Abstract
The addition of palmitoyl moieties to proteins regulates their membrane targeting, subcellular localization, and stability. Dysregulation of the enzymes which catalyzed the palmitoyl addition and/or the substrates of these enzymes have been linked to cancer, cardiovascular, and neurological disorders, implying these enzymes and substrates are valid targets for pharmaceutical intervention. However, current chemical modulators of zDHHC PAT enzymes lack specificity and affinity, underscoring the need for screening campaigns to identify new specific, high affinity modulators. This report describes a mixture based screening approach to identify inhibitors of Erf2 activity. Erf2 is the Saccharomyces cerevisiae PAT responsible for catalyzing the palmitoylation of Ras2, an ortholog of the human Ras oncogene proteins. A chemical library developed by the Torrey Pines Institute for Molecular Studies consists of more than 30 million compounds designed around 68 molecular scaffolds that are systematically arranged into positional scanning and scaffold ranking formats. We have used this approach to identify and characterize several scaffold backbones and R-groups that reduce or eliminate the activity of Erf2 in vitro. Here, we present the analysis of one of the scaffold backbones, bis-cyclic piperazine. We identified compounds that inhibited Erf2 auto-palmitoylation activity using a fluorescence-based, coupled assay in a high throughput screening (HTS) format and validated the hits utilizing an orthogonal gel-based assay. Finally, we examined the effects of the compounds on cell growth in a yeast cell-based assay. Based on our results, we have identified specific, high affinity palmitoyl transferase inhibitors that will serve as a foundation for future compound design.
Keywords: Acyltransferase, bis-cyclic piperazine, Erf2, enzyme inhibitor, high throughput screening (HTS), palmitoylation, zDHHC
1. INTRODUCTION
S-acylation is the enzymatic addition of a fatty acid (acyl) group onto one or more cysteine residues of a protein via a thioester linkage. The bulk of S-acylation involves the addition of a C16:0 carbon palmitoyl moiety and so we will refer to S-acylation as S-palmitoylation. A number of cellular processes involve the regulated addition of palmitate to proteins and includes signal transduction, protein turnover, vesicle fusion, and cell-cell interactions. At the protein level, the addition of palmitate enhances a protein’s membrane affinity as well as distribution in membrane micro-domains, mediates protein-protein interactions, trafficking, stability, and aggregation state [1]. While other protein lipidations, such as prenylation and myristoylation, are physiologically irreversible, the formation of the thioester linkage indicative of protein palmitoylation is reversible, and has led to a proposal that repeated rounds of acylation and de-acylation regulate substrate activity, localization and turn-over [2].
A family of protein acyl transferases (PATs) catalyzes the addition of a palmitoyl moiety to proteins. Genes encoding members of the PAT family have been identified in all sequenced eukaryotic genomes. This family of enzymes catalyze palmitoylation by a two-step reaction [3-6]. The first step, autopalmitoylation, results in the formation of the enzyme-palmitoyl intermediate via a thioester linkage between palmitate, donated from palmitoyl-CoA, and the active site cysteine of the enzyme. The palmitoyl moiety is then transferred from the enzyme to a receiver cysteine of the protein substrate in the second step of the reaction. In the absence of a protein substrate, water attacks the active site causing hydrolysis of the enzyme-palmitoyl complex thioester linkage, thus regenerating the enzyme and producing palmitic acid [3, 4].
Alterations in palmitoylation have been implicated in the etiology of cancer, cardiovascular disease, and neurological disorders [1, 7]. However, there are currently no drugs that target palmitoylation and the limited numbers of inhibitors that do exist exhibit low affinity and lack specificity. The most widely used inhibitor, 2-bromopalmitic acid (2-BP), is a non-metabolizable palmitate analog that elicits pleiotropic effects on cellular metabolism [8]. Despite a recent mass spectrometry study where its preference for palmitoylated substrates or PAT enzymes was not detectable, 2-BP conti-nues to be the primary experimental inhibitor of palmitoylat-ion in part due to the lack of a more suitable alternative [9]. Furthermore, 2-BP also inhibits the depalmitoylating thioesterase, Apt1 [10]. Thus, the need to identify specific, high affinity inhibitors of protein palmitoylation is critical for the progression of palmitoylation research, and for the regulation of palmitoylation for therapeutic intervention.
We have recently described a high-throughput screening technique for quantifying autopalmitoylation and will be applying that assay to a screening campaign for inhibitors of palmitoylation from a unique compound scaffolding chemical library. This approach allows for the interrogation of millions of compounds with only hundreds of reactions [11-13]. In the present study, we describe the use of this assay for the identification of a unique class of compounds, based on a bis-cyclic piperazine scaffold that inhibits the autopalmitoylation activity of the yeast Ras PAT, Erf2.
2. MATERIALS AND METHODS
2.1. Strains, Media, and Yeast Techniques
Yeast growth media were prepared as described previously [14]. Cells were grown in synthetic complete (SC) medium or YPD (1% yeast extract, 2% peptone, and 2% glucose) medium [14]. Induction of GAL1, 10 promoters were achieved by adding 4% galactose to SC medium in the absence of glucose. Yeast transformations were performed using the lithium acetate procedure [15]. Three yeast strains were used for this study: RJY1941 (S288C) MATα leu2-3,112 ura3-52 ade2 ade8 lys2 ras1::HIS3 Ras2(CS-ext) erf2Δ::KANr erg6Δ::TRP1 [YCp52-RAS2], RJY1942 (S288C) MATα leu2-3,112 ura3-52 ade2 ade8 lys2 ras1::HIS3 Ras2(CS-ext) erf2Δ::KANr erg6Δ::TRP1 [YCp52] and RJY1842 (MATa/α ade2-1/ade2-1 leu2-3,112/leu2-3,112 ura3-52/ura3-52 trp1-1/trp1-1 his3-11,15/his3-11,15 can1-100/can1-100 GAL+/GAL+ psi+/psi+ erf4Δ::NATr/erf4Δ::NATr [16].
2.2. Protein Purification
Strain RJY1842 was transformed with pESC(-Leu)-6xHIS-Erf2-(Flag)-Erf4 and grown to 2 x 107 cells/ml in SC(-Leu) medium containing 2% (v/v) ethanol/ 2% (v/v) glycerol at 30°C with shaking. 50 mls (1x109 cells) were added to 1 liter of YEP medium supplemented with 4% galactose for induction. Cells were induced with shaking (230 RPM) for 18 hrs (30°C) and then harvested by centrifugation at 3000xg for 15 mins. The resulting pellet was resuspended in breaking buffer (50 mM Tris-Cl pH 8, 500 ml NaCl, 1 mM EDTA, 1 mM DTT, 1xPIC, 8 µl/ml saturated PMSF/isopropyl alcohol), and the cells were lysed using glass beads (400-600 mesh, Sigma) for 40 mins with 1 min pulses (1 min cooling). The resulting extract was spun at 3000xg for 15 mins to remove cellular debris and unbroken cells, followed by a crude membrane fraction (P13) by centrifugation (13,000xg) for 0.5 hrs at 4°C. The supernatant was discarded and the pellet was resuspended in Tris buffered saline, pH 8, with the aid of a Dounce homogenizer. The resulting extract was adjusted to a final concentration of 1% Triton-X100. To solubilize the membranes, the extract was incubated at 4°C (1.5 hrs). Insoluble material was then removed by centrifugation (13,000xg) for 0.5 hrs at 4°C. The supernatant was incubated with Ni-NTA resin at 4°C for 1 hr. The resin was washed 3x with Solution W (50 mM Tris-HCl, pH 8, 150 mM NaCl and 1% Triton-X100). The protein was eluted with 50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% Triton-X100, 5% glycerol and 250 mM imidazole. Eluates were desalted and concentrated using 50 mM Tris-Cl, pH 8, 150 mM NaCl, 1% Triton-X100 and 5% glycerol. Fractions containing 6xHis-Erf2/FLAG-Erf4 were pooled to obtain approximately 0.2 mg of purified Ras PAT per liter of culture as determined by SDS-PAGE against a standard curve of bovine serum albumin. The complexes were divided into 50 µL aliquots and frozen at -80°C until use.
