Summary
A simple method to detect saxitoxin (STX), one of the main components of the paralytic shellfish poison from red tide, has been developed. By using a next generation dye for double-stranded DNA we were able to differentiate fluorescence from STX-binding aptamers when exposed to different concentrations of STX, suggesting a change in aptamer folding upon target binding. The developed method is extremely rapid, only requiring small sample volumes, with quantitative results in the concentration range of 15 ng/mL to 3 µg/mL of STX, with a detection limit of 7.5 ng/mL.
Key words: red tide, paralytic shellfish poison, aptamers, saxitoxin, high resolution melting
Introduction
Aptamers are short single-stranded nucleic acids that bind to a defined target with high affinity and specificity (1, 2). They are selected from a random library of 1012–1024 oligonucleotides, through a process called SELEX (Systematic Evolution of Ligands by Exponential Enrichment) where target-bound strands are isolated and PCR amplified. The combination of an aptamer specificity and affinity, together with the inherent avoidance of an animal host makes aptamers an extremely attractive alternative to the use of antibodies as analyte recognition elements (3, 4). Aptamers are highly cost-effective, highly stable and are routinely produced by easier and more consistent manufacturing methods than antibodies.
Aptamers have been developed for a wide range of different molecules (5–8), and considerable effort has been placed on engineering and formatting selected aptamers into analytical devices. Since aptamers are a class of nucleic acid molecules, they are inherently much more flexible than antibodies and have been used in various formats not amenable to antibodies, including displacement assays (9, 10), apta-PCR (11, 12) and molecular aptamer beacons (13, 14). The arrangement of an aptamer as a sensing platform is often dependent on its interaction with the target (15–19). Some assays are designed similarly to those with antibodies, based on strategic labelling/immobilization of the participant molecules. Other common formats measure changes in surface properties when an attached molecule binds a free counterpart and recent developments rely on the structural switch that the aptamer itself undergoes when binding its target. In particular, the tertiary structural switch of aptamers that bind small molecules is often significant, and can be measured by a number of strategies (20–23). Molecular aptamer beacons are of particular interest as they allow assays to be performed in a single experimental step that does not require washing or further developing, with the only required end user intervention being sample addition. Successful examples are fluorophore-quencher, dual-pyrene and quantum dot interactions (24–28).
Here, we exploit a previously discovered aptamer (29), which we adapted as a label-free fluorescent sensor for the detection of saxitoxin (STX), a main component of the paralytic shellfish poison (PSP), concentrated by bivalve molluscs when they consume toxic dinoflagellates from a harmful algal bloom (HAB). Contaminated mollusc consumption can be fatal and HABs are thus not only a problem for public health, but also for mollusc fisheries, as well as tourism since they affect living resources in coastal systems (30–32). PSP toxicity detection is typically performed through a mouse bioassay, with obvious disadvantages, and several bioanalytical systems are currently being proposed to replace it (33–35). Herein, we detail the use of the reported STX-binding aptamer as a facile and economically viable molecular analytical tool for the sensitive and specific detection of PSP toxin, exploiting the fluorescence detection of the conformational change of aptamer structure upon binding with its cognate target.
Materials and Methods
Single-stranded DNA corresponding to the aptamer reported to have been selected against STX (29) (STXH aptamer: GGT ATT GAG GGT CGC ATC CCG TGG AAA CAT GTT CAT TGG GCG CAC TCC GCT TTC TGT AGA TGG CTC TAA CTC TCC TCT) as well as the random DNA strand used by the same authors as a negative control (RAN: GGT ATT GAG GGT CGC ATC TAG TAG AAA AGT GCT GAG TAG TTT TAC CTG GTA GAT ATG CGA TGG CTC TAA CTC TCC TCT) were synthesized by and purchased from Integrated DNA Technologies (Hayward, CA, USA).
