Abstract
In this Perspective, we expand the notion of temporal regulation of RNA in the brain and propose that the qualitative nature of RNA and its metabolism, together with RNA abundance, are essential for the molecular mechanisms underlying experience-dependent plasticity. We discuss emerging concepts in the newly burgeoning field of epitranscriptomics, which are predicted to be heavily involved in cognitive function. These include activity-induced RNA modifications, RNA editing, dynamic changes in the secondary structure of RNA, and RNA localization. Each is described with an emphasis on its role in regulating the function of both protein-coding genes, as well as various noncoding regulatory RNAs, and how each might influence learning and memory.
A remarkable feature of the adult brain is its plasticity in response to experience. It is widely accepted that, to have a lasting impact on behavior, activity-induced gene expression followed by protein synthesis in specialized regions of the brain is required for learning and the formation of long-term memory1. However, as time scales for experience-dependent transcription (minutes to hours) differ greatly from those for learning and for the consolidation and maintenance of memory (days to years), a simple, straightforward relationship between gene expression and behavioral adaptation is unlikely2. Moreover, postmitotic neurons transduce signals in mere microseconds using both chemical and electrical processes, through which a myriad of extremely fast-acting signal transduction mechanisms control ion flux, metabolic transformation of small molecules and chemical transformation of macromolecules such as proteins3. Thus, the temporal discordance between activity-induced gene expression and the real-time firing patterns of neurons underlying memory formation raises questions about the link between gene expression, protein synthesis and behavior.
The activity-induced readout of the neuronal transcriptome is dynamic, and factors such as the temporal integration of transcription rate, RNA processing and RNA degradation, as well as variations in the relative contribution of each, can obscure the linear trajectory from transcription to translation4. This is best exemplified by the fact that in many instances messenger RNA (mRNA) and protein levels do not align5. For example, it has been reported that only 40% of the variance in protein levels can be directly attributed to RNA abundance6. When translation rate constants are included the correlation is much stronger, which suggests that the rate of translation, and not necessarily the overall levels of mRNA in a cell, may be a dominant process that controls protein expression6. However, this view has recently been challenged by the observation that the kinetics of both synthesis and degradation of RNA and protein are equally involved in regulating cellular homeostasis in response to an acute stressor7. Jovanovic et al.8 note that RNA abundance contributes significantly to protein levels at steady state in immune cells and that this relationship is even more pronounced following stimulation, in accordance with the idea that translation and degradation might predominate, although in a context-specific manner9. If one factors in cell type and the rate of cell proliferation, the relationship becomes even more complex. Therefore, a consideration of the potential factors underlying the discordance between mRNA and protein levels in the adult brain is crucial for understanding experience-dependent plasticity, particularly with respect to activated postmitotic neurons engaged in the formation of a memory trace.
The understanding of the relationship between learning-induced mRNA expression and protein synthesis, and its role in cognition, is currently undergoing a renaissance as novel modes of gene regulation are being integrated into this conceptual framework. For example, epigenetic mechanisms, including DNA methylation and post-translational histone modifications, are involved in driving experience-dependent gene expression underlying the formation and maintenance of memory10,11. These chemical reactions can proceed within microseconds, and chemical modifications of proteins through post-translational modifications can have profound effects on chromatin structure and function and on subsequent gene expression. The rapid modification of cellular macromolecules is therefore highly relevant for activity-dependent molecular processes required for memory formation.
Like the epigenetic code surrounding DNA modification, there is also an emergent layer of chemistry that can profoundly influence the life of RNA (Fig. 1). For example, RNA methylation in the form of N6-methyladenosine (m6A) and N1-methyladenosine (m1A) is critical for controlling RNA steady-state levels and even the rate and fidelity of protein synthesis12,13. Another highly abundant RNA modification, pseudouridine (Ψ), has recently been shown to be dynamic and responsive to different stimuli14. Changes in RNA editing and RNA structure represent even more sophisticated layers of chemically mediated regulation of RNA, which can alter the protein code of a gene and even control post-transcriptional interactions such as protein binding affinity and microRNA targeting15,16. It is important to note that each of these chemical modifications is catalyzed by enzymatic reactions such as those that occur on DNA or histones and that, in some cases, the same critical cofactors, such as S-adenosyl methionine, are used17.
Figure 1.
