Summary
3-Hydroxypropionic acid (3-HP) is a novel antimicrobial agent against foodborne pathogens like Salmonella and Staphylococcus species. Lactobacillus reuteri converts glycerol into 3-HP using a coenzyme A-dependent pathway, which is encoded by propanediol utilization operon (pdu) subjected to catabolite repression. In a catabolite-repression-deregulated L. reuteri RPRB3007, quantitative PCR revealed a 2.5-fold increase in the transcripts of the genes pduP, pduW and pduL during the mid-log phase of growth. The production of 3-HP was tested in resting cells in phosphate buffer and growing batch cultures in MRS broth of various glucose/glycerol ratios. Due to the upregulation of pathway genes, specific formation rate of 3-HP in the mutant strain was found to be enhanced from 0.167 to 0.257 g per g of cell dry mass per h. Furthermore, formation of 3-HP in resting cells was limited due to the substrate inhibition by reuterin at a concentration of (30±5) mM. In batch cultures, the formation of 3-HP was not observed during the logarithmic and stationary phases of growth of wild-type and mutant strains, which was confirmed by NMR spectroscopy. However, the cells collected in these phases were found to produce 3-HP after washing and converting them to resting cells. Lactate and acetate, the primary end products of glucose catabolism, might be the inhibiting elements for 3-HP formation in batch cultures. This was confirmed when lactate (25±5 mM) or acetate (20±5 mM) were added to biotransformation medium, which prevented the 3-HP formation. Moreover, the removal of sodium acetate and glucose (carbon source for lactic acid production) was found to restore 3-HP formation in the MRS broth in a similar manner to that of the phosphate buffer. Even though the genetic repression was circumvented by the up-regulation of pathway genes using a mutant strain, 3-HP formation was further limited by the substrate and catabolite inhibition.
Key words: 3-hydroxypropionic acid, catabolite inhibition, catabolite repression, Lactobacillus reuteri, biotransformation
Introduction
3-Hydroxypropionic acid (3-HP) is a C3 β-hydroxy carboxylic acid and a structural isomer of lactic acid. It is one of the novel antimicrobial compounds against foodborne pathogens like Salmonella and Staphylococcus species (1). Apart from direct food preservative applications, it has got appreciable nematicidal activity against parasites and saprophytes of staple food crops (2). Currently, biological production of 3-HP is under research and is largely focused on the production of green chemicals like acrylic acid (3). For food and unique biomedical applications, GRAS (Generally Regarded as Safe) production of 3-HP is more suitable and acceptable for regulatory clearances unlike the current methods employed in C3 chemical synthesis (4). Lactobacillus reuteri is an attractive host for 3-HP synthesis as it is known for the production of its precursor molecule, 3-hydroxypropanaldehdye (3-HPA/reuterin), from glycerol. Metabolic pathway of 3-HP in L. reuteri is provided with an ATP-producing step (5) which is necessary for export of weak acids like 3-HP out of the cell (6).
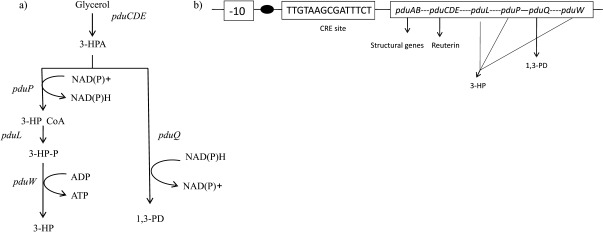
A detailed metabolic pathway (5) is outlined in Fig. 1a and its genetic arrangement is shown in Fig. 1b (7). The previously conducted metabolite characterisation studies on L. reuteri using batch cultures and chemostat cultivations have confirmed the absence of β-hydroxy acid during fermentation (8). The lack of 3-HP formation in the earlier reports might be due to the catabolite repression element (CRE) (9) in the upstream of 3-HP pathway genes. The recombineering approaches in L. reuteri have led to the development of CRE mutants (7) and had not been studied for 3-HP formation, until the recent flux analysis was reported in resting cells (10).
Fig. 1.