2.3. Coupled PAT Assay
The HTS application of protein palmitoylation was recently described [13], but was adjusted for this study. The production of NADH was monitored with a Biotek Mx fluorimeter (Biotek, Winooski, VT) using 340 nm excitation/ 465 nm emission. The 50 µl reaction contained 2 mM 2-Oxoglutarate (α-ketoglutamic acid), 0.25 mM NAD+, 0.2 mM Thiamine Pyrophosphate, 0.5 µg of purified 6xHIS-Erf2/FLAG-Erf4, 1 mM EDTA, 1 mM dithiothreitol, 8 mU 2-oxogluarate dehydrogenase (α-ketoglutarate dehydro-genase), 50 mM sodium phosphate, pH 6.8, and 0 - 100 µM inhibitor in 5% DMF. The reaction was initiated by the addition of 40 μM palmitoyl-CoA and monitored for 30 mins at 30°C. The first 10 mins of the reaction was analyzed to determine the initial rates of CoASH release. The PAT specific activity was determined from a standard curve of NADH production with different CoASH amounts. In these reactions, CoASH was added to the standard PAT reaction mixture (without Erf2/Erf4 complex or palmitoyl-CoA) and the reaction was allowed to proceed to equilibrium before fluorescence was measured.
2.4. Scaffold Ranking Library
The scaffold ranking library contains one sample for each of the 68 positional scanning libraries tested. Each of these samples contains an approximate equal molar amount of each compound in that library. So, for example, the sample 2160 in the scaffold ranking library contains 45,864 compounds in approximately equal molar amounts. These samples can be prepared by mixing the cleaved products of the complete positional scanning library, as was the case for sample 2160, or they can be synthesized directly as a single mixture [12, 17]. Refer to supplemental Material for a thorough description of the synthesis of the scaffold ranking library and subsequent individual compounds.
2.5. Positional Scanning Library 2160
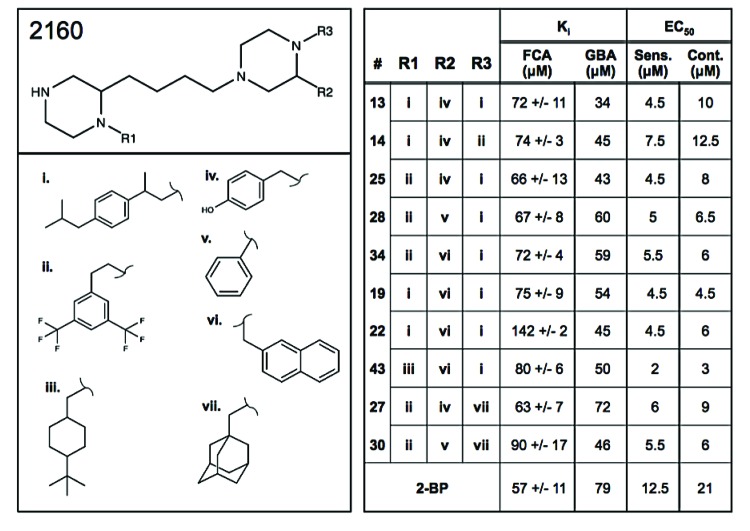
Positional scanning library 2160 was synthesized using the scheme described in Fig. (S1). The positional scanning library incorporates both individual and mixtures of amino acids (R2) and carboxylic acid (R1 and R3). The synthetic technique facilitates the generation of information regarding the likely activity of individual compounds from the screening of the library [11, 18, 19]. Equimolar isokinetic ratios have been previously determined and calculated for each of the amino and carboxylic acids utilized for the respective mixtures [17, 20]. The bis-piperazine library 2160 has a total diversity of 45,864 compounds (42x26x42= 45,864). The R1 and R3 positions as shown in Fig. (S1) each consist of 42 carboxylic acids and the R2 position contains 26 amino acids.
2.6. BODIPY®-C12:0 Autopalmitoylation Assay
BODIPY®-C12:0-CoA (40 µM final, unless specified otherwise) was added to a 50 µl reaction containing approximately 0.5 µg enzyme (6xHIS-Erf2/FLAG-Erf4) and 100 µM inhibitors in 5% DMF in 50 mM sodium phosphate buffer, pH 6.8. The reactions were incubated 10 mins with inhibitor, and then the reaction was initiated with the addition of BODIPY®-C12:0-CoA, and incubated 15 mins at 30°C. The reaction was terminated by the addition of 5x non-reducing protein loading buffer. Each reaction was heated at 65°C for three mins and then subjected to SDS-PAGE (12%). The gel was washed three times in ddH2O and visualized on the Typhoon 9410 Variable Mode Imager (GE Healthcare, Piscataway, NJ) for BODIPY® fluorescence (ex. 488 nm/em. 532 nm) to visualize co-migration of the BODIPY® signal with 6xHIS-Erf2/FLAG-Erf4. The amount of 6xHIS-Erf2/FLAG-Erf4 was determined empirically using SDS-PAGE analysis under reducing conditions against a standard curve of bovine serum albumin.
2.7. Growth Inhibition Assay
The in vivo effect of the inhibitors on Ras2 palmitoylation was investigated by comparing the growth of S. cerevisiae strains previously described for our complementation assay [16]. Briefly, the cells contain a defective allele of RAS2 that is balanced by an episomal copy of RAS2 linked to URA3. Under these conditions, the yeast strain cannot grow unless the episomal copy of RAS2 is palmitoylated. Varying concentrat-ions of the inhibitors were added to 200 µl volumes of the yeast cells at an OD600 between 0.8 and 1.2 in a 96-well plate format. The OD600 was observed every 30 mins for 24 hrs and EC50 values were determined by graphing rate of growth against concentration of inhibitor for each inhibitor.
2.8. Yeast Cell Spot Assay
Following 24 hrs incubation with varying concentrations of the inhibitors in 1% DMF, S. cerevisiae strains RJY1941 and RJY1942 were then diluted 1/100, and spotted onto SC-Ura plates with 2% glucose. Cytotoxicity data was obtained by detecting the colony growth, following 48 hrs incubation at 30°C, with white light detection on the Bio-Rad Molecular Imager® ChemiDocTM XRS+ (Hercules, CA) and performing densitometry with Bio-Rad ImageLabTM software (Hercules, CA). Triplicate reactions were plated in triplicate. Values were normalized to vehicle control (1% DMF) for each plate, and then the averages of each reaction were compared for statistical analysis.
3. RESULTS
3.1. Inhibition of Erf2 Autopalmitoylation using a Fluorescence-Based Coupled Assay
The identification of palmitoylation inhibitors has been hampered by the lack of assays amenable to high throughout screening applications. Previously, we validated an assay that monitors the rate of autopalmitoylation by measuring the production of CoASH generated from the reduction of palmitoyl-CoA [13]. This assay has a Z'-value of 0.87 [13]. The intention of this assay is to couple the amount of CoASH formed (and hence, enzyme:palmitoyl intermediate formed) to the production of NADH, which is fluorescent. This assay can be used to monitor autopalmitoylation in real time or as an end point assay, thus providing flexibility when screening small molecule compounds. Compounds with internal fluorescence or that affect the assay components are easily identified and excluded, allowing for the rapid identification of the compounds that inhibit or activate PAT autopalmitoylation activity.
