It was just after 6 AM on October 7, 2015. A first cup of coffee penetrated my morning fog while I was listening to the BBC news. After the usual worrisome headlines, the announcer concluded with “And this year’s Nobel Prize in Chemistry is shared by three individuals working in the field of DNA repair: Tomas Lindahl, Paul Modrich and Aziz Sancar.” I immediately ran downstairs to write a short congratulatory email to Aziz, knowing full well his ‘inbox’ would be packed and he would have little time to reply as his world would soon get incredibly busy; he actually responded with a kind thank you within a week.
As I sat there staring at the computer screen before pressing “send,” I was transported from Pittsburgh back to the time from January 1985 to the Fall of 1988 when I worked in Aziz’s laboratory in Chapel Hill, NC. Together with Intisar Husain and Dave Thomas, we were Aziz’s first crop of postdocs. I had just completed my PhD in James Regan’s laboratory at the University of Tennessee’s Oak Ridge School of Biomedical Sciences located at Oak Ridge National Laboratory (ORNL). Due to its proximity to nuclear reactors capable of making radioisotope-labeled macromolecule precursors, the Biology Division of ORNL was a hot-bed for molecular biological research. Jim had worked previously with Richard Setlow to show that cells from patients with xeroderma pigmentosum (XP) have a defect in the removal of cyclobutane pyrimidine dimers (CPD) [1], a confirmation of James Cleaver’s earlier result showing that XP cells are defective in repair replication after UV irradiation [2]. Using alkaline sucrose gradients, Jim Regan and Richard Setlow showed that XP cells lack the initial incision step seen in normal cells after irradiation with 254 nm light (UVC) [3]. They also developed a powerful technique using bromodeoxyuridine flash photolysis to measure the patch sizes of DNA repair helping to establish that different patch sizes exist for base excision repair (BER) versus nucleotide excision repair (NER) [4].
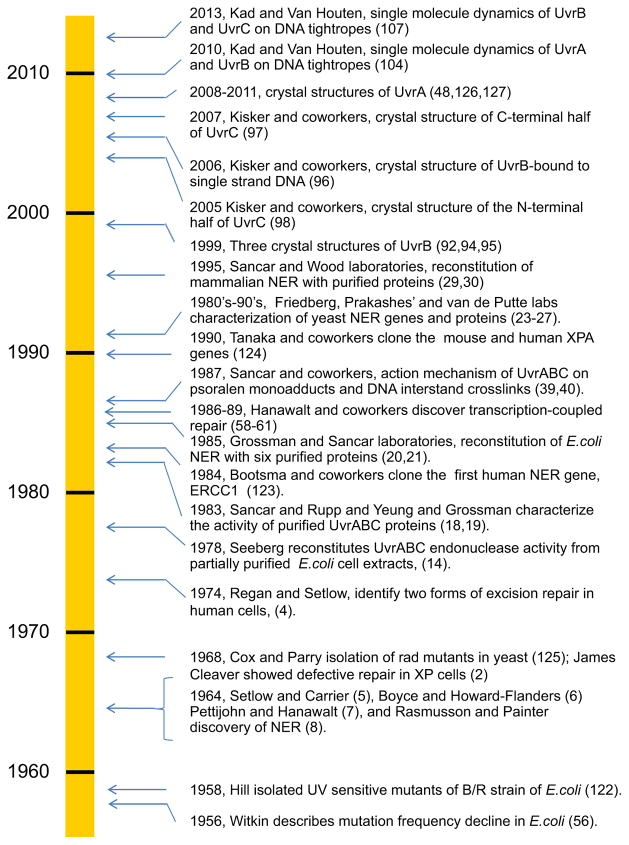
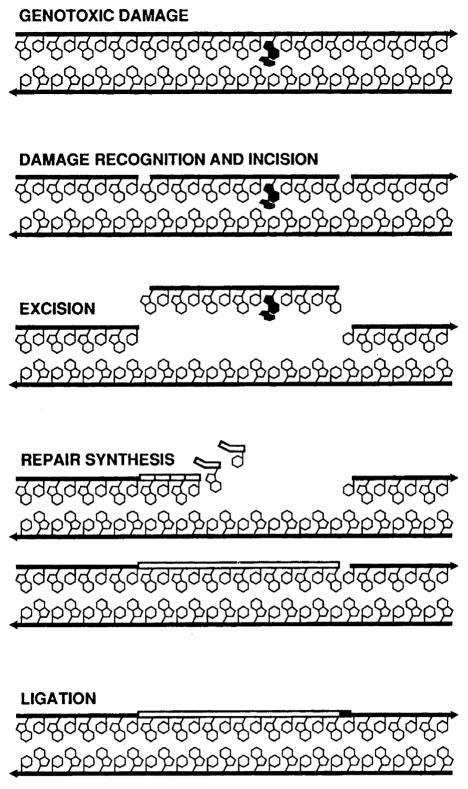
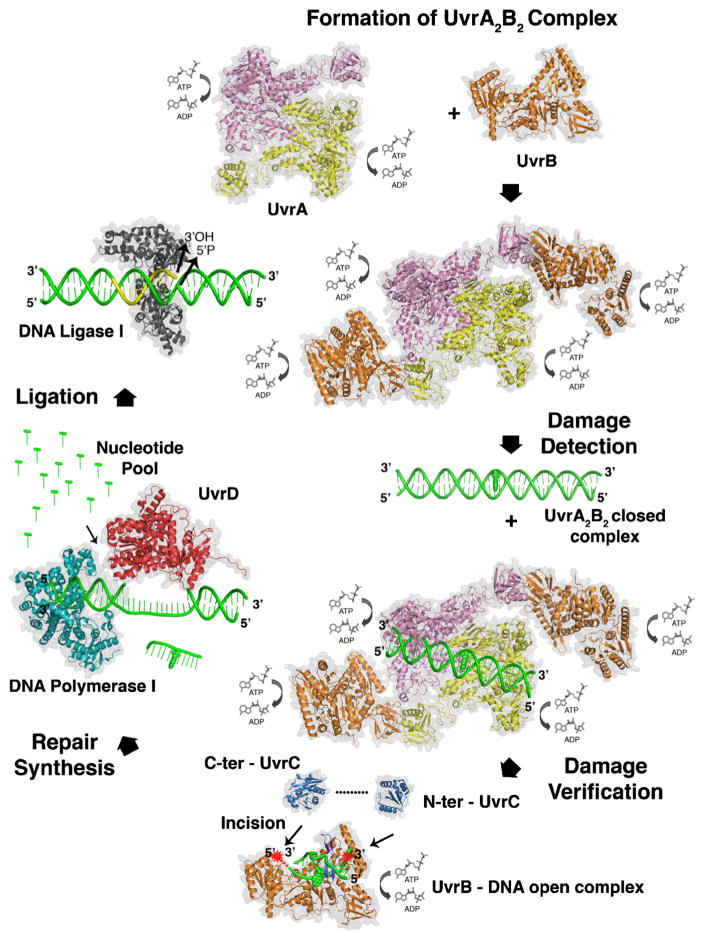
These studies followed the remarkable discovery of NER in 1964 (see timeline in Fig. 1). Bill Carrier and Richard Setlow at ORNL [5], as well as, Richard Boyce and Paul Howard-Flanders at Yale [6], independently discovered an active process by which cyclobutane thymine dimers are removed from Escherichia coli. Concurrently, David Pettijohn and Phil Hanawalt at Stanford demonstrated repair replication the non-conservative incorporation of radioactive nucleotides into parental DNA strands during NER [7]. In the same year, Ronald Rasmussen and Bob Painter at the NASA Ames Research Center found that tritiated-thymidine was incorporated into DNA in non-S-phase mammalian cells after UV irradiation in what they called ‘unscheduled DNA synthesis’ as evidence for NER in mammalian cells [8]. At this time, the model for NER was damage recognition, incision/excision, repair synthesis and DNA ligation (see Fig. 2). However, the incision mechanism was not understood, so it was unclear whether the repair process occurred through a ‘cut and patch’ or a ‘patch and cut’ mechanism [9].
Fig. 1.
Timeline of key events in NER. Bacterial NER is placed into larger perspective of NER; due to space limitations many important contributions are not shown during this 60 year period [122–127].
Fig. 2.
