Abstract
Objective
To evaluate whether endometriosis-associated genetic variation affects risk of ovarian cancer
Design
Pooled genetic analysis
Setting
Research unit in a university hospital
Patients/Animals
Genetic data from 46,176 participants (15,361 ovarian cancer cases and 30,815 controls) from 41 ovarian cancer studies
Intervention(s)
None
Main Outcome Measure(s)
Endometriosis-associated genetic variation and ovarian cancer
Results
There was significant evidence of an association between endometriosis-related genetic variation and ovarian cancer risk, especially for the high-grade serous and clear cell histotypes. Overall, we observed 15 significant burden statistics, which was three times more than expected.
Conclusion
By focusing on candidate regions from a phenotype associated with ovarian cancer, we have shown a clear genetic link between endometriosis and ovarian cancer that warrants further follow-up. The functional significance of the identified regions and SNPs is presently uncertain, though future fine mapping and histotype-specific functional analyses may shed light on the etiologies of both gynecologic conditions.
Keywords: endometriosis, ovarian cancer, genetic variation, SNPs
Capsule
There is a clear genetic link between endometriosis and risk of ovarian cancer, especially for the high-grade serous and clear cell histotypes.
Introduction
Ovarian carcinoma (ovarian cancer) is the most fatal malignancy in the female reproductive system, accounting for more than 140,000 deaths annually worldwide (1). Endometriosis, the presence of ectopic endometrial glands and tissue in the peritoneum, is a common gynecologic condition, occurring in 6 to 10% of the general female population (2). Studies that have controlled for parity have shown that endometriosis is a well-established ovarian cancer risk factor, especially for the endometrioid and clear cell histotypes (3). Although the etiology of endometriosis remains enigmatic, it is influenced by genetic factors, with an estimated heritability of 51% and an incidence that is approximately seven times higher in relatives of women with endometriosis than in women without such family history (4,5). To date, seven variants reaching genome-wide significance have been identified in association with risk of endometriosis (6–8). In addition, the most recent meta-analysis by Nyholt et al has identified multiple additional variants associated with risk, albeit some at a sub-genome-wide significance level (P≤1 × 10−5) (9).
Genetics also plays a role in the etiology of ovarian cancer as women with first-degree family histories of the disease have over a two-fold increased risk (10). High-penetrance susceptibility genes, such as BRCA1 and BRCA2, as well as 18 published common variants identified through genome-wide association studies (GWAS) account for a substantial portion of ovarian cancer’s familial risk, but at least 60% remains unexplained (11–19). Lee and colleagues have previously shown that the cumulative effect of many risk variants contribute to disease heritability (20) and hence, it is likely that the germline genetic contributions to ovarian cancer are not limited to GWAS-identified variants.
Given the important role genetics plays in the etiologies of both endometriosis and ovarian cancer and the consistent epidemiologic evidence of their association with one another, the two gynecologic conditions may also have a similar genetic profile. Hence, based on the results presented by Nyholt and colleagues of their endometriosis GWAS meta-analysis, we present the first report that evaluates whether variation in the 18 regions harboring the top 38 endometriosis-associated SNPs is associated with risk of ovarian cancer.
Materials and Methods
All studies included in this report obtained institutional ethics committee approval and all participating subjects provided written informed consent.
Study Populations
Our analysis included 41 studies participating in the Ovarian Cancer Association Consortium (OCAC), an international collaboration of ovarian cancer studies founded in 2005. In total, 20 studies were conducted in Europe, 19 in North America, and two in Australia. Only participants of European ancestry were included, as was determined from the program LAMP (Local Ancestry in Admixed Populations) (see Statistical Analysis below). We used a combined total of 46,176 participants (15,361 ovarian cancer cases and 30,815 controls) in our analyses; borderline tumors were excluded. Details regarding sample quality control have been previously published (15). Supplementary Table 1 provides an overview of each study’s characteristics and the numbers of subjects included.
Genotyping and Imputation Analyses
The genetic data for our analyses came from three population-based ovarian cancer GWAS, which comprised 2,162 cases and 2,564 controls from a GWAS in North America (“US GWAS”) (21), 1,763 cases and 6,118 controls from a GWAS in the United Kingdom (“UK GWAS”) (11), and 443 cases and 441 controls from a second GWAS in North America. In addition, 11,030 cases and 21,693 controls were genotyped using the iCOGS array, which was a large-scale genotyping project by the Collaborative Oncological Gene-environment Study (COGS) (15). The US and UK GWAS included several independent case-control studies, and samples from these studies were also genotyped using the iCOGS array. All duplicates were removed from the analyses, resulting in genetic data for 15,361 women diagnosed with ovarian cancer and 30,815 controls. Study sets were created based on the scope of genotyping information (GWAS versus COGS) available for imputation. These sets are indicated in Supplementary Table 1. Details regarding the genotyping platform for each dataset have been published previously (15).
Imputation of the entire scope of genetic variation in the genome was carried out separately for iCOGS samples and each of the GWAS. We imputed variants by combining all available genotype data with information from the April 2012 release of the 1000 Genomes Project using the program IMPUTE2 (22). In addition, all data were pre-phased using the software SHAPEIT in order to improve computation efficiency (23).
SNP Selection
The SNPs evaluated here were based on the results of Nyholt et al’s genome-wide association meta-analysis (9), which included two large endometriosis GWAS: one conducted in a Japanese sample obtained from BioBank Japan (BBJ) (7) and the other in an European sample from Australia and the UK by the International Endogene Consortium (IEC) (6). Nyholt et al found six SNPs (rs7521902, rs12700667, rs10859871, rs4141819, rs1537377, rs7739264) that reached genome-wide significance (P≤5 × 10−8) in their analysis as well as an additional SNP (rs13394619) that reached genome-wide significance when combined with the results from a previous meta-analysis of two Japanese case-control cohorts (8). These seven SNPs, as well as an additional 31 SNPs that showed suggestive evidence of an association with endometriosis (P≤1 × 10−5), were included in our analyses for a total of 38 index SNPs. A list of these SNPs is provided in Table 1.