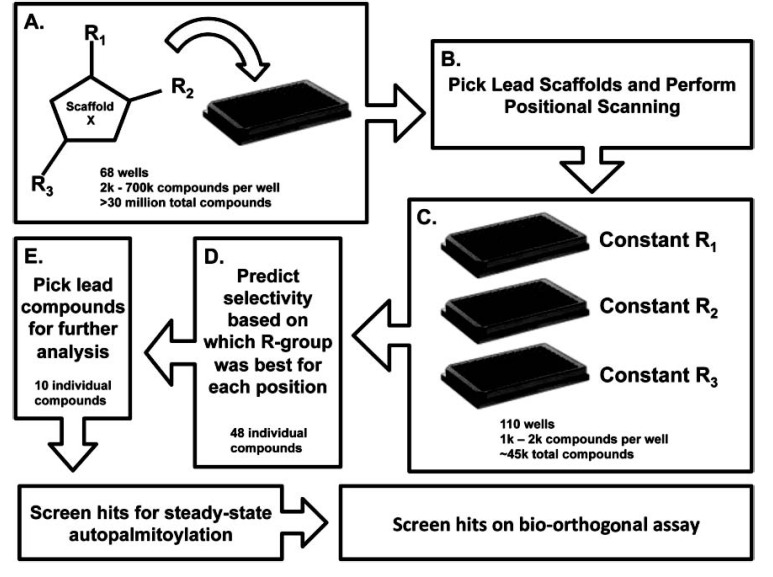
As an initial step in our screening campaign, we interrogated a scaffold ranking library developed by the Torrey Pines Institute for Molecular Studies [11-13]. This strategy allows for the evaluation of >30 million synthetic compounds while screening exponentially fewer reactions. This is achieved by organizing a large number of chemically diverse compounds into 68 core scaffolds. Each scaffold contains between 2,000 to 700,000 unique compounds at approximately equal molar concentrations. It is predicted that structural similarities will dictate additive effects increasing the chance that individual modulators will be detected despite being between nanomolar to sub-nanomolar concentrations [12, 21]. The vast number of structurally similar compounds in each scaffold library sample increases the probability of identifying a compound with useful chemical characteristics and properties [22]. Fig. (1) highlights a flow diagram of the stages involved in the identification and optimization of the compounds. We first screened the 68 scaffolds to determine the optimal scaffold structure (Fig. 1A). Once identified, positional scanning libraries of the lead scaffolds, which are organized by R-groups at each position around a given scaffold, are screened (Fig. 1B). This allows for the prediction of optimal R-groups for each position on the core scaffold (Fig. 1C). Based on the optimized scaffold and knowledge of the active R-groups, individual compounds can be designed and synthesized to determine the positional effects of the R-groups on activity (Fig. 1D). Finally, based on the level of inhibition, we selected a subset of lead compounds for further analysis (Fig. 1E). This approach allows us to expedite the screening process of millions of compounds and to generate structure activity relationship (SAR) information earlier in the screening process. A description of the library construction and use can be found in recent reviews [11, 22-25].
Fig. (1).
Schematic representation of the chemical library screen. A, The Torrey Pines Institute for Molecular Studies scaffold library approach starts with >30 million compounds that are organized by 68 core scaffolds with 2,000 – 700,000 different compounds per scaffold. B, A lead scaffold is selected for the positional scanning screen. C, All of the compounds in the positional scanning screen contain the same core scaffold structure and are organized by the R-groups at each position. Each of the plates contains the same compounds organized by the different R-group position to determine if a particular functional group is optimal at one of the positions. The positional scanning library screened in this study contained 110 positional scanning samples that each comprise of 1,000 – 2,000 individual compounds for a total diversity of 45864 individual compounds. D, The selectivity for the different R-groups is predicted for each position based on the positional scanning results. In this study, 48 individual compounds were synthesized. E, Of the 48 individual compounds synthesized ten were selected for further analysis in additional assays. In total 226 samples were tested; 68 Scaffolds, 110 positional scanning scaffolds, 48 individual compounds.
3.2. Scaffold Ranking of the Erf2 Autopalmitoylation Inhibitors
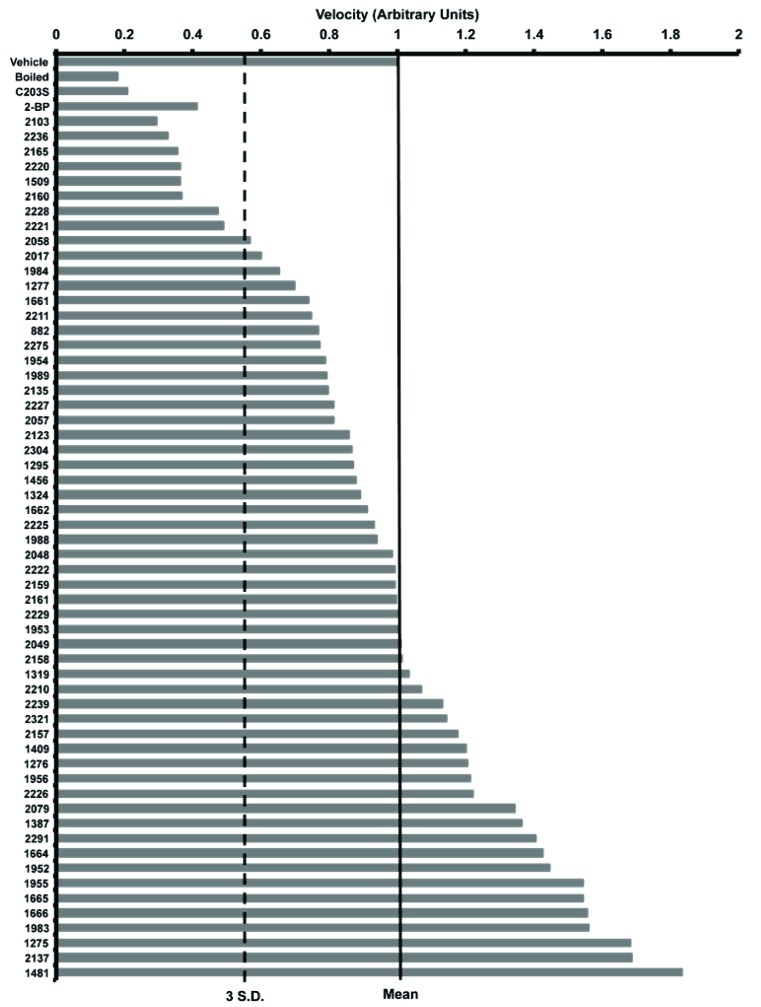
We screened 68 scaffold ranking samples, each at a concentration of 100 μg/ml/scaffold (Fig. 2). Each scaffold sample was pre-incubated with Erf2 enzyme for 10 mins at 30°C before initiating the reaction with the addition of palmitoyl-CoA. We monitored the production of CoASH for 30 mins to ensure a linear response over time and determined the rate of the reaction for each scaffold sample. Incubation of the enzyme with several of the scaffold samples resulted in a reduction in the production of CoASH. As an initial criterion, we defined inhibition as a reduction in activity greater than three standard deviations from the mean activity value. We identified eight of the scaffold libraries that fit this definition. Scaffolds 2103, 2236, 2165, 2220, 1509, 2160, 2228, and 2221 caused at least a 50% reduction in Erf2 autopalmitoylation activity. In contrast, scaffold 1509 appeared to react with the assay components and was excluded from further analysis. The data from two of the scaffolds, 2165 and 2228, were not reproducible and these scaffolds were also excluded (data not shown). We established a baseline of inhibition using boiled and catalytically inactive enzyme.
Fig. (2).
Results of the screen for inhibition of Erf2 auto-palmitoylation. The scaffold ranking library screen of 68 scaffolds was screened at 100 μg/ml, and the average velocity of Erf2 auto-palmitoylation of three reactions is presented as a fraction of vehicle control (1% DMF). The velocity of Erf2 auto-palmitoylation was detected as an increase in fluorescence over time. No effect on Erf2 auto-palmitoylation would fall at 1 arbitrary unit (solid line). Scaffolds that resulted in a reduction in Erf2 auto-palmitoylation 3 standard deviations (dashed line) or greater were considered hits of this assay. A heat inactivated Erf2 (Boiled) and a catalytically inactive mutant of Erf2 (C203S) represent baseline activity in this assay. 100 μM 2-BP is a control for inhibition of Erf2 auto-palmitoylation.
