Summary
A new optimized system for the surface display and secretion of recombinant proteins is described, termed MATE (maximized autotransporter-mediated expression). It is based on an artificial gene consisting of the coding region for the signal peptide of CtxB, a multiple cloning site for passenger gene insertion, flanked by coding sequences for linear epitopes for monoclonal antibodies and OmpT, and factor Xa protease cleavage sites followed by a codon-optimized DNA sequence of the linker and the β-barrel of the type V autotransporter EhaA from Escherichia coli under control of an IPTG-inducible T5 promoter. The MATE system enabled the continuous secretion of recombinant passenger mCherry via OmpT-mediated cleavage, using native OmpT protease activity in E. coli when grown at 37 °C. It is the first example to show that native OmpT activity is sufficient to facilitate the secretion of a correctly folded target protein in preparative amounts obtaining 240 µg of purified mCherry from 800 mL of crude culture supernatant. Because the release of mCherry was achieved by a simple transfer of the encoding plasmid from an OmpT-negative to an OmpT-positive strain, it bears the option to use surface display for screening purposes and secretion for production of the selected variant. A single plasmid could therefore be used for continuous secretion in OmpT-positive strains or surface display in OmpT-negative strains. In conclusion, the MATE system appears to be a versatile tool for the surface display and for the secretion of target proteins in E. coli.
Key words: surface display, secretion, autotransporter, mCherry, OmpT, EhaA, MATE
Introduction
Surface expression of a certain protein or peptide has many advantages compared to intracellular protein expression. The surface-expressed protein is freely accessible for binding studies or enzyme activity assays. Cell-attached proteins are often more stable than those in a solution, similar to the stability obtained through immobilization on a matrix. Secretion of the target protein outside the cell can also have many advantages, such as avoiding the time and cost associated with protein purification. The cells can be quickly removed by a simple centrifugation step. Because Escherichia coli does not naturally secrete proteins at high levels, the secreted target protein can be easily separated from the less complex background (1).
Different secretion mechanisms evolved in Gram- -negative bacteria for translocation of proteins across the inner and outer membrane. There are currently eight types of secretion systems (type I–VIII) and the chaperone-usher pathway is known to act in different ways to cross both membrane barriers. The autotransporter pathway, also known as type Va pathway, is one of the simplest and widespread secretion systems for the translocation of proteins across the cell envelope of Gram-negative bacteria. Like other type V pathways, the Va secretion system is dependent on the Sec machinery (2, 3). A popular representative of the Va secretion pathway is the AIDA-I autotransporter, which is known to mediate epithelial cell adhesion (4). Wells et al. (5) described the novel type Va autotransporter EhaA which contributes to adhesion, colonisation and biofilm formation by enterohemorrhagic E. coli O157:H7. Many of the characterised autotransporters are associated with virulence. The surface-exposed passenger domains are extremely diverse and the functions of most autotransporters remain unknown (6). All autotransporters possess common features necessary for transport to the cell surface: (i) an N-terminal signal peptide that enables the transport across the inner membrane, (ii) a passenger domain that is either surface displayed or released into the extracellular milieu, (iii) a linker for a complete surface exposure, and (iv) a β-barrel domain that is inserted into the outer membrane (7, 8). Unlike the original belief that autotransporters possess all properties needed for their autonomous transport across the cell envelope, there are accessory proteins that conduct the proper transport and integration into the outer cell membrane: after recognition of the signal peptide by the Sec translocon, the translocation of the polypeptide chain takes place in its unfolded form and the signal peptide is cleaved by signal peptidase during translocation (9, 10). The β-barrel assembly machinery (also termed Bam complex) and further periplasmic chaperones, such as Skp, SurA and DegP, build a stable intermediate with the polypeptide chain after its transport through the inner membrane (11, 12). A β-barrel is formed out of the corresponding polypeptide domain and somehow integrated into the outer membrane. The β-barrel possesses a hydrophilic pore through which the linker with the passenger can be traversed to the cell surface (2, 13). The secretion mechanism of autotransporters could be exploited for the transport of a recombinant passenger by simply replacing the coding region of the innate passenger through the sequence of the desired recombinant passenger sequence (7). Release of the passenger is finally conducted either through endogenous host proteases such as OmpT (14-16), or through self-proteolysis, as it is the case for the so--called serine protease autotransporters of Enterobacteriaceae (SPATE) (17). By replacing the natural passenger with the target protein, this system has recently been utilized for secretion of the red fluorescent protein mCherry (18).
OmpT is an endoprotease localized in the outer membrane of E. coli. It cleaves preferentially between two basic residues, and its physiological function remains unclear (19). The display of the cholera toxin B subunit (CtxB) with AIDA-I autotransporter led to the release of the passenger by OmpT (14-16). OmpT recognition sites within the autodisplayed passenger CtxB were not cleaved (16). Jose et al. (20) showed the secretion of dimeric adrenodoxin (Adx) in E. coli strain UT2300 by cleavage of host OmpT protease within the AIDA-I linker region. However, the released amount of free Adx protein was low and a large portion of the protein remained associated with the outer membrane. As the release of the passenger by OmpT is undesirable for surface expression, surface display has typically been carried out in OmpT-negative E. coli strains such as E. coli UT5600 or BL21 (20). Although OmpT-mediated cleavage of passenger proteins has been shown several times for PAGE analysis (15, 16, 20-22), the feasibility of OmpT-mediated secretion on lab scale has not led to sufficient results by utilizing the AIDA-I translocon.
We therefore designed a new system, designated as MATE (maximized autotransporter-mediated expression), that can be simultaneously used for surface expression and secretion of preparative amounts of a protein of interest in E. coli. The MATE system utilizes the endogenous E. coli protease OmpT for continuous secretion in order to obtain preparative amounts of secreted protein for analysis. The system is based on an artificial gene harbouring the coding sequence for the CtxB signal peptide, a multiple cloning site for passenger gene insertion, flanked by coding sequences for linear epitopes for monoclonal antibodies and OmpT and factor Xa cleavage sites, followed by a codon-optimized nucleotide sequence encoding the C-terminal part of EhaA autotransporter for the translocation of the passenger across the cell envelope. It has recently been shown that EhaA is suitable for surface display not only in E. coli (23), but also in Zymobacter palmae and Zymomonas mobilis (24). Whether the protein is secreted or surface displayed depends only on the strain of E. coli in which it is expressed. We could demonstrate the usability of the MATE system for surface display of a small 6xHis peptide, an active esterase, and the red fluorescent protein mCherry in an OmpT-negative host strain, and at the same time apply it for secretion and purification of mCherry using an OmpT-positive background.
Material and Methods
Sequence analyses
For phylogenetic analysis, sequences were accessed from UniProt (25) or NCBI GenBank (26). Homologous sequences were obtained via the SIMAP database (27). Alignments were conducted using the software Mega v. 6.0 (28), and the ClustalW algorithm with gap opening penalties of 10 to 20, and gap extension penalties of 0.1 to 0.2. Phylogenetic analysis was also performed with Mega, using the maximum likelihood algorithm and the JTT matrix (29).