Table 1.
Association between the top 38 SNPs identified through endometriosis genome-wide scans and ovarian cancer risk
| Region* | Chromosome | Position† | SNP | Endometriosis** | Ovarian Cancer‡ | ||
|---|---|---|---|---|---|---|---|
| OR | p-value | OR | p-value | ||||
| A | 1p36 | 22473410 | rs7515106a | 1.18 | 2.1 × 10−6 | 1.09 | 4.9 × 10−6 |
| A | 1p36 | 22490724 | rs7521902a | 1.18 | 4.6 × 10−8 | 1.08 | 7.3 × 10−6 |
| A | 1p36 | 22501846 | rs742356 | 1.15 | 3.1 × 10−6 | 1.05 | 0.004 |
| B | 1p31 | 80278733 | rs12744944a | 1.15 | 9.6 × 10−6 | 1.00 | 0.80 |
| C | 2p25 | 11725241 | rs12470971 | 1.14 | 8.1 × 10−6 | 0.97 | 0.062 |
| C | 2p25 | 11727507 | rs13394619 | 1.15 | 6.1 × 10−8 | 0.98 | 0.23 |
| D | 2p14 | 67864675 | rs4141819a | 1.16 | 4.0 × 10−7 | 1.00 | 0.86 |
| E | 3p24 | 24927203 | rs10510551 | 1.14 | 9.6 × 10−6 | 1.01 | 0.54 |
| F | 3p24 | 25075167 | rs4858692 | 1.16 | 1.3 × 10−6 | 0.96 | 0.025 |
| F | 3p24 | 25075530 | rs1603995 | 1.16 | 1.6 × 10−6 | 0.96 | 0.017 |
| G | 3p14 | 55216779 | rs9311552 | 1.13 | 7.4 × 10−6 | 1.00 | 0.85 |
| H | 3q13 | 104575864 | rs12635480 | 1.34 | 8.6 × 10−6 | 0.99 | 0.78 |
| I | 4q12 | 55931246 | rs4241991 | 1.16 | 4.6 × 10−6 | 0.95 | 0.018 |
| J | 6p22 | 19706761 | rs1016251 | 1.14 | 6.3 × 10−6 | 1.02 | 0.154 |
| J | 6p22 | 19708481 | rs9366312 | 1.14 | 4.3 × 10−6 | 1.02 | 0.14 |
| J | 6p22 | 19729003 | rs9356708a | 1.16 | 3.1 × 10−7 | 1.02 | 0.132 |
| J | 6p22 | 19761215 | rs6916251 | 1.14 | 3.4 × 10−6 | 1.02 | 0.13 |
| J | 6p22 | 19785588 | rs7739264a | 1.16 | 1.3 × 10−7 | 0.98 | 0.13 |
| J | 6p22 | 19790809 | rs2223361 | 1.15 | 7.3 × 10−7 | 1.03 | 0.093 |
| J | 6p22 | 19803768 | rs6907340 | 1.15 | 5.9 × 10−7 | 1.03 | 0.045 |
| K | 6q21 | 113111808 | rs9487982 | 1.22 | 8.2 × 10−6 | 0.99 | 0.68 |
| L | 6q25 | 152638209 | rs1890100 | 1.16 | 8.1 × 10−6 | 1.01 | 0.70 |
| M | 6q25 | 158144402 | rs16900375 | 1.29 | 4.4 × 10−6 | 1.01 | 0.84 |
| N | 7p15 | 25860812 | rs7798431a | 1.16 | 2.1 × 10−6 | 0.98 | 0.28 |
| N | 7p15 | 25871109 | rs1055144a | 1.18 | 5.6 × 10−7 | 0.97 | 0.19 |
| N | 7p15 | 25873110 | rs10282436a | 1.18 | 7.4 × 10−7 | 0.97 | 0.18 |
| N | 7p15 | 25901639 | rs12700667a | 1.22 | 9.3 × 10−10 | 0.97 | 0.072 |
| O | 7q33 | 134607676 | rs7809057 | 1.15 | 2.3 × 10−6 | 1.00 | 0.81 |
| O | 7q33 | 134618710 | rs6973420 | 1.14 | 4.7 × 10−6 | 1.00 | 0.87 |
| P | 9p21 | 22169700 | rs1537377a | 1.14 | 2.5 × 10−6 | 1.03 | 0.071 |
| Q | 10q26 | 117325021 | rs1572396 | 1.16 | 1.5 × 10−6 | 1.02 | 0.24 |
| Q | 10q26 | 117393524 | rs2769422a | 1.14 | 8.2 × 10−6 | 1.01 | 0.55 |
| Q | 10q26 | 117396288 | rs2804250 | 1.15 | 7.6 × 10−6 | 1.01 | 0.54 |
| Q | 10q26 | 117398195 | rs2769417 | 1.15 | 6.9 × 10−6 | 1.01 | 0.55 |
| R | 12q22 | 95574831 | rs10777670a | 1.19 | 1.6 × 10−7 | 1.05 | 0.021 |
| R | 12q22 | 95631276 | rs10859856 | 1.15 | 4.8 × 10−7 | 1.01 | 0.39 |
| R | 12q22 | 95702385 | rs11107973 | 1.14 | 2.1 × 10−6 | 1.01 | 0.36 |
| R | 12q22 | 95711876 | rs10859871a | 1.18 | 5.5 × 10−9 | 1.03 | 0.088 |
| Burden statistic (all SNPs): | 0.001 | ||||||
| Burden statistic (excluding Region A SNPs): | 0.052 | ||||||
Note: Significant p-values at P≤0.05 for ovarian cancer are indicated in bold.
The 38 SNPs represent 18 distinct regions of linkage disequilibrium (Regions A-R).
Based on Build 37.
Odds ratios and p-values reported in Nyholt et al (9) for the association between the SNP and risk of endometriosis.
Odds ratios and p-values for the association between the SNP and risk of ovarian cancer.