Bioconversion pathway of glycerol to 3-hydroxypropionic acid (3-HP): a) pduCDE=vitamin B12-dependent glycerol dehydratase, pduP=coenzyme A- and NAD(P)+-dependent propanaldehyde dehydrogenase, pduL=phosphotransacylase, pduW=propionate kinase and pduQ=NAD(P)H-dependent 1,3-propanediol (1,3-PD) dehydrogenase genes; b) an overview of 1,2-propanediol utilization operon (pdu) showing upstream transcriptional regulatory element, a catabolite repression site. In the catabolite repression element (CRE) mutant, CRE site was modified into TGAATTCCGATTTCT using recombineering approach (7). The black dot indicates the transcriptional start site
Overexpression of the pathway genes either by transcriptional deregulation or a plasmid-borne expression of homologous genes is a widely employed technique to enhance the feasibility of producing a compound in natural hosts. In this study, the 3-HP metabolic pathway of CRE mutant RPRB3007 was analysed quantitatively at transcript levels and the effect of mutation over 3-HP formation was studied in the resting cells and growing batch cultures.
Materials and Methods
Strains and media
Lactobacillus reuteri strain ATCC PTA 6475 (wild-type) and its CRE mutant strain RPRB3007, obtained from Prof. Robert A. Britton (Michigan State University, East Lansing, MI, USA), were propagated anaerobically in MRS complex medium containing 20 g/L of glucose (11). The growth was monitored by measuring the absorbance at 600 nm, where A600 nm=1 was equivalent to 0.33 g of cell dry mass per L of medium.
Transcript analysis using quantitative PCR
The transcript analysis of genes involved in 3-HP metabolic pathway (Fig. 1a), namely pduP, pduL and pduW, was done using a quantitative PCR approach. Once the cells were sampled, they were treated with RNAprotect Bacteria Reagent (Qiagen, Valencia, CA, USA) and stored at –80 °C until the RNA extraction was done using on-column DNase digestion with RNase-free DNase. As it is a Gram-positive bacterium, its cell lysis was supplemented using 50 units of mutanolysin and 25 mg/mL of lysozyme at 37 °C for 20 min during RNA extraction. The synthesis of cDNA using M-MuLV reverse transcriptase and the real-time PCR analysis with SYBR® Green Master Mix (Life Technologies, Carlsbad, CA, USA) were carried out further as described previously (12). For ΔCT calculations, 16S ribosomal RNA was used as reference gene and ΔΔCT was calculated based on the difference between the ΔCT of a gene in the mutant and in the wild-type strains. Transcripts were compared using the fold change, 2–ΔΔCT.
Propanaldehyde dehydrogenase activity in the wild-type and mutant strains
In order to ascertain the effect of CRE mutation on the enzyme activities, one of the pathway enzymes, pduP (Coenzyme A- and NAD(P)+-dependent propionaldehyde dehydrogenase) was assayed using the method described by Leal et al. (13) with certain modifications as follows. Briefly, the enzyme assay reaction mixture in 50 mM potassium phosphate buffer (pH=8) comprised 5 mM NAD+, 100 µM coenzyme A (CoA), 3 mM propanaldehyde, and the total cell lysate (55 µg/mL of total protein). After 10 min of incubation at 37 °C, the amount of produced NADH was measured by checking the absorbance at 340 nm. One unit (U) of enzyme activity was defined as the amount of enzyme producing 1 µmol of NADH per min.
Biotransformation of glycerol in resting cells and supernatant analysis
MRS broth containing 100 mM glycerol was inoculated with 5% cells grown overnight and was cultivated at 37 °C and 100 rpm. Anaerobic conditions were maintained by flushing sterile nitrogen in rubber stopper vials at the beginning of cultivation. In the mid-log phase (6 h), resting cells were prepared as described earlier (14) with following modifications. Cells were washed twice at room temperature using 50 mM phosphate buffer (pH=7.4) and resuspended in the same buffer containing 150 mM glycerol until A600 nm=20 was obtained. Biotransformation was carried out at 37 °C under static conditions and subsequently 3-HP synthesis was monitored using a HPLC (Shimadzu LC-10AT VP, Tokyo, Japan) that was equipped with a refractive index detector (RID) and an Aminex HPX-87H column (300 mm×78 mm, Bio-Rad, Hercules, CA, USA). The mobile phase was 35% acetonitrile in 5 mM sulphuric acid. Elution was carried out at room temperature with RID kept at 50 °C. Other metabolites like lactate, acetate, glycerol, 1,3-propanediol (1,3-PD), and 3-hydroxypropanaldehyde (3-HPA) were also quantified using HPLC as described in our earlier study (11).