STX and gonyautoxin 2/3 (GTX 2/3) preserved in 0.001 M HCl were lyophilized and later resuspended in RNAse- and DNase-free purified water to constitute 80 µg/mL of stock concentration.
The high quantum yield fluorescent dye Evagreen was purchased from Biotium, Inc. (Coralville, KY, USA). Evagreen uses a ‘release on demand’ mechanism, which greatly diminishes background noise, emitting fluorescence upon switching configuration when binding to double-stranded DNA.
Saxitoxin detection was performed in a 10-µL reaction volume containing 10 µM of the STX aptamer STXH in purified water and Evagreen dye at 5× final concentration. The fluorescence was measured by performing high resolution melting (HRM) analyses in a real time thermocycler Eco Illumina (Illumina, San Diego, CA, USA) with excitation/emission of 500/530 nm. The HRM denaturation graphics were monitored in order to verify the change from dsDNA to ssDNAs with increasing temperature, while the fluorescence value taken at 30 °C was used as a baseline value to establish the relation between STX concentration and fluorescence. The HRM protocol included a denaturation step of 5 s at 95 °C, followed by renaturation at 30 °C and a melting step from 30 to 95 °C at a ramp rate set at 0.08 °C/s (36). All measures were carried out in triplicate and the melting analysis was based on the derivative of the fluorescence signal normalized to a control that did not contain STX.
Goodness-of-fit for correlations (R2 values) and their significance (p-values calculated from an F-test) were performed using GraphPad Prism v. 5 for Windows (San Diego, CA, USA).
Results
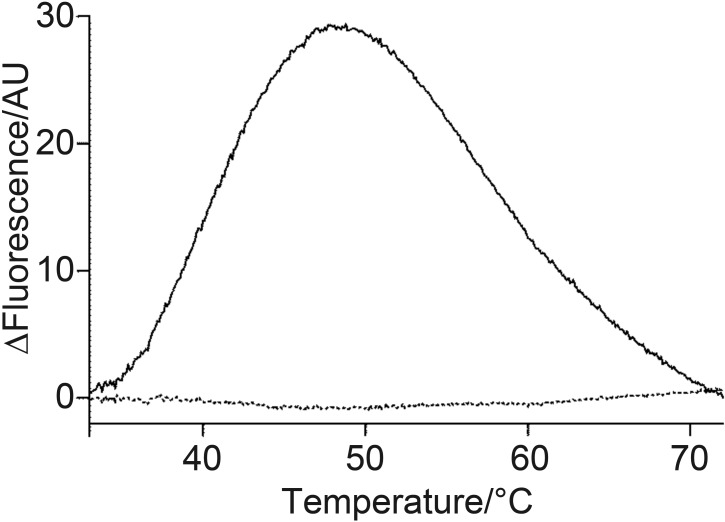
The normalized derivative melting profile of the aptamer bound to STX can be seen in Fig. 1, where the configuration change of the aptamer with 5 µM STX (full line) is clear when normalized to the control signal (dotted line), suggesting a change in the secondary structure stability upon target binding. This melting profile is consistent in all experiments performed with a range of toxin concentrations.
Fig. 1.
High resolution melting (HRM) profile of the saxitoxin- -binding aptamer (STXH) in the presence (full line) and absence (dotted line) of STX
Since the mouse bioassay has a detection limit of approx. 1 µM STX (34), two independent assays were performed around that value. The fluorescence signal of Evagreen bound to the STXH aptamer was tested for ten different concentrations of the toxin between 50 nM and 1 µM and between 1 and 10 µM, yielding a significant linear correlation of 0.821 (p=0.01) and 0.885 (p<0.001) respectively, while the same assays applied on the randomly generated ssDNA sequence RAN yielded correlations of 0.040 and 0.198 respectively, which were not significant (p>0.1).
The lowest amount of STX that influenced the melting profile of STXH aptamer was observed to be 25 nM. At that concentration, the profiles are significantly distinguishable from the melting profile of the unbound aptamer. However, the linearity of the relation concentration/fluorescence greatly increases only when points over 50 nM are considered.