SNP was genotyped.
In addition, we evaluated genetic variation in the surrounding regions of each of the 38 index SNPs. Initially, these regions were defined as the areas 25kb up- and downstream of each index SNP. All variants in these 50kb regions were analyzed and the SNP with the strongest ovarian cancer association in each region was identified. The final regions, which we have labelled as Regions A to R, were defined as including all SNPs with an r2≥0.2 with the most significantly associated ovarian cancer SNPs. Based on each SNP’s linkage disequilibrium pattern, a total of 18 regions of varying sizes was identified as some of the 38 SNPs were strongly correlated with each other. Only SNPs with an imputation r2≥0.5 and a minor allele frequency (MAF) ≥0.05 were considered. In total, 6,981 SNPs (including the original 38 SNPs) were assessed.
Statistical Analysis
We used LAMP to assign intercontinental ancestry based on genotype frequencies in European populations according to HapMap (24). Subjects were classified as European if they had 90% or more European ancestry. Principal components analysis (PCA) to control for population substructure was also performed using a set of 37,000 unlinked markers as well as an in-house program written in C++ that used the Intel MKL library for eigenvectors (http://ccge.medschl.cam.ac.uk/software/pccalc).
We carried out unconditional logistic regression analyses, adjusting for the first five eigenvalues from the PCA for European ancestry, to determine the association between each SNP and risk of ovarian cancer. For all ovarian cancer cases combined and for high-grade serous cases, the analyses were carried out within each study set and the set-specific results were summarized using a fixed-effects meta-analysis approach. For the less common histotypes (mucinous, endometrioid, clear cell), the cases and controls were pooled across all study sets, and study set was included as a term in the model. A log-additive mode of inheritance was used, with each SNP modeled as an ordinal variable. Hence, the effect estimates reflect per-allele odds ratios (ORs). All p-values reported herein are two-sided.
To evaluate whether overall genetic variation implicated in risk of endometriosis also plays a role in risk of ovarian cancer, we calculated burden statistics using the admixture likelihood (AML) method (25). We used the AMLcalc program to accomplish this using 1000 simulations with the maximum proportion of associated SNPs set to 0.2 on the genotyped and imputed data and adjusting for the first five ancestry principal components. P-values for the AML trend test are provided. We calculated burden statistics across the 38 SNPs identified by Nyholt et al as well as across each of the 18 regions for ovarian cancer overall risk and for the four main histotypes. This allowed us to take a global approach to assess whether the combined variation that exists among the 38 endometriosis-associated SNPs or across the 18 regions plays a role in ovarian cancer risk after accounting for the correlation between SNPs.
Manhattan plots were generated by plotting the −log P versus the chromosomal position using GraphPad Prism 6 to provide a picture of the distribution of p-values by histotype across each region. This plotting was done for all regions except Region A since it was presented in a recent meta-analysis by OCAC and the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) (19). Linkage disequilibrium plots depicting pairwise correlation data from the 1000 Genomes CEU (Utah residents (CEPH) with Northern and Western European ancestry) population were generated using HaploView. Epigenomic data made available from the ENCODE and Roadmap Epigenomics Consortia were obtained and visualized in the UCSC Genome Browser.
Results
In total, 38 SNPs with a P≤1 × 10−5 spanning 18 unique, uncorrelated regions were identified from the endometriosis GWAS meta-analysis carried out by Nyholt and colleagues (see Table 1) (9). Table 1 presents the association between each of those 38 SNPs and risk of ovarian cancer. Eight SNPs (rs7515106, rs7521902, rs742356, rs4858692, rs1603995, rs4241991, rs6907340, rs10777670) from five different regions (Regions A, F, I, J, R) showed statistically significant associations with ovarian cancer risk (P≤0.05), with the most significantly associated SNP being rs7515106 from Region A on chromosome 1 (P=4.9 × 10−6). When considering all 38 SNPs together, the calculated AML burden statistic showed significant evidence of an association with ovarian cancer risk (P=0.001); it slightly attenuated when the SNPs from Region A were excluded (P=0.052).
Given that each region was defined by the area containing SNPs with an r2≥0.2 with the SNP most significantly associated with ovarian cancer risk, Table 2 presents the coordinates as well as the number of SNPs with genotyped or imputed data available for each region. Overall, a total of 6,981 SNPs across 18 regions were evaluated. Region O was the largest, spanning almost 500 kb, but Region R included the largest number of SNPs with a MAF≥0.05 and an imputation r2≥0.8 (n=1,309).
Table 2.