NER in the mid-1980s. By 1985 NER was known to proceed as a dynamic process of damage recognition and incision involving endonucleolytic incisions flanking the damaged site, excision of the oligonucleotide, simultaneously with repair synthesis. Finally, restoration of the repair patch into full length DNA was achieved with a ligase. The molecular steps were known, but the action of each protein was not.
Part of my own research at ORNL, in collaboration with Warren Masker, was to partially purify the UvrABC proteins for use in a modified endonuclease sensitive site (ESS) assay. The ESS assay was developed to follow the removal of the CPD lesion, and involved pre-labeling the mammalian DNA with either 14C- or 3H-labeled thymidine and running the DNA on alkaline sucrose gradients with and without treatment with a CPD-specific endonuclease isolated from Micrococcus luteus [10,11]. Our goal was to detect the formation and removal of highly carcinogenic (anti-) and relatively less carcinogenic (syn-) diastereomer benzo[a]pyrene diol epoxide (BPDE) adducts from mammalian cells. These studies were focused on testing the hypothesis that the less carcinogenic syn-BPDE lesions are repaired more quickly than the more carcinogenic anti-BPDE adducts [12]. We therefore had to generalize the ESS assay by using the UvrABC nuclease system, instead of the CPD-specific M. luteus enzyme [13]. We were following the protocols of Erling Seeberg, one of the pioneers in bacterial NER enzymology. He had developed a method for partial purification of the UvrABC proteins, and had shown that their associated ATP-dependent activities [14,15] could incise DNA containing several different types of “bulky lesions,” including those produced by BPDE [16]. As a graduate student, I was struggling with a question that turned into a life-long quest: how do NER enzymes interrogate DNA as they probe for lesions, and what motifs do they use to sense the structural and conformational changes induced by DNA damage?
When I was finishing my PhD, Warren suggested I write to Aziz Sancar about the possibility of postdoctoral training as he had recently set up his own laboratory just east over the Smokey mountains from Oak Ridge, at the University of North Carolina at Chapel Hill. As a postdoc in Dean Rupp’s laboratory, Aziz had developed a unique detection scheme for identifying plasmid encoded proteins [17]. Aided by this approach he purified the UvrA, UvrB and UvrC proteins to homogeneity and shown that the proteins worked in an ATP-dependent manner to incise plasmid DNA containing either UV-induced photoproducts, cisplatin adducts or psoralen adducts [18]. Furthermore, by comparing the incision sites of the CPD-specific T4 endonuclease V to the incision positions produced by the UvrABC proteins, he was able to infer that the UvrABC proteins worked like an “excinuclease” to incise the eighth phosphodiester bond 5′ and the fourth or fifth phosphodiester bond 3′ to a TC, CC or CT CPD, resulting in the release of a 12 or 13 base oligonucleotide containing the damage [18]. During the same year, using highly purified UvrA, UvrB and UvrC proteins, and similar approaches, these results were confirmed by Tony Yeung working in Larry Grossman’s laboratory [19]. Together these two studies supported a cut and patch model of NER (Fig. 2).
I was very lucky to be accepted into the Sancar lab, and despite the excellent bench training I received at ORNL, I still wanted to hone my biochemical and molecular biology skills. I remember the first agarose gel I ran in Aziz’s lab in January of 1985, which slipped out of the gel holder and into the sink, its shattered wreckage a disquieting sign I might be in over my head if not under water as I developed more sophisticated approaches! It was a remarkable time in Aziz’s laboratory. Dave Thomas, a postdoc who was an expert in protein purification, was making large batches of highly purified UvrA, UvrB and UvrC proteins. This was a huge task as we did not have tagged proteins and several of the protein purifications took five to six different chromatography steps. Intisar Husain seemed to have his hands on every project in the lab, and set out to sequence the uvrA gene, while I was there to help see how these proteins worked.
People often ask me what was it like to work in Aziz’s laboratory, and the first line of Charles Dickens’ novel, A Tale of Two Cities comes to mind: “It was the best of times, it was the worst of times…” The best of times, as we were able to do highly innovative experiments that led to a molecular understanding of how E. coli NER proteins process DNA damage. The worst of times because of the extreme pressure to produce results. Aziz and his wife, Gwen Sancar, worked amazingly long hours and we were expected to match or exceed their shift in the laboratory. Aziz took me in with little molecular biology or biochemical experience and I owe my career to his outstanding training. We worked extremely hard, and ironically both the postdocs and graduate students enjoyed lingering after Aziz and Gwen had left. When in their absence, the lab took on an informal atmosphere of camaraderie. The saying in the lab was, “see you today” as we would often depart after midnight in an effort to finish one set of experiments in preparation for another set the next day. We actually had a television in the laboratory, which allowed us to check in on sporting events, and watch Miami Vice on Friday nights before going out for a well-deserved drink. Gwen and Aziz usually worked 9 AM to 9 PM every day of the week except for Sunday, when Aziz would only work “a half a day” so that he could go home to watch the Dallas Cowboys’ football games. Thomas Edison has been quoted as saying that genius is 99% perspiration and 1% inspiration. In the halo of Aziz’s brilliance, we all worked at a higher level and surpassed our wildest expectations. Like an astute coach constantly challenging us to work harder and smarter, Aziz pushed us past our limitations. A tremendous work ethic, the ability to synthesize voluminous literature, the skill to develop highly productive collaborations, and the fearlessness to take on any project are some of the qualities that helped to earn Aziz a Nobel Prize. Aziz has a voracious hunger for knowledge that he tries to satiate by reading huge numbers of papers. This essential ingredient, I believe, contributed greatly to his success because it facilitated tremendous insights as new ideas would bubble up from his subconscious brain to his lips and out onto the bench.
The first task that we three postdocs undertook was to try to reconstitute the bacterial NER pathway with six purified proteins: UvrA, UvrB, UvrC, UvrD, DNA polymerase 1 (Pol 1), and DNA ligase. Our laboratory was in direct competition with Larry Grossman’s group at Johns Hopkins University. They beat us by two months, but fortunately both groups were able to publish in the Proceedings of the National Academy of Sciences [20,21]. These two papers showed that the activity of the UvrA, UvrB and UvC proteins led to a stoichiometric number of incisions, and it was not until we added both Pol I and UvrD, a DNA helicase, that a true turnover of the UvrABC proteins occurred. We found that the combined action of UvrD and Pol I were necessary and sufficient to dissociate the post-incision complex of the UvrBC proteins and allow gap filling by Pol I. The final stage was sealing of the repair patch by DNA ligase. We later showed that if DNA ligase was present, the repair patch filled in by Pol I was quickly ligated close, and greater than 85% of the repair patches were exactly 13–14 nucleotides in length [22].
My own good fortune was further enhanced in 1985, when Aziz shared an email with me from John Hearst at UC Berkeley, who thought it would be interesting to see how the UvrABC system processed psoralen monoadducts and interstrand cross-links. A brilliant postdoc in his laboratory, Howard Gamper, had just come up with a way to make short oligonucleotides with specific psoralen adducts at defined sites. Howard and I met in the spring of 1986 at a Mammalian DNA repair Gordon conference and during a magnificent afternoon by the beach, we outlined what would become three years of exciting experiments, which are described in more detail below. It was during the same meeting that I had lunch with Larry Grossman and Errol Friedberg. It was a memorable occasion for several reasons, including an incredible story Larry told over dessert about having to ditch his P51 Mustang fighter plane after being shot down over the Pacific Ocean during World War II, eventually being rescued as he floated alone in a small inflatable raft. At this time, in 1986, Errol’s laboratory, as well as the labs of Louise and Satya Prakash, were cloning and characterizing the genes and proteins responsible for mediating NER in the budding yeast, Saccharomyces cerevisiae. Later in the 1990s, these two teams went on to characterize the key steps in eukaryotic NER at the molecular level [23–27]. Because the genes and proteins in yeast share high sequence identity with the human repair proteins, their pioneering efforts supported the subsequent reconstitution of the mammalian NER pathway by Aziz’s group [28,29], as well as by Rick Wood’s laboratory [30] (in conjunction with Jean Marc Egly and Jan Hoejimakers) reviewed in [31–34]. This was a truly remarkable accomplishment, as human cells employ ~30 proteins in NER in contrast to bacterial cells, which require only six.