Bacterial strains and growth conditions
E. coli Stellar (F–, endA1, supE44, thi-1, recA1, relA1, gyrA96, phoA, Φ80d lacZΔ M15, Δ[lacZYA-argF] U169, Δ[mrr-hsdRMS-mcrBC], ΔmcrA, λ–) was used for cloning purposes (Clontech Laboratories Inc, a TaKaRa Bio Company, Mountain View, CA, USA). E. coli UT5600 (FŻ, ara- -14, leuB6, secA6, lacY1, proC14, tsx-67, Δ[ompT-fepC]266, entA403, trpE38, rfbD1, rpsL109[Strʳ], xyl-5, mtl-1, thi-1), E. coli UT2300 (FŻ, ara-14, leuB6, secA6, lacY1, proC14, tsx--67, entA403, trpE38, rfbD1, rpsL109[Strʳ], xyl-5, mtl-1, thi--1) and E. coli BL21 (B, FŻ, dcm, ompT, lon, hsdS[rBŻ mBŻ], gal) were used for expression experiments (30). The cells were typically grown in lysogeny broth (LB) containing 60 µg/mL of kanamycin. Cultures for protein expression were inoculated with 0.01% (by volume) of an overnight pre-culture, and grown until A600 nm=0.5 at 37 °C and 200 rpm. Protein expression was induced with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG), and the cells were incubated for another 1.5 h. Cultures for purification of secreted protein were set up as follows: 800 mL of LB medium containing 60 µg/mL of kanamycin was inoculated with 0.01% (by volume) of an overnight culture, and grown until A600 nm=0.5 at 37 °C and 200 rpm. A concentration of 1 mM of IPTG was added, and the cells were incubated for 24 h.
Plasmid construction and expression
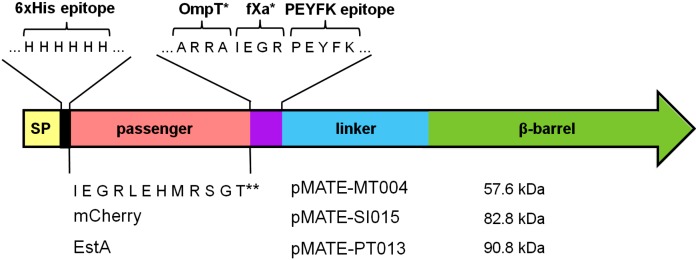
The MATE autotransporter domain was derived from the C-terminal domain (linker and β-barrel) of EhaA from E. coli strain O157:H7 [GenBank Accession No. Q8X6C1, (5)]. The pMATE-MT004 plasmid encoding the autotransporter fusion protein with 6xHis as a passenger was synthesized commercially in the pJExpress-401 vector backbone (DNA2.0, Menlo Park, CA, USA). The MATE fusion protein included an N-terminal signal peptide from the cholera toxin B subunit (CtxB), a 6xHis affinity tag, a multiple cloning site, and the codon-optimized autotransporter domain, which consists of a linker and a β-barrel region. Protease cleavage sites included a factor Xa site between the N-terminal 6xHis and the multiple cloning site, and both (for factor Xa and OmpT) were incorporated after the C-terminal side of the multiple cloning site. An epitope (PEYFK) for a monoclonal antibody (Dü142) (16) was inserted after the protease cleavage sites. The pJExpress-401 plasmid backbone contained coding sequences for rrnB1/B2 terminator, kanamycin resistance gene, LacI repressor, pUC ori, rpn and bla terminators and IPTG-inducible T5 promoter.
For construction of pMATE-SI015, pMATE-PT013 and pAIDAI-PT014, standard In-Fusion® techniques were used as described by the manufacturer (Eco-Dry kit, Clontech Laboratories Inc) and transferred into E. coli Stellar chemically competent cells. A detailed construction strategy including the primer sequences used for amplification of the respective fragments is provided in Fig. 1 and Supplementary Table 1 (available at www.ftb.com.hr).
Fig. 1.
Structure of encoded fusion proteins and plasmid construction scheme used in this study. Plasmid sizes are depicted below the plasmid names. Structure of the encoded fusion proteins are shown schematically, with CtxB signal peptide in yellow, 6xHis epitope in black, passenger domains in light red, epitope and protease cleavage site in violet, EhaA autotransporter in blue/green, and AIDA-I autotransporter in orange/pink. FXa protease recognition site is indicated with an asterisk (*). Two asterisks (**) indicate the amino acid sequence encoded by the multiple cloning site. 1=Commercial synthesis in the pJExpress backbone by DNA2.0 (Menlo Park, CA, USA), 2=amplification of pMATE-MT004 backbone using SI020 and PQ019 primers, 3=amplification of mCherry from pEF1a-mCherry-C1 using SI021 and PQ024 primers, 4=amplification of pMATE-SI015 backbone using PT-MCE-7f and PT-MCE-8r primers, 5=amplification of AIDA-I β-barrel and linker region from pES01 using PT-MCE-5f and PT-MCE6r primers, 6=amplification of pMATE-MT004 backbone using PT-MCE-3r and PT-MCE-4f primers, 7=amplification of estA from pES01 using PT-MCE1f and PT-MCE2r primers. The respective PCR fragments were combined using In-Fusion cloning technique (Clontech Laboratories Inc, Mountain View, CA, USA). pES01 has been described previously by Schultheiss et al. (31)
Supplementary Table 1. Sequences of primers used for the construction of plasmids pMATE-MT004, pMATE-SI015, pMATE-PT013 and pAIDAI-PT014.
| Primer name | Primer sequence (5´ to 3´) |
|---|---|
| SI020 | GCTCGTCGTGCTATTGAGGGCCGCATCCC |
| PQ019 | ATGATGGTGATGGTGGTGGGTGATGTTCTG |
| PQ024 | CACCATCACCATCATATGGTGAGCAAGGGCGAGGAGGATAACATG |
| SI021 | AATAGCACGACGAGCCTTGTACAGCTCGTCCATGCCGCCGGTGG |
| PT-MCE-1f | CTCGAGGGCGGCGGTGACGAC |
| PT-MCE-2r | GGTACCCTTGGTGACGCCGGC |
| PT-MCE-3r | ACCGCCGCCCTCGAGATGATGGTGATGGTGGTGGGTGATGTTCTG |
| PT-MCE-4f | GTCACCAAGGGTACCGCTCGTCGTGCTATTGAGGGCCGC |
| PT-MCE-5f | CTTAATCCTACAAAAGAAAGTGCAGG |
| PT-MCE-6r | GAAGCTGTATTTTATCCCCAGTGCTC |
| PT-MCE-7f | GATAAAATACAGCTTCCTCGAGCCCCAAGGGCGACACCCCCTAAT |
| PT-MCE-8r | CTTTTGTAGGATTAAGTTTAAAGTATTCCGGGATGCGGCCCTCAATAG |
Differential cell fractionation
Overnight cultures of E. coli BL21, UT2300 or UT5600 containing the plasmid of interest were used to inoculate 40 mL of LB medium (1:40 dilution), which was incubated at 37 °C and 200 rpm until the cultures reached A600 nm=0.5. Protein expression was induced by the addition of 1 mM IPTG, and the cells were harvested 1.5 h after induction. For isolation of soluble protein fractions, the pellet was resuspended in 1 mL of Tris/HCl buffer (0.2 M, pH=8.0) and subjected to 7 cycles of sonication (60 s each at 50% of maximum output, UP200S Ultrasonic Processor, Hielscher Ultrasonics GmbH, Teltow, Germany). Membrane proteins were removed by centrifugation at 20 000×g for 30 min at 4 °C. The supernatant contained the soluble proteins and was used for further experiments. Outer membrane proteins were prepared according to a modified rapid isolation protocol as described earlier (32, 33). Briefly, the cell pellet was resuspended in 1.5 mL of Tris/HCl buffer (0.2 M, pH=8.0), followed by the addition of 0.1 mL of saccharose (1 M in water), 0.1 mL of ethylenediaminetetraacetic acid (10 mM in water), 0.1 mL of lysozyme (10 mg/mL in water), and 3.2 mL of water. After 10 min of incubation at room temperature, following reagents were added: 50 µL of phenylmethanesulphonylfluoride (100 mM in n-propanol), 10 µL of aprotinin (10 mg/mL in 10 mM HEPES buffer, pH=8.0), 5 mL of extraction buffer (2% Triton X-100, 10 mM MgCl2 in 50 mM Tris/HCl, pH=8.0) and 0.1 mL of DNAse (1 mg/mL). The suspension was thoroughly mixed and incubated for 30 min on ice. Subsequently, cell debris was removed by centrifugation at 3000×g for 5 min, and the supernatant was centrifuged at 38 700×g for 10 min. The resulting protein pellet was washed twice with 10 mL of ice-cold water.
