15020. doi: 10.1038/nplants.2015.20. [DOI] [PubMed] [Google Scholar]
- 20.Lippman ZB, Zamir D. Heterosis: Revisiting the magic. Trends Genet. 2007;23(2):60–66. doi: 10.1016/j.tig.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Fujimoto R, Taylor JM, Shirasawa S, Peacock WJ, Dennis ES. Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc Natl Acad Sci USA. 2012;109(18):7109–7114. doi: 10.1073/pnas.1204464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer RC, et al. Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J. 2012;71(4):669–683. doi: 10.1111/j.1365-313X.2012.05021.x. [DOI] [PubMed] [Google Scholar]
- 23.Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457(7227):327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore S, Lukens L. An evaluation of Arabidopsis thaliana hybrid traits and their genetic control. G3-Genes Genom Genet. 2011;1(7):571–579. doi: 10.1534/g3.111.001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groszmann M, et al. Intraspecific Arabidopsis hybrids show different patterns of heterosis despite the close relatedness of the parental genomes. Plant Physiol. 2014;166(1):265–280. doi: 10.1104/pp.114.243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dapp M, et al. Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids. Nat Plants. 2015;1(7):15092. doi: 10.1038/nplants.2015.92. [DOI] [PubMed] [Google Scholar]
- 27.Greaves IK, et al. Epigenetic changes in hybrids. Plant Physiol. 2015;168(4):1197–1205. doi: 10.1104/pp.15.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo X, Qiu Z, Li R. PeiAi64S, a dual–purpose sterile line whose sterility is induced by low critical temperature. Hybrid Rice. 1992;7(1):27–29. [Google Scholar]
- 29.Dai Z, et al. Yangdao 6-a new cultivar with fine quality and high yield developed from the strains of rice irradiated by 60Co-γ rays. Acta Agriculturae Nucleatae Sinica. 1999;13(6):377–379. [Google Scholar]
- 30.Lv C, Zou J. Theory and practice on breeding of two-line hybrid rice, Liangyoupeijiu. Scientia Agricultura Sinica. 2016;49(9):1635–1645. [Google Scholar]
- 31.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296(5565):79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 32.Wei G, et al. A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proc Natl Acad Sci USA. 2009;106(19):7695–7701. doi: 10.1073/pnas.0902340106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D, Xia Y, Li X, Hou L, Yu J. The Rice Genome Knowledgebase (RGKbase): An annotation database for rice comparative genomics and evolutionary biology. Nucleic Acids Res. 2013;41(Database issue) D1:D1199–D1205. doi: 10.1093/nar/gks1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xin Y, Yuan L. Heterosis loci and QTL of super hybrid rice Liangyoupeijiu yield by using molecular marker. Scientia Agricultura Sinica. 2014;47(14):2699–2714. [Google Scholar]
- 35.Zou J, et al. Breeding of two-line hybrid rice variety “Liang you pei jiu” and preliminary studies on its cultivation characters. Scientia Agricultura Sinica. 2003;36(8):869–872. [Google Scholar]
- 36.Gao ZY, et al. Dissecting yield-associated loci in super hybrid rice by resequencing recombinant inbred lines and improving parental genome sequences. Proc Natl Acad Sci USA. 2013;110(35):14492–14497. doi: 10.1073/pnas.1306579110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, et al. Construction of chromosomal segment substitution lines and genetic dissection of introgressed segments associated with yield determination in the parents of a super-hybrid rice. Plant Breed. 2016;135(1):63–72. [Google Scholar]
- 38.Ikeda K, Sunohara H, Nagato Y. Developmental course of inflorescence and spikelet in rice. Breed Sci. 2004;54(2):147–156. [Google Scholar]
- 39.Tsuji H, Taoka K, Shimamoto K. Regulation of flowering in rice: Two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol. 2011;14(1):45–52. doi: 10.1016/j.pbi.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 40.Sentoku N, Kato H, Kitano H, Imai R. OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy. Mol Genet Genomics. 2005;273(1):1–9. doi: 10.1007/s00438-004-1093-6. [DOI] [PubMed] [Google Scholar]
- 41.Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL. Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem Biophys Res Commun. 2007;360(1):251–256. doi: 10.1016/j.bbrc.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 42.Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL. Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa. J Plant Physiol. 2008;165(8):876–885. doi: 10.1016/j.jplph.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 43.Guo M, et al. Cell number regulator1 affects plant and organ size in maize: Implications for crop yield enhancement and heterosis. Plant Cell. 2010;22(4):1057–1073. doi: 10.1105/tpc.109.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan WH, et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant. 2011;4(2):319–330. doi: 10.1093/mp/ssq070. [DOI] [PubMed] [Google Scholar]
- 45.Wei X, et al. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153(4):1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai X, et al. LHD1, an allele of DTH8/Ghd8, controls late heading date in common wild rice (Oryza rufipogon) J Integr Plant Biol. 2012;54(10):790–799. doi: 10.1111/j.1744-7909.2012.01166.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, et al. Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 2015;208(4):1056–1066. doi: 10.1111/nph.13538. [DOI] [PubMed] [Google Scholar]
- 48.Jones J. Hybrid vigor in rice. J Am Soc Agron. 1926;18(5):423–428. [Google Scholar]
- 49.Yuan L, Virmani SS. 1988. Status of hybrid rice research and development. Hybrid rice. Proceedings of the International Symposium on Hybrid Rice (International Rice Research Institute, Manila, Philippines)
- 50.Pan X. Reflection of rice development in Yangtze river basin under the new situation. Hybrid Rice. 2015;30(6):1–5. [Google Scholar]
- 51.Zhou G, et al. Genetic composition of yield heterosis in an elite rice hybrid. Proc Natl Acad Sci USA. 2012;109(39):15847–15852. doi: 10.1073/pnas.1214141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Yuan L. Hybrid rice: Genetics, breeding, and seed production. In: Janick J, editor. Plant Breeding Reviews. Vol 17 John Wiley & Sons, Inc.; Oxford, UK: 2000. [Google Scholar]
- 53.Wan J. Chinese Rice Breeding and Cultivar Pedigree (1986–2005) China Agriculture Press; Beijing: 2010. [Google Scholar]
- 54.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 55.Dan Z, et al. Balance between a higher degree of heterosis and increased reproductive isolation: A strategic design for breeding inter-subspecific hybrid rice. PLoS One. 2014;9(3):e93122. doi: 10.1371/journal.pone.0093122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu J, et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant. 2015;8(10):1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 57.Duan P, et al. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat Plants. 2015;2:15203. doi: 10.1038/nplants.2015.203. [DOI] [PubMed] [Google Scholar]
- 58.Che R, et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants. 2015;2:15195. doi: 10.1038/nplants.2015.195. [DOI] [PubMed] [Google Scholar]
- 59.Yuan L. 2014. Progress in breeding of super hybrid rice. Public-private partnership for hybrid rice. Proceedings of the 6th International Hybrid Rice Symposium, ed Xie F HB (International Rice Research Institute, Manila, Philippines)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.