Burden statistics for the association between the 18 genomic regions, harboring SNPs associated with endometriosis, and risk of ovarian cancer by histotype
| Region | Chr. | Region coordinates* | Region size (kb) |
Coverage | # of SNPs in region† |
Burden statistics** | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Overall | High- grade serous |
Mucinous | Endomet- rioid |
Clear cell | ||||||
| A | 1p36 | 22347396 – 22531542 | 184.146 | 0.99 | 594 | 5.0 × 10−5 | 1.0 × 10−4 | 0.78 | 0.35 | 0.007 |
| B | 1p31 | 80152740 – 80338250 | 185.510 | 0.99 | 607 | 0.71 | 0.057 | 0.26 | 0.93 | 0.015 |
| C | 2p25 | 11661140 – 11712075 | 50.935 | 0.98 | 107 | 0.041 | 0.70 | 0.11 | 0.23 | 0.014 |
| D | 2p14 | 67983706 – 68019054 | 35.348 | 0.97 | 77 | 0.044 | 0.21 | 0.16 | 0.43 | 0.54 |
| E | 3p24 | 24940850 – 25060867 | 120.017 | 0.96 | 390 | 0.67 | 0.85 | 0.28 | 0.57 | 0.65 |
| F | 3p24 | 25035804 – 25075645 | 39.841 | 0.96 | 168 | 0.15 | 0.32 | 0.17 | 0.036 | 0.28 |
| G | 3p14 | 55181646 – 55221542 | 39.896 | 0.94 | 113 | 0.30 | 0.32 | 0.79 | 0.54 | 0.68 |
| H | 3q13 | 104584856 – 104828081 | 243.225 | 0.99 | 846 | 0.84 | 0.95 | 0.86 | 0.34 | 0.48 |
| I | 4q12 | 55988082 – 56027573 | 39.491 | 0.96 | 135 | 0.011 | 0.010 | 0.23 | 0.19 | 0.018 |
| J | 6p22 | 19567942 – 19632693 | 64.751 | 0.97 | 194 | 0.61 | 0.54 | 0.20 | 0.23 | 0.29 |
| K | 6q21 | 112966971 – 113226112 | 259.141 | 0.97 | 550 | 0.15 | 0.63 | 0.19 | 0.67 | 0.82 |
| L | 6q25 | 152544371 – 152677815 | 133.444 | 1.00 | 370 | 0.26 | 0.39 | 0.75 | 0.26 | 0.070 |
| M | 6q25 | 158184000 – 158192932 | 8.932 | 0.94 | 19 | 0.23 | 0.50 | 0.075 | 0.48 | 0.61 |
| N | 7p15 | 25888067 – 25897388 | 9.321 | 0.95 | 15 | 0.16 | 0.38 | 0.79 | 0.22 | 0.12 |
| O | 7q33 | 134455276 – 134936517 | 481.241 | 0.98 | 1082 | 0.54 | 0.96 | 0.70 | 0.12 | 0.94 |
| P | 9p21 | 22140648 – 22201586 | 60.938 | 0.96 | 184 | 0.002 | 0.026 | 0.63 | 0.17 | 0.14 |
| Q | 10q26 | 117345263 – 117407423 | 62.160 | 0.99 | 204 | 0.82 | 0.56 | 0.13 | 0.70 | 0.27 |
| R | 12q22 | 95304634 – 95709315 | 404.681 | 0.98 | 1309 | 0.011 | 0.025 | 0.53 | 0.15 | 0.28 |
| Global | -- | -- | -- | -- | -- | 0.002 | 0.002 | 0.61 | 0.20 | 0.039 |
Note: Significant p-values at P≤0.05 are indicated in bold. “Global” refers to when all SNPs across all regions are considered.
Defined as including all SNPs with an r2≥0.2 with the most significant ovarian cancer-associated SNP (when considering the areas 25kb up and downstream of the index SNP as presented in Table 1).
Based on r2≥0.5 and MAF≥0.05.
Took into account the first five ancestry principal components; p-values for trend reported.
Table 2 also presents the burden statistics for each region for all ovarian cancers combined and for the four main histotypes. In total, there were 15 significant burden statistics at a P≤0.05 level, covering eight different regions. Of these, the most significant burden statistic was for Region A (P=5.0 × 10−5 for overall). However, there was a total of six regions (A, C, D, I, P, R) that showed a significant association with ovarian cancer overall and among them, five regions also had significant histotype-specific burden statistics. High-grade serous and clear cell cancers had the greatest number of significant burden statistics, each with four and both sharing Region A. When burden statistics were calculated globally across all 18 regions, significant evidence of an association was found between overall genetic variation and risk of all ovarian cancers combined (P=0.002) as well as risk of the high-grade serous and clear cell histotypes (P=0.002 and P=0.039, respectively).
ORs and 95% CIs for the most significant SNPs associated with ovarian cancer are presented in Table 3. Region A contained the most significant SNP, rs10917151 (OR=1.11, P=4.0 × 10−7), but Regions L, P, and R also had SNPs of notable significance (P=3.3 × 10−4 for rs73007780, P=2.5 × 10−4 for rs1333052, P=1.2 × 10−5 for rs7397212, respectively). Supplementary Tables 2 and 3 present the most significant SNPs for each region by histotype (high-grade serous and mucinous in Supplementary Table 2, endometrioid and clear cell in Supplementary Table 3). Similar to the results for ovarian cancer overall, Region A for high-grade serous cancer contained the most significant SNP, rs3754496 (P=5.3 × 10−8), while Region R for high-grade serous cancer harbored the largest number of significant SNPs (n=327) when looking across the four histotypes. Other SNPs of notable significance were found in Region R for high-grade serous cancer (rs6538605, P=1.2 × 10−4) and endometrioid cancer (rs11107893, P=4.3 × 10−4) as well as in Regions A (rs4654785, P=1.3 × 10−4) and L (rs71575922, P=1.4 × 10−4) for clear cell cancer.
Table 3.
Odds ratios and 95% confidence intervals for the most significant SNP in each of the 18 genomic regions for ovarian cancer overall
| Region | # of significant SNPs |
Most significant SNP |
OR* | 95% CI | P-value |
|---|---|---|---|---|---|
| A | 192 | rs10917151 | 1.11 | 1.07 – 1.15 | 4.0 × 10−7 |
| B | 2 | rs28556754 | 0.96 | 0.93 – 0.99 | 0.017 |
| C | 53 | rs142034631 | 1.05 | 1.01 – 1.10 | 0.004 |
| D | 14 | rs17034382 | 0.94 | 0.91 – 0.98 | 0.004 |
| E | 5 | rs62228500 | 0.94 | 0.89 – 1.00 | 0.033 |
| F | 22 | rs1846555 | 1.04 | 1.01 – 1.08 | 0.015 |
| G | 10 | rs17054879 | 0.94 | 0.90 – 0.98 | 0.005 |
| H | 6 | rs114063826 | 1.08 | 1.02 – 1.14 | 0.007 |
| I | 87 | rs12331507† | 0.95 | 0.92 – 0.98 | 1.0 × 10−3 |
| J | 1 | rs9717730 | 1.07 | 1.02 – 1.13 | 0.012 |
| K | 59 | rs4437516 | 1.04 | 1.01 – 1.07 | 0.018 |
| L | 24 | rs73007780 | 0.88 | 0.83 – 0.95 | 3.3 × 10−4 |
| M | 2 | rs7742463 | 0.94 | 0.90 – 0.98 | 0.002 |
| N | 1 | rs73093677 | 0.93 | 0.88 – 0.99 | 0.023 |
| O | 0 | N/A | -- | -- | -- |
| P | 68 | rs1333052 | 1.06 | 1.03 – 1.09 | 2.5 × 10−4 |
| Q | 2 | rs2804241 | 1.05 | 1.00 – 1.11 | 0.041 |
| R | 256 | rs7397212 | 1.13 | 1.07 – 1.20 | 1.2 × 10−5 |
Note: “Significant” reflects a P≤0.05. "N/A" means there are no significant SNPs in that region.