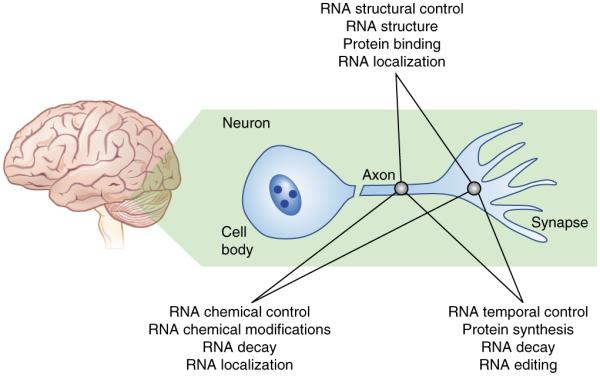
In different local environments of the neuron, epitranscriptomic mechanisms can be employed independently and bidirectionally to regulate the qualitative state of RNA and effect experience-dependent changes in neuronal function in the brain. The intersection of various aspects of RNA control, within differing regions of the same cell, affords both mechanistic and spatial control over RNA metabolism. RNA trafficking within axons to the synapse is controlled by RNA-binding proteins, which recognize unique structural and sequence elements in RNAs. Protein synthesis is controlled temporally to guide protein abundance and synapse formation. Checkpoint mechanisms may be brought about by RNA modifications, which have already been demonstrated to regulate RNA decay. Each of these are interconnected and therefore substantially increase the complexity of RNA regulation in neurons.
Changes in the metabolic state of RNA also occur on similar time scales to those for neuronal firing and depolarization; therefore, it is plausible that these processes may have evolved to be just as important for determining localized genetic flow as other activity-induced epigenetic mechanisms but with the added advantage of exerting their effects without the need to signal back to the nucleus and, in some instances, effectively bypassing the requirement for a linear relationship between mRNA and protein levels in the brain. These regulatory mechanisms represent an elusive additional hidden layer of control in the brain that is likely to be intimately involved in the molecular transactions underlying learning and memory. Advances in our understanding of the epitranscriptome and emerging technologies that can be used to unravel the complex nature of experience-dependent gene regulation in the brain are discussed below.
RNA modification
It has been known for at least half a century that RNA is subject to chemical modification, with more than 140 marks identified to date18. These post-transcriptional ‘epitranscriptomic’ modifications, which direct the functional readout of nascent RNAs in a highly structured and coordinated manner, have recently been found to occur on many classes of RNA beyond ribosomal RNA (rRNA), transfer RNA (tRNA) and snoRNA (small nucleolar RNA). The list now includes mRNAs, as well as short and long noncoding RNAs19. In addition, depending on the locus, chemical modifications on RNA can dictate patterns of alternative splicing and degradation20 and influence secondary structure21. Perhaps most importantly with respect to experience-dependent effects in the brain, RNA modification can modulate the rate of translation22. These mechanisms may therefore serve as an epigenetic code for fine-tuning activity-dependent changes in the state of RNA, imparting functional diversity without the need for further increased levels of transcription. However, beyond the transcriptome-wide mapping of the most prevalent marks and the initial identification of their readers, writers and erasers, these are early days for the study of RNA modification23. Important next steps will be to define the functional relevance of these marks and the precise upstream signals that engage their respective regulatory mechanisms in the brain and to consider these features in a cell-type-specific manner under baseline and activated conditions (Box 1). It is evident that much more effort is required to better define how different epitranscriptomic mechanisms play context-dependent roles in RNA metabolism, particularly in the brain, and to what extent they contribute to learning and memory.
Box 1. Future directions.
Regardless of the rapid advances that have been made in the past few years with respect to our appreciation of RNA modification in the brain, much more work is needed in this field, especially to achieve the following:
A deeper understanding of the diversity of RNA modifications across subcellular compartments, cell types, tissues, brain regions and development and neuronal states.
Direct quantification of the temporal and spatial dynamics of RNA modifications and metabolism and how they relate to transcript and protein levels and to the dynamics of neuronal activity, plasticity and transmission.
Insight into the molecular mechanisms that transduce cellular and neuronal activity into locus-specific changes in RNA and how these persist to maintain a specific cellular or subcellular (for example, synaptic) state.
Determination of the functional relevance of different RNA species, structures and modifications to cognition and memory, which will require innovative new methods for temporally precise and spatially restricted locus-specific causal manipulations.
N6-methyladenosine
The RNA modification N6-methyladenosine (m6A), of which there are a number of readers and writers24, is abundant throughout the mammalian transcriptome and appears to be involved in a variety of biological processes12,25–27, including RNA translation, degradation, localization, splicing and RNA-induced structural states. In the mouse brain, m6A is developmentally regulated and increases in adulthood26, which suggests a role in the post-transcriptional regulation of RNA associated with neural plasticity and behavioral adaptation. Activity-dependent changes in m6A in the mammalian transcriptome have recently been observed in response to heat-shock stress22. We have discovered that m6A is also dynamic in the adult brain, which is reflected by widespread learning-induced, locus-specific accumulation of m6A in RNA derived from the prefrontal cortex28. Furthermore, when the accumulation of m6A is amplified following knockdown of the RNA demethylase FTO, memory is enhanced, an effect that is accompanied by reduced stability of target mRNAs. Hess et al.29 found that RNA hypermethylation is associated with increased levels of target mRNAs but decreased levels of proteins in FTO knockout mice. The findings suggest that m6A is a critically important epitranscriptomic modification associated with behavioral adaptation, although its relationship with RNA expression is not so straightforward. The differential effects of m6A may depend on distinct cis-acting elements that are present on the RNA molecule and which may interact with m6A, including microRNAs30.