The mouse bioassay is meant to quantify saxitoxin within a rough shellfish extract (37). Therefore, the detection system developed here was tested using this matrix. The linear correlation of Evagreen fluorescence and the toxin in concentrations between 1 and 10 µM, while significant (p<0.01), was considerably affected by the compounds of the extract (R2=0.618).
Potential cross-reactivity of the STXH aptamer was then tested with a range of concentrations of GTX 2/3, a similar toxin to that of STX, but with an OSO3– group instead of H group at one of the radical groups (38). No significant change in fluorescence of the Evagreen bound to the aptamer was observed between 1 and 10 µM (R=0.074; p>0.1), again highlighting the specificity of this fluorescence detection method.
Discussion
Most advances on label-free aptasensor development have been based on strategies where the target analyte sequesters the aptamer from a competitive binding molecule, often a complementary DNA strand (39–41). Since in these cases fluorescence is emitted by a dsDNA-binding dye, it is inversely proportional to analyte concentration. Double-stranded DNA is formed with fluorescently labelled DNA that is complementary to the aptamer, and a fluorescent signal is observed, with a quantitative decrease in fluorescence observed upon displacement of the aptamer from its labelled complementary sequence. The sensitivity of this strategy depends on the affinity of the aptamer for its target, which competes with the Watson- -Crick interaction between the aptamer and its complementary strand.
Here, a novel strategy was developed by directly measuring the change in fluorescence of a dsDNA-binding fluorophore, Evagreen, due to a modification in aptamer structure upon target binding, a phenomenon that has commonly been described for aptamers that bind to small molecules (5, 19, 42). With the STX-binding aptamer, the analyte contributes to secondary structure stability, resulting in a different double-stranded configuration compared to that of the free aptamer. Even though the unbound aptamer has a distinct denaturation profile, which indicates that without the target, other double- -stranded configurations might be formed (43–45), the amount of dsDNA area increases with target concentration. The increased stability of the aptamer bound to the toxin was verified by the distinct melting profiles observed in the HRM assays. This is the first report of a fluorescent aptasensor for the detection of saxitoxin.
However, while significant correlation was obtained under simulated field conditions, a weaker correlation compared to that of the laboratory conditions suggests an aptasensor of low robustness. Apparently, STXH aptamer performance might be very sensitive to external conditions, which might explain the poor results of surface plasmon resonance (SPR) analysis published previously (29) and reviewed in our facilities (∆RU<50). Possibly, small steric effects caused by STXH aptamers attached to the SPR chip surface might impede proper STX binding (46). Moreover, when STXH aptamer was tested with STX for competition with its complementary strand, inverse correlation between STX concentration and fluorescence was not present at 30 °C (data not shown). Thus, STXH binding affinity towards its target, not reported in the original publication (29), might not be high enough to overcome Watson-Crick interaction, as opposed to what has been described for other aptamers (39–41).
In this work, the successful detection of STX was performed by taking advantage of STXH configuration change upon binding its target, which increased the amount of dsDNA area. Indeed, aptamer-based sensors are designed specifically depending on the 3D structure and folding of each particular aptamer, which is usually modified when the aptamer binds its target (47–49). Usually, reporter molecules are added so they transduce the configuration change as a measurable signal. Hence, extensive efforts have been directed towards understanding the folding configuration of every developed aptamer. This can be estimated roughly through folding algorithms (50), but they typically have to be refined in order to obtain an accurate tertiary structure. Characterization of folding/unfolding configurations has been performed mainly through nuclear magnetic resonance analysis (51, 52), but also through mutational analysis (43, 53) and recently through the analysis of stretching force by optical tweezers (54, 55).