OR = Per allele odds ratio, adjusted for the first five principal components; based on study set-specific ORs that were meta-analyzed.
SNP was genotyped.
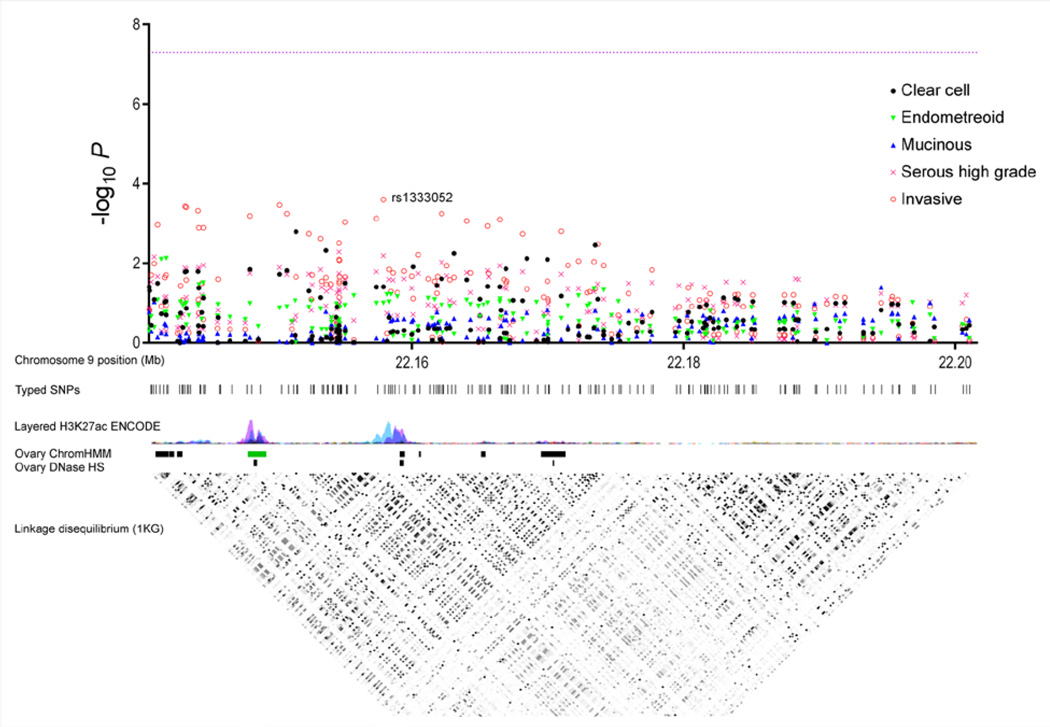
Figure 1 presents the Manhattan and linkage disequilibrium plots for Region P, the region showing the most significant burden statistic across all 18 regions excluding Region A (P=0.002 for ovarian cancer overall). A clear elevation of p-values is present, especially when looking at ovarian cancer overall, with the most significant SNP rs1333052 (P=2.5 × 10−4) indicated. In addition, Supplementary Figure 1 presents the Manhattan plots for the remaining regions with significant burden statistics whereas Supplementary Figure 2 presents these plots for the regions that did not have significant burden statistics. With the exception of Region L showing some elevation of p-values for clear cell cancer, reasonable given its borderline significant burden statistic (P=0.07), all plots in Supplementary Figure 2 look relatively flat.
Figure 1. Manhattan and linkage disequilibrium plots for Region P.
These plots depict the results for all 184 SNPs in Region P (chromosome 9p21, position 22140648 – 22201586). The Manhattan plot includes ovarian cancer overall (“invasive”) and its four histotypes, with the x-axis corresponding to the chromosomal position (in Mb), the y-axis to the –log P, and the line to P=5.0 × 10−8. Rs1333052, the most significant SNP in the region (P=2.5 × 10−4 for ovarian cancer overall), is indicated. The linkage disequilibrium plot depicts pairwise correlation data from the 1000 Genomes CEU population. Epigenomic data from the ENCODE and Roadmap Epigenomics Consortia were obtained and visualized in the UCSC Genome Browser.
Discussion
We took a novel approach toward evaluating endometriosis-associated SNPs with ovarian cancer risk and found appreciable support for a shared genetic etiology between these two gynecologic conditions. Across the 18 regions harboring putative endometriosis SNPs, we calculated a total of 15 significant burden statistics for ovarian cancer risk compared to approximately 5 expected by chance at the P≤0.05 level (i.e., 18 regions × 5 types of ovarian cancer (overall, high-grade serous, mucinous, endometrioid, clear cell) = 90 total), a conservative estimate due to the strong correlation between overall and high-grade serous.
Endometriosis and ovarian cancer were first linked because of their frequent co-occurrence in surgical specimens. Most recently, a pooled analysis of 13 case-control studies by Pearce et al showed that after adjusting for oral contraceptive use and parity, women with a history of endometriosis were 46% more likely to develop ovarian cancer, with the association primarily restricted to the endometrioid and clear cell histotypes (3). Endometriosis may be a precursor lesion for endometrioid and clear cell ovarian cancer, but the process of its malignant transformation is not well-understood (26,27). The two gynecologic conditions are likely to have a shared pathophysiology given evidence from pathology case series reporting endometrioid and clear cell ovarian cancers arising from endometriotic foci as well as epidemiologic studies that highlight their similar hormone-related risk factors, such as nulliparity (27). In addition, both traits appear to thrive in similar hormonal and immune environments, highlighting a possible shared inflammatory etiology.