Because of the versatility of m6A in regulating a plethora of RNA functions, the phenotypic consequence of FTO perturbation in mice is likely to be complicated by multiple regulatory effects exerted by m6A. Although the functions of m6A in alternative splicing, translational dynamics and mRNA transport in vivo remain to be investigated, it is also possible that one function of learning-induced m6A is to constrain the sorting efficiencies or turnover of nascent mRNAs. As indicated, the accumulation of m6A has been shown to mark its target mRNAs for both degradation and translation in the cytoplasm22,31. With FTO knockdown, the increased targeting of plasticity-related genes by m6A may allow them to be efficiently translated and then rapidly degraded to bring the mRNA pool down to a minimum, thereby reducing ‘transcriptional noise’ in neurons32.
Regardless of these interesting threads, many questions remain about the functional relevance of RNA methylation. For example, it was recently discovered that N1-methyladenosine (m1A) represents a distinct RNA modification that exerts its influence on RNA metabolism independent of m6A. Although both marks are highly conserved and have been shown to be dynamic, m6A appears accumulate preferentially at the stop codon and along the 3′ untranslated region (UTR)25, whereas m1A is found selectively at the start codon13. The limitation of these findings is that they, like much of the research to date, used antibody-based immunoprecipitation approaches, which cannot distinguish between m6A and m1A. Moreover, current methods such as photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) or high-throughput sequencing of RNA isolated by crosslinking and immunoprecipitation (HITS-CLIP) are notoriously inefficient, resulting in just 1–5% yields33, with significant biases reported toward highly abundant and stable transcripts such as rRNA or tRNA. Furthermore, these methods are also limited by the amount of input RNA required, which is prohibitive when considering select cell populations derived from specific brain regions. Recent innovations in base resolution approaches such as chemogenetic tagging followed by sequencing and PCR stop analysis or long-read, single-molecule, real-time sequencing will help to clarify where, when and at what position adenosine becomes methylated. This information will then need to be overlaid with a direct readout of RNA abundance, translation (or degradation), splicing and/or localization within the cell, which will be essential for unraveling the functional role of RNA methylation in learning and memory formation.
Pseudouridine
The post-transcriptional modification of uracil is by far the most abundant of all RNA modifications34. Although the presence of Ψ is well established, relatively little is known about the function of this unusual modification, particularly in the context of brain function. There is a sharp increase in the abundance of Ψ residues in eukaryotes compared to bacteria (four- to eightfold more Ψ sites), suggesting a greater demand for Ψ in higher organisms34. Indeed, in mammals, rRNA contains more than 100 Ψ residues per ribosome, while each tRNA molecule itself contains at least three Ψ sites35. Recent evidence indicates the presence of many potentially functional Ψ sites in coding and noncoding transcripts that are enriched in the brain36. The ubiquitous presence of this modification therefore raises the intriguing possibility of its involvement in basic neuronal processes, particularly given that Ψ is responsive to environmental stimuli, including stress37.
Most studies to date have focused on the physiological properties of Ψ in an attempt to determine its function. Ψ is formed post-transcriptionally by pseudouridine synthases (PUS), which act on maturing RNA molecules by isomerizing the uracil base moiety along its N3–C6 axis and forming a glycosidic carbon–carbon bond without additional energy requirements38. Based on this chemistry, Ψ was thought to contribute primarily to the flexibility of the RNA molecule39. However, contrary to the concept of Ψ as a free nucleoside, spectroscopy and molecular simulations suggest that Ψ improves local RNA stacking and interactions with RNA-binding proteins, enhances Ψ duplex formation and stabilizes the overall structure of RNAs40–43.
