Summary
Lactococcus lactis ssp. lactis BGBM50, a producer of lactococcin G and aggregation-promoting factor, was isolated from selected lactic acid bacteria taken from semi-hard cheese traditionally produced in the village Žanjic, Montenegro. Strain BGBM50 harbours a number of plasmids of different sizes. Plasmid curing experiments showed that genes for bacteriocin production are located on pBM140, a plasmid 140 kb in length. PCR analysis with primers specific for lactococcin Q and G genes gave fragment of the expected size. In addition, after plasmid curing of strain BGBM50, different derivatives with altered phenotypes were obtained, among them BGBM50-34 strain, which retained bacteriocin synthesis but had enhanced aggregation ability.
Key words: bacteriocin lactococcin G, aggregation-promoting factor, plasmid curing
Introduction
Some strains of Gram-positive and Gram-negative bacteria have the ability to produce bacteriocins. Bacteriocins are ribosomaly synthesised antimicrobial peptides that generally inhibit the growth of closely related microbes, with a narrow spectrum of activity (1). Many bacteriocins from Gram-positive bacteria show inhibitory activity not only within the species of the producer, but also against species or genera different from the bacteriocin producer, including foodborne pathogens and spoilage microorganisms such as Listeria monocytogenes, Bacillus cereus, Staphylococcus aureus and Clostridium tyrobutyricum (2). Based on structure, mode of action and biochemical and genetic characteristics, bacteriocins from Gram-positive bacteria can be divided into four main groups: (i) lantibiotics, (ii) non-modified heat-stable bacteriocins, (iii) large heat-stable bacteriocins, and (iv) cyclic bacteriocins (3). Bacteriocins produced by lactic acid bacteria (LAB) are of special interest for many reasons, one of the most important being that they are generally regarded as safe (GRAS) for human consumption, since they are found or used in fermented food and feed products (3). LAB are used as starter cultures for a wide variety of fermented dairy and meat products. Among them, Lactococcus lactis strains are the most commonly used by the dairy industry for the manufacture of different types of cheese (4). Bacteriocins from all four main groups have been isolated from different strains of L. lactis. In the last two decades, the interest in the application of bacteriocinogenic LAB and their bacteriocins to food preservation has increased. During this period, several new bacteriocins have been characterised, but only nisin and pediocin are permitted for application in the production of cheese and some meat products (5). The chances of isolating bacteriocinogenic strains from cheese produced from raw milk have increased since this is a natural environment for the growth and development of LAB. In traditionally manufactured homemade cheese, without the use of commercial starter cultures, non-starter lactic acid bacteria (NSLAB) usually become the dominant cheese microbiota that contribute to the texture, flavour and quality of the product (6). The exact role of NSLAB strains is not fully clear, but they certainly influence cheese ripening. On the other hand, they can be the source of a variety of different new, and some old preserved, properties of LAB, which the industrial strains have lost over years of use. Besides the production of antimicrobial substances, aggregation ability could play the main role in the adhesion of bacteria, colonisation ability and coaggregation with other bacterial strains (7, 8). The only aggregation-promoting factor described so far in lactococci is the one characterised in L. lactis ssp. lactis BGKP1 strain (9). The aim of this work is the isolation of natural strains of lactococci from homemade cheese, manufactured without using a starter culture, with the ability to produce bacteriocin(s) and to aggregate, in order to identify a strain with new properties.
Materials and Methods
Isolation of bacteriocin-producing lactococci
The bacterial strains used in this study are listed in Table 1 (10–13). Semi-hard cheese was produced in a traditional way in the village of Žanjic, Montenegro, without the addition of any starter cultures. Cheese sample (20 g) was homogenised with 180 mL of peptone water (0.1% by mass per volume), submitted to serial tenfold dilutions in peptone water and spread on the surface of M17 plates (Merck GmbH, Darmstadt, Germany) supplemented with (by mass per volume): glucose 0.5% and GM17 agar 1.5%. After aerobic incubation at 30 °C for 48 h, colonies were randomly selected and transferred to GM17 broth and incubated at 30 °C. For detection of bacteriocin activity, an agar well diffusion assay was used (14). Lactococcus lactis ssp. lactis BGMN1-596 was used as an indicator strain. Aliquots (50 µL) of the supernatant of 15-hour overnight cultures of tested strains were poured into the wells (Ř 5 mm) made in GM17 soft agar. To confirm the production of antimicrobial compounds of proteinaceous nature, crystals of pronase E (P5147, Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany), proteinase K (P8044, Sigma-Aldrich Chemie GmbH), trypsin (T1005, Sigma- -Aldrich Chemie GmbH) and pepsin (P7000, Sigma-Aldrich Chemie GmbH) were placed close to the edge of the wells. The plates were incubated overnight at 30 °C. A clear zone of inhibition around the well, but not near the pronase E, proteinase K, trypsin and pepsin, was taken as a positive signal for bacteriocin production. For further work, one strain, designated BGBM50, was chosen.