Cofermentation and two-stage fermentation of glycerol
Cofermentation was done anaerobically in 5-mL vials containing different initial glucose/glycerol ratios (in g/g), namely 20:5, 20:10, 20:15, 20:20 and 20:25. Batch cultures were inoculated with 5% inoculum in multiple vials, and every 3 h a vial was used for 3-HP detection. A two-stage fermentation was carried out in a 1.4-litre working volume fermentor (Bioengineering AG, Wald, Switzerland) sparged with nitrogen, in which glycerol was added in the second step after the exhaustion of glucose or sucrose, resulting in biomass formation in the first stage. A feed solution of 40% glycerol was used at a feed rate of 10–30 mL/h. Every two hours, samples were prepared for HPLC analysis as described in an earlier study (11).
NMR analysis using 13C3-glycerol in batch cultures
To ensure the absence of 3-HP in batch cultures, unlabelled glycerol was replaced by 13C3-glycerol in cofermentation studies for NMR analysis, which was performed in duplicates. In the mid-log and late exponential phases of growth, cells were discarded and the supernatants were concentrated by lyophilization. The residue was redissolved in 500 µL of deuterated water to record the NMR spectra at 125.77 MHz on a Bruker AV III 500 MHz FT spectrometer (Bremen, Germany) with a broadband gradient probe head (5 mm) maintained at room temperature. Chemical shifts were assigned with the aid of previously published data available at Spectral Database for Organic Compounds (SDBS), National Institute of Advanced Industrial Science and Technology (AIST), Tokyo, Japan. A unique carboxylic acid signal for 3-HP was searched using previously published 3-HP NMR spectra (1).
Effect of 3-HPA and glycolytic end products on biotransformation of glycerol
To understand the inhibitory effect of the substrate, 3-HPA, and end products of glucose metabolism on 3-HP formation, biotransformation medium was supplemented with 3-HPA and primary end products of glycolysis. The addition of glucose, lactic acid, acetic acid and reuterin to biotransformation medium of resting cells was carried out up to a final concentration of 50 mM individually by the use of appropriate stock solutions in duplicates. Since reuterin was not commercially available, it was prepared using previous methods (8, 11). HPLC analysis was carried out to track the metabolism of the added compounds.