Isomerization of uridine is catalyzed by two separate sets of enzymes with distinct mechanisms for identifying pseudouridylation targets. The first uses one of several H/ACA box snoRNAs that guide the enzyme to appropriate target residues via direct base-pairing of the snoRNA guide strand with the target RNA44. The H/ACA snoRNA ribonucleoprotein complex is responsible for pseudouridylation of several small RNAs and long noncoding RNAs (lncRNAs) within the nucleus. H/ACA snoRNAs can also contain Ψ sites and allow modification of other snoRNAs. In contrast, PUS proteins act independently of H/ACA snoRNAs, with each PUS protein containing a consensus sequence that binds directly to target RNAs, thereby conferring specificity. Seven independent PUS proteins have been identified; they are conserved from yeast to humans, and a recent sequencing analysis of Ψ sites within both species suggests that independent PUS proteins are responsible for pseudouridylation in all classes of RNA14. This is particularly intriguing considering that several of these enzymes, including TruB, the PUS protein responsible for the universally conserved U55 modification in tRNA, have high levels of expression and an enrichment of targets in the brain. Finally, it has been suggested that transglycosylation, a process that is required for the formation of Ψ, might also lead to a novel form of editing reflected by an unusual U-to-A conversion in RNA45. This is an interesting hypothesis and, if proven true, could have a very significant impact on the dynamic coding potential of the transcriptome given that activity-induced Ψ-mediated RNA base-flipping in the brain would then represent a novel mode of creating RNA diversity. At this stage, however, the functional role of Ψ in the brain, and any potential downstream editing events associated with the formation of Ψ, remain to be determined.
RNA editing
The conversion of adenosine to inosine residues by base deamination, or A-to-I editing, leads to qualitatively different proteins, promotes functional diversity and serves to fine-tune the genomic response to rapidly changing environmental demands46. There has been an extraordinary expansion in the discovery of RNA editing sites in the human brain47. Through the use of emerging technology, ~1.4 million editing sites have been identified, with the majority occurring in Alu repeats48, and over 100 million are now predicted to occur in the mammalian brain49. These findings strongly suggest a role for this epitranscriptomic process in the evolution of cognitive function50. RNA editing is mediated by two major classes of enzymes; the first, comprising a group of adenosine deaminases called ADARs, exhibits tissue-specific and context-dependent patterns of expression51–53. The second, the vertebrate-specific APOBEC family, promotes C-to-U editing by cytosine deamination. Interestingly, APOBEC3 is primate-specific, again hinting at a relationship between RNA editing and cognitive evolution as suggested by Barry and Mattick50. With respect to A-to-I conversion, ADAR1 and ADAR2 promote the conversion of glutamine to arginine (Q/R site) within the 5-HT2C subunit of serotonin receptors and within the GluR2 subunit of AMPA receptors, as well as voltage-gated calcium channels, and can even alter the structure of the synapse, which is interesting given that each of these targets is known to affect learning and memory (for a comprehensive review of RNA editing in the brain, see ref. 16).
Emerging evidence suggests that methyladenosine is a direct target for deamination by the editing enzyme activation-induced cytidine deaminase (ref. 54), highlighting the potentially interconnected nature of chemically modified RNA and RNA editing. A key unresolved issue is how nascent or constitutively expressed transcripts are localized in the cell or targeted for RNA editing. How would one go about quantifying this process in the brain? Our recent study on learning-induced accumulation of m6A in the adult brain28 suggests that RNA modification may hold the answer. We found that in 20% of all transcripts where m6A accumulates in an experience-dependent manner, this epitranscriptomic mark is present in mRNA encoding proteins that are subsequently localized to the synapse. To address this question in a robust, quantitative manner, a method that enables the capture of high-quality synaptically localized RNA must be developed (Box 2). Ideally, this approach would also differentiate between activated and quiescent synapses.
Box 2. Emerging technologies.
Methods for understanding the inner workings of postmitotic neurons are undergoing a revolution due to the merger of classical biochemical techniques with transcriptomics. These approaches are being employed to gain a holistic view of how RNA molecules are controlled from transcription to decay. RNA translation within neurons is controlled in space and time. A new method called ribosome profiling allows the transcriptome-wide analysis of ribosome footprints73 and has also been performed in subcellular compartments, thereby permitting the analysis of translation in space74,75. An application of ribosome profiling in different segments within neurons (cell body, axon, dendrite, etc.) permits tracking RNA movement and examining control of translation at the transcriptome level. If performed in select cell populations following a learning event, such an approach would provide an analysis of experience-dependent translation, which would represent a significant advance over current total RNA-seq approaches.
The ability to understand how RNA modification influences RNA metabolism will only be as good as the precision at which RNA modification can be controlled and assayed. Clustered, regularly interspaced, short palindromic repeats (CRISPR) proteins have been shown to be amenable to engineering and depositing chemical modifications on genomic DNA to better understand how such marks control transcription76. The recent discovery that CRISPR–Cas effector systems can be employed to target cellular RNA opens the window for employing this targeted approach for similar analyses of RNA modification77,78. For example, engineering Cas9-C2c2-PUS fusions may enable pseudouridylation to be directed to predicted sites on RNA. The same could be performed with other RNA-editing and RNA-modifying enzymes such as ADAR and the RNA methyltransferases. This would facilitate high-resolution interrogation of the causal relationships between single marks at individual sites and how they influence RNA biology in the brain.