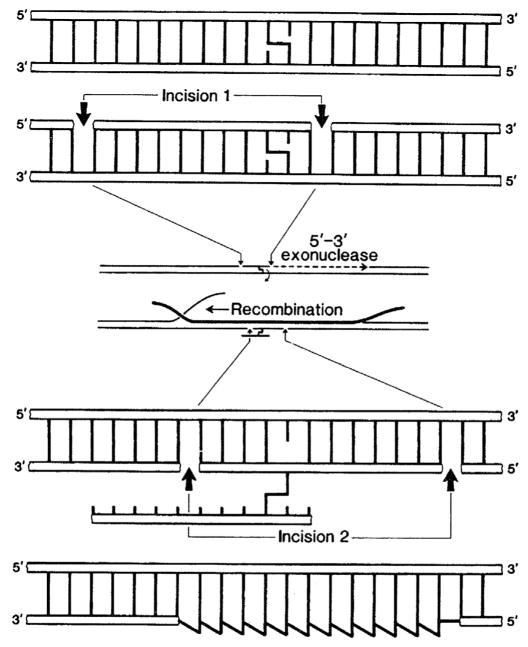
Working first with Howard Gamper and later with Suzanne Cheng (a graduate student in John Hearst’s laboratory at UC Berkeley) we ligated psoralen containing oligos into larger duplexes. Using these substrates and the purified Uvr protein system, we were able to describe each step of the bacterial NER pathway: from damage recognition, to verification, and then incision (see Fig. 2). In this way we could precisely map at the nucleotide level where the UvrABC system would incise the damage-containing DNA strand at the eighth phosphodiester bond 5′ and the fourth or fifth phosphodiester bond 3′ to a psoralen-thymine monoadduct, confirming Aziz’s early work from 1983 [18]. Using DNAase I footprinting, we were able to determine the binding affinity of the UvrA and the UvrAB complexes. Moreover, we found that UvrA bound to a region of 33 bp and made strong contact with the non-damaged strand, but surprisingly the footprint shrank down to just 19 bp when we added UvrB [35]. What we didn’t appreciate at the time, until Dave Orren, a graduate student in the lab, later showed that once the UvrAB complex finds a lesion, UvrA positions UvrB at the site of the damage and then dissociates, leaving behind a very stable UvrB-DNA complex [36,37]. Thus, Dave helped to elucidate the molecular nature of the salt-insensitive recognition complex characterized by filter binding assays as reported several years earlier by Larry’s group [19]. In another exciting project in Aziz’s lab, we were able to investigate how the UvrABC proteins process inter-strand cross-links using an “incision/recombination/incision” pathway—a repair model first suggested by Ron Cole [38]. We were able to show at the molecular level that UvrABC cuts only one side of a psoralen inter-strand adduct [39], while RecA helps to insert a complementary strand into the gap to generate a three stranded DNA structure [40]. This intermediate is then further processed by UvrABC in a second round of incisions that attacks the other psoralen-modified strand, which is cross-linked to the excised oligo (see Fig. 3).
Fig. 3.
Working model for the repair of inter-strand DNA cross-links in bacteria. The UvrABC proteins were shown to incise on one side of a psoralen cross-link. This gapped substrate was processed by a RecA filament to generate a three-stranded intermediate. In a second round of incisions, UvrABC was shown to act on the psoralen-cross-linked-excised oligonucleotide. Gap filling by DNA polymerase I and ligation complete the repair process. This incision/recombination/incision model is based on a previous model of Cole [38]. From Ref. [76] with permission.
There were several times during my stay in Aziz’s lab when I felt I was given lucky breaks. For example, Aziz does not like to travel and he was asked to give a talk at a big meeting in the Netherlands and at the last minute decided not to go. Over the last four decades, the Netherlands has had an incredibly strong program in DNA repair, first under the direction of Dirk Bootsma and now Jan Hoejimakers and Wim Vermeulen in Rotterdam, and Paul Lohman and Leon Mullenders in Leiden. So attending this extremely important and exciting meeting was a ‘big deal’, and as fate would have it, the organizing committee called me and asked me if I could present the laboratory’s work on the biochemistry of bacterial NER. Having never been to Europe I of course jumped at the chance to go. Since my last name is Dutch, I received a very warm introduction, to which I spontaneously said at the beginning of my lecture, “on behalf of my genes it’s good to be home again”. Many people bought me beers that night, and the presentation of our data was a big success; it truly helped launch my career. I am very lucky that Aziz doesn’t like to attend meetings.
While we were characterizing the molecular activities of the proteins, we were also busily sequencing the E. coli uvrA, and uvrB genes. Gwen and Aziz had started sequencing uvrC while still in Dean Rupp’s laboratory, with the task finally completed in 1984 [41]. Recall that although entire bacterial genomes currently may be sequenced in just days, in the mid 1980s a single gene took many months of hard work. We were not able to get dideoxy sequencing to work efficiently in the lab, so we settled for the classical Maxim-Gilbert sequencing (Walter Gilbert shared the Nobel Prize in Chemistry in 1980 with Fredrick Sanger and Paul Berg) in which we used the chemical reactivity of specific reagents for particular bases to facilitate nucleotide-specific cleavage, which we could read on huge hand-poured sequencing gels. We sequenced the uvrA gene by taking DNA fragments of it, labeling both 5′ ends with 32P, cutting each fragment with a restriction enzyme, then sequencing as far from each end as we could, hoping to encounter more restriction enzyme sites that would allow us to progress further. Remarkably, UvrA apparently evolved via gene duplication [42], encompassing two putative ATP binding sites and several putative zinc fingers [43], the presence of which were definitively established by Gary Myles, a graduate student in the lab [44–46]. The functional ATP cassettes were also confirmed by Larry’s group in a similar way [47]. However, it was many years later, after the UvrA crystal structure was solved, when it was determined that UvrA’s ATPase domains comprised an unusual intramolecular interaction in which the Walker A domain from one region interacted with the Walker B domain of the corresponding duplicated region [48]. Another important research group studying bacterial NER in the 1980s and 90s was Pieter van de Putte’s laboratory at the Laboratory of Molecular Genetics at the University of Leiden. This group, which included Nora Goosen (who went on to run her own highly successful laboratory investigating the UvrABC machinery), made many important contributions to the field of NER, including the first direct evidence of UvrA loading of UvrB using gel mobility shift assays [49]. Nora’s group proved that nuclease centers responsible for the 3′ and 5′ incisions are housed in the N- and C-terminus of UvrC, respectively [50]. These studies extended the previous work by Jing-Jer Lin and Aziz who identified of the catalytic triad of UvrC responsible for the 5′ incision [51]. Additionally, Nora’s lab discovered that E. coli and some other bacteria contain a homolog of UvrC, Cho that lacks the N-terminus and incises 10 nucleotides away from the lesion on the 3′ side of the damage [52,53] (Fig. 4).
Fig. 4.
Bacterial NER in the 1990s. Many of the key steps mediated by the UvrA, UvrB and UvrC proteins were becoming known: UvrA and UvrB work together to recognize the lesion. In an ATP-dependent step, UvrB engages the damaged nucleotide causing strand opening allowing UvrA to dissociate. Recruitment of UvrC causes dual incisions 5′ and 3′ to the damaged site. The dual action of UvrD and Pol I are necessary for gap filling and turnover of UvrB and UvrC from the post incision complex. Finally DNA ligase works to seal the newly completed repair patch. From Ref. [76] with permission.
It was during the intense period of DNA sequencing of the uvrA and uvrB genes that we gained some insights into Aziz’s early years growing up in Savur, Turkey, when a friend and colleague of his, Erlap Arikan, visited the lab to help sequence the uvrB gene [54]. His amicable smile and jolly persona affected all of us in a positive way. He arrived in the lab with rudimentary English language skills, but his enthusiasm for learning and speaking English brought the whole group together. This was especially true in the evenings as we were drying down sequencing gels and waiting to put them down on X-ray films for overnight exposures. As he practiced his English he told us that he knew Aziz from school, where Aziz preferred reading books rather than running around and playing soccer with his classmates. Aziz’s thirst for knowledge was truly unquenchable; this in turn, impelled him to press us to work hard; he knew what everyone was doing each day. He loved to pour over data with us, and together we would discuss the next steps, and ultimately, the resultant manuscript—he understood the imperative to publish research in a timely fashion.
The intense working environment of Aziz’s laboratory prompted the need to escape the daily burden of churning out data. Many of us would go to the gym over dinner time, and we would suffer Aziz’s disapprobation as we would leave the lab. “You know, your heart only has so many beats – when you go work out you are using them up.” Of course Aziz knew that was nonsense, I believe it was his way of saying, “please stay here and keep working—I need to know more.” Sometimes in Aziz’s lab when we were particularly productive, we would drink shots of whiskey as a group every time we got a paper accepted—a monthly event.
