Abstract
Accumulating evidence indicates that miRNAs, a class of small non-coding RNAs, are implicated in the pathogenesis of various diseases such as cancer and intervertebral disc degeneration. The aim of this study was to investigate the expression and the biological function of microRNA-34a in intervertebral disc degeneration. In this study, microRNA-34a expression was assessed in nucleus pulposus specimens and in IL-1β-stimulated nucleus pulposus cells by real-time polymerase chain reaction. microRNA-34a functions were investigated by using gain and loss of function experiments in nucleus pulposus cells and a dual luciferase reporter assay in 293T cells. microRNA-34a was dramatically up-regulated in degenerative nucleus pulposus tissues and in IL-1β-stimulated nucleus pulposus cells when compared with controls. Furthermore, growth differentiation factor 5 was identified as a target of microRNA-34a. Aberrant expression of microRNA-34a inhibited growth differentiation factor 5 expression by direct binding to its 3′-untranslated region. This inhibition was abolished by mutation of the microRNA-34a binding sites. In addition, microRNA-34a silencing reversed IL-1β-induced decrease in type II collagen and aggrecan expression in nucleus pulposus cells. This effect was substantially suppressed by growth differentiation factor 5 silencing. Our results suggested that microRNA-34a inhibition prevents IL-1β-induced extracellular matrix degradation in human nucleus pulposus by increasing growth differentiation factor 5 expression. microRNA-34a inhibition may be a novel molecular target for intervertebral disc degeneration treatment through the prevention of nucleus pulposus extracellular matrix degradation.
Keywords: Extracellular matrix degradation, growth differentiation factor 5, IL-1β, intervertebral disc degeneration, microRNA-34a, nucleus pulposus cell
Introduction
Currently, lower back pain is a common medical problem and epidemiologic studies show that its incidence is increasing every year.1 Lower back pain seriously influences human health and results in enormous socioeconomic burdens.2 Although complex and multiple factors are involved in triggering lower back pain, it is generally believed to be associated with intervertebral disc degeneration (IDD). Numerous studies indicate that the etiology and pathogenesis of IDD involve various factors, including age, gene, smoking, trauma, hyper-physiological loading, or overload.3–7 However, the underlying mechanisms of IDD remain unknown. Recently, there has been accumulating evidence showing that the predominant changes in IDD correlate with intervertebral disc (IVD) and extracellular matrix (ECM) degeneration.8,9 It is well established that collagens and proteoglycans (PGs) such as type II collagen and aggrecan are crucial for proper disc function, particularly in the nucleus pulposus (NP), and the loss of type II collagen and aggrecan plays a pivotal role in the development of disc degeneration.10,11 During IDD, NP cells function abnormally with decreased synthesis of normal IVD matrix components and increased production of ECM degradative enzymes, thereby leading to the degeneration of the ECM and the loss of the normal homeostatic metabolism in the IVD.12,13 There is accumulating evidence indicating that inflammatory cytokines such as IL-1β are increased in IDD and can mediate ECM degradation in the IVD.14–16 Therefore, the use of biological agents that protect NP cells from inflammatory cytokine-induced ECM degradation might become one of the therapeutic strategies for IDD treatment.
Recent evidence shows that growth differentiation factor 5 (GDF5) plays an essential role in suppressing the ECM degeneration in IVD cells. GDF5 is expressed in normal, moderate, and severe degeneration of IVD, particularly in cells of the NP.17 However, a small decrease in the number of GDF5 is observed during degeneration.18 In GDF5-deficient mice, type II collagen and aggrecan expression is down-regulated in IVD cells and the PGs content in discs is decreased. Treatment of disc cells from the GDF5-deficient mice with recombinant GDF5 resulted in the up-regulation of the type II collagen and aggrecan genes.19 Moreover, GDF5 treatment up-regulated the expression of type II collagen and aggrecan in NP cells derived from degenerative IVDs and induced greater production of PGs.18 In addition, IL-1β down-regulates GDF-5 expression in human disc cells and the suppression of GDF-5 expression might down-regulate ECM anabolism in the IVD to accelerate the process of IL-1β-induced ECM degeneration.20 Therefore, GDF5 overexpression in human NP cells may block IL-1β-induced ECM degeneration in IDD.
Accumulating evidence shows that multiple cellular processes, including cell proliferation, differentiation, and apoptosis, are regulated by the newly defined small non-coding RNAs, miRNAs.21–23 miRNAs control gene expression primarily by binding to their target mRNAs 3′-untranslated regions (3′-UTRs) and triggering either translation repression or RNA degradation.21 Several miRNAs exhibit a tissue-specific or developmental stage-specific expression pattern and have been implicated in diverse pathological conditions such as cancer, osteoarthritis, neurodegeneration, and cardiovascular disease.23–26 Currently, accumulating evidence indicates that miRNAs might play a role in IDD pathogenesis. The aberrant expression of miRNAs is involved in proliferation, apoptosis, and ECM degradation in human NP cells.27–29
MiRNA-34a (miR-34a), a tumor suppressor miRNA transcriptionally activated by p53, is associated with a variety of diseases.30,31 Current evidence indicates that miR-34a plays a role in the regulation of cartilage degradation. Moreover, miR-34a silencing significantly prevented IL-1-induced down-regulation of type II collagen as well as IL-1-induced up-regulation of iNOS in chondrocytes.23 Additionally, miR-34a silencing could reduce the cartilage endplate chondrocyte apoptosis induced by Fas, reversed Fas-induced decrease of type II collagen and aggrecan mRNA levels, and increased MMP-3 and MMP-13 mRNA levels.28 Given that miR-34a is crucially involved in ECM degradation, we hypothesized that miR-34a might play a role in the process of IDD. However, to date, there is no report regarding the functional activity of miR-34a in NP cells.