Genomic technologies have also been used to study RNA localization. However, they are limited by low-resolution fractionation methods. Attempts at isolating organelles (greater cellular resolution) with high purity or preserving spatial relationships have proven much less fruitful. The isolation of these compartments for study relies on centrifugation gradients, which can often lead to high false-positive rates due to lysis79,80. Preserving the spatial organization of a cell before lysis would have a significant impact on how RNA localization is assayed. There has been some work toward this goal with proteins. Engineered ascorbate peroxidase (APEX)-generated molecular labeling can provide information regarding intracellular localization of proteins. In this technique, tyrosyl radicals are generated to make protein cross-links in a distance-dependent manner81,82. This approach has not yet been shown to work on RNA, but it is possible that RNAs associated with labeled proteins could be purified and sequenced to reveal their spatial location.
Finally, assaying and understanding RNA content inside cells is also critical for addressing another major outstanding question in biology: what dictates neuronal specification? This is a significant problem for neuroscientists, who do not have a clear picture of how many unique cell types exist in the brain. One way to overcome this barrier would be to better understand the gene expression profiles and RNA metabolism, and even translation, of single cells. This will require major technological advances and bioinformatics development. Some headway has been made in this regard as it has been shown that single-cell RNA sequencing can reveal the expression patterns of a multitude of different neuronal subtypes in the human cortex83. A next critical experiment will be to profile RNA expression in each of these neuronal subtypes following learning, and to devise new technology to capture nascent transcripts from these cells at different time points in the same cell, in vivo. Another benchmark would be to quantify translation at the single-cell or single-RNA level. Morisaki et al.84 have begun to tackle this issue with a method they call nascent chain tracking. They use multi-epitope tags and antibody-based fluorescent probes to visualize and measure the translation dynamics of individual RNAs in vivo in real time. Such advances represent important progress in understanding RNA biology and will be integral to elucidating the contribution of the qualitative state of RNA in individual neurons to memory formation and experience-dependent change across the lifespan.
RNA structure
Neurons respond to dynamic switches in the cellular environment. As such, the molecular components that regulate this responsiveness must also be malleable and controlled by modular components that can react quickly to stimuli and drive gene expression. The structure of RNA provides a modifiable context in which this can occur. Indeed, RNA structural elements respond to ion concentrations, metabolite flux, RNA-binding proteins and even changes due to RNA–RNA interactions55,56. As such, RNA structure may serve as a central conduit controlling signal transduction pathways that are critical for single-neuron or even high-order cerebral function. This role of RNA as a molecular sensor requires that RNA structures be (i) highly specific, so that distinct RNA structures can respond to specific cellular stimuli, and (ii) highly dynamic, so that the cellular response can be rapid. Below we elaborate on a few examples that demonstrate the specific and dynamic character of RNA structures and how identifying such structures in a transcriptome-wide manner can enhance our understanding of RNA function in the brain.
The lncRNA MALAT1 has emerged as a prototypical example of a brain-enriched lncRNA that is subject to chemical and structural modification. It is found in nuclear paraspeckles within hippocampal neurons, where one of its reported roles is to regulate the splicing of genes related to synaptogenesis57, as well as within cortical neurons, where it appears to have a cis-regulatory function58. Recent evidence indicates that m6A can predict whether an individual RNA will change its secondary structure21 and that, when m6A accumulates in MALAT1, this modification alters the interaction of MALAT1 with its RNA-binding proteins15. Furthermore, we have recently shown the presence of dynamic, experience-dependent lncRNA expression (including MALAT1) in the mammalian brain59,60, and the majority of these transcripts harbor potential m6A and Ψ sites. It remains to be determined whether modification at these leads to functionally relevant m6A- or Ψ-mediated effects on activity-induced lncRNAs in the brain and whether they contribute substantially to learning.
Chemical modifications are also known to affect the folding of RNA and are critical for determining its secondary and tertiary structures, which can influence the function of RNA inside the cell21,61. RNA structure has recently been linked to learning and memory. A stem-loop structure in the 3′ UTR of BDNF, a key neurotropic factor for learning and memory, has been shown to be calcium-dependent and necessary for RNA stabilization62. In addition, others have shown that the G-quadruplex RNA structure is required for localization of Ca2+/ calmodulin-dependent protein kinase II (CAMK2α) and postsynaptic density protein (PSD-95) to neurites, both of which are essential for the synaptic plasticity required for learning63. Alternative splicing of exon 10 of tau, a protein well known for its role in neurodegenerative disorders, is regulated by a stem loop induced by a particular RNA helicase64. Furthermore, the RNA editing capacity of ADAR1 is regulated by RNA structure and favored in a Z-RNA confirmation65.