It is with very fond memories that I look back on our long discussions with Gary, Dave, and a fellow graduate student, Gillian Payne who helped workout the action mechanism of DNA photolyase in another exciting and collaborative project with Gwen Sancar’s laboratory. I wouldn’t trade one day of my three and a half years in Aziz’s lab for any other; in spite of the grueling pace, and perhaps because of it—it was truly the best of times.
With the help and support from our chair of Biochemistry at UNC, Mary Ellen Jones, I moved on to take my first faculty position at the University of Vermont and Aziz’s group went on to purify and characterize the transcription repair coupling factor (TRCF) encoded by the mfd gene [55]. This genetic locus was first discovered by Evelyn Witkin, who was awarded the 2015 Lasker prize for her studies on damage-inducible genes. Evelyn had discovered mutation frequency decline, a process in which bacterial cells starved of nutrients following UV-irradiation had lower levels of UV-induced mutagenesis [56,57]. Transcription-coupled repair was first identified in mammalian cells by Vilhelm Bohr, Allen Smith and Diane Okumoto working with Phil Hanawalt [58,59] during the same period while we were working out the steps in bacterial NER.
A postdoc in Phil’s lab, Isabel Mellon, suggested that the translocating RNA polymerase might be the sensor of DNA lesions, and then she proved that the transcribed strand is repaired more rapidly than the non-transcribed strand in mammalian cells [60,61], as well as in bacteria [59] Later, in her own laboratory, she ascertained that transcription coupled repair in E. coli was under the direct control of the MFD protein [62,63]. During transcription-coupled NER the arrested RNA polymerase at the site of a DNA lesion helps to recruit other repair factors to the site of damage on the transcribed strand. Through some impressive detective work, Aziz, working with Chris Selby, was able to identify and purify the coupling factor, TRFC, and show that it is the product of the mfd gene, and that it assisted in the removal of stalled RNA polymerase from the DNA. They speculated that it might recruit UvrABC to the site of the damage [55,64–67]. It was later shown by several groups who helped solve the structure and function of TRCF/MFD that this protein shares a UvrB-homology domain to which UvrA binds [68–70]. It is believed that once RNA polymerase stalls at a damaged site, binding of MFD triggers opening of this UvrA interacting domain to facilitate pushing RNA polymerase off the damaged site through its translocase activity while simultaneously recruiting UvrA to the site [71–75].
Once at the University of Vermont, I invited several scientists working in the field of DNA repair to give lectures about their work. During his visit, Errol Friedberg suggested that I write a comprehensive review of the bacterial nucleotide excision repair system. What a delightful, if not daunting task, as I contemplated the diverse repertoire of substrates upon which the UvrABC system acted [76]. Unlike the glycosylase initiators of BER (see [129]), which sense highly specific structural features of DNA (i.e., damaged or incorrect bases), NER proteins dance a tango with DNA to sense dynamic changes in its motion and stability caused by DNA lesions. In my 1990 review, I was accorded the space and the time to explore the question that I began to ponder as a graduate student, “how do NER proteins work together to probe DNA for imperfections?” This monograph was a labor of love, and for a brief spell I was able to familiarize myself with almost every key piece of data about bacterial NER and critically analyze the merits of each study [76] to develop a working model of NER, see Fig. 3. I didn’t realize at the time how much I was channeling Aziz as I read and digested several hundred papers that punctuated the field of bacterial NER.
The process of writing a comprehensive review forged an arc of discovery that has propelled my group forward for more than two decades. This is a great lesson for new faculty members: work with a member of your new laboratory to write an exhaustive review on the field you are hoping to pursue for the years to come. I am still riding this spectacular surge of discovery that started when I was a graduate student with Jim Regan and swelled to a huge wave during my time in Aziz’s lab, which my group and I continue to surf in our efforts to understand what molecular details DNA repair enzymes detect in DNA lesions as they probe for DNA damage.
Once settled at the Univeristy of Vermont, with great mentorship of several colleagues who soon became my scientific siblings, Nick Heintz, Wah Kow, Sriram Krishnaswamy, Nick Farrell, Brooke Mossman and Susan Wallace, my research program became established. Susan, Wah and I organized a meeting on DNA Damage Recognition [77] where we learned from John Tainer about the first resolved structure of a DNA repair enzyme, exonuclease III [78,79], and were trained in the molecular dynamics of DNA by Suse Broyde, New York University [80]. Suse offered a fantastic tutorial on how to use a silicon graphics work-station to model the vibrations of DNA, as it was being probed by DNA repair enzymes. The intervening years have born witness to the huge increase in computational power that now allows us to do such types of imaging using Pymol running on laptops and even smart phones! Together, she and Nick Geacintov have contributed a series of beautiful papers that have helped to reveal what NER enzymes see as they probe the DNA for damage-induced chemical and conformational changes [81–83]. Their most recent study shows in great detail how XPC uses its beta-hairpins to probe for DNA damage [84]. They have taught me that damage recognition during NER is highly dynamic, and that while crystal structures are informative, they are only snap-shots of the process; it is the miraculous dance of repair proteins along the undulating fibers of DNA that allows molecular recognition of structural perturbations induced by a large array of damaging agents acted on by NER. I feel so privileged to be a student of NER and to work in this exciting field with such brilliant colleagues.
After a productive period in Vermont, I was lucky enough to be recruited by Sam Wilson when he assembled a gifted group of researchers studying genome stability at the University of Texas Medical Branch (UTMB) in Galveston, Texas. In addition to Sam Wilson and me, the group consisted of Louise and Sayta Prakash, Patrick Sung, Sankar Mitra (whom I knew and admired from ORNL for is work on replication and then BER), and Stephen Lloyd (who worked out the action mechanism of the CPD-specific T4 lyase). The Sealy Center for Molecular Science was a powerful engine of discovery in all aspects of genome stability and it was our year of glory when Science magazine declared DNA repair enzymes the “molecule of the year” in 1994 [85]. Under Sam’s mentorship, and the collective help of the faculty at the Sealy Center for Molecular Science, my research program blossomed in two directions: we continued to make important inroads into understanding the UvrABC system [86–89] and we initiated investigations into the nature of mitochondrial DNA damage and repair [90,91].
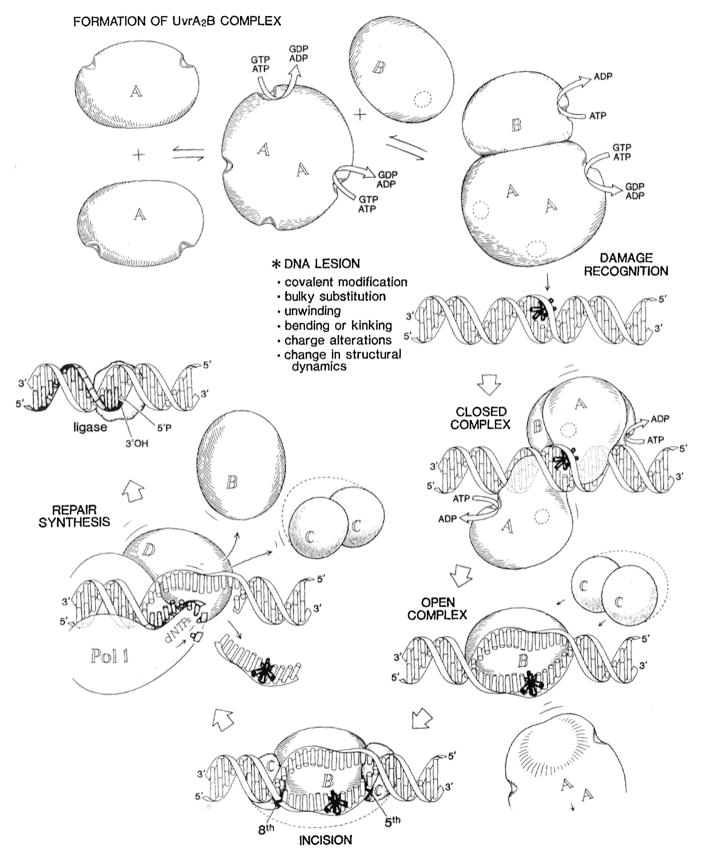
Several years later Sam Wilson was recruited to the position of Deputy Director at the National Institute of Environmental Health Sciences (NIEHS), NIH, and he hired me to run an intramural lab, as well as to direct the Program Analysis Branch in the extramural division of the institute. It was during these years at NIEHS that I had the fortune of beginning a highly productive collaboration with Caroline Kisker at SUNY Stony Brook and subsequently, when she moved to the University of Würzburg. Together we contributed one of three new crystal structures of UvrB [92–95], which has a helicase fold, as well as, the first structure of UvrB bound to DNA [96], and two nuclease domains of UvrC (Fig. 5) [97,98]. We discovered that UvrB interrogates and verifies the presence of a DNA lesion by inserting a beta-hairpin into the DNA and that it uses key hydrophobic residues to distinguish the damaged structure from undamaged DNA [99–101]. My first graduate student, Amanda Snowden (McCullough), and I had first proposed such a detection mechanism in a review some 13 years earlier [102]. Knowing that stacking interactions of the bases governs the configuration of B-form DNA, we envisioned that aromatic side chains might intercalate between the bases to measure “the closeness of fit to the Watson and Crick structure,” as had been suggested by Phil Hanawalt and Bob Haynes in 1965, when they reported that, in addition to UV induced CPDs, chemical adducts to DNA were subject to excision repair, and that the system must be able to deal with a wide variety of different types of alterations [103].