Table 1. Bacterial strains and plasmids.
| Bacterial strain | Description | Source or reference |
|---|---|---|
| Lactococcus lactis | ||
| BGBM50 | Natural isolate from semi-hard cheese, Bac+, Bacim, Agg+/– | This work |
| BGBM50-8 | Derivative of BGBM50, Bac+,Bacim, Agg+/– | This work |
| BGBM50-34 | Derivative of BGBM50-8, Bac+, Bacim, Agg+ | This work |
| BGBM50-11 | Derivative of BGBM50-8, Bac+, Bacim, Agg+/– | This work |
| BGBM50-13 | Derivative of BGBM50-11, Bac+, Bacim, Agg+/– | This work |
| BGBM50-14 | Derivative of BGBM50-11, Bac–, Bacs, Agg+/– | This work |
| BGBM50-25 | Derivative of BGBM50-11, Bac+, Bacim, Agg+/– | This work |
| BGBM50-62 | Derivative of BGBM50-11, Bac+, Bacim, Agg+/– | This work |
| BGMN1-5 | Natural isolate from semi-hard cheese, Bac+(lactococcin B, lsbB producer) Bacim, Agg+ | (10) |
| BGMN1-501 | Derivative of BGBM1-5, Bac+ (lactococcin B producer) lacBim | (10) |
| BGMN1-596T | Derivative of BGBM1-5, Bac+ (lsbB producer), Bacim, Agg– | (10) |
| MG7284/pAZIL-lsbB | Derivate of MG7284 with pAZIL-lsbB, (lsbB producer) lsbBim | (11) |
| BGMN1-596 | Plasmid-free derivative of BGMN1-5, Prt-, Bacim, Agg– | (10) |
| BGBU1-4 | Natural isolate from semi-hard cheese, Bac+ (unknown bacteriocin producer), Bacim | (12) |
| BGSM1-19 | Natural isolate from semi-hard cheese, Bac+ (lactin RM, lactococcin B producer), Bacim | (13) |
Bac+=bacteriocin producer, Bac–=non-bacteriocin producer, Bacs=sensitivity to bacteriocin, Bacim=immunity to bacteriocin, Agg+=aggregation, Agg+/–=slow aggregation
Molecular determination of the bacteriocin producer
Preliminary determination of strain BGBM50 was performed with classical microbiological and biochemical techniques. Fermentation ability was determined by API 50CHL (API System S.A., bioMérieux, Marcy l’Etoile, France). Total DNA from pure cultures was extracted by the method of Hopwood et al. (15), modified as follows: a pellet of log culture (A600 nm=0.6–0.8) was washed in 0.5 mL of TEN buffer (50 mM Tris-HCl, pH=8, 10 mM EDTA, pH=8, 50 mM NaCl) and resuspended in 0.5 mL of buffer (0.5 M sucrose, 40 mM ammonium acetate and 10 mM magnesium acetate, pH=7). The concentration of lysozyme was 8 mg/mL. Cells were incubated at 37 °C for 30 min, and then 250 µL of 2% sodium dodecyl sulphate (SDS) were added and vortexed for 1 min. Following cell lysis, phenol-chloroform extraction was performed until the solution was almost protein free. DNA was concentrated with 3 M sodium acetate, pH=4.8 (1/10 of total volume) and isopropanol (the same volume as the DNA water solution). After centifugation (13 000 rpm for 5 min), the pellet was washed with ethanol (75%, 1 mL), centrifuged again (13 000 rpm for 5 min), air dried and resuspended in 50 µL of double distilled water. Final species identification based on 16S rDNA sequencing was performed as described previously by Golic et al. (16). The 16S rDNA was sequenced (Macrogen, Amsterdam, The Netherlands) with UNI16SF and UNI16SR (17) primers.