OmpT protease activity assays
To measure OmpT activity in whole cells and membrane samples, we adopted the fluorescence resonance energy transfer (FRET) method that Kramer et al. (34) used to measure the activity of purified OmpT. The peptide substrate Abz-Ala-Arg-Arg-Ala-Tyr(NO2)-NH2 was synthesized commercially (Bachem, Bubendorf, Switzerland) and dissolved in FRET assay buffer (0.5 M Tris/HCl, pH=7.0) to a concentration of 8 mM. Continuous assays using a final substrate concentration of 0.16 mM were conducted using solutions of whole E. coli cells or isolated outer membrane proteins. For the preparation of whole E. coli cell solutions, LB medium was inoculated with a pre-culture of E. coli UT2300 or E. coli UT5600 as described above. Cells were grown at 37 °C to A600 nm=0.5, harvested and washed with assay buffer (0.5 M Tris/HCl, pH=7.0), and subsequently resuspended in the same buffer to A600= 0.5. Fluorescence was measured using a microplate reader (Mithras 940, Berthold, Bad Wildbad, Germany) at an excitation of (355±20) nm and an emission of (420±5) nm.
SDS-PAGE and Western blot
Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) of proteins isolated from the outer membrane was conducted as described elsewhere (35) with stacking (4% T) and resolving (10% T) gels (Mini- -Protean III, Bio-Rad, Hercules, CA, USA). Protein samples were diluted 1:2 in sample buffer consisting of Tris/HCl (100 mM, pH=6.8), sodium dodecyl sulphate (4%), Bromophenol Blue (0.2%), dithiothreitol (200 mM) and glycerol (20%) and heated to 100 °C for 15 min before electrophoresis. Proteins were then detected with Coomassie Brilliant Blue R-250. PageRuler prestained protein ladder from Fermentas (St. Leon-Rot, Germany) served as a molecular mass marker.
For Western blotting, proteins were transferred onto a nitrocellulose membrane using standard electroblotting techniques (Mini-trans blot, Bio-Rad). The blot was then blocked with phosphate-buffered saline (PBS, pH=7.4) containing 3% bovine serum albumin (BSA), and incubated with a 1:1000 dilution in PBS of the primary antibody against 6xHis (mouse anti-6-His mAb IgG1, Dianova, Hamburg, Germany). The blot was then washed in PBS, and incubated for 2 h with a 1:6000 dilution in PBS of the secondary antibody (horseradish peroxidase-conjugated anti-mouse IgG; antibodies-online.com, Atlanta, GA, USA). The blot was then washed and incubated with Pierce enhanced chemiluminescent (ECL) substrate (Thermo Scientific, Waltham, MA, USA). Chemiluminescence was detected with a chemiluminescent reader (ChemoCam ECL imager, Intas, Göttingen, Germany).
Secretion and IMAC purification of mCherry
A volume of 800 mL of LB medium was inoculated with 8 mL of an overnight culture of either E. coli UT2300 or E. coli UT5600 harbouring pMATE-SI015 plasmid, and grown until A600 nm=0.5 at 37 °C and 200 rpm. A concentration of 1 mM of IPTG was added, and the cells were incubated for 24 h. The cells were then removed by centrifugation (10 000×g for 20 min at 4 °C).
The mCherry protein secreted into the LB medium was supposed to contain a polyhistidine tag towards the N-terminus, and could therefore be purified by immobilized metal affinity chromatography (IMAC). After the removal of the cells, the complete volume of cell-free growth medium was filtered through a 0.45-µm filter unit and loaded on a column containing Ni-NTA medium that had been pre-equilibrated with binding buffer (20 mM sodium phosphate, 0.5 M sodium chloride and 20 mM imidazole, pH=7.0). The column was washed with 10 column volumes of binding buffer followed by 10 column volumes of washing buffer (binding buffer+50 mM imidazole). The protein was eluted with 10 mL of elution buffer (binding buffer+200 mM imidazole), and the eluate was collected in five fractions. The eluted protein fractions were concentrated in a centrifugal filter device with a cut--off size of 10 kDa (Amicon Ultra-4, Merck Millipore, Darmstadt, Germany). All purification steps were carried out at 4 °C. Protein concentration of mCherry was determined by measuring the absorption at 280 nm using a nanophotometer (Implen P300, Implen GmbH, München, Germany) with a molar absorption coefficient of 34 380 L/(M·cm) and a protein mass of 28.4 kDa. Additionally, protein concentration was determined by means of Roti®- -Quant Bradford assay according to the manufacturer’s protocol (Carl Roth GmbH & Co. KG, Karlsruhe, Germany). Briefly, 200 µL of 5×-staining solution were mixed with the protein sample diluted in 800 µL of Tris/HCl buffer (0.2 M, pH=8.0) and absorption was measured after 10 min of incubation at 595 nm. BSA served as a reference. Both methods provided very similar protein concentration values.
Flow cytometry
Cell cultivation and protein expression were carried out as described, and the culture was centrifuged and stored overnight in PBS. The cells were washed twice with PBS (filter-sterilized) and diluted to A600 nm=0.4. A volume of 1 mL of the cell suspension was then pelleted and resuspended in 100 µL of PBS containing mouse anti--6xHis antibody (1:50, THE™ HisTag, GenScript, Piscataway, NJ, USA). After incubation for 30 min at room temperature, the washing step was repeated three times, and the cells were incubated for 30 min in the dark with a goat anti-mouse IgG antibody which was coupled to DyLight 633 (1:50, Thermo Scientific, USA). After washing again, the cells were analyzed by means of a FACSAria III flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) using an excitation wavelength of 633 nm for DyLight 633, and an excitation wavelength of 561 nm for mCherry.
Esterase activity assay
Esterase activity assay was performed as described previously (36), with minor modifications: cells were grown as described above and protein expression was induced. After cell harvest and a washing step with 100 mM Tris/HCl buffer (pH=7.0), the absorbance was adjusted to A600 nm=1. A volume of 40 µL of this cell suspension was added to a 160-µL assay solution composed of 100 mM Tris/HCl buffer (pH=7.0) with 3.2 mM p-nitrophenyl acetate as substrate. Increase of absorption at 405 nm due to release of p-nitrophenol was measured spectrophotometrically in a microplate reader (Mithras LB 940, Berthold, Germany).
Results
Identification of the EhaA autotransporter domain used in the MATE system
The EhaA autotransporter belongs to the type Va secretion group and shows a phenotype of cell aggregation when expressed with the native passenger domain. As with other autotransporters such as the AIDA-I from E. coli, replacing the passenger domain with a protein of interest facilitates surface display. Although a number of EhaA amino acid sequences in public databases are labelled AIDA-I-like, Celik et al. (6) placed EhaA within the large IcsA/VirG subfamily (Group 12), rather than the neighbouring ‘long-type’ (Group 13) that included AIDA--I. In the C-terminal region we found that EhaA had a slightly higher sequence identity to IcsA of Shigella flexneri than to AIDA-I (data not shown), supporting the classification of Celik et al. (6). However, homology searches revealed a large number of diverse sequences that were more similar to EhaA than to either AIDA-I or IcsA (data not shown). This suggests that EhaA-like sequences form their own subgroup within the autotransporter family. The EhaA-like sequences (>60% amino acid identity) were present in a large number of Gram-negative strains (data not shown), suggesting that the EhaA autotransporter domain could be useful for expression in a variety of Gram-negative bacteria.