Here, for the first time, we characterized an aptamer and its interaction with the target by its unfolding speed in relation to small increments of temperature in HRM assays. HRM has been used so far to distinguish among different PCR amplicons in population studies, where a given DNA fragment can be identified by a particular denaturation profile, even when its sequence differs by a single base pair, applying constant temperature increments homogeneously among different samples (56, 57). The technique is performed by most nowadays real-time thermocyclers and it is considered one of the most precise, inexpensive and fastest methods to distinguish base--pair binding differentials (58, 59). The STXH aptamer showed a distinct HRM profile compared to the RAN sequence (data not shown) and additionally, dramatically changed its HRM profile upon interacting with STX. HRM assays could then be used as a simple and quick system to characterize aptamer configuration and to evaluate its response to its target, as well as to structural modifications or changes in the reaction media.
Conclusions
Here the feasibility of quantifying saxitoxin (STX) with specific aptamers was shown through the use of Evagreen, a dsDNA-binding fluorophore. When the aptamers are in a solution containing STX, they present a distinguishable melting profile, which is probably given by specific dsDNA areas that are formed when the target stabilizes the structure. Additionally, at a given temperature, fluorescence of the Evagreen bound to the dsDNA can be correlated with STX concentration. The method developed here is inexpensive, easy to prepare, fast and sensitive. However, correlation falls dramatically when trying to quantify STX in a rough shellfish extract. This is probably caused by the relatively low affinity of the aptamer for STX, thus, future developments towards high affinity aptamers could lead to a robust aptamer-mediated method to quantify STX in field samples.
Acknowledgements
Support for the work was provided by FONDECYT grant 11110050 and PIA grant ACT1108 from CONICYT Chile. The authors would also like to acknowledge the help provided by Alessandro Pinto, Ioanis Katakis and Juan Kuznar.
References
- 1.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–83. 10.1146/annurev.med.56.062904.144915 [DOI] [PubMed] [Google Scholar]
- 2.O’Sullivan CK. Aptasensors – the future of biosensing? Anal Bioanal Chem. 2002;372:44–8. 10.1007/s00216-001-1189-3 [DOI] [PubMed] [Google Scholar]
- 3.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–50. [PubMed] [Google Scholar]
- 4.Tombelli S, Minunni M, Mascini M. Analytical applications of aptamers. Biosens Bioelectron. 2005;20:2424–34. 10.1016/j.bios.2004.11.006 [DOI] [PubMed] [Google Scholar]
- 5.Cho EJ, Lee JW, Ellington AD. Applications of aptamers as sensors. Annu Rev Anal Chem (Palo Alto Calif). 2009;2:241–64. 10.1146/annurev.anchem.1.031207.112851 [DOI] [PubMed] [Google Scholar]
- 6.Kang KN, Lee YS. RNA aptamers: a review of recent trends and applications. In: Zhong J, editor. Advances in biochemical engineering/biotechnology, Vol. 131: Future trends in biotechnology. Berlin, Germany: Springer-Verlag; 2013. pp. 153–69. http://dx.doi.org/ 10.1007/10_2012_136 [DOI] [PubMed] [Google Scholar]