In this report, we demonstrated that miR-34a is highly expressed in human degenerative NP tissues and in IL-1β-stimulated NP cells. Moreover, we validated GDF5 as a direct target of miR-34a and miR-34a negatively regulated GDF5 by binding to its 3′-UTR, leading to the inhibition of GDF5 expression. In addition, our results suggest that miR-34a acts as a novel regulator of NP ECM homeostasis and silencing miR-34a expression might significantly prevents IL-1β-induced ECM degradation in human NP via increasing GDF5 expression.
Materials and methods
Ethics statement, patients, and sample collection
This study was approved by the Clinical Research Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology. The written informed consents were obtained from all patients or their parents (on behalf of their children).
Eight normal lumbar NP specimens were obtained from four patients with idiopathic scoliosis (average age 17.8, range 16–20 years) who had no history of IDD. Ten degenerated lumbar NP specimens were obtained from 10 IDD patients (average age 48.5, range 34–65 years) undergoing discectomy. The surgical inclusion criteria were as follows: (1) conservative treatment fail and (2) progressive neurologic deficits such as progressive motor weakness or cauda equine syndrome. The exclusion criteria were as follows: (1) lumbar stenosis; (2) ankylosing spondylitis; (3) degenerative scoliosis; and (4) degenerative or spondylolysis spondylolisthesis. The degree of disc degeneration was determined according to a modified Pfirrmann classification.32 All normal disc specimens from idiopathic scoliosis patients were classified as grade II. Of them, four NP specimens were used as controls for real-time quantitative polymerase chain reaction (qRT-PCR) to examine miRNA expression levels. The others were used for NP cell isolation and culture. Five degenerated disc specimens from IDD patients were classified as grade IV and the others were grade V. All these degenerated specimens were used for qRT-PCR to examine miRNA expression levels.
Isolation and culture of human NP cells
NP cells were isolated as previously described.33,34 Four normal tissue specimens were obtained from four patients with idiopathic scoliosis (average age 17.8, range 16–20 years) and washed twice in phosphate-buffered saline (PBS) and cut into pieces (2–3 mm3). The NP cells, which were chondrocyte-like cells, were isolated by enzymatic digestion for 8 h at 37℃ in Dulbecco’s modified Eagle medium (DMEM; Gibco, Grand Island, NY, USA) with 0.25 mg/mL type II collagenase (Invitrogen, Carlsbad, CA, USA). After isolation, the NP cells were plated and expanded for three weeks in DMEM containing 15% fetal bovine serum (FBS; Gibco), and 1% penicillin/streptomycin (Invitrogen) at 37℃ in a 5% CO2 incubator. The culture medium was replaced twice a week with the exception that the primary cells were allowed more time (6.7 ± 1.4 days) to adhere prior to the first change. The second passage cells were used for subsequent experiments.
Cell transfection
The miR-34a (mimic), anti-miR-34a (inhibitor), GDF5 siRNA, and their negative controls were synthesized by GenePharma (Shanghai, China). Human NP cells (1.5 × 105 cells/well) were seeded in 24-well plates in 250 µL of culture medium. When cultured NP cells were grown to 80% confluence, all transfections were performed using the Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 24 h of transfection, cells were cultured in serum-free medium for 12 h and then stimulated with IL-1β (10 ng/mL) (Sino Biological Inc., North Wales, PA, USA) for 24 h at 37℃ under 5% CO2. Cells were harvested for RNA isolation and Western blot. To knock down miR-34a, anti-miR-34a (150 nM) or its negative control was transfected into NP cells. GDF5 knockdown in cultured NP cells was achieved by transfection of a selective GDF5 siRNA (150 nM) after validation of its inhibition efficacy. For each cell transfection, three replicate experiments were performed.
qRT-PCR
Total RNA was extracted from the NP tissues and cells with TRIzol reagent (Invitrogen) following the manufacturer’s instructions. After determination of RNA concentration using spectrophotometry, RNA was reverse transcribed with a PrimeScript™ RT Master Mix (TakaRa, Dalian, China) according to manufacturer's instructions. Real-time PCR was performed to quantify the miR-34a, GDF5, collagen type II, and aggrecan expression levels. Real-time PCR was conducted using One Step SYBR® PrimeScript™ RT-PCR Kit (TakaRa, China). U6 served as the reference gene to normalize the expression of miR-34a and β-actin served as the reference gene to normalize the expression of GDF5, collagen type II, and aggrecan. PCR was performed with the following primers: hsa-miR-34a Forward primer (F) 5-TGC GCT GGC AGT GTC TTA GCT-3, Reverse primer(R) 5-CCA GTG CAG GGT CCG AGG TAT T-3; GDF5 F: 5-CGA TAA GAC CGT GTA TGA GT-3, R: 5-CTC GCA GTG GAA AGC CTC GT-3; Collagen F: 5-TCC AGA TGA CCT TCC TAC GC-3, R: 5-GGT ATG TTT CGT GCA GCC AT-3; Aggrecan F: 5-TGA GCG GCA GCA CTT TGA C-3, R: 5-TGA GTA CAG GAG GCT TGA G-3; U6 F: 5-CGC TTC GGC AGC ACA TAT AC-3, R: 5-AAA TAT GGA ACG CTT CAC GA-3; β-actin F:5-AGC GAG CAT CCC CCA AAG TT-3,R:5-GGG CAC GAA GGC TCA TCA TT-3.The expression levels of mRNA and miRNA were calculated by the 2−ΔΔCt method relative to β-actin or U6.35 All experiments were carried out at least in triplicate.