Some of these results are contrary to what we expected. Consistent with our hypothesis, we calculated a significant global burden statistic for clear cell ovarian cancer when considering all endometriosis SNPs together. However, we did not see this association for the endometrioid histotype despite its well-established association with endometriosis. In addition, we observed a link with the high-grade serous histotype, which previous epidemiologic studies have not found to be associated with endometriosis; we did not observe any associations with the low-grade serous histotype (data not shown due to small numbers). Our findings suggest that while endometriosis is not a precursor lesion for high-grade serous ovarian cancer, the genetic pathways related to risk of these two diseases are shared. This finding paves the way for interesting lines of inquiry related to shared pathways. Perhaps women with endometriosis are more likely to have endosalpingiosis, which is common but infrequently noted in pathology reports, and which may predispose to high-grade serous ovarian cancer.
Our ovarian cancer susceptibility GWAS have identified thousands of nominally significant associations, but most are due to chance. Deciphering the noise from true signals is difficult despite our large sample size. In this analysis, we took a novel two-pronged approach, first selecting candidate regions from a phenotype linked to ovarian cancer and then assessing whether the burden of associations in these regions was significant. The burden statistics implicate some of these regions in risk of ovarian cancer although the exact relevance of the regions remains unknown. Region A includes the most significantly associated SNP among all 6,981 considered, rs3754496 (P=5.4 × 10−8 for high-grade serous), an imputed SNP located near WNT4, a gene involved in steroidogenesis and in the development of the ovarian follicle and the female reproductive tract, biological functions that make its role in the development of both endometriosis and ovarian cancer compelling (28). Our data from OCAC for Region A have previously been combined with those of CIMBA and a genome-wide significant association was observed (19).
In addition, Table 1 shows that an association with the remaining regions may exist even after excluding Region A (P=0.052). Only three of the 15 significant burden statistics presented in Table 2 are found in Region A, with the other 12 spanning seven other regions. Region P had the most significant burden statistic (P=0.001 for ovarian cancer overall) with rs1333052 as its most significant SNP (P=2.5 × 10−4). This SNP is located adjacent to CDKN2B, a tumor suppressor gene whose methylation has been commonly seen in ovarian carcinogenesis (29). In addition, rs1333052 lies within a binding site for GATA2, one of six factors that constitute the GATA family of transcriptional regulatory proteins which has been shown to play a role in ovarian function; it was identified by genome-wide ChIP-sequencing (30). Rs12331507 in Region I, which, other than Region A, had the greatest number of significant burden statistics, lies in an intergenic region approximately 20 kb upstream of KDR, a gene that encodes one of the two receptors for vascular endothelial growth factor (VEGF); VEGF has been shown to be expressed by tumor cells in ovarian cancer (31). Interestingly, the most significant SNP in Region C, rs142034631, is located in GREB1, an estrogen-responsive gene that modulates tumor progression in models of ovarian cancer (32).
While the specific relevance of these genes and regions as well as the functional significance of their most significantly associated SNPs cannot be determined at this time, these results suggest that additional real associations exist between ovarian cancer risk and variation in the endometriosis-related regions. More importantly, they highlight the presence of a genetic commonality that high-grade serous and clear cell ovarian cancers and endometriosis are likely to share.
In conclusion, we have shown significant evidence of a genetic link between endometriosis and ovarian cancer that warrants further follow-up. Whether the association between these two gynecologic conditions is causal remains unknown. However, fine mapping studies of the regions we have identified will greatly contribute to our knowledge regarding the etiologies of both diseases as well as shed some light on their likely shared pathophysiology. The clinical implications of these results remain to be established, but next steps would include fine mapping of the regions with significant burden statistics (Regions A, B, C, D, F, I, P, R) and functional analyses of the most likely causal SNP(s).
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Cancer Institute [K07-CA095666, K07-CA80668, K07-CA143047, K22-CA138563, N01-CN55424, N01-PC67001, N01-PC067010, N01-PC035137, P01-CA017054, P01-CA087696, P30-CA072720, P30-CA15083, P30-CA008748, P50-CA159981, P50-CA105009, P50-CA136393, R01-CA149429, R01-CA014089, R01-CA016056, R01-CA017054, R01-CA049449, R01-CA050385, R01-CA054419, R01-CA058598, R01-CA058860, R01-CA061107, R01-CA061132, R01-CA063678, R01-CA063682, R01-CA067262, R01-CA071766, R01-CA074850, R01-CA076016, R01-CA080978, R01-CA083918, R01-CA087538, R01-CA092044, R01-CA095023, R01-CA122443, R01-CA112523, R01-CA114343, R01-CA126841, R01-CA136924, R03-CA113148, R03-CA115195, U01-CA069417, U01-CA071966 and Intramural research funds]; the European Commission's Seventh Framework Programme [agreement number 223175 HEALTH F2 2009-223175]; a National Cancer Institute, Cancer Post-GWAS Initiative [U19-CA148112]; the Genetic Associations and Mechanisms in Oncology (GAME-ON); the Ovarian Cancer Research Fund (thanks to donations by the family and friends of Kathryn Sladek Smith) [PPD/RPCI.07]; the National Institute of Environmental Health Sciences [T32ES013678 to A.W.L]; National Health and Medical Research Council [to G.C.T.]; American Cancer Society Early Detection Professorship [SIOP-06-258-01-COUN to B.K.]; National Center for Advancing Translational Sciences (NCATS) [UL1TR000124 to B.K.]; California Cancer Research Program [00-01389V-20170, N01-CN25403, 2II0200]; the Canadian Institutes of Health Research [MOP-86727]; Cancer Australia; Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK [C490/A6187, C490/A10119, C490/A10124]; the Danish Cancer Society [94-222-52]; the ELAN Program of the University of Erlangen-Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Helse Vest; the Norwegian Cancer Society; the Norwegian Research Council; the Ovarian Cancer Research Fund; Nationaal Kankerplan of Belgium; the L & S Milken Foundation; the Polish Ministry of Science and Higher Education [4 PO5C 028 14, 2 PO5A 068 27]; the Roswell Park Cancer Institute Alliance Foundation; the National Institutes of Health/National Center for Research Resources/General Clinical Research Center [MO1-RR000056]; the US Army Medical Research and Material Command [DAMD17-01-1-0729, DAMD17-02-1-0666, DAMD17-02-1-0669, W81XWH-07-0449, W81XWH-10-1-02802]; the U.S. Public Health Service [PSA-042205]; the National Health and Medical Research Council of Australia [199600, 400281]; the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research [01GB 9401]; the State of Baden-Wurttemberg through Medical Faculty of the University of Ulm [P.685]; the German Cancer Research Center; the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Lon V. Smith Foundation [LVS-39420]; the Oak Foundation; the Oregon Health and Science University (OHSU) Foundation; the Mermaid I project; the Rudolf-Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital “Womens Health Theme” and the Royal Marsden Hospital; and WorkSafeBC 14.