Results and Discussion
Transcript analysis and enzyme activity of pduP
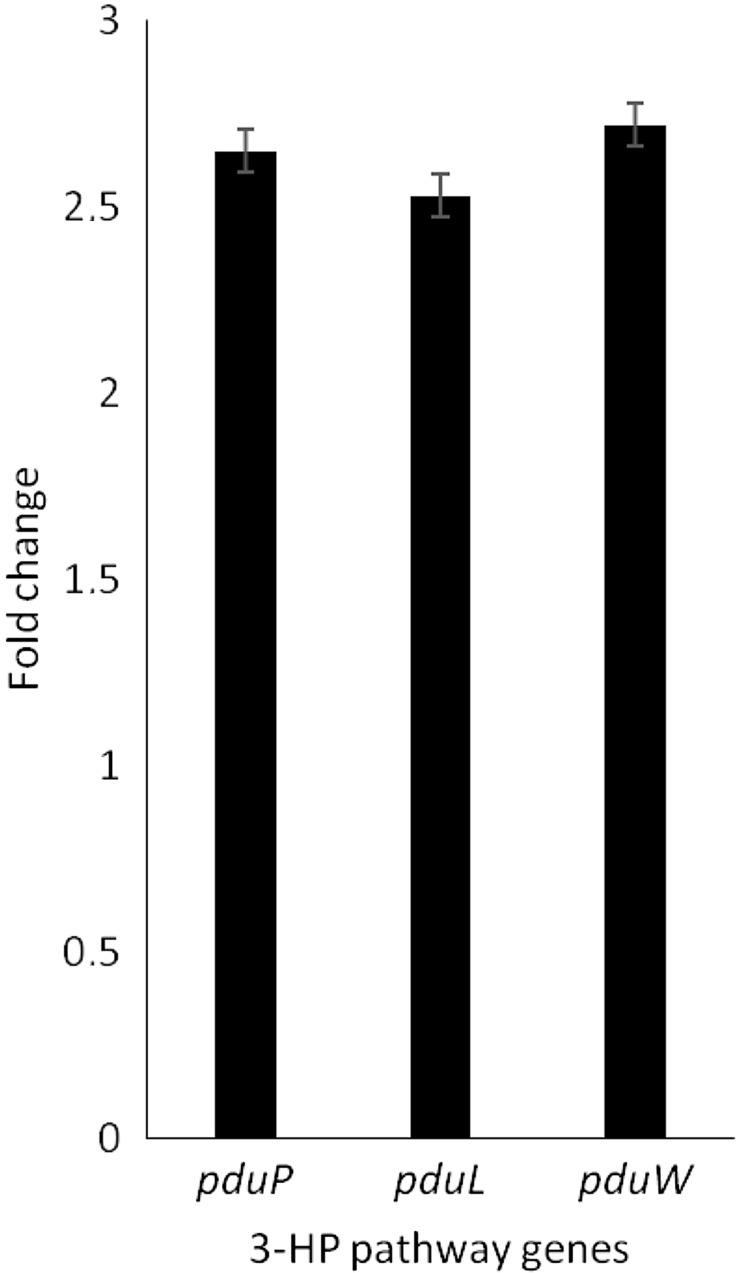
The deregulation of catabolite repression of pdu operon has resulted in enhancement of transcripts when grown in MRS broth with 100 mM glycerol. A comparative transcript analysis between the wild-type and mutant strains relevant for 3-HP pathway is shown in Fig. 2. The observed 2.5-fold increase in the mRNA levels from single transcription unit (15) was found to be consistent when the residual glucose concentration was between 60 and 80 mM. The enhancement in the levels of the transcript was revealed in the measurable in vitro enzyme activities. For instance, propanaldehdye dehydrogenase activity was marked to be upregulated from 0.42 to 0.87 U/mg.
Fig. 2.
Comparison of transcripts of 3-hydroxypropionic acid (3-HP) pathway genes between the wild-type and catabolite repression element (CRE) mutant in mid-log phase of growth, when resting cells were prepared. GACGGTGGGATGGTTATGTATG and AGGGTGAGCACCAAAGTAAAG, CGTTCACACTCACAGGTAGAAA and GCCATCAAGATCACCAGACA, GGAACGCGATGTGGAGATATT and AAGCCCTGAGTCTTCATTCATT were used as forward and reverse primers in Q-PCR for the amplification of pduP, pduL and pduW genes, respectively
Production of 3-HP in the resting cells
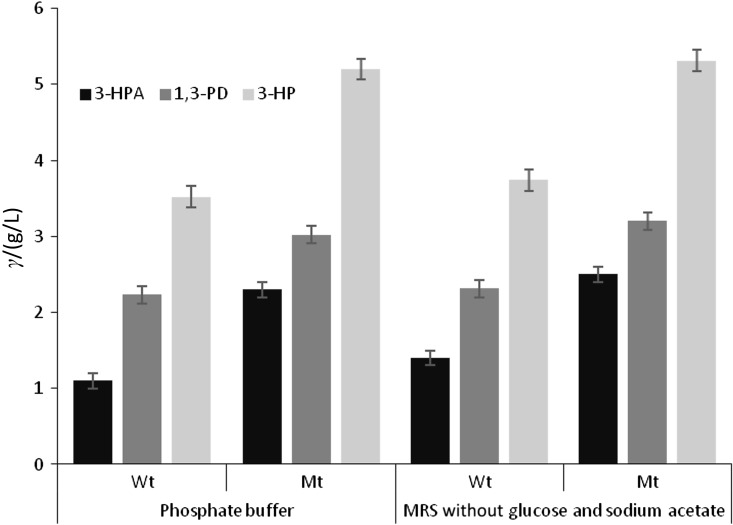
A comparison of the glycerol biotransformation in the wild-type and mutant strains is shown in Fig. 3. Specific productivity of 3-HP by the mutant increased from 0.167 to 0.257 g per g of cell dry mass per h. Furthermore, 52% more glycerol was consumed. Increase in the oxidation rate of glycerol to 3-HP was supported by simultaneous increase in the glycerol to 1,3-PD reduction rate from 0.7 to 1.12 g/(L·h). The observed upregulation in glycerol bioconversion was corroborated in a recent metabolic flux analysis (10). In the wild-type and mutant resting cells, addition of (30±5) mM 3-HPA at 0 h has shown to completely inhibit 3-HP synthesis, similar to the previous enzyme assays (5). Hence, after the accumulation of reuterin to nearly inhibitory concentration, there was no further 3-HP production in the biotransformation medium.