Plasmid constructs
The wild-type pmiR-GDF5 plasmid was generated using the following primers: sense 5′-GCG CTC GAGTGC CAA CAA CGT GGT GTA T-3′, antisense 5′-AAT GCG GCC GCC ACA GTT TTA GGC ACA GTT-3′. The amplified sequences were inserted into pmiR-RB-Report vector (RiboBio Co., Guangzhou, China) within the XhoI and NotI restriction sites. With wild-type pmiR-GDF5 as a template, the mutation on miR-34a binding sites in human GDF5 3′-UTR was generated by performing site-directed mutagenesis (primers: Forward: ACA GGT GCG TGA CGG TCC TCA AAT CAC ATT TGT and Reverse: TTT GAG GAC CGT CAC GCA CCT GTG GCT CTC CTG). All constructs were verified by sequencing.
Dual luciferase assays
Human embryonic kidney (HEK) 293 cells were used for luciferase activity analysis. HEK293 cells are a specific cell line originally derived from human embryonic kidney cells grown in tissue culture. They are very easy to grow and transfect. They have been widely used in dual luciferase assays for many years.36,37 HEK293 cells were cultured in 48-well plates, and co-transfected with wild-type pmiR-GDF5 plasmid or mutant-type pmiR-GDF5 plasmid (WT and Mut, respectively) and miR-34a or control mimic with Lipofectamine 2000. Twenty-four hours later, cells were harvested and luciferase activity was examined by Dual-Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Firefly luciferase activity was normalized to Renilla luciferase activity. All experiments were carried out three times.
Western blot
Western blot analysis was performed in some experiments in this study. The type of cells used for Western blot analysis varied depending on the experiment. In brief, untransfected NP cells and IL-1β-stimulated transfected or untransfected NP cells were used for Western blot analysis in the first set of experiments. Normal NP cells transfected with miR-34a/miR-NC or anti-miR-34a/anti-miR-NC were used for Western blot analysis in the second set of experiments. IL-1β-stimulated NP cells untransfected or co-transfected with the miRNA inhibitor along with siRNA and their negative controls were used for Western blot analysis in the third set of experiments. Western blot analysis was performed following standard methods. The culture supernatants were collected and the cells were lysed for 20 min in cold radioimmunoprecipitation (RIPA) lysis buffer (Beyotime, Beijing, China), and protein concentrations were measured by the Enhanced BCA Protein Assay Kit (Beyotime, China). Equal amounts of proteins were separated by using 10% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking with TBST containing 5% non-fat milk, the membranes were incubated with primary antibodies against anti-GDF5 (ab93855, Abcam, Cambridge, UK), anti-aggrecan (ab3778, Abcam), anti-collagen type II (ab34712, Abcam), and anti-β-actin (ab8227, Abcam) overnight at 4℃. After washes in TBST, the immobilized primary antibodies were detected using a horseradish peroxidase (HRP)-conjugated secondary goat anti-rabbit IgG antibody and were visualized with an ECL Chemiluminescence kit (Thermo, Waltham, MA, USA). The experiment was performed at least in triplicate.
Immunofluorescence staining
Cultured cells were rinsed three times in PBS, fixed with 4% paraformaldehyde for 15 min at room temperature, and blocked with 3% BSA for 1 h. Subsequently, cells were incubated overnight at 4℃ with an anti-type II collagen (Abcam; 1:200 dilution). After three washes with PBS, cells were treated for 1 h with a Cy3-conjugated goat anti-rabbit IgG antibody (fluorescently labeled; GE Healthcare, UK; 1:100 dilution) at room temperature. The samples were then incubated with 0.1 g/mL DAPI for 3 min. After the final round of washes, fluorescence images were acquired using a laser-scanning confocal microscope (LSM 710, Carl Zeiss, Oberkochen, Germany).
Statistical analysis
All data are expressed as the mean ± standard deviation (SD). The statistical analysis was performed using the GraphPad Prism 6.0 (San Diego, CA, USA) with the Student's t-test or one-way ANOVA. The level of significance was set at P < 0.05.