Ovarian Cancer Association Consortium: Roberta B. Ness, Agnieszka Dansonka-Mieszkowska, Aleksandra Gentry-Maharaj, Alexander Hein, Alice S. Whittemore, Allan Jensen, Andreas du Bois, Angela Brooks-Wilson, Anja Rudolph, Anna Jakubowska, Anna H. Wu, Argyrios Ziogas, Arif B. Ekici, Arto Leminen, Australian Cancer Study (Ovarian Cancer), Australian Ovarian Cancer Study Group, Barry Rosen, Beata Spiewankiewicz, Beth Y. Karlan, Britton Trabert, Brooke L. Fridley, C. Blake Gilks, Camilla Krakstad, Catherine M. Phelan, Cezary Cybulski, Christine Walsh, Claus Hogdall, Daniel W. Cramer, David G. Huntsman, Diana Eccles, Diether Lambrechts, Dong Liang, Douglas A. Levine, Edwin S. Iversen, Elisa V. Bandera, Elizabeth M. Poole, Ellen L. Goode, Els Van Nieuwenhuysen, Estrid Hogdall, Fiona Bruinsma, Florian Heitz, Francesmary Modugno, Graham G. Giles, Harvey A. Risch, Helen Baker, Helga B. Salvesen, Heli Nevanlinna, Hoda Anton-Culver, Honglin Song, Iain McNeish, Ian G. Campbell, Ignace Vergote, Ingo B. Runnebaum, Ingvild L. Tangen, Ira Schwaab, Jacek Gronwald, James Paul, Jan Lubinski, Jennifer A. Doherty, Jenny Chang-Claude, Jenny Lester, Joellen M. Schildkraut, John R. McLaughlin, Jolanta Lissowska, Jolanta Kupryjanczyk, Jonathan Tyrer, Joseph L. Kelley, Joseph H. Rothstein, Julie M. Cunningham, Karen Lu, Karen Carty, Kathryn L. Terry, Katja K.H. Aben, Kirsten B. Moysich, Kristine G. Wicklund, Kunle Odunsi, Lambertus A. Kiemeney, Lara Sucheston-Campbell, Lene Lundvall, Leon F.A.G. Massuger, Liisa M. Pelttari, Linda E. Kelemen, Linda S. Cook, Line Bjorge, Lotte Nedergaard, Louise A. Brinton, Lynne R. Wilkens, Malcolm C. Pike, Marc T. Goodman, Maria Bisogna, Mary Anne Rossing, Matthias W. Beckmann, Matthias Dürst, Melissa C. Southey, Melissa Kellar, Michelle A.T. Hildebrandt, Nadeem Siddiqui, Natalia Antonenkova, Natalia Bogdanova, Nhu D. Le, Nicolas Wentzensen, Pamela J. Thompson, Patricia Harrington, Penelope M. Webb, Peter A. Fasching, Peter Hillemanns, Philipp Harter, Piotr Sobiczewski, Rachel Palmieri Weber, Ralf Butzow, Robert P. Edwards, Robert A. Vierkant, Rosalind Glasspool, Sandra Orsulic, Sandrina Lambrechts, Sara H. Olson, Shan Wang-Gohrke, Shashi Lele, Shelley S. Tworoger, Simon A. Gayther, Stacey A. Missmer, Steven A. Narod, Susan J. Ramus, Susanne K. Kjaer, Tanja Pejovic, Thilo Dörk, Ursula Eilber, Usha Menon, Valerie McGuire, Weiva Sieh, Xifeng Wu, Yukie Bean, Yurii B Shvetsov
We thank all the individuals who took part in this study and all the researchers, clinicians and technical and administrative staff who have made possible the many studies contributing to this work. In particular, we thank: D. Bowtell, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward and D. Whiteman (AUS); G. Peuteman, T. Van Brussel and D. Smeets (BEL); the staff of the genotyping unit, S LaBoissiere and F Robidoux (Genome Quebec); L. Gacucova (HMO); P. Schurmann, F. Kramer, W. Zheng, T. W. Park, Simon, K. Beer-Grondke and D. Schmidt (HJO); S. Windebank, C. Hilker and J. Vollenweider (MAY); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, HI, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WYL (NHS); L. Paddock, M. King, L. Rodriguez-Rodriguez, A. Samoila, and Y. Bensman (NJO); M. Sherman, A. Hutchinson, N. Szeszenia-Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao and M. Stagner (POL); C. Luccarini, P. Harrington the SEARCH team and ECRIC (SEA); I. Jacobs, M. Widschwendter, E. Wozniak, N. Balogun, A. Ryan, C. Karpinskyj, and J. Ford (UKO); Carole Pye (UKR); A. Amin Al Olama, J. Dennis, E. Dicks, K. Michilaidou, K. Kuchenbaker (COGS); G. Montgomery.