Fig. 5.
Current state of bacterial NER in 2015. Crystal structures for several key intermediates of bacterial NER are shown. The dynamics of the interactions and the structures of several key intermediates are not yet known.
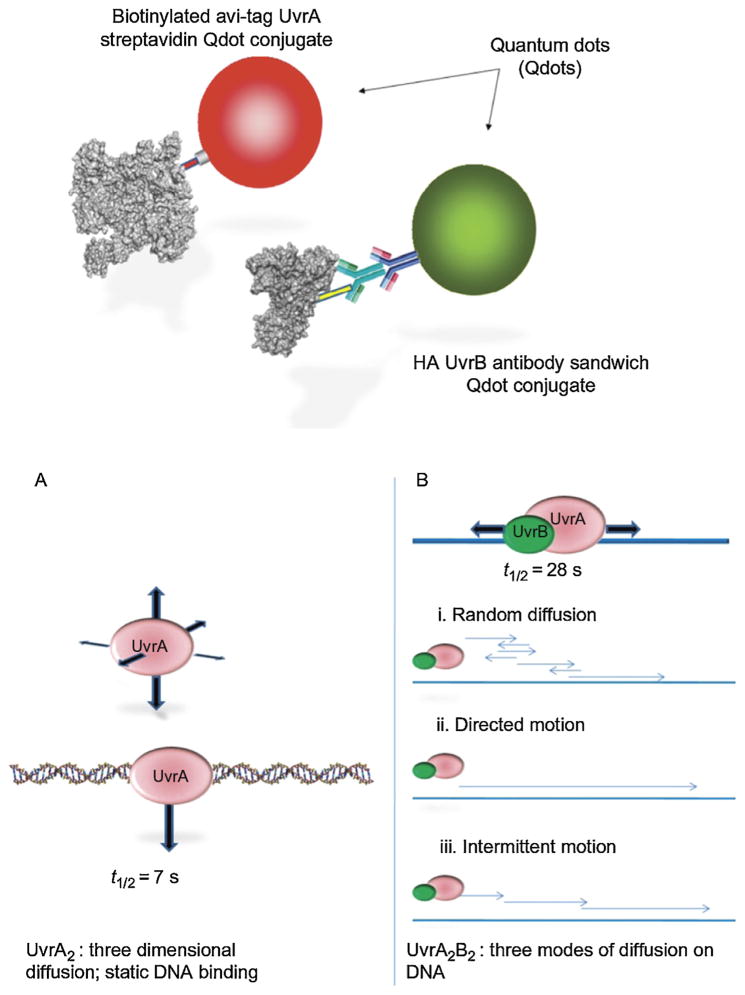
I moved to the University of Pittsburgh in 2008, and thanks to a superb collaboration with Neil Kad (University of Kent), David Warshaw (University of Vermont), and Hong Wang (a talented postdoc in my laboratory, now at North Carolina State running her own productive lab) we were able to achieve a lifelong goal of mine: to actually watch individual molecules of UvrA and UvrB search for DNA damage. Using a DNA tightrope assay developed by Neil, we observed quantum-dot-labeled UvrA and UvrAB complexes interrogate DNA for lesions in real time (see Fig. 6) [104,105]. We made the astonishing discovery that UvrA scans for damage by a relatively inefficient three-dimensional approach; however, the addition of UvrB to UvrA causes a collapse in the search apparatus from a three-dimensional to a one-dimensional sliding mechanism. It is worthy of note that Larry Grossman had postulated such a sliding mechanism no less than 15 years earlier [106]. Neil and his group have also found that UvrB and UvrC can combine on DNA and slide—something we never envisaged during my tenure years ago in Aziz’s lab [107]. Caroline’s focus has moved to eukaryotic NER, and the structure and function of XPD, a DNA helicase [108], which we published contemporaneously with the results from two other groups [109,110]. This protein works as part of the seven-membered TFIIH core complex to help authenticate DNA lesions during eukaryotic NER. Thus XPD shares a similar function as UvrB for damage verification [111].
Fig. 6.
Single molecule analysis of UvrA and UvrB search strategies. UvrA and UvrB differentially labeled with two differently colored Qdots were observed moving on DNA together. UvrA does a three-dimensional search and the addition of UvrB causes one-dimensional sliding of the UvrAB complex.
Recently, Aziz and his group have characterized key proteins that control DNA damage responses [112,113] and have discovered that DNA excision repair efficiency in mice and humans exhibits a circadian rhythm [114–116], and that this may have important implications for the etiology of some environmentally-induced cancers [115,117,118], as well as translational value for chemotherapeutic approaches to treat cancer [119]. I am confident that the enormous influence NER exerts on the molecular biology of the cell will become increasingly evident as researchers discover new roles for NER proteins and establish how components of the NER machinery participate in other cellular functions, such as transcription, replication, damage signaling, cell cycle control and even nuclear architecture involving actin filaments [120]. Aziz continues at the forefront of such research, as shown by his recent study that employed a high throughput sequencing approach (coined XR-seq) to analyze excised nucleotide fragments and create a genome-wide map of DNA repair at the nucleotide level in human cells [121]. Knowing Aziz, his laboratory will continue blazing new paths that we will all race to follow. The wondrous thing about science is that the next big discovery always lies in front of us.
Acknowledgments
I am indebted to Aziz Sancar for his mentoring and for giving me the great opportunity to work in his laboratory. I also acknowledge the advice and important collaborative interactions with Jim Regan, Bill Carrier, Warren Masker, Julian Preston, Nick Heintz, Brooke Mossman, Susan Wallace, Sam Wilson, and last but not least, my outstanding students and postdocs who reverse mentor me to work harder and smarter. During my career, including my period in Aziz’s laboratory, I have been amazingly lucky to have enjoyed the opportunity to work with a huge number of talented people, many of whom I have highlighted in this perspective. It was impossible to include everyone, and for those of you who have not been cited in this brief essay for lack of time and space, please forgive my sin of omission. I greatly appreciate the helpful comments and editing by Bill Cannon, Caroline Kisker, Victoria Woshner, Neil Kad, Muwen Kong and Emily Beckwitt. My laboratory is currently supported by the University of Pittsburgh Cancer Institute, and NIH Grants, 1R01ES019566; 1R21/33ES025606, and 1R01CA174713.