Surface display of a polyhistidine tag, mCherry and esterase EstA
The C-terminal linker and β-barrel domains of an autotransporter protein are required for transport to the cell surface. We defined the start of the EhaA linker region based on a pairwise alignment with a C-terminal fragment of AIDA-I that had previously been used for surface display (16). EhaA also contains a proline-rich 31 amino acid insert that is lacking in AIDA-I; however, most of the remaining sequence could be well aligned. The amino acid identity of linker and β-barrel regions of the two proteins was 37% excluding gaps. The codons of the DNA sequence encoding the fusion protein were optimized according to the codon usage of E. coli proteins expressed under conditions of physiological stress (37). The codon usage was further altered to remove selected restriction sites from the open reading frame, and to minimize the predicted secondary structure in the region encoding the CtxB signal peptide. To transport the fusion protein across the inner membrane, we added the N-terminal CtxB signal peptide (16).
The plasmid pMATE-MT004 encoded the autotransporter fusion protein with a simple 6xHis, followed by a multiple cloning site as the passenger, pMATE-SI015 encoded mCherry, and pMATE-PT013 the esterase EstA from B. gladioli as passenger domain (Fig. 2). The correct fusions were controlled by restriction analyses and DNA sequencing.
Fig. 2.
Scheme of the used MATE autotransporter fusion proteins. The fusion proteins include an N-terminal signal peptide from the cholera toxin B subunit (CtxB), a 6xHis epitope, the respective passenger domains, and the C-terminal autotransporter domain, which consists of a linker and β-barrel region. The protein sequence contains factor Xa and OmpT protease cleavage sites, which were incorporated after the passenger domains. Protease recognition sites in the expressed protein are indicated with an asterisk (*). Two asterisks (**) indicate the amino acid sequence of the small passenger consisting of a further fXa cleavage site (IEGR) and the peptide product of the multiple cloning site. A PEYFK epitope for a monoclonal antibody (Dü142) was inserted after the protease cleavage sites. The predicted size of the encoded proteins after cleavage of the signal peptide is indicated
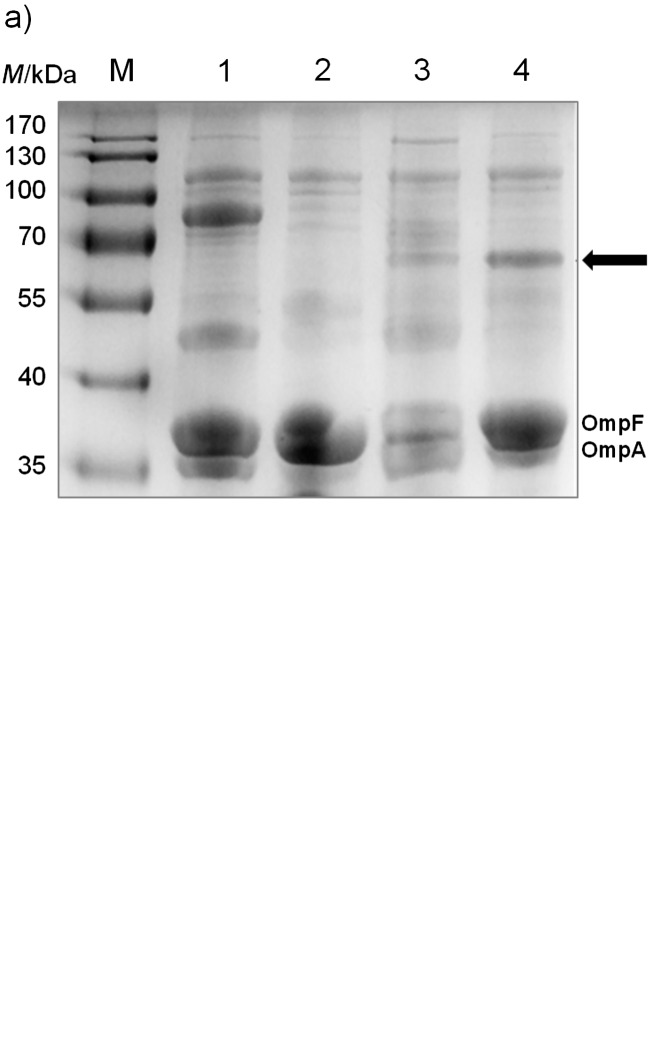
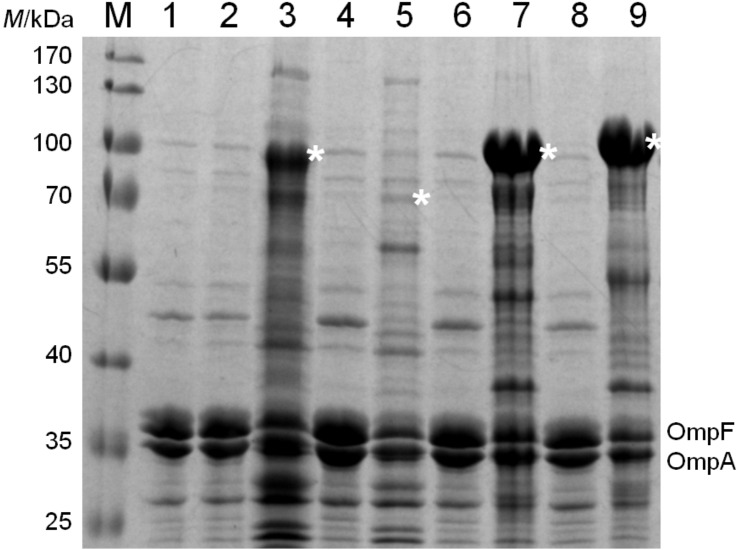
It was first tested whether the EhaA domain used in the MATE system could transport a small peptide to the outer membrane of E. coli. After transforming the plasmid pMATE-MT004 into E. coli UT5600 and inducing protein expression with IPTG, SDS-PAGE and Western blot revealed a protein of 60–65 kDa in the outer membrane. The predicted size of the autotransporter fusion protein with 6xHis as a passenger was 57.6 kDa after cleavage of the N-terminal signal peptide. The fusion protein was visible in both cases as a single band after SDS-PAGE (Fig. 3; 31,37). Interestingly, a much higher amount of fusion protein could be found in the outer membrane of E. coli when mCherry or EstA were inserted as passenger domains (Fig. 4). Obviously, the translocation efficiency strongly depends on the passenger sequence itself, whereby its length does not seem to be the crucial parameter. To compare the transport efficiencies of MATE and the previously quite frequently applied AIDA-I (7, 39), we constructed pAIDAI-PT014, in which the EhaA translocation unit from pMATE-SI015 is substituted by that of AIDA-I. According to the band intensities in the SDS-PAGE, MATE- -mCherry is obviously present in considerably larger amounts in the outer membrane than AIDA-I-mCherry (Fig. 4).
Fig. 3.
Utilization of the MATE system for the transport of a small peptide with an N-terminal 6xHis to the outer membrane. E. coli BL21 cells were grown in LB medium to A600 nm=0.5. Protein expression was induced by the addition of 1 mM IPTG, and the cells were harvested after 1.5 h. Outer membrane proteins were isolated according to the modified method of Park et al. (32): a) SDS-PAGE of outer membrane proteins including the reference outer membrane proteins OmpF and OmpA; b) Western blot of outer membrane proteins with an antibody against 6xHis. M=PageRuler prestained protein marker, 1=cells with negative control plasmid, expressing a 74.5-kDa sized fusion protein containing the AIDA-I autotransporter with a single- -chain antibody fragment as passenger [pST005, (38)], 2=cells with empty vector (pJExpress-401), 3=cells with pMATE-MT004 without the addition of IPTG, 4=cells with pMATE-MT004 with 1 mM IPTG for the induction of protein expression. The arrow indicates the band associated with the MATE fusion protein
Fig. 4.