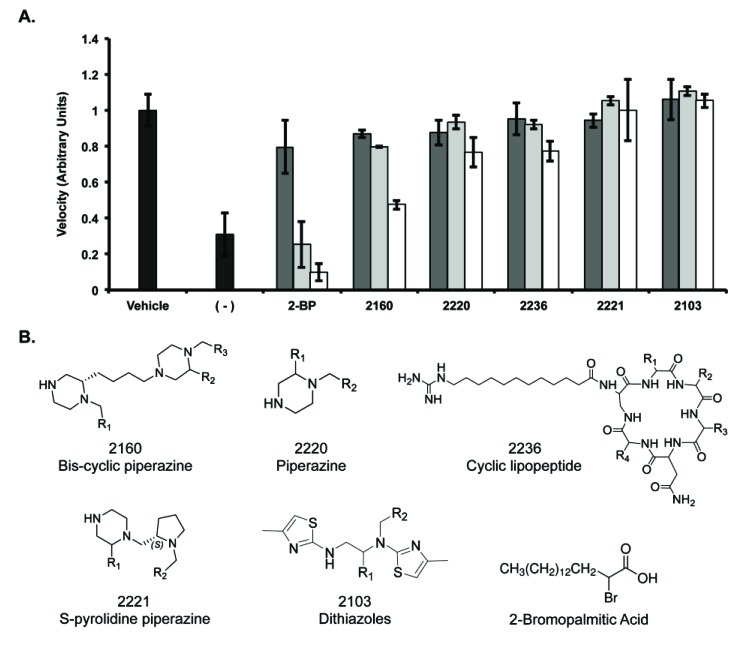
The remaining 5 scaffolds were then screened for dose responsiveness at 200 μg/ml, 100 μg/ml, and 50 μg/ml (Fig. 3A). Similar to the dosage response observed for 2-BP, three of the five scaffold libraries (2160, 2220 and 2236) demonstrated some degree of dosage response at these concentrations. Interestingly, these three scaffold libraries included a piperazine-analog feature. Scaffold 2220 is a piperazine, scaffold 2221 is a pyrolidine piperazine, and scaffold 2160 is a bis-cyclic piperazine (Fig. 3B). Of these, scaffold 2160 reproducibly decreased PAT activity in a dose dependent fashion to the greatest extent. Structurally, scaffold 2160 has a core bis-cyclic piperazine with three R-groups, and a total diversity of 45864 compounds (Fig. 1B).
Fig. (3).
Inhibition of Erf2 auto-palmitoylation by varying the concentration of library samples. A, The velocity of Erf2 auto-palmitoylation on the coupled assay with the lead scaffold ranking library samples (2160, 2220, 2236, 2221, 2103). The velocity of Erf2 auto-palmitoylation was detected as an increase in fluorescence over time. Average values of three reactions are presented as a fraction of vehicle control (1% DMF) +/- standard deviation. Samples were screened at 50 μg/ml (dark grey bars), 100 μg/ml (light grey bars), and 200 μg/ml (white bars) compared to 2-BP at 50 μM (dark grey bars), 100 μM (light grey bars), and 200 μM (white bars). A reaction lacking Erf2 (-) represents baseline activity in the assay. B, Structures of lead scaffolds and 2-Bromopalmitic Acid. Positions for varying functional groups are denoted by R1-R4. Scaffold 2160 contains three R-group positions, scaffolds 2220, 2221, and 2103 each contain two R-group positions, and scaffold 2236 contains four R-group positions.
The 45864 compounds in scaffold 2160 were synthesized into 110 mixture samples organized by R-group (Fig. 1C). At position R1 and R3 there were 42 functionalities derived from different carboxylic acids (samples 2160.001-2160.042 and 2160.069-2160.110, respectively), and 26 derived from amino acids at the R2 position (samples 2160.043-2160.068). Thus, samples organized by R1 or R3 positions contain 1092 (42 x 26) compounds per sample, and samples organized by the R2 position contain 1764 (42 x 42) compounds per sample. The 110 samples were screened for inhibition of Erf2 autopalmitoylation at a concentration of 100 μg/ml. Ninety-nine of the 110 mixture samples had no significant effect on Erf2 autopalmitoylation, where 25% inhibition was considered the cutoff for significance (data not shown). Four functional groups at position R1, three at position R2 and four at position R3, resulted in a greater than 25% reduction and were selected for synthesis of 48 individual compounds.
3.3. Individual Compounds Screened for Inhibition of Erf2 Autopalmitoylation
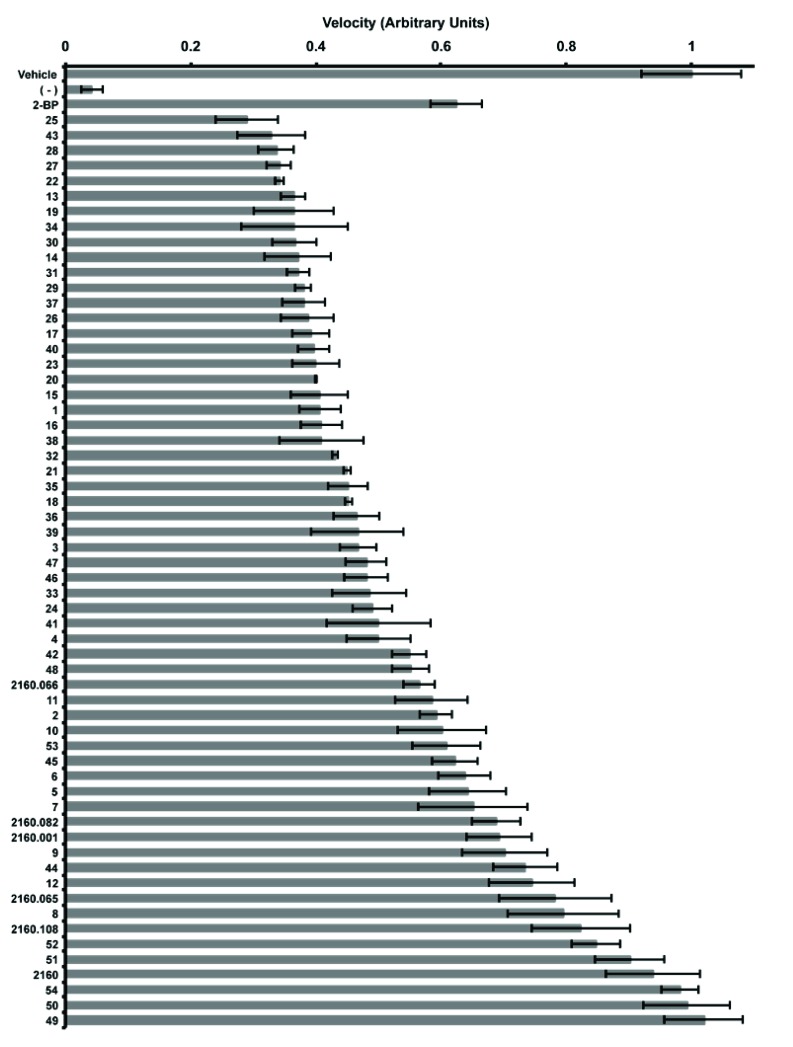
The 48 compounds were screened with five of the hits from the positional scanning samples (2160.066, 2160.082, 2160.001, 2160.065, and 2160.108) and the complete 2160-scaffold sample (Fig. 4). Scaffold 2160 and the positional scanning samples inhibited Erf2 autopalmitoylation consistent with prior screens and demonstrated that as the complexity decreased from 45864 compounds (scaffold 2160), to 1000-2000 compounds (positional scanning), and finally, to individual compounds, there was a concomitant increase in the inhibition of Erf2 autopalmitoylation (Fig. 4). As an additional control, we included non-optimized compounds (compounds 49-54), which were designed with side groups that did not pass the original cutoff in the positional scanning screen. These compounds, as anticipated, demonstrated poor inhibition of Erf2 autopalmitoylation in comparison to the compounds whose design was based on
Fig. (4).