- 7.Long F, Zhu A, Shi H. Recent advances in optical biosensors for environmental monitoring and early warning. Sensors (Basel). 2013;13:13928–48. 10.3390/s131013928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiviyanathan V, Gorenstein DG. Aptamers and the next generation of diagnostic reagents. Proteomics Clin Appl. 2012;6:563–73. 10.1002/prca.201200042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen BT, Anslyn EV. Indicator–displacement assays. Coord Chem Rev. 2006;250:3118–27. 10.1016/j.ccr.2006.04.009 [DOI] [Google Scholar]
- 10.Cruz-Aguado JA, Penner G. Fluorescence polarization based displacement assay for the determination of small molecules with aptamers. Anal Chem. 2008;80(22):8853–5. 10.1021/ac8017058 [DOI] [PubMed] [Google Scholar]
- 11.Svobodova M, Mairal T, Nadal P, Bermudo MC, O’Sullivan CK. Ultrasensitive aptamer based detection of β-conglutin food allergen. Food Chem. 2014;165:419–23. 10.1016/j.foodchem.2014.05.128 [DOI] [PubMed] [Google Scholar]
- 12.Pinto A, Bermudo Redondo MC, Ozalp VC, O’Sullivan CK. Real-time apta-PCR for 20 000-fold improvement in detection limit. Mol Biosyst. 2009;5:548–53. 10.1039/b814398f [DOI] [PubMed] [Google Scholar]
- 13.Mairal T, Ozalp VC, Lozano Sanchez P, Mir M, Katakis I, O’Sullivan CK. Aptamers: molecular tools for analytical applications. Anal Bioanal Chem. 2008;390:989–1007. 10.1007/s00216-007-1346-4 [DOI] [PubMed] [Google Scholar]
- 14.Radi AE, Acero Sánchez JL, Baldrich E, O’Sullivan CK. Reagentless, reusable, ultrasensitive electrochemical molecular beacon aptasensor. J Am Chem Soc. 2006;128:117–24. 10.1021/ja053121d [DOI] [PubMed] [Google Scholar]
- 15.Gopinath SC. Methods developed for SELEX. Anal Bioanal Chem. 2007;387:171–82. 10.1007/s00216-006-0826-2 [DOI] [PubMed] [Google Scholar]
- 16.Song KM, Lee S, Ban C. Aptamers and their biological applications. Sensors (Basel). 2012;12:612–31. 10.3390/s120100612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han K, Liang Z, Zhou N. Design strategies for aptamer- -based biosensors. Sensors (Basel). 2010;10:4541–57. 10.3390/s100504541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tainaka K, Sakaguchi R, Hayashi H, Nakano S, Liew FF, Morii T. Design strategies of fluorescent biosensors based on biological macromolecular receptors. Sensors (Basel). 2010;10:1355–76. 10.3390/s100201355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng AKH, Sen D, Yu HZ. Design and testing of aptamer- -based electrochemical biosensors for proteins and small molecules. Bioelectrochemistry. 2009;77:1–12. 10.1016/j.bioelechem.2009.04.007 [DOI] [PubMed] [Google Scholar]
- 20.Feng C, Dai S, Wang L. Optical aptasensors for quantitative detection of small biomolecules: a review. Biosens Bioelectron. 2014;59:64–74. 10.1016/j.bios.2014.03.014 [DOI] [PubMed] [Google Scholar]
- 21.Giannetti A, Tombelli S, Baldini F. Oligonucleotide optical switches for intracellular sensing. Anal Bioanal Chem. 2013;405:6181–96. 10.1007/s00216-013-7086-8 [DOI] [PubMed] [Google Scholar]
- 22.Pu Y, Liu H, Liu B, Liao J, Liu J, Zhao Z, et al. Development of aptamer-based nanomaterials for biological analysis. Curr Mol Med. 2013;13:681–9. 10.2174/1566524011313040016 [DOI] [PubMed] [Google Scholar]
- 23.Juskowiak B. Nucleic acid-based fluorescent probes and their analytical potential. Anal Bioanal Chem. 2011;399:3157–76. 10.1007/s00216-010-4304-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang CJ, Jockusch S, Vicens M, Turro NJ, Tan W. Light- -switching excimer probes for rapid protein monitoring in complex biological fluids. Proc Natl Acad Sci USA. 2005;102:17278–83. 10.1073/pnas.0508821102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozaki H, Nishihira A, Wakabayashi M, Kuwahara M, Sawai H. Biomolecular sensor based on fluorescence-labeled aptamer. Bioorg Med Chem Lett. 2006;16(16):4381–4. 10.1016/j.bmcl.2006.05.054 [DOI] [PubMed] [Google Scholar]