Results
miR-34a expression levels in NP tissues of patients with IDD and in IL-1β-stimulated NP cells
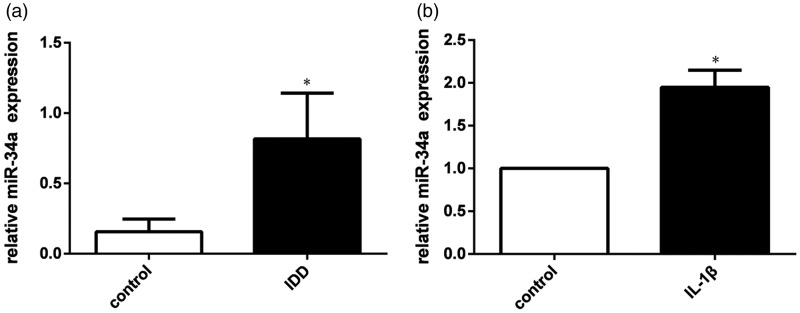
To investigate the regulatory effect of miR-34a in IDD pathogenesis, qRT-PCR was used to examine its expression levels. As shown in Figure 1(a), miR-34a was significantly up-regulated in degenerative NP tissues (n = 10) from patients with IDD compared to non-degenerative NP tissues (n = 4) from patients with idiopathic scoliosis. Non-degenerative NP tissues were used as controls. Subsequently, miR-34a expression in human NP cells stimulated by IL-1β and in un-stimulated NP cells was also analyzed by real-time PCR. Our results demonstrated that miR-34a was significantly up-regulated in IL-1β-stimulated NP cells when compared with non-stimulated NP cells. Non-stimulated NP cells were used as controls. (Figure 1(b)).
Figure 1.
miR-34a expression levels in NP tissues of patients with IDD and in IL-1β-stimulated NP cells. (a) miR-34a was significantly increased in NP tissues from patients with IDD compared to NP tissues from the control group.(b) miR-34a was significantly increased in IL-1β-stimulated NP cells compared to IL-1β unstimulated NP cells. U6 was used as an internal control. Data are presented as mean ± SD. *P < 0.05
Effect of miR-34a silencing on type II collagen and aggrecan expression in IL-1β-stimulated NP cells
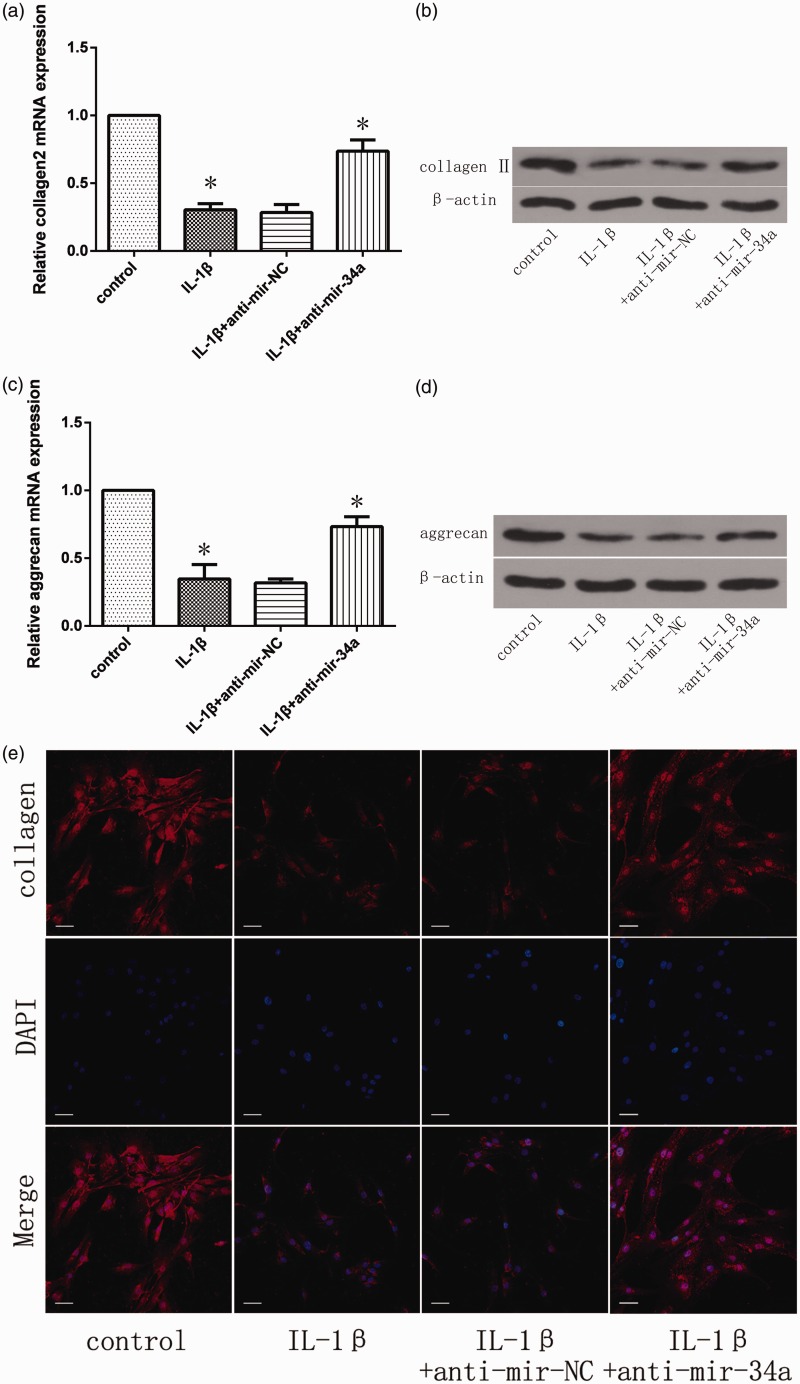
To elucidate the effect of miR-34a silencing in NP cells, NP cells were transfected with miR-34a inhibitor or negative control and then treated with IL-1β 24 h post-miRNA transfection. After IL-1β treatment for 24 h, real-time PCR and Western blot were used to quantify the mRNA and protein expression of type II collagen and aggrecan. IL-1β significantly inhibited the mRNA and protein expression of type II collagen and aggrecan in NP cells (Figure 2(a) to (d)). Conversely, the anti-miR-34a significantly reversed the IL-1β-induced inhibition of type II collagen and aggrecan mRNA expression (Figure 2(a) and (c)). Similarly, miR-34a silencing abrogated the effects of IL-1β and resulted in the preservation of type II collagen and aggrecan content (Figure 2(b) and (d)).