References
- 1.Regan JD, Trosko JE, Carrier WL. Evidence for excision of ultraviolet-induced pyrimidine dimers from the DNA of human cells in vitro. Biophys J. 1968;8:319–325. doi: 10.1016/S0006-3495(68)86490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 3.Setlow RB, Regan JD, German J, Carrier WL. Evidence that xeroderma pigmentosum cells do not perform the first step in the repair of ultraviolet damage to their DNA. Proc Natl Acad Sci U S A. 1969;64:1035–1041. doi: 10.1073/pnas.64.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Regan JD, Setlow RB. Two forms of repair in the DNA of human cells damaged by chemical carcinogens and mutagens. Cancer Res. 1974;34:3318–3325. [PubMed] [Google Scholar]
- 5.Setlow RB, Carrier WL. The disappearance of thymine dimers from DNA: an error-correcting mechanism. Proc Natl Acad Sci U S A. 1964;51:226–231. doi: 10.1073/pnas.51.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyce RP, Howard-Flanders P. Release of ultraviolet light-induced thymine dimers from DNA in E. coli K-12. Proc Natl Acad Sci U S A. 1964;51:293–300. doi: 10.1073/pnas.51.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettijohn D, Hanawalt P. Evidence for repair-replication of ultraviolet damaged DNA in bacteria. J Mol Biol. 1964;9:395–410. doi: 10.1016/s0022-2836(64)80216-3. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen RE, Painter RB. Evidence for repair of ultra-violet damaged deoxyribonucleic acid in cultured mammalian cells. Nature. 1964;203:1360–1362. doi: 10.1038/2031360a0. [DOI] [PubMed] [Google Scholar]
- 9.Hanawalt PC, Haynes RH. The repair of DNA. Sci Am. 1967;216:36–43. doi: 10.1038/scientificamerican0267-36. [DOI] [PubMed] [Google Scholar]
- 10.Buhl SN, Regan JD. Repair endonuclease-sensitive sites in daughter DNA of ultraviolet-irradiated human cells. Nature. 1973;246:484. doi: 10.1038/246484a0. [DOI] [PubMed] [Google Scholar]
- 11.Paterson MC, Lohman PH, Westerveld A, Sluyter ML. DNA repair monitored by an enzymatic assay in multinucleate xeroderma pigmentosum cells after fusion. Nature. 1974;248:50–52. doi: 10.1038/248050a0. [DOI] [PubMed] [Google Scholar]
- 12.Van Houten B. Studies of Ultraviolet Light and Benzo[a]pyrene Diol Epoxide Induced DNA Damage and Repair in Human Cells. Oak Ridge Graduate School of Biomedical Sciences, University of Tennessee; Oak Ridge: 1984. p. 218. [Google Scholar]
- 13.Van Houten B, Masker WE, Carrier WL, Regan JD. Quantitation of carcinogen-induced DNA damage and repair in human cells with the UVR ABC excision nuclease from Escherichia coli. Carcinogenesis. 1986;7:83–87. doi: 10.1093/carcin/7.1.83. [DOI] [PubMed] [Google Scholar]
- 14.Seeberg E. Reconstitution of an Escherichia coli repair endonuclease activity from the separated uvrA+ and uvrB+/uvrC+ gene products. Proc Natl Acad Sci U S A. 1978;75:2569–2573. doi: 10.1073/pnas.75.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seeberg E, Nissen-Meyer J, Strike P. Incision of ultraviolet-irradiated DNA by extracts of E. coli requires three different gene products. Nature. 1976;263:524–526. doi: 10.1038/263524a0. [DOI] [PubMed] [Google Scholar]
- 16.Seeberg E, Steinum AL, Nordenskjold M, Soderhall S, Jernstrom B. Strand-break formation in DNA modified by benzo[alpha]pyrene diolepoxide. Quantitative cleavage by Escherichia coli UvrABC endonuclease. Mutat Res. 1983;112:139–145. doi: 10.1016/0167-8817(83)90036-6. [DOI] [PubMed] [Google Scholar]
- 17.Sancar A, Hack AM, Rupp WD. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979;137:692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sancar A, Rupp WD. A novel repair enzyme: UvrABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 19.Yeung AT, Mattes WB, Oh EY, Grossman L. Enzymatic properties of purified Escherichia coli UvrABC proteins. Proc Natl Acad Sci U S A. 1983;80:6157–6161. doi: 10.1073/pnas.80.20.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caron PR, Kushner SR, Grossman L. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the UvrABC protein complex. Proc Natl Acad Sci U S A. 1985;82:4925–4929. doi: 10.1073/pnas.82.15.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Husain I, Van Houten B, Thomas DC, Abdel-Monem M, Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci U S A. 1985;82:6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Houten B, Gamper H, Hearst JE, Sancar A. Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J Biol Chem. 1988;263:16553–16560. [PubMed] [Google Scholar]
- 23.Friedberg EC. Yeast genes involved in DNA-repair processes: new looks on old faces. Mol Microbiol. 1991;5:2303–2310. doi: 10.1111/j.1365-2958.1991.tb02074.x. [DOI] [PubMed] [Google Scholar]
- 24.Friedberg EC. Eukaryotic DNA repair: glimpses through the yeast Saccharomyces cerevisiae. Bioessays. 1991;13:295–302. doi: 10.1002/bies.950130607. [DOI] [PubMed] [Google Scholar]
- 25.Friedberg EC, Bardwell AJ, Bardwell L, Feaver WJ, Kornberg RD, Svejstrup JQ, Tomkinson AE, Wang Z. Nucleotide excision repair in the yeast Saccharomyces cerevisiae: its relationship to specialized mitotic recombination and RNA polymerase II basal transcription. Philos Trans R Soc Lond Ser B: Biol Sci. 1995;347:63–68. doi: 10.1098/rstb.1995.0010. [DOI] [PubMed] [Google Scholar]
- 26.Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 27.Prakash S, Sung P, Prakash L. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu Rev Genet. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- 28.Mu D, Hsu DS, Sancar A. Reaction mechanism of human DNA repair excision nuclease. J Biol Chem. 1996;271:8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 29.Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 30.Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 31.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 32.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 33.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 34.Wood RD, Araujo SJ, Ariza RR, Batty DP, Biggerstaff M, Evans E, Gaillard PH, Gunz D, Koberle B, Kuraoka I, Moggs JG, Sandall JK, Shivji MK. DNA damage recognition and nucleotide excision repair in mammalian cells. Cold Spring Harb Symp Quant Biol. 2000;65:173–182. doi: 10.1101/sqb.2000.65.173. [DOI] [PubMed] [Google Scholar]
- 35.Van Houten B, Gamper H, Sancar A, Hearst JE. DNase I footprint of ABC excinuclease. J Biol Chem. 1987;262:13180–13187. [PubMed] [Google Scholar]
- 36.Orren DK, Sancar A. The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex. Proc Natl Acad Sci U S A. 1989;86:5237–5241. doi: 10.1073/pnas.86.14.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orren DK, Sancar A. Formation and enzymatic properties of the UvrB.DNA complex. J Biol Chem. 1990;265:15796–15803. [PubMed] [Google Scholar]
- 38.Cole RS. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc Natl Acad Sci U S A. 1973;70:1064–1068. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Houten B, Gamper H, Holbrook SR, Hearst JE, Sancar A. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc Natl Acad Sci U S A. 1986;83:8077–8081. doi: 10.1073/pnas.83.21.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng S, Van Houten B, Gamper HB, Sancar A, Hearst JE. Use of psoralen-modified oligonucleotides to trap three-stranded RecA-DNA complexes and repair of these cross-linked complexes by ABC excinuclease. J Biol Chem. 1988;263:15110–15117. [PubMed] [Google Scholar]
- 41.Sancar GB, Sancar A, Rupp WD. Sequences of the E. coli uvrC gene and protein. Nucleic Acids Res. 1984;12:4593–4608. doi: 10.1093/nar/12.11.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doolittle RF, Johnson MS, Husain I, Van Houten B, Thomas DC, Sancar A. Domainal evolution of a prokaryotic DNA repair protein and its relationship to active-transport proteins. Nature. 1986;323:451–453. doi: 10.1038/323451a0. [DOI] [PubMed] [Google Scholar]
- 43.Husain I, Van Houten B, Thomas DC, Sancar A. Sequences of Escherichia coli uvrA gene and protein reveal two potential ATP binding sites. J Biol Chem. 1986;261:4895–4901. [PubMed] [Google Scholar]
- 44.Myles GM, Sancar A. Isolation and characterization of functional domains of UvrA. Biochemistry. 1991;30:3834–3840. doi: 10.1021/bi00230a005. [DOI] [PubMed] [Google Scholar]
- 45.Myles GM, Hearst JE, Sancar A. Site-specific mutagenesis of conserved residues within Walker A and B sequences of Escherichia coli UvrA protein. Biochemistry. 1991;30:3824–3834. doi: 10.1021/bi00230a004. [DOI] [PubMed] [Google Scholar]