Fig. 3.
Comparison of biotransformation of 150 mM glycerol into 3-HPA, 3-HP and 1,3-PD in the resting cells of wild-type (Wt) and CRE mutant (Mt) in two different biotransformation media, namely phosphate buffer and MRS broth without glucose and sodium acetate. Bioconversion proceeded up to 3 h only, due to the accumulation of inhibitory reuterin
The yield of 3-HP from glycerol was smaller due to the accumulation of coproducts like 3-HPA and 1,3-PD. Since it was an unaerated process, 1,3-PD formation regenerated the NAD(P)+ required for 3-HP synthesis. For high-value and low-volume production, a single purification step such as preparative chromatography is required to get rid of the residual glycerol and co-products. A trade-off between the purification costs and surplus raw material is an essential factor to be considered during scaling up of this process. Nevertheless, the process is superior to that of resting cells of Klebsiella species where glycerol was found to produce additional by-products like lactic acid and ethanol due to parallel oxidation reaction mediated through dihydroxyacetone pathway (16). This sort of biotransformation would become economical if biomass of L. reuteri were obtained from the cast-off of other processes such as large scale mannitol (17) and 1,3- -PD production (4).
Batch cultures in glucose did not produce 3-HP
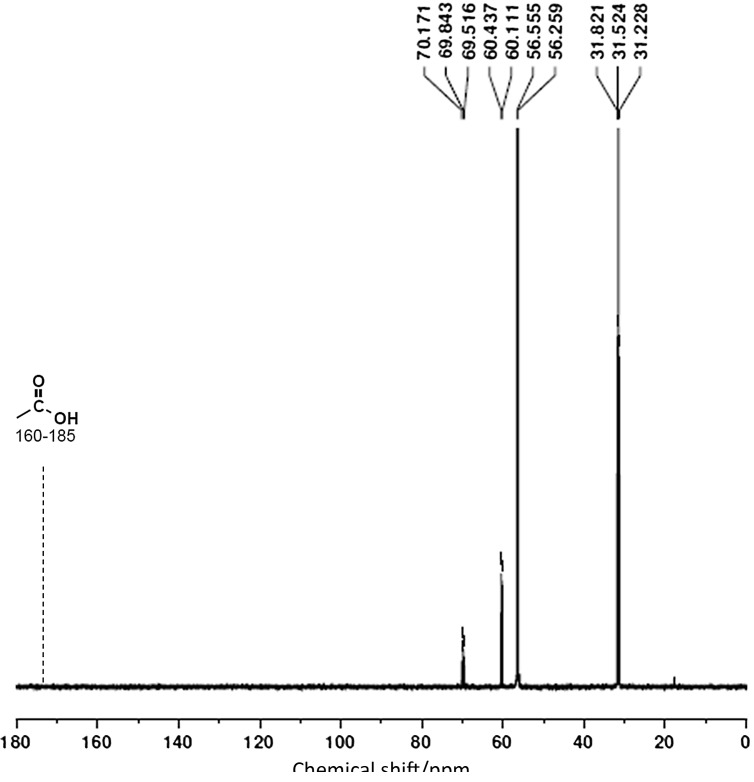
Lactobacillus reuteri does not grow on glycerol as a sole carbon source. Hence, 3-HP formation from glycerol was analysed in the presence of other cofermentable carbon sources like glucose and sucrose. For the various glucose (or sucrose)/glycerol ratios tested in the batch cultivation of both wild-type and mutant, 3-HP formation remained undetected by HPLC. NMR studies confirmed that neither the wild-type nor the mutant strains had chemical shifts between 160 and 185 ppm, corresponding to the carboxylic acid functional group (at position 1) (Fig. 4) at any of the tested sugar/glycerol ratios. Unlike the absence of 3-HP even after the enhanced expression of the pdu genes in mutant strain, both 1,3-PD and 3-HPA formation increased in the batch cultures. For instance, when using the glucose/glycerol ratio of 20:10, 1,3-PD and 3-HPA fractions were found to be 16.54 and 18.72%, respectively, more than in the wild-type after 12 h of cofermentation. Lactic acid and acetic acid were found to be the common end products of glucose and sucrose catabolism. The main differences between the glucose and sucrose metabolism were the formation of mannitol in sucrose-grown cultures and enhanced ethanol formation in the glucose- -grown cultures.