Outer membrane proteins of E. coli UT5600 cells with: 1=empty expression vector pJExpress-401, 2=plasmid encoding for AIDA-I-mCherry, not induced and 3=induced, 4=plasmid encoding for MATE-6xHis, not induced and 5=induced, 6=plasmid encoding for MATE-mCherry, not induced and 7=induced, 8=plasmid encoding for MATE-EstA, not induced and 9=induced. Fusion proteins are marked with an asterisk (*). Sample amounts were normalized to OmpF/OmpA protein bands
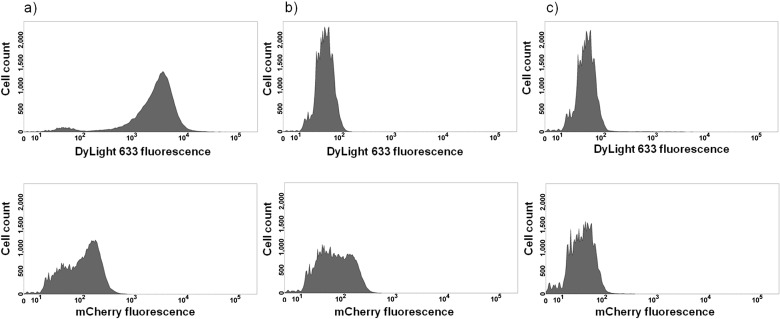
Next, it was investigated if the MATE system enables a stable presentation of mCherry on the cell surface of an OmpT negative strain. For this purpose, pMATE-SI015 was inserted into E. coli UT5600, and protein expression was induced as described. The cells were then incubated with anti-6xHis antibody, followed by incubation with a DyLight 633-coupled second antibody. As antibodies are too large to penetrate the bacterial outer cell membrane, binding to an epitope (in this case the 6xHis epitope at the N-terminus of mCherry) can only occur when the epitope is actually located on the outside of the cell. The same cells, but without antibody labelling (to exclude interference between mCherry and DyLight 633 fluorescence), and cells containing the empty expression vector pJExpress-401 were used as negative controls. As can be seen in the flow cytometry histograms, cells expressing MATE--mCherry (Fig. 5, a above) exert a significantly higher DyLight 633 fluorescence intensity than the control cells without antibody labelling (Fig. 5, b above) and with empty expression vector (Fig. 5, c above), providing evidence that mCherry was displayed on the surface of OmpT-negative E. coli cells. Furthermore, these cells also showed mCherry fluorescence (Fig. 5, a and b below), providing evidence that the protein adopts a functional conformation when displayed with the MATE system. Even with the naked eye, a distinct colouring of these cells on agar plates is recognizable when using a LED illuminator or similar device (data not shown).
Fig. 5.
Surface display of mCherry analyzed by flow cytometry: a) UT5600 cells displaying mCherry; b) UT5600 cells displaying mCherry, without antibody treatment; c) UT5600 control cells with empty expression vector. Above: DyLight 633 fluorescence. Below: mCherry fluorescence. Cells were cultivated and protein expression was induced as described. Cells were then harvested and, except sample b), incubated with anti-6xHis antibody, followed by incubation with DyLight 633 fluorophore-labelled second antibody. Cell count was 50 000 cells for each sample
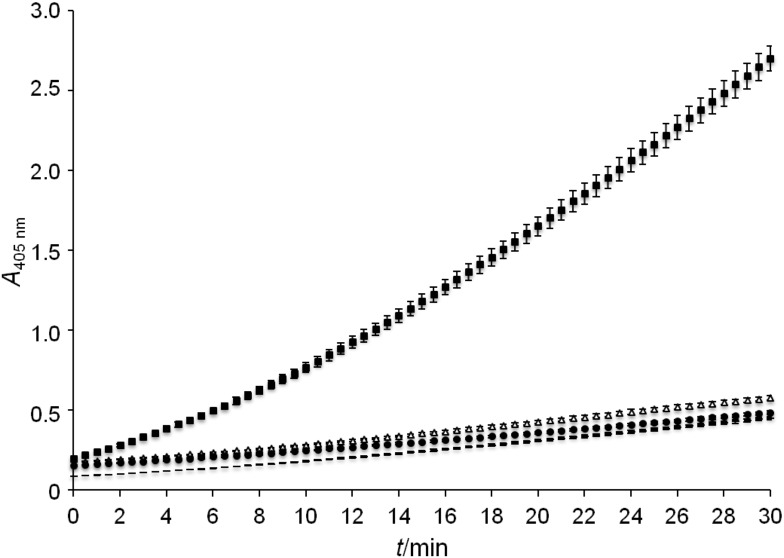
To demonstrate that an enzyme is functional when displayed by means of the MATE system, E. coli UT5600 cells were transformed with plasmid pMATE-PT013, which encodes the esterase EstA from B. gladioli (31) as passenger domain. The presentation of EstA on the cell surface was verified by flow cytometry as described above (data not shown). After inducing protein expression, cells were incubated with p-nitrophenyl acetate, and hydrolysis of this substrate was monitored photometrically at 405 nm. E. coli UT5600 pJExpress-401 and E. coli UT5600 pMATE-SI015 displaying mCherry were used as negative controls. As can be seen in Fig. 6, cells displaying the esterase catalyzed the hydrolysis of the applied substrate, indicating that EstA adopts an active conformation on the bacterial surface after transport with MATE.
Fig. 6.
Esterase activity assay of cells displaying EstA (■), cells displaying mCherry (Δ), cells with empty expression vector (●), and cell-free sample (־־). Cell cultivation and protein expression were performed as described, cells were washed twice, resuspended in 100 mM Tris/HCl buffer, pH=7.0, containing 3.2 mM p-nitrophenyl acetate, and absorption at 405 nm was monitored photometrically for 30 min
Secretion of mCherry
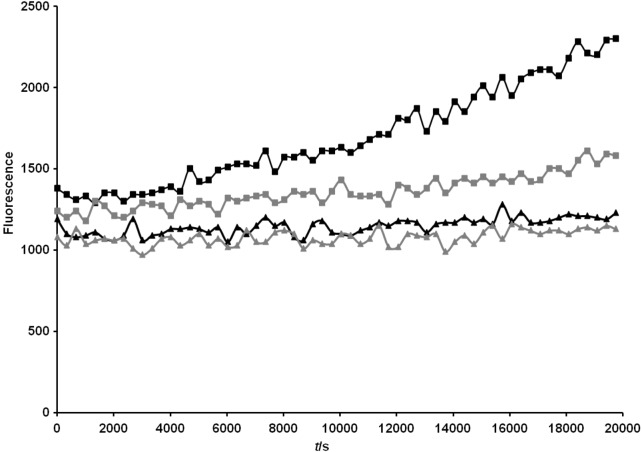
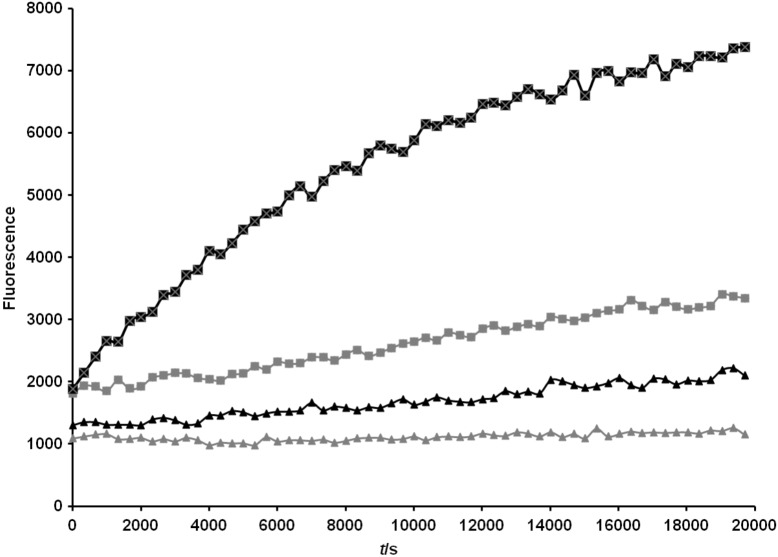
In the next step it was investigated whether the MATE system can be used for secretion of a full-length protein into the medium. In our case it was unknown whether OmpT protease would facilitate cleavage in the native linker. For this reason, we inserted an artificial OmpT cleavage site (Ala-Arg-Arg-Ala) in the C-terminal region of the recombinant passenger. We attempted to measure the activity of the native OmpT protease in whole cells and outer membrane preparations. The assay relies on the decrease in fluorescence resonance energy transfer (FRET) observed upon the hydrolysis of the substrate Abz-Ala-Arg-Arg-Ala-Tyr(NO2)-NH2. In whole cell assays, the measured activity of the OmpT-positive strain E. coli UT2300 was consistently higher, although the substrate was cleaved by proteases in both E. coli UT2300 and UT5600 strains (Fig. 7; available at www.ftb.com.hr). Incubating the protease substrate with isolated membrane proteins resulted in a greater differentiation between the OmpT-positive and -negative strains (Fig. 8; available at www.ftb.com.hr).