- 46.Navaratnam S, Myles GM, Strange RW, Sancar A. Evidence from extended X-ray absorption fine structure and site-specific mutagenesis for zinc fingers in UvrA protein of Escherichia coli. J Biol Chem. 1989;264:16067–16071. [PubMed] [Google Scholar]
- 47.Thiagalingam S, Grossman L. Both ATPase sites of Escherichia coli UvrA have functional roles in nucleotide excision repair. J Biol Chem. 1991;266:11395–11403. [PubMed] [Google Scholar]
- 48.Pakotiprapha D, Inuzuka Y, Bowman BR, Moolenaar GF, Goosen N, Jeruzalmi D, Verdine GL. Crystal structure of Bacillus stearothermophilus UvrA provides insight into ATP-modulated dimerization, UvrB interaction, and DNA binding. Mol Cell. 2008;29:122–133. doi: 10.1016/j.molcel.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visse R, de Ruijter M, Moolenaar GF, van de Putte P. Analysis of UvrABC endonuclease reaction intermediates on cisplatin-damaged DNA using mobility shift gel electrophoresis. J Biol Chem. 1992;267:6736–6742. [PubMed] [Google Scholar]
- 50.Moolenaar GF, Franken KL, Dijkstra DM, Thomas-Oates JE, Visse R, van de Putte P, Goosen N. The C-terminal region of the UvrB protein of Escherichia coli contains an important determinant for UvrC binding to the preincision complex but not the catalytic site for 3′-incision. J Biol Chem. 1995;270:30508–30515. doi: 10.1074/jbc.270.51.30508. [DOI] [PubMed] [Google Scholar]
- 51.Lin JJ, Sancar A. Active site of (A)BC excinuclease. I. Evidence for 5′ incision by UvrC through a catalytic site involving Asp399, Asp438, Asp466, and His538 residues. J Biol chem. 1992;267:17688–17692. [PubMed] [Google Scholar]
- 52.Moolenaar GF, van Rossum-Fikkert S, van Kesteren M, Goosen N. Cho, a second endonuclease involved in Escherichia coli nucleotide excision repair. Proc Natl Acad Sci U S A. 2002;99:1467–1472. doi: 10.1073/pnas.032584099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Houten B, Eisen JA, Hanawalt PC. A cut above: discovery of an alternative excision repair pathway in bacteria. Proc Natl Acad Sci U S A. 2002;99:2581–2583. doi: 10.1073/pnas.062062599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arikan E, Kulkarni MS, Thomas DC, Sancar A. Sequences of the E. coli uvrB gene and protein. Nucleic Acids Res. 1986;14:2637–2650. doi: 10.1093/nar/14.6.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science (New York, NY) 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 56.Witkin EM. Time, temperature, and protein synthesis: a study of ultraviolet-induced mutation in bacteria. Cold Spring Harb Symp Quant Biol. 1956;21:123–140. doi: 10.1101/sqb.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 57.Witkin EM. Mutation frequency decline revisited. Bioessays. 1994;16:437–444. doi: 10.1002/bies.950160613. [DOI] [PubMed] [Google Scholar]
- 58.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 59.Bohr VA, Okumoto DS, Hanawalt PC. Survival of UV-irradiated mammalian cells correlates with efficient DNA repair in an essential gene. Proc Natl Acad Sci U S A. 1986;83:3830–3833. doi: 10.1073/pnas.83.11.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mellon I, Bohr VA, Smith CA, Hanawalt PC. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci U S A. 1986;83:8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 62.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 63.Mellon I, Rajpal DK, Koi M, Boland CR, Champe GN. Transcription-coupled repair deficiency and mutations in human mismatch repair genes. Science. 1996;272:557–560. doi: 10.1126/science.272.5261.557. [DOI] [PubMed] [Google Scholar]
- 64.Selby CP, Sancar A. Transcription-repair coupling and mutation frequency decline. J Bacteriol. 1993;175:7509–7514. doi: 10.1128/jb.175.23.7509-7514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Selby CP, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor, I. Structural domains and binding properties. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 67.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. II. Catalytic properties. J Biol Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 68.Manelyte L, Kim YI, Smith AJ, Smith RM, Savery NJ. Regulation and rate enhancement during transcription-coupled DNA repair. Mol Cell. 2010;40:714–724. doi: 10.1016/j.molcel.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murphy MN, Gong P, Ralto K, Manelyte L, Savery NJ, Theis K. An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd. Nucleic Acids Res. 2009;37:6042–6053. doi: 10.1093/nar/gkp680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Savery NJ. The molecular mechanism of transcription-coupled DNA repair. Trends Microbiol. 2007;15:326–333. doi: 10.1016/j.tim.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 71.Deaconescu AM, Sevostyanova A, Artsimovitch I, Grigorieff N. Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc Natl Acad Sci U S A. 2012;109:3353–3358. doi: 10.1073/pnas.1115105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Howan K, Smith AJ, Westblade LF, Joly N, Grange W, Zorman S, Darst SA, Savery NJ, Strick TR. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012;490:431–434. doi: 10.1038/nature11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monnet J, Grange W, Strick TR, Joly N. Mfd as a central partner of transcription coupled repair. Transcription. 2013;4:109–113. doi: 10.4161/trns.24934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haines NM, Kim YI, Smith AJ, Savery NJ. Stalled transcription complexes promote DNA repair at a distance. Proc Natl Acad Sci U S A. 2014;111:4037–4042. doi: 10.1073/pnas.1322350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Houten B, Kisker C. Transcriptional pausing to scout ahead for DNA damage. Proc Natl Acad Sci U S A. 2014;111:3905–3906. doi: 10.1073/pnas.1402020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990;54:18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wallace SS, Houten Bv, Kow YW. DNA damage: effects on DNA structure and protein recognition. New York Academy of Sciences; New York, NY: 1994. [Google Scholar]
- 78.Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 79.Kuo CF, Mol CD, Thayer MM, Cunningham RP, Tainer JA. Structure and function of the DNA repair enzyme exonuclease III from E. coli. Ann N Y Acad Sci. 1994;726:223–234. doi: 10.1111/j.1749-6632.1994.tb52820.x. discussion 234–225. [DOI] [PubMed] [Google Scholar]
- 80.Singh SB, Li B, Hingerty BE, Shapiro R, Broyde S. Molecular dynamics simulation of a tumorigenic benzo[a]pyrene metabolite bound to DNA at a single strand-double strand junction. Ann N Y Acad Sci. 1994;726:57–70. doi: 10.1111/j.1749-6632.1994.tb52798.x. [DOI] [PubMed] [Google Scholar]
- 81.Lee YC, Cai Y, Mu H, Broyde S, Amin S, Chen X, Min JH, Geacintov NE. The relationships between XPC binding to conformationally diverse DNA adducts and their excision by the human NER system: is there a correlation? DNA Repair. 2014;19:55–63. doi: 10.1016/j.dnarep.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, Geacintov NE. Probing for DNA damage with beta-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair. 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mu H, Kropachev K, Chen Y, Zhang H, Cai Y, Geacintov NE, Broyde S. Role of structural and energetic factors in regulating repair of a bulky DNA lesion with different opposite partner bases. Biochemistry. 2013;52:5517–5521. doi: 10.1021/bi4009177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mu H, Geacintov NE, Zhang Y, Broyde S. Recognition of damaged DNA for nucleotide excision repair: a correlated motion mechanism with a mismatched cis-syn thymine dimer lesion. Biochemistry. 2015;54:5263–5267. doi: 10.1021/acs.biochem.5b00840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koshland DE., Jr Molecule of the year: the DNA repair enzyme. Science. 1994;266:1925. doi: 10.1126/science.7801114. [DOI] [PubMed] [Google Scholar]
- 86.Zou Y, Bassett H, Walker R, Bishop A, Amin S, Geacintov NE, Van Houten B. Hydrophobic forces dominate the thermodynamic characteristics of UvrA–DNA damage interactions. J Mol Biol. 1998;281:107–119. doi: 10.1006/jmbi.1998.1903. [DOI] [PubMed] [Google Scholar]
- 87.Zou Y, Crowley DJ, Van Houten B. Involvement of molecular chaperonins in nucleotide excision repair, Dnak leads to increased thermal stability of UvrA, catalytic UvrB loading, enhanced repair, and increased UV resistance. J Biol Chem. 1998;273:12887–12892. doi: 10.1074/jbc.273.21.12887. [DOI] [PubMed] [Google Scholar]