Fig. 4.
3C13 NMR spectrum of the supernatant from cofermentation of glucose and glycerol. Dashed line (------) indicates the expected carboxylic acid chemical shift for 3-HP and was found to be absent from cofermenting samples of growing batch cultures in various glucose/glycerol ratios
In two-stage fermentation, none of the feeding rates of glycerol was found to produce 3-HP. In contrast, when the cells obtained from the two-stage or cofermentation were washed and resuspended as resting cells in phosphate buffer with 150 mM glycerol, synthesis of 3-HPA, 1,3-PD and 3-HP was observed. It is noteworthy to mention that the expression of pdu genes was ubiquitous and did not require the presence of glycerol or other inducers like 1,2-propanediol (18). Despite this expression, 3-HP was not formed in the batch cultures unless the cells were washed and converted to resting cells.
Since the washing process was found to elicit the formation of 3-HP, components in the fermentation broth might be inhibitory to 3-HP formation. With this regard, when lactate (25±5) mM or acetate (20±5) mM was added to biotransformation medium of the resting cells under buffered conditions separately, 3-HP production was absent. However, 3-HPA and 1,3-PD were formed at 41 and 32%, respectively, at a lower amount in comparison with the control. Addition of lactate resulted in the formation of ethanol and acetic acid. In the samples treated with acetic acid, traces of ethanol were seen. Acetate is a potential substrate for pduW activity because of its conserved motif region (19) and hence a substrate competition between the acetic acid and 3-hydroxypropyonyl phosphate can occur. With regard to lactic acid, the inhibitory mechanism was not clear. In the cofermentation and two-stage fermentation there might be catabolite inhibition due to the presence of end products of glycolysis, especially lactate and acetate, affecting the formation of 3-HP. Ensuring this hypothesis, MRS broth devoid of glucose (carbon source for lactic acid production) and sodium acetate was found to have similar biotransformation profile in comparison with the phosphate buffer (Fig. 3).
The resting cells obtained with glucose to glycerol molar ratio (R) greater than 0.42 did not produce 3-HP (Table 1). Below the molar ratio of 0.42, equimolar formation of 1,3-PD and 3-HP was absent in comparison with the control without glucose. It is well known that L. reuteri uses glycerol as an electron acceptor (20) and hence at higher glucose/glycerol ratios, reduction of glycerol to 1,3-PD is more favourable than oxidation to 3-HP. At lower molar ratios of glucose/glycerol, even though the thermodynamic feasibility of 3-HP formation was greater, it might be prevented by 3-HPA accumulation and glycolytic end products.