Inhibition of Erf2 auto-palmitoylation by individual compounds derived from the 2160 scaffold. Relative velocity of Erf2 auto-palmitoylation with 100 μg/ml of optimized individual compounds (1-48), non-specific individual compounds (49-54), positional scanning samples (2160.066, 2160.082, 2160.001, 2160.065, and 2160.108), and lead scaffold ranking library sample (2160). The velocity of Erf2 auto-palmitoylation was detected as an increase in fluorescence over time. Average values of three reactions presented as a fraction of vehicle control (1% DMF) +/- standard deviation. A reaction lacking Erf2 (-) represents baseline activity in the assay. 50 μM 2-BP is a control for inhibition of Erf2 auto-palmitoylation.
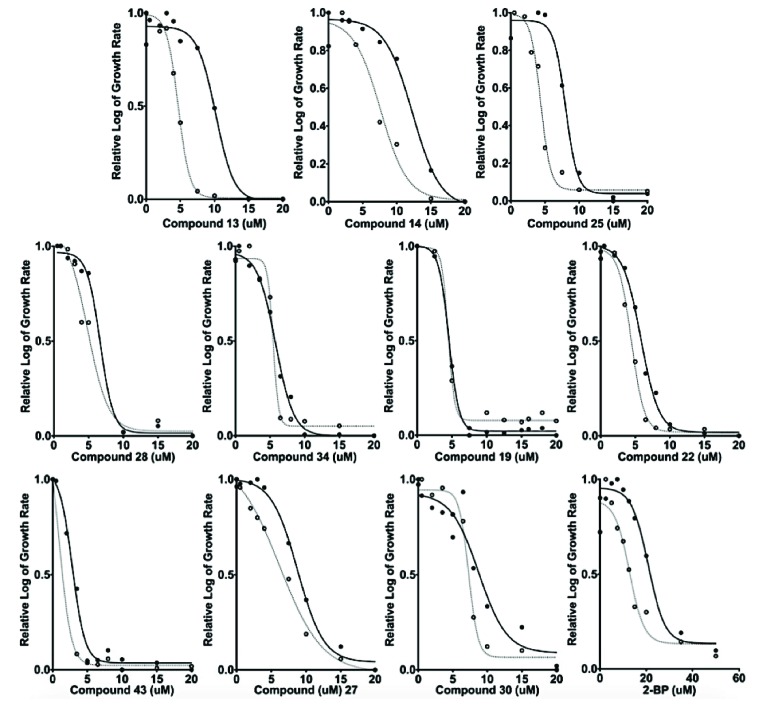
the positional scanning results. We determined the inhibition constant (Ki) for the top ten compounds by determining the rate of autopalmitoylation activity for varying amounts of inhibitor compound (data not shown). The autopalmitoylat-ion inhibition constants ranged from ~63-142 µM for the ten compounds identified in this study, and 59 μM for 2-BP (Fig. 5, right panel).
There did not appear to be a preference within the top 40 compounds for the R2 position amongst the four possibilities; hydroxybenzyl, phenyl, and (S or R) naphthymethyl groups (Fig. 5). There were three possibilities at the R3 position, and it was at this position that the greatest preference was observed. 2-(4-isobutyl-phenyl)-propyl was the optimal group at this position and is present in seven of the top ten hits. 2-(3,5-bis-trifluoromethyl-phenyl)-ethyl is the most prevalent in the next 15, and the completely aliphatic functional group, adamantan-1-yl-methyl, is in this position for a majority of the remaining top 40 individual
Fig. (5).
Structure of the scaffold 2160 derived compounds. The structure for scaffold 2160, bis-cyclic piperazine has three R-group positions designated R1, R2, and R3. The lead functional groups are 2-(4-Isobutyl-phenyl)-propyl (i), 2-(3,5-bis-trifluoromethyl-phenyl)-ethyl (ii), 4-tert-butyl-cyclohexyl-methyl (iii), S-4-hydroxybenzyl (iv), S-phenyl (v), S-2-naphthylmethyl (vI), and adamantan-1-yl-methyl (vii). The curved lines on the functional group structures designate where they will share a bond with the core scaffold. Positions for each functional group are described in the right panel. Ki values for the fluorescence-based coupled assay of the rate of Erf2 autopalmitoylation (FCA) and the orthogonal gel-based assay of steady-state Erf2 autopalmitoylation (GBA) are listed for each compound (14, 13, 25, 28, 34, 19, 22, 43, 27, and 30) as well as EC50 values obtained from the growth curve experiments in S. cerevisiae RJY1942 (Sens.) and RJY1941 (cont.). All values are represented in μM concentrations. The increased detection sensitivity of the FCA allowed for the determination of standard deviation.
compounds. The R1 position also showed similar specificity for the functional groups utilized in the individual compounds. At the R1 position, nearly all of the top 30 compounds have either 2-(3,5-bis-trifluoromethyl-phenyl)-ethyl or 2-(4-isobutyl-phenyl)-propyl, the same top two groups for the R3 position. A >50% reduction in Erf2 autopalmitoylation was observed for most of the optimized individual compounds, but only the top ten were selected for additional analysis.
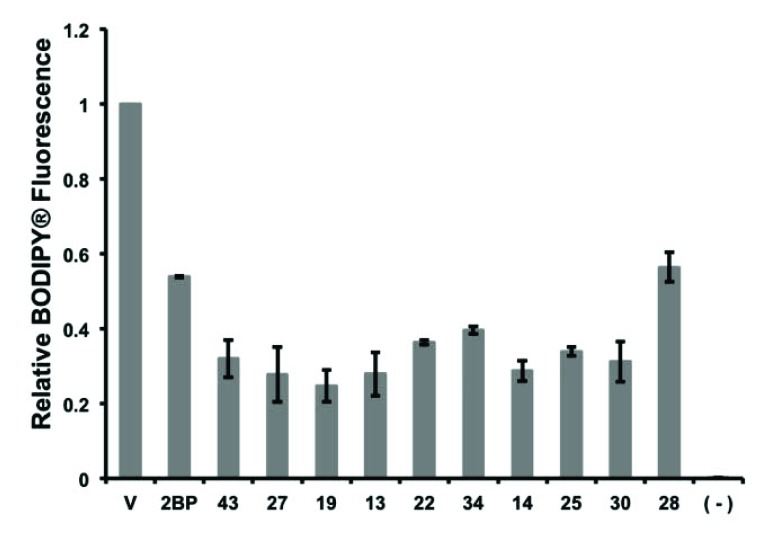
To determine if the ten compounds identified in this study were inhibiting autopalmitoylation, and not targeting hydrolysis of the enzyme, steady-state autopalmitoylation was evaluated at 100 μM of each compound compared to 100 μM 2-BP and DMF (vehicle) alone. This was performed in a gel-based reaction using BODIPY®-C12-CoA [26]. The reactions were separated by SDS-PAGE and steady-state autopalmitoylation was determined by the relative BODIPY® fluorophore that co-migrated with Erf2, demonstrating that Erf2 was acylated by BODIPY®-C12 [26]. At 100 μM, the ten compounds each resulted in a 50% or greater decrease in steady-state Erf2 autopalmitoylation compared to vehicle alone. From these screens we observed a decrease in BODIPY® fluorescence at the apparent
molecular weight of Erf2. This does not confirm that the inhibitors are directly interacting with Erf2 as they could be reducing the stability of the Erf2-Erf4 complex. Taking that into consideration, all of the compounds inhibited autopalmitoylation to either an equal or greater extent compared to 2-BP (Fig. 6). Dose response curves of steady-state autopalmitoylation activity were examined to determine the Ki for each compound (data not shown). The steady-state autopalmitoylation inhibition constants ranged from ~34-72 µM for the ten compounds identified in this study, and 79 μM for 2-BP (Fig. 5, right panel).
Fig. (6).