Biochemical characterisation of bacteriocin produced by L. lactis ssp. lactis BGBM50
Cell-free culture supernatant of strain BGBM50 was incubated for 15 min at 40, 60, 80 and 100 °C. Then, the samples were cooled to room temperature and the remaining bacteriocin activity was determined. To determine the effect of pH on bacteriocin activity, the pH of the bacteriocin samples was adjusted to pH=2, 7 and 10 with 1 M HCl or 1 M NaOH. Samples were incubated for 1 h at 30 °C and bacteriocin activity was determined using the agar well diffusion test. The supernatant of the non-bacteriocin-producing strain BGMN1-596 was pretreated in the same way as the bacteriocin sample and used as a control to eliminate the effect of pH alone on the growth of the indicator strain.
Search for bacteriocin-encoding genes by polymerase chain reaction
Genes coding for the most common bacteriocins produced by L. lactis strains were sought by specific PCR primers (Table 2; 18,19). All amplification reactions were conducted under standard conditions at an annealing temperature of 50 °C. The PCR products were purified, sequenced (Macrogen) and analysed using BLAST algorithm (National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD, USA).
Table 2. Primers used throughout this study.
| Name | Sequence (5`-3`) | Bacteriocin gene | Reference |
|---|---|---|---|
| Lact481-F | CCAATGTCATTGCATCTGCAC | Lacticin 481 gene | (18) |
| Lact481-R | GTCCTTATGTTGCTATTCATC | Lacticin 481 gene | (18) |
| Lcn972-F | TTGTAGCTCCTGCAGAAGGAACATGG | Lactococcin 972 gene | (19) |
| Lcn972-R | GCCTTAGCTTTGAATTCTTACCAAAAG | Lactococcin 972 gene | (19) |
| LactGQ-F | GAAAGAATTATCAGAAAAAG | Lactococcin G and Q genes | (18) |
| LactGQ-R | CCACTTATCTTTATTTCCCTCT | Lactococcin G and Q genes | (18) |
| Lact3147-F | GTCTTTGTGTTGTTTGGAGATG | Lacticin 3147 gene | (18) |
| Lact3147-R | CAACTCCCGAAATAAATCATCG | Lacticin 3147 gene | (18) |
Plasmid curing experiments
Plasmid curing was achieved by growing the cells in the presence of novobiocin at sublethal temperature (20). Briefly, prewarmed GM17 broth (39 °C) containing novobiocin (8 µg/mL) was inoculated with 103 cells per mL. After 2 h of incubation, the cells were collected by centrifugation and resuspended in the same volume of prewarmed novobiocin containing GM17 broth to avoid a bacteriocin-killing effect on the cured (Bac–Bacs) cells. The same step was repeated ten times and the aliquots (0.1 mL) were then plated onto GM17 agar plates, which were then incubated at 30 °C for 48 h. To detect Bac–Bacs derivatives, Petri plates were overlaid with a reconstituted GM17 soft agar containing indicator cells BGMN1-596 and incubated overnight at 30 °C. Colonies that lost the ability to inhibit the indicator strain were taken from the original master plate and used as indicator strains for rechecking their Bac–Bacs phenotype. Plasmid profile and pulsed field gel electrophoresis (PFGE) were used to confirm that the obtained derivatives, after plasmid curing experiments, originated from strain BGBM50. Incorporation of lactococcal bacterial cells into agarose blocks, restriction enzyme treatment and PFGE were conducted as described previously (21).
Visual aggregation assay
Among 50 selected isolates from homemade cheese in the village Žanjic, Montenegro, the strains that exhibited aggregation ability were screened by a visual assay. Aggregation received positive scores when clearly visible sand-like particles, formed by aggregated cells, gravitated to the bottom of the tube, forming a precipitate and leaving a clear supernatant.