Table 1. Effect of glucose on 3-hydroxypropionic acid (3-HP) formation in resting cells of wild-type strain after 3 h of biotransformation. Similar trend was obtained in the catabolite repression element (CRE) mutant also (data not shown).
| R(glucose/glycerol) | γ/(g/L) | ||
|---|---|---|---|
| 3-HPA | 1,3-PD | 3-HP | |
| 0 | 1.10 | 2.23 | 3.50 |
| 0.12 | 1.60 | 1.44 | 0.72 |
| 0.33 | 6.20 | 0.90 | 0.57 |
| 0.42 | 5.58 | 1.78 | 0 |
| 0.66 | 2.91 | 3.15 | 0 |
| 1.33 | 0.95 | 4.22 | 0 |
| 2.66 | 0.47 | 6.50 | 0 |
In the batch culture, preference of glycolysis over 3-HP pathway for regeneration of NAD(P)H might be related to higher affinity of enzymes towards NAD(P)+, as observed in glucose-6-phosphate dehydrogenase (Km= 0.0075 mM) in E. coli (21). In our earlier study on L. reuteri (11), cellular NAD(P)+ level was increased through enhanced 1,3-PD production by means of overexpression of NAD(P)H-dependent alcohol dehydrogenase. During such redox-related imbalance within the glycerol bioconversion pathway, glycolysis remained as an electron sink for NAD(P)H regeneration and absence of cooxidation of glycerol to 3-HP (data not shown). Apart from that, the affinity of an aldehyde dehydrogenase towards 3-HPA was reported to be high (22), which makes it interesting for further studies.
Conclusion
The deregulation of the catabolite repression was quantified at the transcript level, and the enhancement in metabolic activity of glycerol bioconversion to 3-HP and other end products was observed in resting cells. From the NMR profiles in different physiologic states, it was clear that batch cultures did not produce 3-HP, even after the enhanced expression of pathway genes. The underlying reason was hypothesised to be catabolite inhibition due to the end products of glycolysis like acetic acid, apart from the previously reported substrate inhibition in enzyme assays. The conversion of 3-HPA to 3-HP under cofermentation conditions could be done possibly through e.g. enhancement of the affinity of oxidising enzyme for the substrate 3-HPA, rather than mere increase in the expression levels of pathway enzymes. In future, 3-HP production in L. reuteri would be worth exploring by means of overexpression of the gene encoding for an aldehyde dehydrogenase like puuC, whose catabolite regulation and substrate inhibition are still not known.
Acknowledgements
Authors are thankful for the grant within the UGC MRP project 34-265\2008 (SR). First author is thankful to CSIR for the senior research fellowship.
References
- 1.Sebastianes FL, Cabedo N, Aouad NE, Valente AM, Lacava PT, Azevedo JL, et al. 3-Hydroxypropionic acid as an antibacterial agent from endophytic fungi Diaporthe phaseolorum. Curr Microbiol. 2012;65:622–32. 10.1007/s00284-012-0206-4 [DOI] [PubMed] [Google Scholar]
- 2.Schwarz M, Köpcke B, Weber RW, Sterner O, Anke H. 3-Hydroxypropionic acid as a nematicidal principle in endophytic fungi. Phytochemistry. 2004;65:2239–45. 10.1016/j.phytochem.2004.06.035 [DOI] [PubMed] [Google Scholar]
- 3.Jiang X, Meng X, Xian M. Biosynthetic pathways for 3-hydroxypropionic acid. Appl Microbiol Biotechnol. 2009;82:995–1003. 10.1007/s00253-009-1898-7 [DOI] [PubMed] [Google Scholar]
- 4.Jolly J, Hitzmann B, Ramalingam S, Ramachandran KB. Biosynthesis of 1,3-propanediol from glycerol with Lactobacillus reuteri: effect of operating variables. J Biosci Bioeng. 2014;118:188–94. 10.1016/j.jbiosc.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 5.Sabet-Azad R, Linares-Pastén JA, Torkelson L, Sardari RR, Hatti-Kaul R. Coenzyme A-acylating propionaldehyde dehydrogenase (PduP) from Lactobacillus reuteri kinetic characterization and molecular modelling. Enzyme Microb Technol. 2013;53:235–42. 10.1016/j.enzmictec.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 6.van Maris AJ, Konings WN, Van Dijken JP, Pronk JT. Microbial export of lactic and 3-hydroxypropanoic acid: implications for industrial fermentation processes. Metab Eng. 2004;6:245–55. 10.1016/j.ymben.2004.05.001 [DOI] [PubMed] [Google Scholar]