Fig. 7.
OmpT activity in whole cells of E. coli. Cells were grown to A600 nm=0.5 as described in the Materials and Methods. A volume of 100 µL of protein solution was added to the Abz-Ala-Arg-Arg-Ala-Tyr(NO2)-NH2 substrate to a final substrate concentration of 0.16 mM. Fluorescence was recorded over 5 h at 37 °C in intervals of 5 min with an excitation wavelength of 320 nm and an emission wavelength of 420 nm. Designations: outer membrane proteins from OmpT-positive E. coli UT2300 cells with substrate (■), and without substrate (▲); outer membrane proteins from OmpT-negative E. coli UT5600 cells with substrate (■), and without susbstrate (▲). LB medium was inoculated with a pre-culture of E. coli UT2300 or E. coli UT5600 as described above. Cells were grown at 37 °C to A600 nm=0.5. After a centrifugation step, cells were washed in assay buffer (0.5 M Tris/HCl, pH=7.0) and subsequently resuspended to A600 nm=0.5
Fig. 8.
OmpT activity in isolated outer membrane proteins of E. coli. Outer membrane proteins were isolated from a 40-mL culture of cells at A600 nm=0.5 as described in the Materials and Methods and resuspended in 1 mL of assay buffer. A volume of 100 µL of protein solution was added to the Abz-Ala-Arg-Arg-Ala-Tyr(NO2)-NH2 substrate to a final substrate concentration of 0.16 mM. Fluorescence was recorded over 5 h at 37 °C in intervals of 5 min with an excitation wavelength of 320 nm and an emission wavelength of 420 nm. Designations: outer membrane proteins from OmpT-positive E. coli UT2300 cells with substrate (■), and without substrate (▲); outer membrane proteins from OmpT-negative E. coli UT5600 cells with substrate (■), and without substrate (▲)
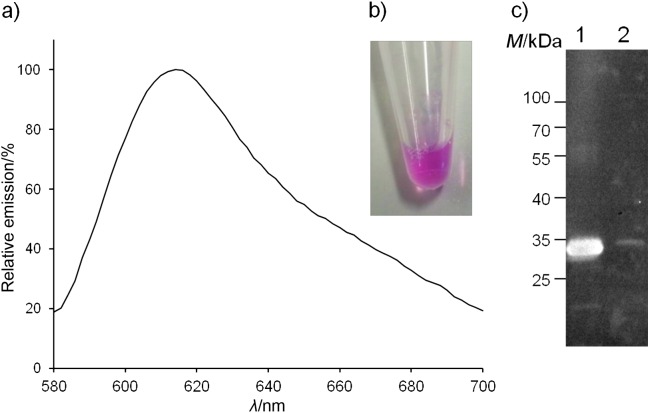
To examine the functionality of the artificial OmpT cleavage site for secretion of mCherry, we transferred the plasmid pMATE-SI015 into the OmpT-positive strain E. coli UT2300 and looked for the presence of mCherry in the growth medium. The OmpT-negative strain E. coli UT5600 served as a negative control. Supernatants from cultures of both E. coli UT2300 and E. coli UT5600 showed no visible colour difference. However, the eluate from the Ni-NTA column showed the distinctive pink colouring of mCherry in the sample of OmpT-positive strain E. coli UT2300. In contrast, the same eluate from OmpT-negative cells was colourless. The secreted mCherry exhibited a characteristic emission peak between 580 and 700 nm, with a maximum at about 615 nm (Fig. 9). The negative control did not show any significant fluorescence at the examined wavelength (data not shown).
Fig. 9.
Emission spectrum of: a) secreted affinity-purified 6xHis--mCherry. The plasmid encoding the MATE fusion protein with mCherry (pMATE-SI015) as a passenger was transformed into the OmpT-positive strain E. coli UT2300. Cells were grown to A600 nm=0.5, protein expression was induced, and the cells cultured at 37 °C and 200 rpm for 24 h. The cells were then removed from the growth medium by centrifugation, and the supernatant was loaded on a Ni-NTA column for purification. The secreted 6xHis containing protein was then eluted with 200 mM imidazole. The eluate was analysed in a fluorescence microplate reader at an excitation wavelength of 550 nm and emission spectra were recorded from 580 to 700 nm. Repeating the purification procedure with the growth medium from an OmpT-negative strain (E. coli UT5600) did not show any significant fluorescence at the examined wavelengths (data not shown); b) characteristic pink colour (see www.ftb.com.hr) of mCherry purified from the supernatant of E. coli UT2300 cells expressing MATE-mCherry; c) Western blot of eluate from Ni- -NTA purification of the supernatant of 1=UT2300 and 2=UT5600 cells expressing MATE-mCherry. A mass of 5 µg of protein was loaded into lane 1
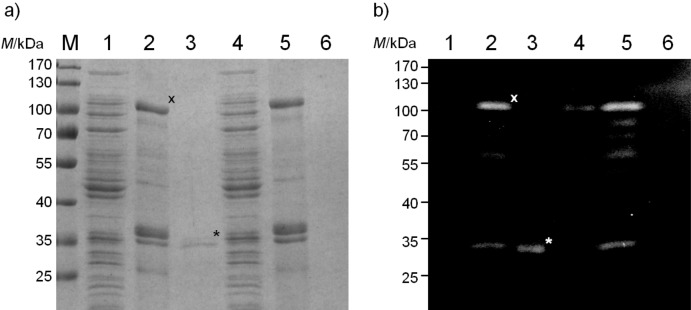
The overall yield of purified 6xHis mCherry was about 240 µg obtained from the 800-mL supernatant of a bacterial culture from E. coli UT2300 harbouring pMATE--SI015. The secreted affinity-purified 6xHis-mCherry from E. coli UT2300 was visible with an apparent molecular mass of approx. 30 kDa after electrophoresis (Fig. 9, c, lane 1). The calculated molecular mass of the secreted 6xHis-mCherry was 28.4 kDa after processing by the signal peptidase [AHG-TP, SignalP 4.0, (40)] and OmpT at the artificial cleavage site (AR-RA). This band was not visible in the OmpT-negative strain E. coli UT5600. There were no 6xHis-containing fragments smaller than 30 kDa, suggesting that there was no undesired cleavage by OmpT in the globular mCherry domain of the fusion protein. To get an impression of the transport and secretion efficiency of the MATE system, soluble protein and outer membrane fractions of E. coli UT2300 and UT5600 pMATE-SI015 cells were also separated by SDS-PAGE (Fig. 10, a) and the intensities of the protein bands assigned to MATE-mCherry and mCherry were analyzed using ImageJ software (41). The volume loaded into each lane was normalized to the number of cells from which the samples were obtained. Only a very small portion of fusion protein could be detected in the soluble protein fractions of both E. coli UT2300 and UT5600 cells (Fig. 10, a, lanes 1 and 4). The corresponding protein band is only visible in the Western blot when applying double volume of the sample, which we have avoided as other protein bands would then be overexposed (data not shown). In both strains, the largest part of expressed protein was located in the outer membrane fraction of the cells (Fig. 10, lanes 2 and 5), indicating that the transport of the fusion protein is nearly complete, and consequently, that the MATE system is very efficient in terms of surface translocation. However, when comparing the band intensities of MATE-mCherry in the outer membrane of E. coli UT2300 cells (Fig. 10, a, lane 2, marked with an x) with those of secreted mCherry in the supernatant (Fig 10, a, lane 3, marked with an asterisk), it becomes clear that only a relatively small portion of expressed MATE-mCherry ended up in the supernatant in form of secreted mCherry. The Coomassie-stained protein bands have a ratio of approx. 1 to 3.7, which would account for approx. 20% secretion. However, secreted mCherry is about threefold smaller than MATE-mCherry, and thus takes up correspondingly less Coomassie Brilliant Blue. Taking this into account, the secretion ratio would be 1 to 1.22, which is equivalent to 45% secretion. Although basically not adequate for quantifying the ratio of proteins of different sizes, Western blot revealed similar band intensity ratios. Obviously, the endogenous OmpT activity of E. coli UT2300 is not sufficient enough to release more of the displayed mCherry from the cell surface.