Figure 2.
Effect of miR-34a silencing on type II collagen and aggrecan expression in IL-1β-stimulated NP cells. NP cells were transfected with miR-34a inhibitor or negative control and then treated with IL-1β 24 h post-miRNA transfection. Type II collagen (a, b) and aggrecan (c, d) expression levels were quantified by qRT-PCR and Western blot. The relative mRNA expression levels in untransfected and non-stimulated NP cells were set to one and used as controls. β-actin was used as an internal control. Data are presented as mean ± SD. *P < 0.05. (e) Type II collagen protein levels were detected by immunofluorescence. Scale bars = 40 µm
In addition, the immunofluorescence experiments showed that type II collagen protein was significantly increased by treatment with the miR-34a inhibitor (Figure 2(e)).
GDF5 is a target of miR-34a
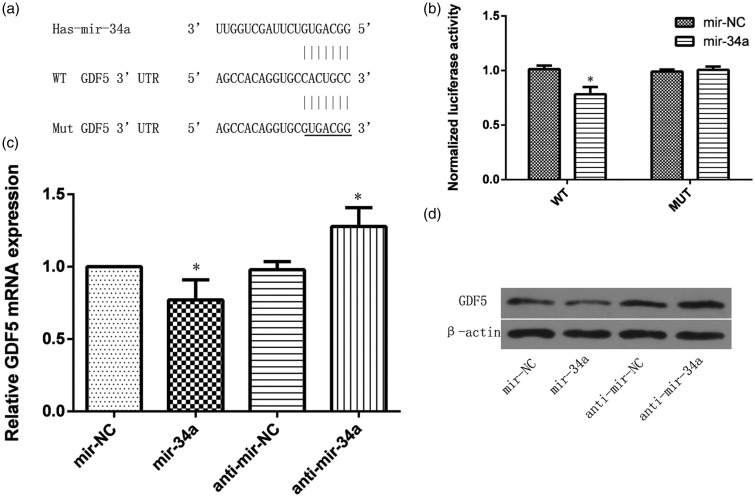
Data mining predicted that GDF5 was a miR-34a target (Figure 3(a)). To validate the functional interaction between miR-34a and GDF5 generated by the target prediction algorithms, we performed luciferase reporter assays with either the wild-type pmiR-GDF5 plasmid or the mutated pmiR-GDF5 plasmid. Luciferase activity assay showed that miR-34a significantly suppressed the luciferase activity of the wild type GDF5 3′-UTR reporter plasmid, but not that of the mutated GDF5 3′-UTR reporter plasmid in HEK293 cells (Figure 3(b)). Furthermore, to further examine whether GDF5 is a target of miR-34a in NP cells, the NP cells were transfected with the miR-34a mimic and anti-miR-34a and their negative controls. miR-34a overexpression significantly suppressed GDF5 mRNA and protein levels, while miR-34a inhibition had the opposite effects (Figure 3(c) and (d))
Figure 3.
GDF5 is a target of miR-34a. (a) Computational analysis showed that miR-34a potentially targeted GDF5. (b) Luciferase reporter assay was used to validate the relationship between miR-34a and GDF5. (c) GDF5 mRNA expression was detected by qRT-PCR in NP cells transfected with miR-34a/miR-NC or anti-miR-34a/anti-miR-NC. (d) Protein level was detected by Western blot in NP cells transfected with miR-34a/miR-NC or anti-miR-34a/anti-miR-NC. β-actin was used as an internal control. Data are presented as mean ± SD. *P < 0.05
miR-34a silencing induces type II collagen and aggrecan up-regulation in NP cells, probably by targeting GDF5
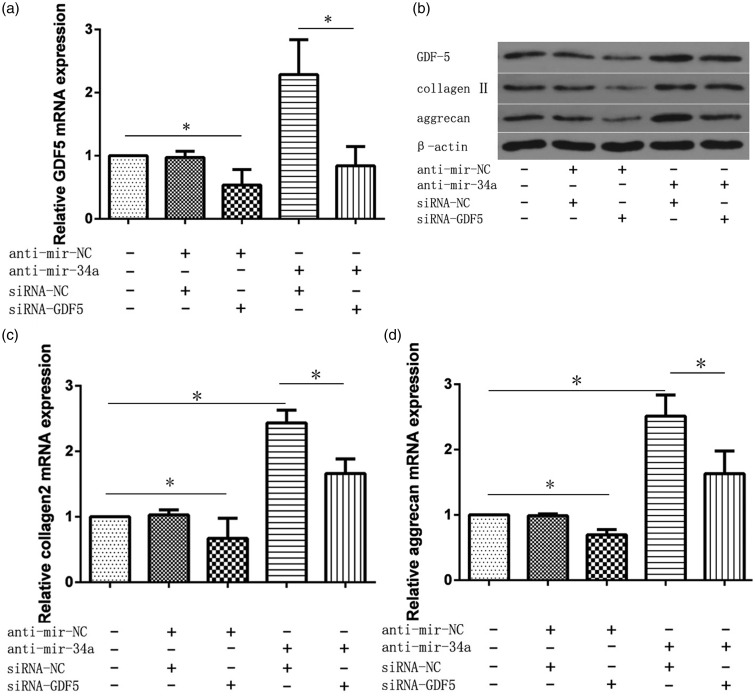
Our studies indicated that miR-34a silencing increases the level of ECM gene expression as well as type II collagen and aggrecan synthesis. Whether GDF5 is involved in the function of miR-34a in NP cells remains unknown. To determine whether the effects of miR-34a silencing involved GDF5, we co-transfected human NP cells with the miR-34a inhibitor along with a GDF5 siRNA and their negative controls and then treated the cells with IL-1β 24 h post-miRNA transfection. After IL-1β treatment for 24 h, real-time PCR and Western blot were performed to quantify the mRNA and protein expression of GDF5, type II collagen, and aggrecan. GDF5 mRNA and protein expression was significantly suppressed by siGDF5. Furthermore, the increased GDF5 level post-miR-34a inhibitor transfection was reduced by co-transfection with siGDF5 (Figure 4(a) and (b)). In addition, comparison of the anti-miR-34a and GDF5 siRNA co-transfection with the transfection with the anti-miR-34a alone demonstrated that the up-regulation of type II collagen and aggrecan mRNA and protein expression, upon silencing of miR-34a, was significantly attenuated by GDF5 silencing. (Figure 4(b) to (d)).