Results
Identification of bacteriocin-producing isolates
Fifty randomly selected colonies, isolated using standard procedures for the isolation of lactic acid bacteria, were collected from semi-hard homemade cheese manufactured in the village of Žanjic and subjected to bacteriocin testing. Two isolates showing inhibitory activity of the growth of indicator strain Lactococcus lactis ssp. lactis BGMN1-596 were identified according to microbiological techniques, the API50 CHL system and by sequencing of the genes for 16S rRNA. According to these results, isolate BGBM50 showed the highest zone of inhibition and aggregation phenotype. It was identified as Lactococcus lactis ssp. lactis and chosen for further study.
Bacteriocin production and its biochemical characterisation
The inhibitory substance produced by isolate BGBM 50 was temporarily designated as bacteriocin BacBM50. It was observed that the production of bacteriocin by strain BGBM50 in GM17 medium depended on the growth phase and reached a maximal plateau when the culture entered the late exponential phase (data not shown). Bacteriocin BacBM50 is a heat-stable molecule; its antimicrobial activity was not affected by treatment at 100 °C for 30 min, but after 60 min, the activity was completely abolished. Present in the cell-free supernatant of the producer strain, bacteriocin BacBM50 retained antimicrobial activity in a pH range between 1 and 12, with maximum activity observed between pH=5 and 7. The inhibitory activity on the growth of sensitive indicator cells was eliminated by treatment with proteolytic enzymes such as pepsin, trypsin, pronase E and proteinase K.
Targeting the bacteriocin-encoding genes by PCR
PCR analysis was undertaken using gene-specific primers to determine genes of the most common lactococcal bacteriocins. Positive amplification was obtained only when gene-specific primers for the genes encoding bacteriocin lactococcin G were used. The amplified fragment was sequenced and it confirmed that strain BGBM50 produces lactococcin G. Interestingly, a cross-immunity test with Lactococcus lactis LMG 2081 strain, a producer of lactococcin G (22), revealed that strain BGBM50 does not inhibit the growth of strain LMG 2081. On the contrary, strain LMG 2081 inhibited the growth of BGBM50 indicating that strain LMG 2081 produces additional bacteriocin(s). In order to determine the antimicrobial spectrum of bacteriocin lactococcin G, cross-immunity tests with other lactococcal bacteriocin-producing strains were performed and the results are presented in Table 3.
Table 3. Antimicrobial activity of bacteriocin-producing Lactococcus lactis strains.
| Bac producer Indicator strain |
BGBM50 | BGMN1-5 | BGMN1-501 | BGMN1-596T | MG7284/pAZIL-lsbB | BGBU1-4 | BGSM1-19 |
|---|---|---|---|---|---|---|---|
| BGBM50 | – | + | – | – | – | + | – |
| BGMN1-5 | + | – | – | – | – | + | – |
| BGMN1-501 | + | + | – | + | + | + | + |
| BGMN1-596T | + | + | + | – | – | + | + |
| MG7284/pAZIL-lsbB | + | + | – | – | – | + | + |
| BGBU1-4 | + | + | – | – | – | – | + |
| BGSM1-19 | – | – | – | – | – | + | – |
| BGMN1-596 | + | + | + | + | + | + | + |
Screening of L. lactis strains for antimicrobial activity using agar well diffusion method was performed in triplicate +=an inhibition zone sensitive to protease; –=without a zone
Localisation of the genes coding for lactococcin G production
Plasmid curing was performed to check whether the genes coding for bacteriocin synthesis and immunity are located on plasmids. In plasmid curing experiments, the derivative BGBM50 that lost its immunity to bacteriocin and the ability of synthesis was obtained. The difference in bacteriocin phenotypes of plasmid-cured derivatives was determined by using the agar well diffusion assay. Two groups of bacteriocinogenic derivates were obtained. Among the cured derivatives it was noticed that one, BGBM50-11, showed much smaller zone (2 mm) of inhibition compared to that of the parental strain (5 mm). The plasmid profile analysis revealed that derivative BGBM- 50-11 lost some plasmids (Fig. 1), but it was not possible to make any correlation with reduced bacteriocin activity. However, one of the most interesting derivatives was found inside the second group. BGBM50-14 did not produce lactococcin G, but was sensitive to it. PFGE analyses revealed that BGBM50-14 lost a plasmid of at least 140 kb (pBM140), indicating that the genes coding for bacteriocin lactococcin G synthesis and immunity are located on plasmid pBM140 in strain BGBM50 (Fig. 2). The appearance of one extra band in PFGE macrorestriction analysis of the derivatives compared to the wild-type strain, without changes in the sizes or pattern of other restriction fragments, strongly indicates that the extra fragment belongs to extrachromosomal element (plasmid). In cases when restriction enzymes with unusual appearance are used for macrorestriction analysis, and good distribution of fragments on the PFGE gel is obtained, it can be postulated that the extra fragments observed represent a complete plasmid (20).