The above observations demonstrate that RNA structural elements are both dynamic and critical for the regulation of many cellular processes. A key challenge for future efforts aimed at elucidating the role of RNA structure in the regulation of neuronal pathways is to obtain a systems-level understanding of RNA structure inside living cells. The functional relevance of modified secondary RNA structures in the context of behavioral adaptation has yet to be explored. This will be challenging due to the spatiotemporal nature of RNA structure state changes in vivo, which will require a method for tagging RNAs in neurons in a freely behaving animal. One such approach may be to employ photoinducible ‘click chemistry’ combined with standard optics platforms, such as those used for optogenetics. In this way, the spatial and temporal integration of the chemogenetic tag in specific cell populations in the brain can be controlled, and these populations could then be purified and sequenced using now-standard transcriptome-wide sequencing platforms. Indeed, recent efforts have focused on transcriptome-wide measurements of RNA structure66–69. These studies, although not yet applied to neurons, have nonetheless revealed general principles of RNA structural regulation and the critical contribution of RNA structural elements to the control of translation site selection, the binding of RNA-binding proteins and even m6A RNA methylation.
RNA localization
Determining how a cell organizes its molecular components is one of the great remaining challenges in neuroscience. Proteins were once thought to be the only molecules with specific localization properties. However, an overwhelming body of evidence has been gathered since the mid-1980s to indicate that cells localize proteins, at least in part, by directing the corresponding mRNAs. The localization of mRNAs to subcellular compartments provides a mechanism for regulating gene expression with exquisite temporal and spatial control. Localization of RNAs is widespread and evolutionarily conserved. RNA localization, whereby mRNAs can be targeted to specific neuronal subcellular domains to enable rapid changes in the spatial proteome through local translation, is a hallmark of neurobiology70.
The vast majority of work in the field has focused on the localization of protein-coding mRNAs; nevertheless, lncRNAs are also often subject to specific subcellular localization. Although focused on non-neurobiological systems, the examples below are worth mentioning as they illustrate that lncRNAs can influence cellular function by acting outside the nucleus. For example, the lncRNA NRON forms a complex with the shuttling protein importin to regulate the subcellular trafficking of nuclear factor of activated T cells (NFAT; ref. 71). The double-stranded RNA–binding protein Staufen1 can be recruited to mRNA targets through Alu elements in cytoplasmic lncRNAs to enhance decay72. Thus the many functions of lncRNAs, which may be dictated by both RNA modification and structural state, from chromatin remodeling to translation control, have prompted the need to develop new biochemical tools for studying lncRNA function in living cells. Overall, these examples demonstrate how RNA interplay and localization outside the nucleus of regulatory RNAs could be controlling gene expression. Although it has been studied for many years, RNA localization within neurons remains a frontier for investigation in the context of the adult brain and experience-dependent plasticity.
Outlook and conclusions
It is evident that the coordination and synchronicity of biochemical processes related to RNA metabolism, including its modification, editing and structural variation, bidirectionally contribute to the language and internal dialog of the cell and are critically important for driving experience-dependent plasticity in the brain and adaptive behavior. However, it is also clear that much more work is needed in this area (Box 1). With this in mind, it may not be beyond the limitations of a neuron to coordinate the regulation of a single RNA from the time of its inception to its degradation. In fact, each of the mechanisms discussed above could be critical for controlling the biology of a single mRNA molecule in real time, without the need to signal back to the nucleus. There are clear overlaps and relationships between RNA chemical modification, editing, structure and localization, which can impact translation (Fig. 2). These mechanisms could be the driving force behind RNA regulation that occurs without a change in transcription signals to and from the nucleus. Once the RNA is at the synapse, it could be temporally edited to alter the coding sequence and give rise to an alternative protein product that has enhanced or reduced efficacy. As RNA methylation has been demonstrated to control RNA decay, temporal changes in the accumulation of m6A may serve as a signal for localized degradation at the synapse. Because these events can be altered and reinitiated in seconds, they can be aligned with neuronal firing and gene responses that may be responsible for memory consolidation and the organizational architecture of neuronal networks.
Figure 2.