Fig. 10.
Differential cell fractionation of E. coli cells expressing MATE-mCherry from either an OmpT-positive strain UT2300 (lanes 1–3) or an OmpT-negative strain UT5600 (lanes 4–6). a) SDS-PAGE stained with Coomassie Brilliant Blue; b) Western blot with antibody against 6xHis epitope. M=PageRuler prestained protein marker. Fractions of E. coli soluble proteins (lanes 1 and 4), outer membrane proteins (lanes 2 and 5) and secreted proteins purified from the growth medium (lanes 3 and 6) are shown. Sample loading was normalized so that the proteins from an equivalent number of cells is given in each lane. The MATE-mCherry fusion protein is marked with an x, the secreted mCherry is marked with an asterisk (*)
Discussion
Autotransporters can be used for either surface display or secretion of recombinant proteins. Secretion of recombinant proteins has several advantages for the large-scale production of pharmaceutical and industrial proteins. Although proteins can be produced at high rates in the cytoplasm of E. coli, there are several problems associated with the accumulation of the proteins within the cell. They can aggregate into insoluble inclusion bodies that require time-consuming refolding procedures. Alternatively, the purification can require multiple chromatography steps. In contrast, secretion into the media efficiently separates the protein of interest from the E. coli proteome. Autotransporters have a unique pathway that may allow the rapid secretion and purification of proteins that are difficult to express in other systems. In this study we describe a new, maximized autotransporter-mediated expression (MATE) based on the codon-optimized EhaA autotransporter of E. coli originally discovered by Wells et al. (5).
Sevastsyanovich et al. (18) recently exploited the autocatalytic cleavage of SPATEs for the secretion of mCherry using an E. coli host. We developed this concept to create a novel system in which a single plasmid vector can be either used for surface display or continuous secretion. EhaA has been previously described as a member of the AIDA-I-type subfamily of autotransporter proteins (3). The expression of EhaA with its native passenger in E. coli resulted in cell aggregation and biofilm formation (5). To obtain the EhaA autotransporter domain, we used a C-terminal region of EhaA that could be aligned with the linker and β-barrel of AIDA-I previously used for surface display (16, 42). This region of EhaA was found to include the minimal translocation domain, as it facilitated the surface display of different passenger proteins, namely a small 6xHis passenger, mCherry and EstA, in E. coli. The respective fusion proteins were found in the outer membrane, and both mCherry and EstA could be shown by flow cytometry to be correctly displayed on the cell surface. Furthermore, both proteins retained their functional conformation on the cell surface. The artificial constructs did not result in cellular aggregation as found with native EhaA, suggesting that the cell-cell interaction observed by Wells et al. (5) is mediated by the EhaA N-terminal domain.
Secretion after surface display with an autotransporter requires endoproteolytic cleavage to release the passenger from β-barrel anchor in the outer membrane. OmpT-mediated cleavage of passenger proteins has been shown several times by utilizing the AIDA-I autotransporter domain (15, 16, 20-22), although the feasibility of OmpT-mediated secretion on preparative protein amounts did not lead to sufficient results. Several systems describe the utilization of OmpT as a processing enzyme for liberation of a desired peptide from a fusion protein. Even whole cells have been used as a source of OmpT protease for the processing of fusion proteins (30, 43). We demonstrate here the OmpT-mediated continuous secretion of mCherry via an artificial OmpT recognition site (Ala-Arg--Arg-Ala) inserted between the autotransporter linker and passenger domain. The MATE system allowed continuous secretion and efficient affinity purification of fluorescent 6xHis-mCherry from the growth medium. Previous reports show the successful secretion of fluorescent dsRed variants such as mCherry through the autotransporter pathway (18, 44). Sevastsyanovich et al. (18) reported yields in the range of 1 mg/L, which is considerably higher than the yield of approx. 0.3 mg/L that we were able to obtain using the MATE system. This discrepancy could in principle originate from differences in the used vector systems (in particular the plasmid replication origin and promoter), which leads to different expression levels of the autotransporter proteins. However, we consider another reason to be more likely: the quantitative comparison between expressed MATE-mCherry and secreted mCherry revealed that only approx. 45% of the expressed MATE-mCherry was actually cleaved and secreted into the growth medium. As nearly all of the expressed protein was located within the outer membrane, the translocation and incorporation of the MATE-mCherry protein into the outer membrane of E. coli can be considered as very efficient. Therefore, the endogenous OmpT activity, rather than MATE expression, translocation, or incorporation, appears to be the limiting factor for the mCherry secretion process. In contrast to the study of Sevastsyanovich et al. (18), in which every SPATE brings along an intrinsic protease functionality, the MATE system depends on the native OmpT activity of the host strain, which has to process a strongly overexpressed protein on the cell surface. Still, the yield obtained using the MATE system is high enough for preparative purposes, and could possibly be further enhanced by using an expression strain with overexpressed OmpT, as it has already been reported (34). In comparison with the utilization of SPATEs for secretion, the MATE system also allows surface display with the same plasmid in OmpT-negative strains.
Two additional OmpT cleavage sites (RR, RK or KR) can be found in mCherry. The lack of degradation products in the Western blot analyses indicated that OmpT did not cleave within the passenger domains. This corresponded well with the results obtained with the surface display of CtxB, where the consensus site in the linker domain was cleaved but not the dibasic residues within CtxB itself (15). An explanation could be that the passenger domains were folded before getting into contact with OmpT, and were therefore inaccessible to the protease. Overall, our results show that continuous secretion of mCherry with the MATE system can be successfully achieved regardless of the presence of predicted OmpT recognition sites within the passenger protein of interest.
Conclusion
In conclusion, the MATE system in Escherichia coli allows continuous secretion or surface display of a protein of interest. As both options can be realized with a single plasmid, this allows high-throughput screening of new protein functions, and the subsequent rapid and inexpensive purification of the improved variants.
Acknowledgements
The support of I.E.P.T. by the Reiner Lemoine foundation is gratefully acknowledged.