Fig. 1.
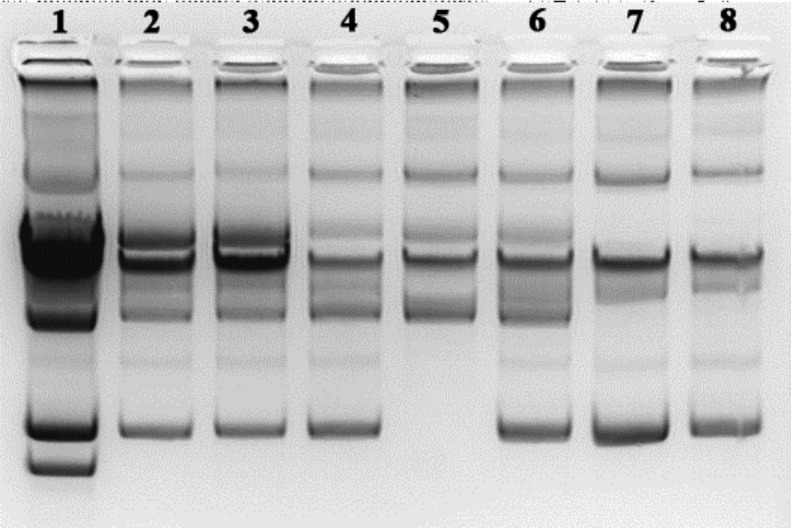
Plasmid profiles of cured derivates of Lactococcus lactis BGBM50. Lane 1=BGBM50, lane 2=BGBM50-8, lane 3=BGBM50--34, lane 4=BGBM50-11, lane 5=BGBM50-13, lane 6=BGBM50-14, lane 7=BGBM50-25, lane 8=BGBM50-62
Fig. 2.
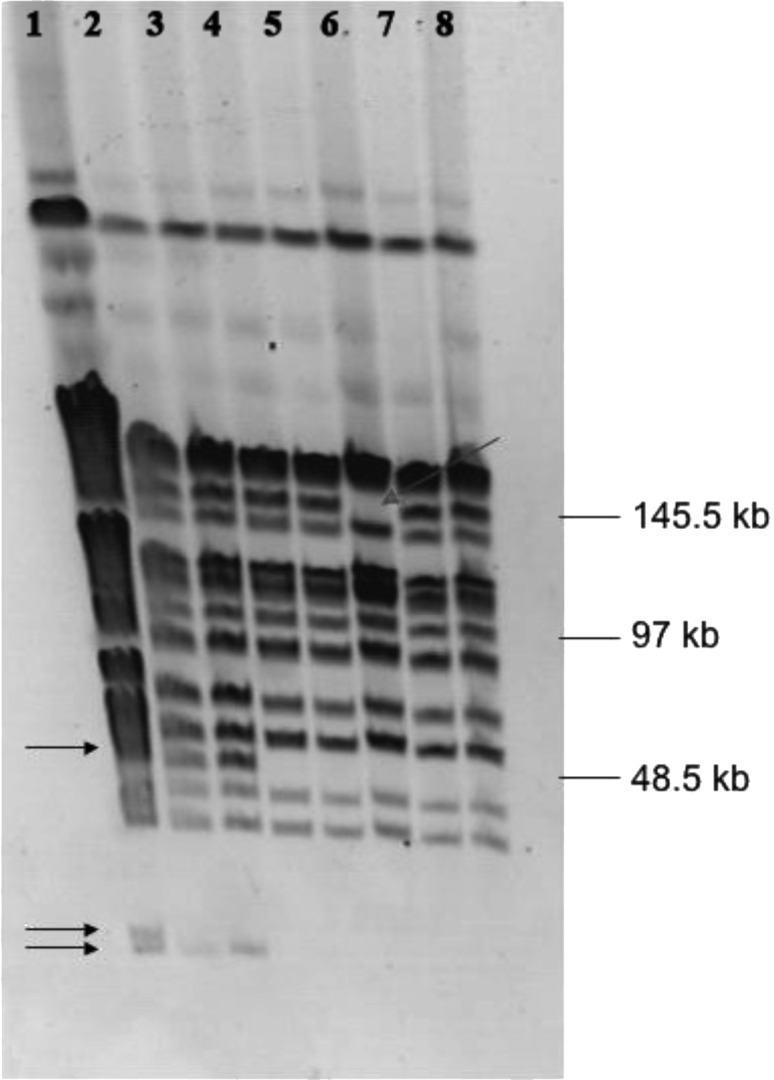
SmaI PFGE restriction pattern of Lactococcus lactis BGBM- 50 and its plasmid cured derivates. Lane 1=BGBM50, lane 2=BGBM- 50-8, lane 3=BGBM50-34, lane 4=BGBM50-11, lane 5=BGBM50- -13, lane 6=BGBM50-14, lane 7=BGBM50-25, lane 8=BGBM50-62. Arrows indicate SmaI DNA fragments missed in cured derivatives
Strain BGBM50 produces aggregation factor similar to AggL
Among cured derivatives we noticed that derivative BGBM50-34 had enhanced aggregation ability compared to the wild-type strain, but it retained bacteriocin production ability. Since AggL is a known lactococcal aggregation-promoting factor (9), we subjected both wild-type strain BGBM50 and derivative BGBM50-34 to Southern hybridisation experiments with an aggL gene probe. It was found that the gene for aggregation-promoting factor in strain BGBM50 shares high identity with the known aggL gene. It is interesting that no differences in size and intensity of hybridisation signals were observed between BGBM50 and derivative BGBM50-34 (data not shown), indicating that some small changes that affect the regulation of aggL gene expression occurred during curing experiment.
Discussion
Lactic acid bacteria (LAB) inhabit different natural niches, ranging from plants to mammalian cavities in which they live among a wide variety of different bacteria. In order to adapt to different environments and bacterial species they encounter, LAB synthesise various compounds, among which are bacteriocins, ribosomally synthesised antimicrobial peptides. Most of the studies on the biotechnological application of diverse bacteriocins have focused on their use as food preservatives, and more recently as preservatives for pharmaceutical industries and as alternatives to antibiotics. An emerging problem all over the world is the quick spread of multidrug resistance among different bacterial species. There is an urgent need for a new approach to the treatment of these bacteria. Synergy between antibiotics and different antimicrobial peptides as well as bacteriocins is becoming a new area of scientific research (23). For that reason, there is a permanent need to search for new LAB strains that synthesise bacteriocins with greater potency. Sources of such LAB strains may include