Inhibition of Erf2 auto-palmitoylation measured by a gel-based assay. An orthogonal screen of steady-state Erf2 auto-palmitoylation with 100 μM of each of the ten individual compounds (43, 27, 19, 13, 22, 34, 14, 25, 30, and 28) was separated by SDS-PAGE. The average relative BODIPY® fluorescence from three reactions that co-migrated with Erf2 is presented as a fraction of vehicle control (V; 5% DMF) +/- standard deviation. A reaction lacking Erf2 (-) represents baseline activity in the assay. 100 μM 2-BP is a control for inhibition of Erf2 steady-state auto-palmitoylation.
3.4. Compounds 13 and 25 Competitively Inhibit Erf2 Autopalmitoylation
The potential for the symmetry of the compounds to play a role in their efficacy is of interest as the same functional groups were identified for the top two leads at both positions R1 and R3, and positions R1 and R3 are located on nitrogens of the opposing piperazine ring structures on the bis-cyclic piperazine scaffold. To interrogate this, the compounds 13, 14, and 25 were selected for further analysis as they represent a unique set of three compounds where all three have the same functional group at position R1, compound 13
has the lead functional group, 2-(3,5-bis-trifluoromethyl-phenyl)-ethyl, at positions R1 and R3, compound 14 also has 2-(3,5-bis-trifluoromethyl-phenyl)-ethyl at position R1, but has the second lead functional group, 4-tert-butyl-cyclo-hexyl-methyl at the R3 position, and compound 25 has the reverse; 4-tert-butyl-cyclohexyl-methyl at the R1 position
and 2-(3,5-bis-trifluoromethyl-phenyl)-ethyl at position R3. Thus, compound 13 represents a compound with symmetry and compounds 14 and 25 are complementary non-symmetrical representatives with the same alternative functional group at opposite positions. Of the ten lead compounds, these three are the only ones to satisfy this scenario.
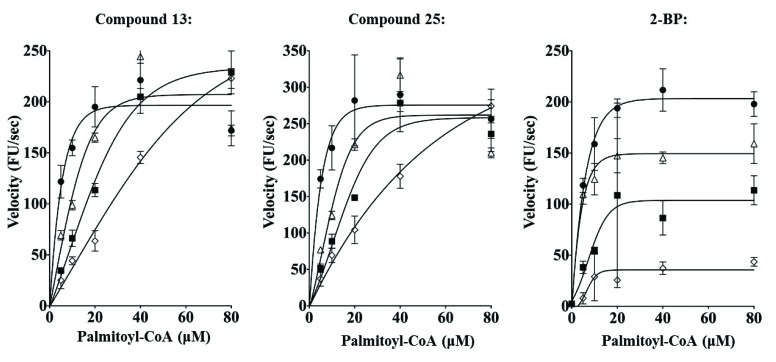
To first elucidate if these compounds were mechanistically different from 2-BP, we performed inhibitor titrations, varying the amount of compound 13 (or compound 25) and palmitoyl-CoA. Over the concentration range tested, compounds 13 and 25, increasing inhibitor concentration increased the KM (palmitoyl-CoA) of the reaction while having little effect on the VMAX. (Fig. 7). It was not possible to increase palmitoyl-CoA concentrations above 80 µM due to the CMC. The KM and VMAX values calculated are summarized in Table 1.
Fig. (7).
Michaelis-Menten plots for Compounds 13 and 25, and 2-BP. The velocity of Erf2 auto-palmitoylation as altered by the addition of the inhibitors at 100 μM (empty diamonds), 50 μM (filled squares), 25 μM (empty triangles), and vehicle control (1% DMF; filled circles) for compounds 13 and 25, and 2-BP. The velocity of Erf2 auto-palmitoylation was detected as an increase in fluorescence over time. VMAX is the maximum velocity of the reaction, and KM is the substrate concentration that allows the reaction to reach a velocity that is 50% of the VMAX. On the Michaelis-Menton Plot, a competitive inhibitor would increase KM without altering VMAX, whereas an uncompetitive inhibitor would result in a decrease of VMAX without altering the KM of the reaction.
Table 1.
The effect of varying the concentration of Compounds 13 and 25, and 2-BP on the Km and Vmax for the Palmitoyl-CoA substrate.
| Compound 13 | KM | VMAX | R2 |
|---|---|---|---|
| (μM) | (μM) | (μM) | |
| 0 | 3 +/- 1 | 210 +/- 11 | 0.9 |
| 25 | 11 +/- 3 | 243 +/- 20 | 0.87 |
| 50 | 38 +/- 6 | 354 +/- 26 | 0.97 |
| 100 | 145 +/- 30 | 631 +/- 94 | 0.98 |
| Compound 25 | KM | VMAX | R2 |
| (μM) | (μM) | (μM) | |
| 0 | 3 +/- 1 | 296 +/- 17 | 0.88 |
| 25 | 11 +/-3 | 307 +/- 30 | 0.83 |
| 50 | 23 +/- 6 | 344 +/- 34 | 0.91 |
| 100 | 79 +/- 15 | 542 +/- 60 | 0.98 |
| 2-BP | KM | VMAX | R2 |
| (μM) | (μM) | (μM) | |
| 0 | 4 +/- 1 | 222 +/-7 | 0.96 |
| 25 | 2 +/-1 | 160 +/-5 | 0.95 |
| 50 | 10 +/-6 | 126 +/- 23 | 0.53 |
| 100 | 14 +/- 7 | 51 +/-8 | 0.71 |
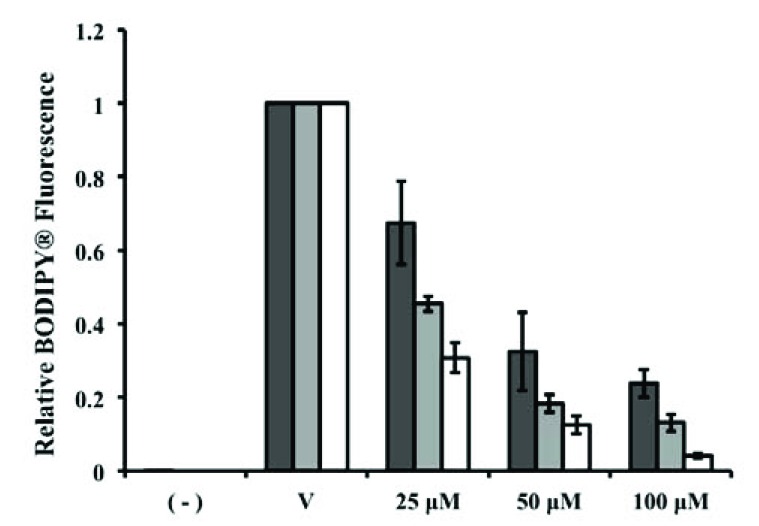
The mode of inhibition was also explored for effects on steady-state autopalmitoylation using the gel-based orthogonal assay. Compound 13 exhibits a greater dosage response for Erf2 autopalmitoylation inhibition at lower BODIPY®-C12-CoA concentrations rather than at higher BODIPY®-C12-CoA concentrations, supporting that compound 13 is acting competitively with BODIPY®-C12-CoA (Fig. 8). Conversely, 2-BP has been previously demonstrated as an uncompetitive or mixed inhibitor of Erf2 autopalmitoylation [13], demonstrating that compounds 13 and 25 utilize a different inhibitory mechanism compared to 2-BP. Thus, we have identified a new class of palmitoylation inhibitors that are structurally distinct and utilize a mode of action different from the currently used palmitoylation inhibitor, 2-BP.
Fig. (8).
Inhibition of palmitoyl-CoA concentration dependent palmitoylation of Erf2 by Compound 13. Orthogonal screen of steady-state Erf2 auto-palmitoylation with compound 13 at 25 μM (dark grey bars), 50 μM (light grey bars) and 100 μM (white bars) was separated by SDS-PAGE. The average relative BODIPY® fluorescence from three reactions that co-migrated with Erf2 is presented as a fraction of vehicle control (VC; 5% DMF) for each concentration of BODIPY®-C12-CoA +/- standard deviation. A reaction lacking Erf2 (-) represents baseline activity in the assay. Prior experiments in this study used 100 μM Compound 13 with 40 μM BODIPY®-C12-CoA.