References
- 1.Jose J. Autodisplay: efficient bacterial surface display of recombinant proteins. Appl Microbiol Biotechnol. 2006;69:607–14. 10.1007/s00253-005-0227-z [DOI] [PubMed] [Google Scholar]
- 2.Gawarzewski I, Smits SH, Schmitt L, Jose J. Structural comparison of the transport units of type V secretion systems. Biol Chem. 2013;394:1385–98. 10.1515/hsz-2013-0162 [DOI] [PubMed] [Google Scholar]
- 3.Wells TJ, Totsika M, Schembri MA. Autotransporters of Escherichia coli: a sequence-based characterization. Microbiology. 2010;156:2459–69. 10.1099/mic.0.039024-0 [DOI] [PubMed] [Google Scholar]
- 4.Benz I, Schmidt MA. Isolation and serologic characterization of AIDA-I, the adhesin mediating the diffuse adherence phenotype of the diarrhea-associated Escherichia coli strain 2787 (O126:H27). Infect Immun. 1992;60:13–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, et al. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol. 2008;10:589–604. 10.1111/j.1462-2920.2007.01479.x [DOI] [PubMed] [Google Scholar]
- 6.Celik N, Webb CT, Leyton DL, Holt KE, Heinz E, Gorrell R, et al. A bioinformatic strategy for the detection, classification and analysis of bacterial autotransporters. PLoS One. 2012;7:e43245. 10.1371/journal.pone.0043245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jose J, Meyer TF. The autodisplay story, from discovery to biotechnical and biomedical applications. Microbiol Mol Biol Rev. 2007;71:600–19. 10.1128/MMBR.00011-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werner J, Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol. 2005;57:1450–9. 10.1111/j.1365-2958.2005.04775.x [DOI] [PubMed] [Google Scholar]
- 9.Peterson JH, Szabady RL, Bernstein HD. An unusual signal peptide extension inhibits the binding of bacterial presecretory proteins to the signal recognition particle, trigger factor, and the SecYEG complex. J Biol Chem. 2006;281:9038–48. 10.1074/jbc.M508681200 [DOI] [PubMed] [Google Scholar]
- 10.Xie K, Dalbey RE. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol. 2008;6:234–44. 10.1038/nrmicro1845 [DOI] [PubMed] [Google Scholar]
- 11.Purdy GE, Fisher CR, Payne SM. IcsA surface presentation in Shigella flexneri requires the periplasmic chaperones DegP, Skp, and SurA. J Bacteriol. 2007;189:5566–73. 10.1128/JB.00483-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J Bacteriol. 2009;191:6571–83. 10.1128/JB.00754-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leyton DL, Rossiter AE, Henderson IR. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol. 2012;10:213–25. 10.1038/nrmicro2733 [DOI] [PubMed] [Google Scholar]
- 14.Klauser T, Pohlner J, Meyer TF. Extracellular transport of cholera toxin B subunit using Neisseria IgA protease beta-domain: conformation-dependent outer membrane translocation. EMBO J. 1990;9:1991–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klauser T, Pohlner J, Meyer TF. Selective extracellular release of cholera toxin B subunit by Escherichia coli: dissection of Neisseria Iga beta-mediated outer membrane transport. EMBO J. 1992;11:2327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maurer J, Jose J, Meyer TF. Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J Bacteriol. 1997;179:794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dautin N. Serine protease autotransporters of enterobacteriaceae (SPATEs): biogenesis and function. Toxins (Basel). 2010;2:1179–206. 10.3390/toxins2061179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sevastsyanovich YR, Leyton D, Wells T, Wardius C, Tveen-Jensen K, Morris F, et al. A generalised module for the selective extracellular accumulation of recombinant proteins. Microb Cell Fact. 2012;11:69. 10.1186/1475-2859-11-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuno K, Yabuta M, Ooi T, Kinoshita S. Utilization of Escherichia coli outer-membrane endoprotease OmpT variants as processing enzymes for production of peptides from designer fusion proteins. Appl Environ Microbiol. 2004;70:76–86. 10.1128/AEM.70.1.76-86.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jose J, Bernhardt R, Hannemann F. Cellular surface display of dimeric Adx and whole cell P450-mediated steroid synthesis on E. coli. J Biotechnol. 2002;95:257–68. 10.1016/S0168-1656(02)00030-5 [DOI] [PubMed] [Google Scholar]
- 21.Hanke C, Hess J, Schumacher G, Goebel W. Processing by OmpT of fusion proteins carrying the HlyA transport signal during secretion by the Escherichia coli hemolysin transport system. Mol Gen Genet. 1992;233:42–8. 10.1007/BF00587559 [DOI] [PubMed] [Google Scholar]
- 22.Gustavsson M, Backlund E, Larsson G. Optimisation of surface expression using the AIDA autotransporter. Microb Cell Fact. 2011;10:72. 10.1186/1475-2859-10-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salema V, Marin E, Martinez-Arteaga R, Ruano-Gallego D, Fraile S, Margolles Y, et al. Selection of single domain antibodies from immune libraries displayed on the surface of E. coli cells with two beta-domains of opposite topologies. PLoS One. 2013;8:e75126. 10.1371/journal.pone.0075126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tozakidis IE, Sichwart S, Jose J. Going beyond E. coli: autotransporter based surface display on alternative host organisms. N Biotechnol. 2015;•••: 10.1016/j.nbt.2014.12.008 [DOI] [PubMed] [Google Scholar]
- 25.The UniProt Consortium . UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–12. 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2015;43:D30–5. 10.1093/nar/gku1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rattei T, Tischler P, Götz S, Jehl MA, Hoser J, Arnold R, et al. SIMAP – a comprehensive database of pre-calculated protein sequence similarities, domains, annotations and clusters. Nucleic Acids Res. 2010;38:D223–6. 10.1093/nar/gkp949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. [DOI] [PubMed] [Google Scholar]
- 30.Grodberg J, Dunn JJ. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988;170:1245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultheiss E, Paar C, Schwab H, Jose J. Functional esterase surface display by the autotransporter pathway in Escherichia coli. J Mol Catal, B Enzym. 2002;18:89–97. 10.1016/S1381-1177(02)00063-2 [DOI] [Google Scholar]
- 32.Park M, Yoo G, Bong JH, Jose J, Kang MJ, Pyun JC. Isolation and characterization of the outer membrane of Escherichia coli with autodisplayed Z-domains. Biochim Biophys Acta. 2015;1848:842–7. [DOI] [PubMed] [Google Scholar]
- 33.Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–92. 10.1007/BF00269672 [DOI] [PubMed] [Google Scholar]
- 34.Kramer RA, Zandwijken D, Egmond MR, Dekker N. In vitro folding, purification and characterization of Escherichia coli outer membrane protease ompT. Eur J Biochem. 2000;267:885–93. 10.1046/j.1432-1327.2000.01073.x [DOI] [PubMed] [Google Scholar]
- 35.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 36.Tozakidis IE, Sichwart S, Teese MG, Jose J. Autotransporter mediated esterase display on Zymomonas mobilis and Zymobacter palmae. J Biotechnol. 2014;191:228–35. 10.1016/j.jbiotec.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 37.Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, et al. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4:e7002. 10.1371/journal.pone.0007002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thömmes S. Development and autodisplay of human antibody libraries: new erythropoietin-binding variants [PhD Thesis]. Münster, Germany: Westphalian Wilhelms-University of Münster; 2013 (in German). [Google Scholar]
- 39.Jose J, Maas RM, Teese MG. Autodisplay of enzymes – molecular basis and perspectives. J Biotechnol. 2012;161:92–103. 10.1016/j.jbiotec.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 40.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 41.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maurer J, Jose J, Meyer TF. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J Bacteriol. 1999;181:7014–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yabuta M, Ochi N, Ohsuye K. Hyperproduction of a recombinant fusion protein of Staphylococcus aureus V8 protease in Escherichia coli and its processing by OmpT protease to release an active V8 protease derivative. Appl Microbiol Biotechnol. 1995;44:118–25. 10.1007/BF00164490 [DOI] [PubMed] [Google Scholar]
- 44.Ko HJ, Park E, Song J, Yang TH, Lee HJ, Kim KH, et al. Functional cell surface display and controlled secretion of diverse agarolytic enzymes by Escherichia coli with a novel ligation--independent cloning vector based on the autotransporter YfaL. Appl Environ Microbiol. 2012;78:3051–8. 10.1128/AEM.07004-11 [DOI] [PMC free article] [PubMed] [Google Scholar]