Figure 4.
Silencing of miR-34a induces type II collagen and aggrecan up-regulation in NP cells, probably by targeting GDF5. The NP cells were co-transfected with the miR-34a inhibitor along with GDF5 siRNA and their negative controls and then treated with IL-1β 24 h post-miRNA transfection. mRNA levels of GDF5 (a), type II collagen (c), and aggrecan (d) were analyzed by real-time PCR. (b) Protein levels of GDF5, type II collagen, and aggrecan were analyzed by Western blot. The relative expression levels of mRNA in untransfected NP cells were set to one, as control. β-actin was used as an internal control. Data are presented as mean ± SD. * P < 0.05
Discussion
Increasing evidence suggests that miRNAs play a crucial role in the pathogenesis of IDD and can be promising therapeutic targets.29,36,37 As a tumor suppressor gene, miR-34a targets many oncogenes related to proliferation, apoptosis, and invasion.30 In addition, miR-34a deregulation plays pivotal roles in cartilage endplate chondrocyte apoptosis and ECM degradation.28 Moreover, miR-34a expression is induced by IL-1β and miR-34a silencing can prevent type II collagen down-regulation induced by IL-1β in chondrocytes.23 However, miR-34a roles in the pathogenesis of IDD, particularly in the NP cell homeostasis, remain largely unknown. In this study, we found that miR-34a was markedly elevated in human degenerated NP tissues and miR-34a expression was significantly up-regulated by IL-1β. Therefore, we hypothesized that miR-34a plays a key role in IL-1β-induced NP ECM degradation.
Consistent with this hypothesis, we first validated miR-34a role in the pathogenesis of IL-1β-induced NP ECM degradation. Our results provide evidence that mir-34a silencing can up-regulate type II collagen and aggrecan expression levels. During disc degeneration, a number of changes are observed within the ECM of the IVD. Many of these are simply due to breakdown of type II collagen and aggrecan components.9 In addition, previous studies indicated that IL-1β can induce a decrease in the gene expression of IVD matrix genes and inhibiting IL-1β could be an important therapeutic target for preventing and reversing disc degeneration.14,15 Thus, our results revealed that miR-34a silencing prevents NP cells from IL-1β-induced ECM degradation and that miR-34a inhibition may represent a novel intervention for IDD treatment through the prevention of NP cells degradation.
We further verified that GDF5 was a direct target of miR-34a in NP cells, as predicted by bioinformatics analysis. A luciferase activity assay was used to explore the relationship between miR-34a and GDF5. Our results show that miR-34a is a repressor of GDF5. In addition, our results indicate that miR-34a negatively regulates GDF5 expression in NP cells, confirming that GDF5 is indeed a target of miR-34a.