various milk products. For the present research, we chose cheese manufactured without the addition of known strains, in order to isolate new strains with bacteriocin production ability. Among a number of isolated bacteriocin-producing strains obtained in previous research (21), BGBM50, with the highest potency, was used for further analysis. Biochemical characterisation of BacBM50 showed similar biochemical characteristics to other previously characterised bacteriocins produced by lactococcal strains (10, 14, 24). The quickest approach to the detection of new bacteriocin genes was a combination of a few powerful techniques: searching for new genes for bacteriocin synthesis by PCR with specific primers for known bacteriocin genes, plasmid curing, and cross-immunity test with known bacteriocin producers. It has been demonstrated that strain BGBM50 produces bacteriocin lactococin G, for which synthesis genes are located on a big plasmid of at least 140 kb. The location of genes for bacteriocin production on such big plasmid has been detected in L. lactis ssp. lactis bv. diacetylactis S50 (20). It is interesting that strain BGBM50 produces only lactococcin G, in contrast to LMG 2081, and can be used in cross-immunity test as a producer of only one bacteriocin. Isolation and characterisation of strain BGBM50 as a producer of only one bacteriocin (lactococcin G) enabled us to establish that the strain LMG 2081 produces more than one bacteriocin, which has not been known until now. In addition, the real inhibitory spectra of bacteriocin producers can be determined only when we are sure that a strain produces only one inhibitory compound. Moreover, strain BGBM50 showed slow aggregation phenotype, with a high identity with the one determined for the L. lactis ssp. lactis BGKP1 strain (9), which was increased in mutant BGBM50-34. This was most probably due to the influence of curing treatment on the genes for the regulation of DNA elements responsible for transcription, not for plasmid replication, or it could be that aggL gene somehow attenuated in the wild-type strain. This work has demonstrated the feasibility of studies aimed at discovering new strains that can express multiple traits important for probiotic activity and biopreservation of foods such as the synthesis of bacteriocins and aggregation-promoting factors. Moreover, L. lactis strains from traditional types of cheese are important source of new properties, which are lost or changed in the industrial strains used for decades in food production.