- 88.Zou Y, Van Houten B. Strand opening by the UvrA(2)B complex allows dynamic recognition of DNA damage. EMBO J. 1999;18:4889–4901. doi: 10.1093/emboj/18.17.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zou Y, Walker R, Bassett H, Geacintov NE, Van Houten B. Formation of DNA repair intermediates and incision by the ATP-dependent UvrB-UvrC endonuclease. J Biol Chem. 1997;272:4820–4827. doi: 10.1074/jbc.272.8.4820. [DOI] [PubMed] [Google Scholar]
- 90.Salazar JJ, Van Houten B. Preferential mitochondrial DNA injury caused by glucose oxidase as a steady generator of hydrogen peroxide in human fibroblasts. Mutat Res. 1997;385:139–149. doi: 10.1016/s0921-8777(97)00047-5. [DOI] [PubMed] [Google Scholar]
- 91.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Theis K, Skorvaga M, Machius M, Nakagawa N, Van Houten B, Kisker C. The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch. Mutat Res. 2000;460:277–300. doi: 10.1016/s0921-8777(00)00032-x. [DOI] [PubMed] [Google Scholar]
- 94.Machius M, Henry L, Palnitkar M, Deisenhofer J. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc Natl Acad Sci U S A. 1999;96:11717–11722. doi: 10.1073/pnas.96.21.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakagawa N, Sugahara M, Masui R, Kato R, Fukuyama K, Kuramitsu S. Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J Biochem. 1999;126:986–990. doi: 10.1093/oxfordjournals.jbchem.a022566. [DOI] [PubMed] [Google Scholar]
- 96.Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C. Structural basis for DNA recognition and processing by UvrB. Nat Struct Mol Biol. 2006;13:360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 97.Karakas E, Truglio JJ, Croteau D, Rhau B, Wang L, Van Houten B, Kisker C. Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad. EMBO J. 2007;26:613–622. doi: 10.1038/sj.emboj.7601497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Truglio JJ, Rhau B, Croteau DL, Wang L, Skorvaga M, Karakas E, DellaVecchia MJ, Wang H, Van Houten B, Kisker C. Structural insights into the first incision reaction during nucleotide excision repair. EMBO J. 2005;24:885–894. doi: 10.1038/sj.emboj.7600568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Skorvaga M, DellaVecchia MJ, Croteau DL, Theis K, Truglio JJ, Mandavilli BS, Kisker C, Van Houten B. Identification of residues within UvrB that are important for efficient DNA binding and damage processing. J Biol Chem. 2004;279:51574–51580. doi: 10.1074/jbc.M409266200. [DOI] [PubMed] [Google Scholar]
- 100.Skorvaga M, Theis K, Mandavilli BS, Kisker C, Van Houten B. The beta-hairpin motif of UvrB is essential for DNA binding, damage processing, and UvrC-mediated incisions. J Biol Chem. 2002;277:1553–1559. doi: 10.1074/jbc.M108847200. [DOI] [PubMed] [Google Scholar]
- 101.Kisker C, Kuper J, Van Houten B. Prokaryotic nucleotide excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012591. doi: 10.1101/cshperspect.a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Van Houten B, Snowden A. Mechanism of action of the Escherichia coli UvrABC nuclease: clues to the damage recognition problem. Bioessays. 1993;15:51–59. doi: 10.1002/bies.950150108. [DOI] [PubMed] [Google Scholar]
- 103.Hanawalt PC, Haynes RH. Repair replication of DNA in bacteria: irrelevance of chemical nature of base defect. Biochem Biophys Res Commun. 1965;19:462–467. doi: 10.1016/0006-291x(65)90147-6. [DOI] [PubMed] [Google Scholar]
- 104.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single-molecule imaging of quantum-dot-labeled proteins. Mol Cell. 2010;37:702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Houten B, Kad N. Investigation of bacterial nucleotide excision repair using single-molecule techniques. DNA Repair. 2014;20:41–48. doi: 10.1016/j.dnarep.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grossman L. Damage recognition by UvrABC. A study of vectorial movement. Ann N Y Acad Sci. 1994;726:252–265. doi: 10.1111/j.1749-6632.1994.tb52823.x. discussion 265–256. [DOI] [PubMed] [Google Scholar]
- 107.Hughes CD, Wang H, Ghodke H, Simons M, Towheed A, Peng Y, Van Houten B, Kad NM. Real-time single-molecule imaging reveals a direct interaction between UvrC and UvrB on DNA tightropes. Nucleic Acids Res. 2013;41:4901–4912. doi: 10.1093/nar/gkt177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wolski SC, Kuper J, Hanzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie AM, Brown SE, Naismith JH, White MF. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuper J, Wolski SC, Michels G, Kisker C. Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 2012;31:494–502. doi: 10.1038/emboj.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lindsey-Boltz LA, Reardon JT, Wold MS, Sancar A. In vitro analysis of the role of replication protein A (RPA) and RPA phosphorylation in ATR-mediated checkpoint signaling. J Biol Chem. 2012;287:36123–36131. doi: 10.1074/jbc.M112.407825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Choi JH, Kim SY, Kim SK, Kemp MG, Sancar A. An integrated approach for analysis of the DNA damage response in mammalian cells: nucleotide excision repair, DNA damage checkpoint, and apoptosis. J Biol Chem. 2015 doi: 10.1074/jbc.M115.690354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci U S A. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kang TH, Reardon JT, Sancar A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res. 2011;39:3176–3187. doi: 10.1093/nar/gkq1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kang TH, Sancar A. Circadian regulation of DNA excision repair: implications for chrono-chemotherapy. Cell Cycle (Georgetown, Tex) 2009;8:1665–1667. doi: 10.4161/cc.8.11.8707. [DOI] [PubMed] [Google Scholar]
- 117.Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci U S A. 2011;108:18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lindsey-Boltz LA, Kemp MG, Reardon JT, DeRocco V, Iyer RR, Modrich P, Sancar A. Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J Biol Chem. 2014;289:5074–5082. doi: 10.1074/jbc.M113.542787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sancar A, Lindsey-Boltz LA, Gaddameedhi S, Selby CP, Ye R, Chiou YY, Kemp MG, Hu J, Lee JH, Ozturk N. Circadian clock, cancer, and chemotherapy. Biochemistry. 2015;54:110–123. doi: 10.1021/bi5007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Belin BJ, Lee T, Mullins RD. DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-(1/2) that promotes efficient DNA repair. eLife. 2015;4:10. doi: 10.7554/eLife.07735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hill RF. A radiation-sensitive mutant of Escherichia coli. Biochim Biophys Acta. 1958;30:636–637. doi: 10.1016/0006-3002(58)90112-4. [DOI] [PubMed] [Google Scholar]
- 123.Westerveld A, Hoeijmakers JH, van Duin M, de Wit J, Odijk H, Pastink A, Wood RD, Bootsma D. Molecular cloning of a human DNA repair gene. Nature. 1984;310:425–429. doi: 10.1038/310425a0. [DOI] [PubMed] [Google Scholar]
- 124.Tanaka K, Miura N, Satokata I, Miyamoto I, Yoshida MC, Satoh Y, Kondo S, Yasui A, Okayama H, Okada Y. Analysis of a human DNA excision repair gene involved in group A xeroderma pigmentosum and containing a zinc-finger domain. Nature. 1990;348:73–76. doi: 10.1038/348073a0. [DOI] [PubMed] [Google Scholar]
- 125.Cox BS, Parry JM. The isolation, genetics and survival characteristics of ultraviolet light-sensitive mutants in yeast. Mutat Res. 1968;6:37–55. doi: 10.1016/0027-5107(68)90101-2. [DOI] [PubMed] [Google Scholar]
- 126.Timmins J, Gordon E, Caria S, Leonard G, Acajjaoui S, Kuo MS, Monchois V, McSweeney S. Structural and mutational analyses of Deinococcus radiodurans UvrA2 provide insight into DNA binding and damage recognition by UvrAs. Structure. 2009;17:547–558. doi: 10.1016/j.str.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 127.Jaciuk M, Nowak E, Skowronek K, Tanska A, Nowotny M. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nat Struct Mol Biol. 2011;18:191–197. doi: 10.1038/nsmb.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kad NM, Van Houten B. Dynamics of lesion processing by bacterial nucleotide excision repair proteins. Prog Mol Biol Transl Sci. 2012;110:1–24. doi: 10.1016/B978-0-12-387665-2.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Friedberg EC. A history of the DNA Repair and Mutagenesis field – The discovery of base excision repair. DNA Repair. 2016;37:A35–A39. doi: 10.1016/j.dnarep.2015.12.003. [DOI] [PubMed] [Google Scholar]