3.5. Inhibition of Ras-Dependent Growth in a Palmitoylation-Sensitive S. cerevisiae Strain
To examine the inhibition of Ras palmitoylation in vivo, we utilized two strains of S. cerevisiae. The yeast strain RJY1942 (palmitoylation sensitive) requires Ras2 palmitoylation for viability [16]. The isogenic wild type control yeast strain (RJY1941) is not dependent on Ras2 palmitoylation for viability. The growth of both strains was monitored in the presence (or absence) of our inhibitors every 30 mins for 24 hrs. EC50 values were calculated using GraphPad Prism® (La Jolla, CA) to extrapolate the effective concentration of each inhibitor that caused a 50% reduction in growth (Fig. 9). EC50 values ranged from 2 μM to 7.5 μM in the sensitive strain, compared to 12.5 μM for 2-BP in the sensitive strain. Compounds 13, 25, and 2-BP show a modest selectivity for the sensitive strain, RJY1942, over the control strain, RJY1941, resulting in a 2-fold greater EC50 value in the control strain as compared to the sensitive strain (values listed in Fig. 5, right panel). It is not clear at this time why only compounds 13 and 25 exhibit selectivity in the cell based assay, whereas there is the others inhibit the purified enzyme to a similar extent. Additional work is needed to determine the mechanism of growth inhibition and selectivity of these compounds.
Fig. (9).
Inhibition of Ras palmitoylation-sensitized yeast. The EC50 for each of the individual compounds was established by graphing the logarithmic growth of S. cerevisiae strain, RJY1941 (solid lines and solid circles) and RJY1942 (hashed lines and open circles), versus the concentration of each compound. The curves were used to estimate the EC50 values are presented for 2-BP and the ten compounds identified in this study. Values were normalized to visually compare relative log of the growth rate of each compound and 2-BP.
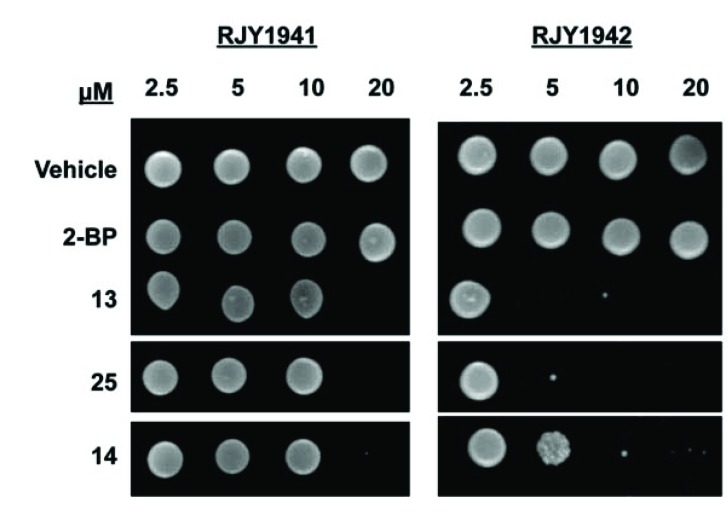
We next compared the ability of either strain to grow following the 24 hrs incubation with Compounds 13, 14, or 25. Serial dilutions of RJY1941 and RJY1942 were spotted on agar plates following incubation for 24 hrs with varying inhibitor concentrations. Compounds 13, 14, and 25 comple-tely inhibited growth of RJY1941 (control) at 20 µM, but had no detectable effect at lower concentrations (Fig. 10). Conversely, compound 13 inhibited (100%) the growth of RJY1942 (sensitized) at 5 µM. Compound 25 also inhibited at 5 μM, albeit at approximately 90% of that observed for compound 13 for the sensitized strain. Compound 14 demonstrated partial inhibition (approximately 10%) at 5 µM, however, total inhibition could be observed for the sensitized strain at higher inhibitor concentrations. 2-BP did not inhibit, most likely due to poor permeability of 2-BP on solid yeast medium.
Fig. (10).
Compounds 13, 25, and 14 inhibit palmitoylated Ras-dependent growth in yeast. A representative (n=3) spot assay of cytotoxic effects in wild type (RJY1941) S. cerevisiae is compared to that of the palmitoylation-dependent (RJY1942) strain. Liquid cultures containing the shown inhibitors were spotted onto selective media and incubated at 30°C for 3 days. The concentrations used in the liquid cultures for compounds 13, 25, 14 and 2-BP were 2.5 μM, 5 μM, 10 μM, and 20 μM in 1% DMF.
4. DISCUSSION
Protein S-palmitoylation is a posttranslational modification that regulates the subcellular localization and activity of a diverse set of signaling and structural proteins. Dysregulation of protein palmitoylation has been linked to a number of diseases. Examples include up-regulation of zDHHC9 in colorectal cancer [27] and down-regulation of zDHHC9 in leukemogenesis [28]; zDHHC3/GODZ is linked to cervical cancer [29] and zDHHC2 mutations have been found in several colorectal cancers [30]. The importance of palmitoylation in physiology and pathophysiology suggests that modulators of catalyzed palmitoyl transfer may play a role in disease treatment. The availability of PAT inhibitors is very limited. 2-Bromo-palmitate (2-BP), cerulenin, and tunicamycin have been reported to inhibit palmitoylation [8, 31-33], but 2-BP is now known to be highly promiscuous, with no preference for CoA-dependent enzymes, including zDHHC PATs [9]. Similarly, cerulenin and tunicamycin inhibit palmitoylation within cells but also inhibit other cellular process including fatty acid synthesis [34] and N-glycosylation [35], respectively. In addition, there are few assays available that can measure palmitoylation [3, 13] in a high throughput platform. There is therefore a need to establish methods to identify small molecule PAT inhibitors for use in vitro and in vivo.
The current lack of high affinity, specific inhibitors is due in part to difficulties purifying biochemical quantities of PATs, limited information on the enzymatic mechanism and the lack of a 3D crystal structure. We have developed methods to purify stable, active zDHHC PATs, along with a
validated HTS method to identify inhibitors. This has allowed the search for chemical modulators of PAT enzymes using HTS coupled with counter screens, orthogonal validation and cell-based assays, to characterize and analyze candidate inhibitors. With these tools in hand we are able to identify compounds that regulate the Ras PAT, Erf2.
Through the use of a scaffold ranking approach to screen for novel inhibitors of Erf2 autopalmitoylation, we have identified a group of inhibitors based on the bis-cyclic piperazine backbone. Piperazine analogues have already been demonstrated to be effective drug-like compounds.
They are currently used as pharmaceutical modulators of GPCR activity and piperazine-like compounds and analogs are also used as pharmaceuticals to treat cancer, behavioral disorders, and insomnia (e.g., imatinib, quetiapine, and eszopiclone) [36-38]. To further explore their specificity for use as modulators of palmitoylation, there is ongoing work to address any inhibitor:enzyme specificity issues by establishing if the inhibitors identified in this study act specifically on Erf2-dependent palmitoylation, or if they act on one or more of the other PAT enzymes. Further analysis will be needed to validate these compounds as pan inhibitors of palmitoylation, which would be beneficial over 2-BP due to the extent of non-specific inhibition that occurs with the use of 2-BP. Specific inhibitors of Erf2 and its human homolog, zDHHC9, will be beneficial for anti-cancer drug therapy interventions. Understanding how the different functional groups lend to their activity and specificity is key to further optimize these compounds. The compounds best align with the Ki and EC50 values from the respective screens when they were organized by the R3 position with consideration of the R1 and R2 positions (Fig. 5). Of the functionalities used at the R1 and R3 positions both 2-(4-Isobutyl-phenyl)-propyl and 2-(3,5-bis-trifluoromethyl-phenyl)-ethyl were the most preferential for both positions. This, along with the apparent symmetry of bis-piperazine, suggests that perhaps these compounds can act as symmetrical entities. The lack of preference, other than for an aromatic group, in the R2 position supports this notion.