Footnotes
Conflict of Interest Statement
Dr. Pharoah reports grants from National Institutes of Health and Cancer Research UK during the conduct of the study.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Bulletti C, Coccia ME, Battistoni S, Borini A. Endometriosis and infertility. J Assist Reprod Genet. 2010;27:441–447. doi: 10.1007/s10815-010-9436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearce CL, Templeman C, Rossing MA, Lee A, Near AM, Webb PM, et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. Lancet Oncol. 2012;13:385–394. doi: 10.1016/S1470-2045(11)70404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Treloar SA, O'Connor DT, O'Connor VM, Martin NG. Genetic influences on endometriosis in an Australian twin sample. sueT@qimr.edu.au. Fertil Steril. 1999;71:701–710. doi: 10.1016/s0015-0282(98)00540-8. [DOI] [PubMed] [Google Scholar]
- 5.Matalliotakis IM, Arici A, Cakmak H, Goumenou AG, Koumantakis G, Mahutte NG. Familial aggregation of endometriosis in the Yale Series. Arch Gynecol Obstet. 2008;278:507–511. doi: 10.1007/s00404-008-0644-1. [DOI] [PubMed] [Google Scholar]
- 6.Painter JN, Anderson CA, Nyholt DR, Macgregor S, Lin J, Lee SH, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Genet. 2011;43:51–54. doi: 10.1038/ng.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uno S, Zembutsu H, Hirasawa A, Takahashi A, Kubo M, Akahane T, et al. A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Genet. 2010;42:707–710. doi: 10.1038/ng.612. [DOI] [PubMed] [Google Scholar]
- 8.Adachi S, Tajima A, Quan J, Haino K, Yoshihara K, Masuzaki H, et al. Meta-analysis of genome-wide association scans for genetic susceptibility to endometriosis in Japanese population. J Hum Genet. 2010;55:816–821. doi: 10.1038/jhg.2010.118. [DOI] [PubMed] [Google Scholar]
- 9.Nyholt DR, Low SK, Anderson CA, Painter JN, Uno S, Morris AP, et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat Genet. 2012;44:1355–1359. doi: 10.1038/ng.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce CL, Rossing MA, Lee AW, Ness RB, Webb PM, for Australian Cancer S, et al. Combined and interactive effects of environmental and GWAS-identified risk factors in ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:880–890. doi: 10.1158/1055-9965.EPI-12-1030-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42:874–879. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Permuth-Wey J, Lawrenson K, Shen HC, Velkova A, Tyrer JP, Chen Z, et al. Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nat Commun. 2013;4:1627. doi: 10.1038/ncomms2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45:362–370. 70e1–70e2. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen H, Fridley BL, Song H, Lawrenson K, Cunningham JM, Ramus SJ, et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat Commun. 2013;4:1628. doi: 10.1038/ncomms2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–384. 84e1–84e2. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolton KL, Ganda C, Berchuck A, Pharaoh PD, Gayther SA. Role of common genetic variants in ovarian cancer susceptibility and outcome: progress to date from the Ovarian Cancer Association Consortium (OCAC) J Intern Med. 2012;271:366–378. doi: 10.1111/j.1365-2796.2011.02509.x. [DOI] [PubMed] [Google Scholar]
- 19.Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet. 2015;47:164–171. doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SH, Harold D, Nyholt DR, Consortium AN, International Endogene C, Genetic, et al. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer's disease, multiple sclerosis and endometriosis. Hum Mol Genet. 2013;22:832–841. doi: 10.1093/hmg/dds491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Permuth-Wey J, Kim D, Tsai YY, Lin HY, Chen YA, Barnholtz-Sloan J, et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 2011;71:3896–3903. doi: 10.1158/0008-5472.CAN-10-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sankararaman S, Sridhar S, Kimmel G, Halperin E. Estimating local ancestry in admixed populations. Am J Hum Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyrer J, Pharoah PD, Easton DF. The admixture maximum likelihood test: a novel experiment-wise test of association between disease and multiple SNPs. Genet Epidemiol. 2006;30:636–643. doi: 10.1002/gepi.20175. [DOI] [PubMed] [Google Scholar]
- 26.Nezhat F, Datta MS, Hanson V, Pejovic T, Nezhat C, Nezhat C. The relationship of endometriosis and ovarian malignancy: a review. Fertil Steril. 2008;90:1559–1570. doi: 10.1016/j.fertnstert.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Ness RB. Endometriosis and ovarian cancer: thoughts on shared pathophysiology. Am J Obstet Gynecol. 2003;189:280–294. doi: 10.1067/mob.2003.408. [DOI] [PubMed] [Google Scholar]
- 28.Franco HL, Dai D, Lee KY, Rubel CA, Roop D, Boerboom D, et al. WNT4 is a key regulator of normal postnatal uterine development and progesterone signaling during embryo implantation and decidualization in the mouse. FASEB J. 2011;25:1176–1187. doi: 10.1096/fj.10-175349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozdemir F, Altinisik J, Karateke A, Coksuer H, Buyru N. Methylation of tumor suppressor genes in ovarian cancer. Exp Ther Med. 2012;4:1092–1096. doi: 10.3892/etm.2012.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viger RS, Guittot SM, Anttonen M, Wilson DB, Heikinheimo M. Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol Endocrinol. 2008;22:781–798. doi: 10.1210/me.2007-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boocock CA, Charnock-Jones DS, Sharkey AM, McLaren J, Barker PJ, Wright KA, et al. Expression of vascular endothelial growth factor and its receptors flt and KDR in ovarian carcinoma. J Natl Cancer Inst. 1995;87:506–516. doi: 10.1093/jnci/87.7.506. [DOI] [PubMed] [Google Scholar]
- 32.Laviolette LA, Hodgkinson KM, Minhas N, Perez-Iratxeta C, Vanderhyden BC. 17beta-estradiol upregulates GREB1 and accelerates ovarian tumor progression in vivo. Int J Cancer. 2014;135:1072–1084. doi: 10.1002/ijc.28741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.