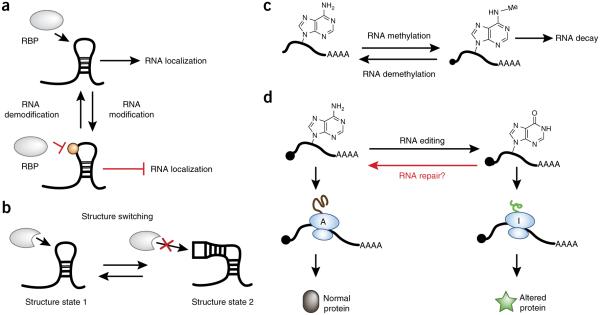
Projected mechanisms that can control the life of a single RNA gene, perhaps at a single-molecule level. (a) RNA-binding proteins control many facets of RNA biology. RNA–protein interactions can be controlled by chemical modifications (m6A, for example). The interplay between physical changes to RNA and protein binding is therefore complex and affords many opportunities for potential regulation (RBP: RNA-binding protein). (b) Structure switching is a key mechanism that can either inhibit or enhance protein binding. (c) Schematic demonstrating N6-methylation of adenosine, which has been shown to lead to RNA decay. (d) RNA editing can result in the expression of an altered protein. This figure demonstrates A-to-I editing, which can alter codon identity, leading to a protein with an altered sequence.
If we are to fully understand how RNA biology is being controlled in individual neurons or cellular subtypes within the brain, we need to further expand our armamentarium of biochemical methods. There are now many RNA-centric methodologies (Box 2) that could be explored to further understand the qualitative nature of RNA and its metabolism, together with RNA abundance, as key features of the functional diversity of RNA in the adult brain. Armed with new sequencing technologies and cell-type-specific profiling, the field is ready to evolve beyond linear, unidirectional relationships between gene transcription and protein synthesis and to accept the challenge of elucidating the fundamental features of epitranscriptomic mechanisms in the brain and their role in regulating learning and memory.
ACKNOWLEDGMENTS
The authors thank R. Tweedale for editing the manuscript and gratefully acknowledge grant support from the NIH (5R01MH105398-T.W.B.), NIH (1R01MH109588-01-T.W.B. and R.C.S.), NIGMS (1DP2GM119164-01-R.C.S.) and the NHMRC (APP1033127-T.W.B.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Alberini CM, Kandel ER. The regulation of transcription in memory consolidation. Cold Spring Harb. Perspect. Biol. 2014;7:a021741. doi: 10.1101/cshperspect.a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGaugh JL. Memory–a century of consolidation. Science. 2000;287:248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- 3.Sweatt JD. Neural plasticity & behavior–sixty years of conceptual advances. J. Neurochem. 2016;2016:14. doi: 10.1111/jnc.13580. [DOI] [PubMed] [Google Scholar]
- 4.Rabani M, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Z, et al. Differential dynamics of the mammalian mRNA and protein expression response to misfolding stress. Mol. Syst. Biol. 2016;12:855. doi: 10.15252/msb.20156423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jovanovic M, et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347:1259038. doi: 10.1126/science.1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JJ, et al. System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ. 2014 doi: 10.7717/peerj.270. https://peerj.com/articles/270/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker-Andresen D, Ratnu VS, Bredy TW. Dynamic DNA methylation: a prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci. 2013;36:3–13. doi: 10.1016/j.tins.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Lopez-Atalaya JP, Barco A. Can changes in histone acetylation contribute to memory formation? Trends Genet. 2014;12:529–539. doi: 10.1016/j.tig.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 13.Dominissini D, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behm M, Öhman M. RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet. 2016;32:165–175. doi: 10.1016/j.tig.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Liu N, Pan T. N6-methyladenosine–encoded epitranscriptomics. Nat. Struct. Mol. Biol. 2016;23:98–102. doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- 18.Machnicka MA, et al. MODOMICS: a database of RNA modification pathways— 2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saletore Y, et al. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13:175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K, et al. Structural and functional characterization of the proteins responsible for N6-methyladenosine modification and recognition. Curr. Protein Pept. Sci. 2015 [PubMed] [Google Scholar]
- 21.Spitale RC, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 2016;6:365–372. doi: 10.1038/nrg.2016.47. [DOI] [PubMed] [Google Scholar]
- 24.Roundtree IA, He C. RNA epigenetics--chemical messages for posttranscriptional gene regulation. Curr. Opin. Chem. Biol. 2016;30:46–51. doi: 10.1016/j.cbpa.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 26.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Widagdo J, et al. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 2016;36:6771–6777. doi: 10.1523/JNEUROSCI.4053-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hess ME, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 2013;16:1042–1048. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- 30.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siciliano V, et al. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat. Commun. 2013;4:2364. doi: 10.1038/ncomms3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 2002;514:17–25. doi: 10.1016/s0014-5793(02)02305-0. [DOI] [PubMed] [Google Scholar]