GDF5, as a key regulator of chondrogenesis, plays an essential role in IDD pathogenesis. GDF5 (−/−) mice presented with significantly lower T2-weighted signal intensity in the central region of their lumbar discs, and disc histology revealed the loss of the normal lamellar architecture of the annulus fibrosus and a shrunken, disorganized NP. GDF5 deficiency significantly reduced type II collagen and aggrecan mRNA expression and PG content.19 GDF5 gene mutation not only changed the growth and differentiation of NP cells, reduced the synthesis of ECM, and induced disc degeneration, but also affected the disc vertebral height or length of the long bones, causing the height variation.38 GDF5 overexpression can significantly enhance cells proliferation and increase the expression of genes encoding ECM proteins such as type II collagen and aggrecan in IVD cells, whereas GDF5 down-regulation attenuates these processes.39,40 Thus, GDF5 gene deficiency, mutation, or abnormal expression can cause variable changes in cell growth and ECM synthesis and promote the development of IDD. Increasing evidence suggests that GDF5 aids in maintaining the structural integrity of the IVD and effectively restores disc physiological function, thereby delaying or even reversing IDD.39,41 In some GDF5-injected discs, the expansion of inner annular fibrochondrocyte populations into the nucleus and an increase in the synthesis of ECM were observed.41 Moreover, recombinant GDF5 treatment of disc cells, isolated from the GDF5-deficient mice, actively expressed aggrecan and type II collagen mRNA.19 In our study, the inhibition of IL-1β-induced down-regulation of aggrecan and type II collagen by miR-34a silencing was attenuated by GDF5 silencing. This experimental evidence suggests that miR-34a inhibition reverses IL-1β-induced NP ECM degradation by increasing GDF5 expression.
In conclusion, this study provides the first evidence that miR-34a expression is up-regulated in NP tissues from patients with IDD and can be induced by IL-1β in human NP cells and that GDF5 is a direct target of miR-34a. Finally, miR-34a silencing contributes to reversing ECM degradation in human NP via increasing GDF5 expression. Our findings shed new light on the role of miR-34a in the pathogenesis of IDD and provide new therapeutic targets for inhibiting ECM degradation in IDD.
Acknowledgments
This work was supported by National Natural Science Foundation of China (grant no.81272025).
Authors’ contributions
WL contributed to study conception and design, data collection, and analysis, contributed to manuscript writing, and had final approval of the manuscript. YKZ contributed to study conception and design, and in critical revision and review of the manuscript. XTF, SL, SHY, and ZWS analyzed the data and reviewed the manuscript. YG, KW, YS, and JT performed the experiments. CY secured funding and contributed final manuscript preparation. All authors read and approved the final manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Cassidy JD, Côté P, Carroll LJ, Kristman V. Incidence and course of low back pain episodes in the general population. Spine 2005; 30: 2817–23. [DOI] [PubMed] [Google Scholar]
- 2.Mehra M, Hill K, Nicholl D, Schadrack J. The burden of chronic low back pain with and without a neuropathic component: a healthcare resource use and cost analysis. J Med Econ 2012; 15: 245–52. [DOI] [PubMed] [Google Scholar]
- 3.Boos N, Weissbach S, Rohrbach H, Weiler C, Spratt KF, Nerlich AG. Classification of age-related changes in lumbar intervertebral discs: Volvo Award in basic science. Spine 2002; 27: 2631–44. [DOI] [PubMed] [Google Scholar]
- 4.Solovieva S, Lohiniva J, Leino-Arjas P, Raininko R, Luoma K, Ala-Kokko L, Riihimäki H. Intervertebral disc degeneration in relation to the COL9A3 and the IL-1ss gene polymorphisms. Eur Spine J 2006; 15: 613–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pye SR, Reid DM, Adams JE, Silman AJ, O'Neill TW. Influence of weight, body mass index and lifestyle factors on radiographic features of lumbar disc degeneration. Ann Rheum Dis 2007; 66: 426–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guehring T, Omlor GW, Lorenz H, Bertram H, Steck E, Richter W, Carstens C, Kroeber M. Stimulation of gene expression and loss of annular architecture caused by experimental disc degeneration – an in vivo animal study. Spine 2005; 30: 2510–5. [DOI] [PubMed] [Google Scholar]
- 7.Stokes IA, Iatridis JC. Mechanical conditions that accelerate inter-vertebral disc degeneration: overload versus immobilization. Spine 2004; 29: 2724–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crean JK, Roberts S, Jaffray DC, Eisenstein SM, Duance VC. Matrix metalloproteinases in the human intervertebral disc: role in disc degeneration and scoliosis. Spine 1997; 22: 2877–84. [DOI] [PubMed] [Google Scholar]
- 9.Le Maitre CL, Pockert A, Buttle DJ, Freemont AJ, Hoyland JA. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans 2007; 35: 652–5. [DOI] [PubMed] [Google Scholar]
- 10.Sivan SS, Wachtel E, Roughley P. Structure, function, aging and turnover of aggrecan in the intervertebral disc. Biochim Biophys Acta 2014; 1840: 3181–9. [DOI] [PubMed] [Google Scholar]
- 11.Kluba T, Niemeyer T, Gaissmaier C, Gründer T. Human anulus fibrosis and nucleus pulposus cells of the intervertebral disc: effect of degeneration and culture system on cell phenotype. Spine 2005; 30: 2743–8. [DOI] [PubMed] [Google Scholar]
- 12.Kluba T, Niemeyer T, Gaissmaier C, Gründer T. Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J 2013; 13: 331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Maitre CL, Freemont AJ, Hoyland JA. Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc. J Pathol 2004; 204: 47–54. [DOI] [PubMed] [Google Scholar]