- 26.Freeman R, Girsh J, Willner I. Nucleic acid/quantum dots (QDs) hybrid systems for optical and photoelectrochemical sensing. ACS Appl Mater Interfaces. 2013;5:2815–34. 10.1021/am303189h [DOI] [PubMed] [Google Scholar]
- 27.Lee JH. Conjugation approaches for construction of aptamer-modified nanoparticles for application in imaging. Curr Top Med Chem. 2013;13:504–12. 10.2174/1568026611313040010 [DOI] [PubMed] [Google Scholar]
- 28.Meyer M, Scheper T, Walter JG. Aptamers: versatile probes for flow cytometry. Appl Microbiol Biotechnol. 2013;97:7097–109. 10.1007/s00253-013-5070-z [DOI] [PubMed] [Google Scholar]
- 29.Handy SM, Yakes BJ, DeGrasse JA, Campbell K, Elliott CT, Kanyuck KM, et al. First report of the use of a saxitoxin–protein conjugate to develop a DNA aptamer to a small molecule toxin. Toxicon. 2013;61:30–7. 10.1016/j.toxicon.2012.10.015 [DOI] [PubMed] [Google Scholar]
- 30.Heisler J, Glibert PM, Burkholder JM, Anderson DM, Cochlan W, Dennison WC, et al. Eutrophication and harmful algal blooms: a scientific consensus. Harmful Algae. 2008;8:3–13. 10.1016/j.hal.2008.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James KJ, Carey B, O’Halloran J, van Pelt FN, Skrabakova Z. Shellfish toxicity: human health implications of marine algal toxins. Epidemiol Infect. 2010;138:927–40. 10.1017/S0950268810000853 [DOI] [PubMed] [Google Scholar]
- 32.Shumway SE. A review of the effects of algal blooms on shellfish and aquaculture. J World Aquacult Soc. 1990;21:65–104. 10.1111/j.1749-7345.1990.tb00529.x [DOI] [Google Scholar]
- 33.Sellner KG, Doucette GJ, Kirkpatrick GJ. Harmful algal blooms: causes, impacts and detection. J Ind Microbiol Biotechnol. 2003;30:383–406. 10.1007/s10295-003-0074-9 [DOI] [PubMed] [Google Scholar]
- 34.Alexander J, Benford D, Cockburn A, Cravedi JP, Dogliotti E, Domenico AD, et al. Scientific opinion of the panel on contaminants in the food chain – marine biotoxins in shellfish – saxitoxin group. The EFSA J. 2009;1019:1–76. [Google Scholar]
- 35.Levin RE. Paralytic shellfish toxins: their origin, characteristics and methods of detection: a review. J Food Biochem. 1991;15:405–17. 10.1111/j.1745-4514.1991.tb00425.x [DOI] [Google Scholar]
- 36.Kalthoff D, Beer M, Hoffmann B. High resolution melting analysis: rapid and precise characterisation of recombinant influenza A genomes. Virol J. 2013;10:284. 10.1186/1743-422X-10-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truman P, Lake RJ. Comparison of mouse bioassay and sodium channel cytotoxicity assay for detecting paralytic shellfish poisoning toxins in shellfish extracts. J AOAC Int. 1996;79:1130–3. [PubMed] [Google Scholar]
- 38.Patockaa J, Stredab L. Brief review of natural nonprotein neurotoxins. ASA Newsl. 2002;89:16–24. [Google Scholar]
- 39.Li D, Song S, Fan C. Target-responsive structural switching for nucleic acid-based sensors. Acc Chem Res. 2010;43:631–41. 10.1021/ar900245u [DOI] [PubMed] [Google Scholar]
- 40.Choi MS, Yoon M, Baeg JO, Kim J. Label-free dual assay of DNA sequences and potassium ions using an aptamer probe and a molecular light switch complex. Chem Commun (Camb). 2009;47:7419–21. 10.1039/b915994k [DOI] [PubMed] [Google Scholar]