Conclusions
In conclusion, a bacteriocin-producing strain, Lactococcus lactis ssp. lactis BGBM50, was selected from a collection of 50 strains. All strains originated from traditional semi-hard cheese made from starter-free raw milk. This strain produced lactococcin G, which we characterised both on molecular and biochemical level. According to these results, we propose this bacteriocin for potential use as a biopreservative in the food industry. Moreover, a derivative of this strain expresses increased aggregation phenotype, indicating its probiotic potential.
Acknowledgements
The Ministry of Education, Science and Technological Development of the Republic of Serbia, Republic of Serbia, (Grant nos. 173019 and 046010) supported this work.
References
- 1.Cotter PD, Hill C, Ross P. Developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–88. 10.1038/nrmicro1273 [DOI] [PubMed] [Google Scholar]
- 2.Gálvez A, Lopez RL, Abriouel H, Valdivia E, Omar NB. Application of bacteriocins in the control of foodborne pathogenic and spoilage bacteria. Crit Rev Biotechnol. 2008;28:125–52. 10.1080/07388550802107202 [DOI] [PubMed] [Google Scholar]
- 3.Nes IF, Yoon SS, Diep DB. Ribosomally synthesized antimicrobial peptides (bacteriocins) in lactic acid bacteria: a review. Food Sci Biotechnol. 2007;16:675–90. [Google Scholar]
- 4.Limsowtin GKY, Powell IB, Parente E. Dairy starter cultures. New York, NY, USA: VCH Publishers; 1995. [Google Scholar]
- 5.Fadda S, López C, Vignolo G. Role of lactic acid bacteria during meat conditioning and fermentation: peptides generated as sensorial and hygienic biomarkers. Meat Sci. 2010;86:66–79. 10.1016/j.meatsci.2010.04.023 [DOI] [PubMed] [Google Scholar]
- 6.Terzic-Vidojevic A, Vukasinovic M, Veljovic K, Ostojic M, Topisirovic L. Characterization of microflora in homemade semi-hard white Zlatar cheese. Int J Food Microbiol. 2007;114:36–42. 10.1016/j.ijfoodmicro.2006.10.038 [DOI] [PubMed] [Google Scholar]
- 7.Boonaert CJP, Rouxhet G. Surface of lactic acid bacteria: relationships between chemical composition and physicochemical properties. Appl Environ Microbiol. 2000;66:2548–54. 10.1128/AEM.66.6.2548-2554.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schachtsiek M, Hammes WP, Hertel C. Characterization of Lactobacillus coryniformis DSM 20001T surface protein Cpf mediating coaggregation with and aggregation among pathogens. Appl Environ Microbiol. 2004;70:7078–85. 10.1128/AEM.70.12.7078-7085.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojic M, Jovcic B, Strahinic I, Begovic J, Lozo J, Veljovic K, et al. Cloning and expression of novel lactococcal aggregation factor from Lactococcus lactis subsp. lactis BGKP1. BMC Microbiol. 2011;11:265. 10.1186/1471-2180-11-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gajic O, Kojic M, Banina A, Topisirovic L. Characterization of natural isolate Lactococcus lactis subsp. lactis BGMN1-5, a strain producing two bacteriocins, cell wall-associated proteinase and showing clumping phenotype. Arch Biol Sci. 1999;51:69–78. 10.2298/ABS1004889T [DOI] [Google Scholar]