- 14.Lee JM, Song JY, Baek M, Jung HY, Kang H, Han IB, Kwon YD, Shin DE. Interleukin-1β induces angiogenesis and innervation in human intervertebral disc degeneration. J Orthop Res 2011; 29: 265–9. [DOI] [PubMed] [Google Scholar]
- 15.Le Maitre CL, Freemont AJ, Hoyland JA. The role of interleukin-1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res Ther 2005; 7: R732–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyland JA, Le Maitre C, Freemont AJ. Investigation of the role of IL-1 and TNF in matrix degradation in the intervertebral disc. Rheumatology 2008; 47: 809–14. [DOI] [PubMed] [Google Scholar]
- 17.Li YF, Tang XZ, Liang CG, Hui YM, Ji YH, Xu W, Qiu W, Cheng LM. Role of growth differentiation factor-5 and bone morphogenetic protein type II receptor in the development of lumbar intervertebral disc degeneration. Int J Clin Exp Pathol 2015; 8: 719–26. [PMC free article] [PubMed] [Google Scholar]
- 18.Le Maitre CL, Freemont AJ, Hoyland JA. Expression of cartilage-derived morphogenetic protein in human intervertebral discs and its effect on matrix synthesis in degenerate human nucleus pulposus cells. Arthritis Res Ther 2009; 11: R137–R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Leo BM, Beck G, Balian G, Anderson GD. Collagen and proteoglycan abnormalities in the GDF-5-deficient mice and molecular changes when treating disk cells with recombinant growth factor. Spine 2004; 29: 2229–34. [DOI] [PubMed] [Google Scholar]
- 20.Gruber HE, Hoelscher GL, Ingram JA, Bethea S, Hanley EN., Jr Growth and differentiation factor-5 (GDF-5) in the human intervertebral annulus cells and its modulation by IL-1β and TNF-α in vitro. Exp Mol Pathol 2014; 96: 225–9. [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 22.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004; 303: 83–6. [DOI] [PubMed] [Google Scholar]
- 23.Abouheif MM, Nakasa T, Shibuya H, Niimoto T, Kongcharoensombat W, Ochi M. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology 2010; 49: 2054–60. [DOI] [PubMed] [Google Scholar]
- 24.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–8. [DOI] [PubMed] [Google Scholar]
- 25.Ponomarev ED, Veremeyko T, Barteneva NS. Visualization and quantitation of the expression of microRNAs and their target genes in neuroblastoma single cells using imaging cytometry. BMC Res Notes 2011; 4: 517–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aghabozorg ASS, Ghaderian SM. The role of microRNAs in cardiovascular disease. Int J Mol Cell Med 2013; 2: 50–7. [PMC free article] [PubMed] [Google Scholar]
- 27.Jing W, Jiang W. MicroRNA-93 regulates collagen loss by targeting MMP3 in human nucleus pulposus cells. Cell Prolif 2015; 48: 284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Wang J, Hu B, Wu X, Chen Y, Li R, Yuan W. MiR-34a promotes Fas-mediated cartilage endplate chondrocyte apoptosis by targeting Bcl-2. Mol Cell Biochem 2015; 406: 21–30. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X, Qiu G. MicroRNA-10b promotes nucleus pulposus cell proliferation through RhoC-Akt pathway by targeting HOXD10 in intervertebral disc degeneration. PLoS One 2013; 8: e83080–e83080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Li XJ, Ren ZJ, Tang JH. MicroRNA-34a: a potential therapeutic target in human cancer. Cell Death Dis 2014; 5: e1327–e1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanette DL, Rivadavia F, Molfetta GA, Barbuzano FG, Proto-Siqueira R, Silva-Jr WA, Falcão RP, Zago MA. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res 2007; 40: 1435–40. [DOI] [PubMed] [Google Scholar]
- 32.Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine 2001; 26: 1873–78. [DOI] [PubMed] [Google Scholar]
- 33.Wang F, Wu XT, Zhuang SY, Wang YT, Hong X, Zhu L, Bao JP. Ex vivo observation of human nucleus pulposus chondrocytes isolated from degenerated intervertebral discs. Asian Spine J 2011; 5: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Shen J, Wu WK, Yu X, Liang J, Qiu G, Liu J. The role of leptin on the organization and expression of cytoskeleton elements in nucleus pulposus cells. J Orthop Res 2013; 31: 847–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–8. [DOI] [PubMed] [Google Scholar]
- 36.Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis D, Jia LT, Wu SX, Huang J, Chen J, Luo ZJ. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J Pathol 2011; 225: 232–42. [DOI] [PubMed] [Google Scholar]
- 37.Liu G, Cao P, Chen H, Yuan W, Wang J, Tang X. MiR-27a regulates apoptosis in nucleus pulposus cells by targeting PI3K. PLoS One 2013; 8: e75251–e75251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanna S, Jackson AU, Nagaraja R, et al. Common variants in the GDF5- BFZB region are associated with variation in human height. Nat Genet 2008; 2: 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui M, Wan Y, Anderson DG, Shen FH, Leo BM, Laurencin CT, Balian G, Li X. Mouse growth and differentiation factor-5 protein and DNA therapy potentiates intervertebral disc cell aggregation and chondrogenic gene expression. Spine J 2008; 8: 287–95. [DOI] [PubMed] [Google Scholar]
- 40.Liang H, Ma SY, Feng G, Shen FH, Joshua Li X. Therapeutic effects of adenovirus-mediated growth and differentiation factor-5 in a mice disc degeneration model induced by annulus needle puncture. Spine J 2010; 10: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh AJ, Bradford DS, Lotz JC. In vivo growth factor treatment of degenerated intervertebral discs. Spine 2004; 29: 156–163. [DOI] [PubMed] [Google Scholar]