- 41.Lv Z, Chen AL, Liu JC, Guan Z, Zhou Y, Xu SY, et al. A simple and sensitive approach for ochratoxin A detection using a label-free fluorescent aptasensor. PLoS One. 2014;9:e85968. 10.1371/journal.pone.0085968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwalbe H, Buck J, Fürtig B, Noeske J, Wöhnert J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew Chem Int Ed Engl. 2007;46:1212–9. 10.1002/anie.200604163 [DOI] [PubMed] [Google Scholar]
- 43.Stojanovic MN, de Prada P, Landry DW. Aptamer-based folding fluorescent sensor for cocaine. J Am Chem Soc. 2001;123:4928–31. 10.1021/ja0038171 [DOI] [PubMed] [Google Scholar]
- 44.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287(5454):820–5. 10.1126/science.287.5454.820 [DOI] [PubMed] [Google Scholar]
- 45.Nutiu R, Li Y. Structure-switching signaling aptamers. J Am Chem Soc. 2003;125:4771–8. 10.1021/ja028962o [DOI] [PubMed] [Google Scholar]
- 46.Tang Q, Su X, Loh KP. Surface plasmon resonance spectroscopy study of interfacial binding of thrombin to antithrombin DNA aptamers. J Colloid Interface Sci. 2007;315:99–106. 10.1016/j.jcis.2007.06.040 [DOI] [PubMed] [Google Scholar]
- 47.Fang X, Sen A, Vicens M, Tan W. Synthetic DNA aptamers to detect protein molecular variants in a high-throughput fluorescence quenching assay. ChemBioChem. 2003;4:829–34. 10.1002/cbic.200300615 [DOI] [PubMed] [Google Scholar]
- 48.Frauendorf C, Jaschke A. Detection of small organic analytes by fluorescing molecular switches. Bioorg Med Chem. 2001;9:2521–4. 10.1016/S0968-0896(01)00027-X [DOI] [PubMed] [Google Scholar]
- 49.Rajendran M, Ellington AD. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal Bioanal Chem. 2008;390:1067–75. 10.1007/s00216-007-1735-8 [DOI] [PubMed] [Google Scholar]
- 50.Zuker M, Mathews DH, Turner DH. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark BFC, editors. RNA biochemistry and biotechnology. NATAO Science Series: Series 3: High technology. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1999;70:11–43. http://dx.doi.org/ 10.1007/978-94-011-4485-8_2 [DOI] [Google Scholar]
- 51.Patel DJ. Structural analysis of nucleic acid aptamers. Curr Opin Chem Biol. 1997;1:32–46. 10.1016/S1367-5931(97)80106-8 [DOI] [PubMed] [Google Scholar]
- 52.Dieckmann T, Suzuki E, Nakamura GK, Feigon J. Solution structure of an ATP-binding RNA aptamer reveals a novel fold. RNA. 1996;2:628–40. [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Z, Szostak JW. Evolution of aptamers with a new specificity and new secondary structures from an ATP aptamer. RNA. 2003;9:1456–63. 10.1261/rna.5990203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319(5863):630–3. 10.1126/science.1151298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodside MT, Garcia-Garcia C, Block SM. Folding and unfolding single RNA molecules under tension. Curr Opin Chem Biol. 2008;12:640–6. 10.1016/j.cbpa.2008.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittwer CT. High-resolution DNA melting analysis: advancements and limitations. Hum Mutat. 2009;30:857–9. 10.1002/humu.20951 [DOI] [PubMed] [Google Scholar]
- 57.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50:1748–54. 10.1373/clinchem.2003.029751 [DOI] [PubMed] [Google Scholar]
- 58.Vossen RH, Aten E, Roos A, den Dunnen JT. High-resolution melting analysis (HRMA) – more than just sequence variant screening. Hum Mutat. 2009;30:860–6. 10.1002/humu.21019 [DOI] [PubMed] [Google Scholar]
- 59.Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8:597–608. 10.2217/14622416.8.6.597 [DOI] [PubMed] [Google Scholar]