- 7.Van Pijkeren JP, Neoh KM, Sirias D, Findley AS, Britton RA. Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered. 2012;3:209–17. 10.4161/bioe.21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Ziney MG, Arneborg N, Uyttendaele M, Debevere J, Jakobsen M. Characterization of growth and metabolite production of Lactobacillus reuteri during glucose/glycerol co-fermentation in batch and continuous cultures. Biotechnol Lett. 1998;20:913–6. 10.1023/A:1005434316757 [DOI] [Google Scholar]
- 9.Sriramulu DD, Liang M, Hernandez-Romero D, Raux-Deery E, Lünsdorf H, Parsons JB, et al. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J Bacteriol. 2008;190:4559–67. 10.1128/JB.01535-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dishisha T, Pereyra LP, Pyo SH, Britton RA, Hatti-Kaul R. Flux analysis of the Lactobacillus reuteri propanediol-utilization pathway for production of 3-hydroxypropionaldehyde, 3-hydroxypropionic acid and 1,3-propanediol from glycerol. Microb Cell Fact. 2014;13:76. 10.1186/1475-2859-13-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaidyanathan H, Kandasamy V, Ramakrishnan GG, Ramachandran KB, Jayaraman G, Ramalingam S. Glycerol conversion to 1,3-propanediol is enhanced by the expression of a heterologous alcohol dehydrogenase gene in Lactobacillus reuteri. AMB Express. 2011;1:37. 10.1186/2191-0855-1-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kandasamy V, Vaidyanathan H, Djurdjevic I, Jayamani E, Ramachandran KB, Buckel W, et al. Engineering Escherichia coli with acrylate pathway genes for propionic acid synthesis and its impact on mixed-acid fermentation. Appl Microbiol Biotechnol. 2013;97:1191–1200. 10.1007/s00253-012-4274-y [DOI] [PubMed] [Google Scholar]
- 13.Leal NA, Havemann GD, Bobik TA. PduP is a coenzyme-a-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch Microbiol. 2003;180:353–61. 10.1007/s00203-003-0601-0 [DOI] [PubMed] [Google Scholar]
- 14.Lüthi-Peng Q, Dileme FB, Puhan Z. Effect of glucose on glycerol bioconversion by Lactobacillus reuteri. Appl Microbiol Biotechnol. 2002;59:289–96. 10.1007/s00253-002-1002-z [DOI] [PubMed] [Google Scholar]
- 15.MetaCyc. SRI International, Menlo Park, CA. http://metacyc.org/.
- 16.Ashok S, Mohan Raj S, Ko Y, Sankaranarayanan M, Zhou S, Kumar V, et al. Effect of puuC overexpression and nitrate addition on glycerol metabolism and anaerobic 3-hydroxypropionic acid production in recombinant Klebsiella pneumoniae ΔglpKΔdhaT. Metab Eng. 2013;15:10–24. 10.1016/j.ymben.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 17.Ortiz ME, Fornaguera MJ, Raya RR, Mozzi F. Lactobacillus reuteri CRL 1101 highly produces mannitol from sugarcane molasses as carbon source. Appl Microbiol Biotechnol. 2012;95:991–9. 10.1007/s00253-012-3945-z [DOI] [PubMed] [Google Scholar]
- 18.Saulnier DM, Santos F, Roos S, Mistretta T, Spinler JK, Molenaar D. Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS One. 2011;6:e18783. 10.1371/journal.pone.0018783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motif Finder. PROSITE database. http://www.genome.jp/tools/ motif/.
- 20.Talarico TL, Axelsson LT, Novotny J, Fiuzat M, Dobrogosz WJ. Utilization of glycerol as a hydrogen acceptor by Lactobacillus reuteri: purification of 1,3-propanediol:NAD+ oxidoreductase. Appl Environ Microbiol. 1990;56:943–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.BRENDA. TU, Braunschweig, Germany. http://www.brenda--enzymes.info/.
- 22.Jo JE, Mohan Raj S, Rathnasingh C, Selvakumar E, Jung WC, Park S. Cloning, expression, and characterization of an aldehyde dehydrogenase from Escherichia coli K-12 that utilizes 3-hydroxypropionaldehyde as a substrate. Appl Microbiol Biotechnol. 2008;81:51–60. 10.1007/s00253-008-1608-x [DOI] [PubMed] [Google Scholar]