- 35.Roovers M, et al. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res. 2006;34:4293–4301. doi: 10.1093/nar/gkl530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015;8:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 37.Wu G, Huang C, Yu YT. Pseudouridine in mRNA: Incorporation, Detection, and Recoding. Methods Enzymol. 2015;560:187–217. doi: 10.1016/bs.mie.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preumont A, Snoussi K, Stroobant V, Collet JF, Van Schaftingen E. Molecular identification of pseudouridine-metabolizing enzymes. J. Biol. Chem. 2008;283:25238–25246. doi: 10.1074/jbc.M804122200. [DOI] [PubMed] [Google Scholar]
- 39.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 40.Neumann JM, Bernassau JM, Guéron M, Tran-Dinh S. Comparative conformations of uridine and pseudouridine and their derivatives. Eur. J. Biochem. 1980;108:457–463. doi: 10.1111/j.1432-1033.1980.tb04742.x. [DOI] [PubMed] [Google Scholar]
- 41.Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 42.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu YT, Meier UT. RNA-guided isomerization of uridine to pseudouridine– pseudouridylation. RNA Biol. 2014;11:1483–1494. doi: 10.4161/15476286.2014.972855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge J, Yu YT. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 2013;38:210–218. doi: 10.1016/j.tibs.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia GA, Kittendorf JD. Transglycosylation: a mechanism for RNA modification (and editing?) Bioorg. Chem. 2005;33:229–251. doi: 10.1016/j.bioorg.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li JB, Church GM. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat. Neurosci. 2013;16:1518–1522. doi: 10.1038/nn.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramaswami G, et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bazak L, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barry G, Mattick JS. The role of regulatory RNA in cognitive evolution. Trends Cogn. Sci. 2012;10:497–503. doi: 10.1016/j.tics.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 51.Paupard M-C, O’Connell MA, Gerber AP, Zukin RS. Patterns of developmental expression of the RNA editing enzyme rADAR2. Neuroscience. 2000;95:869–879. doi: 10.1016/s0306-4522(99)00431-5. [DOI] [PubMed] [Google Scholar]
- 52.Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl. Acad. Sci. USA. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cattenoz PB, Taft RJ, Westhof E, Mattick JS. Transcriptome-wide identification of A > I RNA editing sites by inosine specific cleavage. RNA. 2013;19:257–270. doi: 10.1261/rna.036202.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franchini DM, et al. Processive DNA demethylation via DNA deaminase-induced lesion resolution. PLoS One. 2014;9:e97754. doi: 10.1371/journal.pone.0097754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011;12:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 57.Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang B, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barry G, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry. 2014;19:486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 60.Spadaro PA, et al. Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biol. Psychiatry. 2015;78:848–859. doi: 10.1016/j.biopsych.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fukuchi M, Tsuda M. Involvement of the 3′-untranslated region of the brain-derived neurotrophic factor gene in activity-dependent mRNA stabilization. J. Neurochem. 2010;115:1222–1233. doi: 10.1111/j.1471-4159.2010.07016.x. [DOI] [PubMed] [Google Scholar]
- 63.Subramanian M, et al. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kar A, et al. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol. Cell. Biol. 2011;31:1812–1821. doi: 10.1128/MCB.01149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koeris M, Funke L, Shrestha J, Rich A, Maas S. Modulation of ADAR1 editing activity by Z-RNA in vitro. Nucleic Acids Res. 2005;33:5362–5370. doi: 10.1093/nar/gki849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 67.Pop C, et al. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014;10:770. doi: 10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kubota M, Tran C, Spitale RC. Progress and challenges for chemical probing of RNA structure inside living cells. Nat. Chem. Biol. 2015;11:933–941. doi: 10.1038/nchembio.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lipshitz HD, Smibert CA. Mechanisms of RNA localization and translational regulation. Curr. Opin. Genet. Dev. 2000;10:476–488. doi: 10.1016/s0959-437x(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 71.Willingham AT, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 72.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- 74.Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nelles DA, et al. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell. 2016;165:488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abudayyeh OO, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016 doi: 10.1126/science.aaf5573. http://dx.doi.org/10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riley KJ, Yario TA, Steitz JA. Association of Argonaute proteins and microRNAs can occur after cell lysis. RNA. 2012;18:1581–1585. doi: 10.1261/rna.034934.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hung V, et al. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell. 2014;55:332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee SY, et al. APEX fingerprinting reveals the subcellular localization of proteins of interest. Cell Rep. 2016;15:1837–1847. doi: 10.1016/j.celrep.2016.04.064. [DOI] [PubMed] [Google Scholar]
- 83.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morisaki T, et al. Real-time quantification of single RNA translation dynamics in living cells. Science. 2016;352:1425–1429. doi: 10.1126/science.aaf0899. [DOI] [PubMed] [Google Scholar]