- 11.Uzelac G, Kojic M, Lozo J, Aleksandrzak-Piekarczyk T, Kristensen T, Nes IF, et al. Zn-dependent metallopeptidase is responsible for sensitivity to LsbB, a class II leaderless bacteriocin of Lactococcus lactis subsp. lactis BGMN1-5. J Bacteriol. 2013;195:5614–21. 10.1128/JB.00859-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golić N, Cadez N, Terzic-Vidojevic A, Suranska H, Beganovic J, Lozo J, et al. Evaluation of lactic acid bacteria and yeast diversity in traditional white pickled and fresh soft cheeses form mountain regions of Serbia and lowland regions of Croatia. Int J Food Microbiol. 2013;166:294–300. 10.1016/j.ijfoodmicro.2013.05.032 [DOI] [PubMed] [Google Scholar]
- 13.Strahinic I, Cvetanovic D, Kojic M, Fira D, Tolinacki M, Topisirovic L. Characterization and antimicrobial activity of natural isolate Lactococcus lactis subsp. lactis BGSM1-19. Acta Vet (Beogr). 2007;57:509–21. 10.2298/AVB0706509S [DOI] [Google Scholar]
- 14.Kojic M, Lozo J, Begovic J, Jovcic B, Topisirovic L. Charcterization of lactococci isolated from homemade kefir. Arch Biol Sci. 2007;59:13–22. 10.2298/ABS0701013K [DOI] [Google Scholar]
- 15.Hopwood DA, Bibb MJ, Chater KF, Keiser T, Bruton CJ, Keiser HM, et al. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, UK: John Innes Foundation; 1985. http://dx.doi.org/ 10.1016/0307-4412(86)90228-1 [DOI] [Google Scholar]
- 16.Golic N, Strahinic I, Terzic-Vidojevic A, Nikolic M, Tolinacki M, Topisirovic L. Molecular diversity among natural populations of Lactobacillus paracasei and Lactobacillus plantarum/ paraplantarum strains isolated from autochthonous dairy products. Eur Food Res Technol. 2012;234:627–38. 10.1007/s00217-012-1670-6 [DOI] [Google Scholar]
- 17.Jovcic B, Begovic J, Lozo J, Topisirovic L, Kojic M. Dynamic of sodium dodecyl sulfate utilization and antibiotic susceptibility of strain Pseudomonas sp. ATCC19151. Arch Biol Sci. 2009;61:159–65. 10.2298/ABS0902159J [DOI] [Google Scholar]
- 18.Alegría A, Delgado S, Roces S, Lopez B, Mayo B. Bacteriocins produced by wild Lactococcus lactis strains isolated from traditional, starter-free cheeses made of raw milk. Int J Food Microbiol. 2010;143:61–6. 10.1016/j.ijfoodmicro.2010.07.029 [DOI] [PubMed] [Google Scholar]
- 19.Martínez B, Fernandez M, Suarez JE, Rodriguez A. Synthesis of lactococcin 972, a bacteriocin produced by Lactococcus lactis IPLA 972, depends on the expression of a plasmid-encoded bicistronic operon. Microbiol SGM. 1999;145:3155–61. 10.1099/00221287-145-11-3155 [DOI] [PubMed] [Google Scholar]
- 20.Kojic M, Strahinic I, Topisirovic L. Proteinase PI and lactococcin A are located on the largest plasmid in Lactococcus lactis subsp. lactis biovar. diacetylactis S50. Can J Microbiol. 2005;51:305–14. 10.1139/w05-009 [DOI] [PubMed] [Google Scholar]
- 21.Kojic M, Strahinic I, Fira D, Jovcic B, Topisirovic L. Plasmid content and bacteriocin production by five strains of Lactococcus lactis isolated from semi-hard homemade cheese. Can J Microbiol. 2006;52:1110–20. 10.1139/w06-072 [DOI] [PubMed] [Google Scholar]
- 22.Nissen-Meyer J, Holo H, Hĺverstein LS, Sletten K, Nes IF. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992;174:5686–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mataraci E, Dosler S. In vitro activities of antibiotics and antimicrobial cationic peptides alone and in comination against methicillin-resistant Staphylococcus aureus biofilms. Antimicrob Agents Chemother. 2012;56:6366–71. 10.1128/AAC.01180-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kojic M, Svircevic J, Banina A, Topisirovic L. Bacteriocin-producing strain of Lactococcus lactis subsp. lactis biovar. diacetylactis S50. Appl Environ Microbiol. 1991;57:1835–7. [DOI] [PMC free article] [PubMed] [Google Scholar]