CONCLUSION
We have identified a novel class of bis-cyclic piperazines that inhibit Erf2 autopalmitoylation utilizing a fluorescence-based coupled assay, orthogonal gel-based assay and cell growth assay. A subset of these bis-cyclic piperazines shows Ras-dependent inhibition of yeast cells in vivo.
ACKNOWLEDGeMENTS
We thank Krishna D. Reddy for critical reading of the manuscript. This work was funded in part through the Florida Drug Discovery Acceleration Program by the State of Florida Department of Health, and NIH CA502-17 and a Fred Wright Jr. Cancer Research Endowment.
ABBREVIATIONS
- 2-BP
2-bromopalmitic acid
- APT
Acyl protein thioesterase
- CoA
Coenzyme A
- DCM
Dichloromethane
- DIC
Diisopropylcarbodiide
- DIEA
Diisopropylethylamine
- DMF
Dimethylformamide
- HOBt
1-hydroxybenzotriazole hydrate
- HTS
High throughput screen
- MBHA
p-Methylbenzyhydrylamine
- MEOH
Methanol
- PAT
Protein acyl transferase
- TFA
Trifluoroacetic acid
- THF
Tetrahydrofuran
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Chamberlain L.H., Shipston M.J. The physiology of protein S-acylation. Physiol. Rev. 2015;95:341–376. doi: 10.1152/physrev.00032.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rocks O., Peyker A., Kahms M., Verveer P.J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., Bastiaens P.I. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell D.A., Mitchell G., Ling Y., Budde C., Deschenes R.J. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J. Biol. Chem. 2010;285:38104–38114. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jennings B.C., Linder M.E. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J. Biol. Chem. 2012;287:7236–7245. doi: 10.1074/jbc.M111.337246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohno Y., Kihara A., Sano T., Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. 2006. [DOI] [PubMed]
- 6.Roth A.F., Wan J., Bailey A.O., Sun B., Kuchar J.A., Green W.N., Phinney B.S., Yates J.R., III, Davis N.G. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeste-Velasco M., Linder M.E., Lu Y.J. Protein S-palmitoylation and cancer. Biochim. Biophys. Acta. 1856;2015:107–120. doi: 10.1016/j.bbcan.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Webb Y., Hermida-Matsumoto L., Resh M.D. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 9.Davda D., El Azzouny M.A., Tom C.T., Hernandez J.L., Majmudar J.D., Kennedy R.T., Martin B.R. Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem. Biol. 2013;8:1912–1917. doi: 10.1021/cb400380s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedro M.P., Vilcaes A.A., Tomatis V.M., Oliveira R.G., Gomez G.A., Daniotti J.L. 2-Bromopalmitate reduces protein deacylation by inhibition of acyl-protein thioesterase enzymatic activities. PLoS One. 2013;8:e75232. doi: 10.1371/journal.pone.0075232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houghten R.A., Pinilla C., Giulianotti M.A., Appel J.R., Dooley C.T., Nefzi A., Ostresh J.M., Yu Y., Maggiora G.M., Medina-Franco J.L., Brunner D., Schneider J. Strategies for the use of mixture-based synthetic combinatorial libraries: scaffold ranking, direct testing in vivo, and enhanced deconvolution by computational methods. J. Comb. Chem. 2008;10:3–19. doi: 10.1021/cc7001205. [DOI] [PubMed] [Google Scholar]
- 12.Santos R.G., Appel J.R., Giulianotti M.A., Edwards B.S., Sklar L.A., Houghten R.A., Pinilla C. The mathematics of a successful deconvolution: a quantitative assessment of mixture-based combinatorial libraries screened against two formylpeptide receptors. Molecules. 2013;18:6408–6424. doi: 10.3390/molecules18066408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamel L.D., Deschenes R.J., Mitchell D.A. A fluorescence-based assay to monitor autopalmitoylation of zDHHC proteins applicable to high throughput screening. Anal. Biochem. 2014;460:1–8. doi: 10.1016/j.ab.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherman F., Fink G.R., Hicks J.B. Laboratory Course Manual: Methods in Yeast Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 15.Ito H., Fukada Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartels D.J., Mitchell D.A., Dong X., Deschenes R.J. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6775–6787. doi: 10.1128/mcb.19.10.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostresh J.M., Winkle J.H., Hamashin V.T. Houghten, R.A. Peptide libraries: determination of relative reaction rates of protected amino acids in competitive couplings. Biopolymers. 1994;34:1681–1689. doi: 10.1002/bip.360341212. [DOI] [PubMed] [Google Scholar]
- 18.Houghten R.A., Pinilla C., Appel J.R., Blondelle S.E., Dooley C.T., Eichler J., Nefzi A., Ostresh J.M. Mixture-based synthetic combinatorial libraries. J. Med. Chem. 1999;42:3743–3778. doi: 10.1021/jm990174v. [DOI] [PubMed] [Google Scholar]
- 19.Pinilla C., Appel J.R., Blanc P., Houghten R.A. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques. 1992;13:901–905. [PubMed] [Google Scholar]
- 20.Acharya A.N., Ostresh J.M., Houghten R.A. Determination of isokinetic ratios necessary for equimolar incorporation of carboxylic acids in the solid-phase synthesis of mixture-based combinatorial libraries. Biopolymers. 2002;65:32–39. doi: 10.1002/bip.10206. [DOI] [PubMed] [Google Scholar]
- 21.Santos R.G., Giulianotti M.A., Dooley C.T., Pinilla C., Appel J.R., Houghten R.A. Use and implications of the harmonic mean model on mixtures for basic research and drug discovery. ACS Comb. Sci. 2011;13:337–344. doi: 10.1021/co100065a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos R.G., Giulianotti M.A., Houghten R.A., Medina-Franco J.L. Conditional probabilistic analysis for prediction of the activity landscape and relative compound activities. J. Chem. Inf. Model. 2013;53:2613–2625. doi: 10.1021/ci400243e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minond D., Cudic M., Bionda N., Giulianotti M., Maida L., Houghten R.A., Fields G.B. Discovery of novel inhibitors of a disintegrin and metalloprotease 17 (ADAM17) using glycosylated and non-glycosylated substrates. J. Biol. Chem. 2012;287:36473–36487. doi: 10.1074/jbc.M112.389114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reilley K.J., Giulianotti M., Dooley C.T., Nefzi A., McLaugh-lin J.P., Houghten R.A. Identification of two novel, potent, low-liability antinociceptive compounds from the direct in vivo screening of a large mixture-based combinatorial library. AAPS J. 2010;12:318–329. doi: 10.1208/s12248-010-9191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J., Zhang Y., Maida L.E., Santos R.G., Welmaker G.S., LaVoi T.M., Nefzi A., Yu Y., Houghten R.A., Toll L., Giulianotti M.A. Scaffold ranking and positional scanning utilized in the discovery of nAChR-selective compounds suitable for optimization studies. J. Med. Chem. 2013;56:10103–10117. doi: 10.1021/jm401543h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell D.A., Hamel L.D., Ishizuka K., Mitchell G., Schaefer L.M., Deschenes R.J. The Erf4 subunit of the yeast Ras palmitoyl acyltransferase is required for stability of the Acyl-Erf2 intermediate and palmitoyl transfer to a Ras2 substrate. J. Biol. Chem. 2012;287:34337–34348. doi: 10.1074/jbc.M112.379297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansilla F., Birkenkamp-Demtroder K., Kruhoffer M., Sorensen F.B., Andersen C.L., Laiho P., Aaltonen L.A., Verspaget H.W., Orntoft T.F. Differential expression of DHHC9 in microsatellite stable and instable human colorectal cancer subgroups. Br. J. Cancer. 2007;96:1896–1903. doi: 10.1038/sj.bjc.6603818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuiffo B., Ren R. Palmitoylation of oncogenic NRAS is essential for leukemogenesis. Blood. 2010;115:3598–3605. doi: 10.1182/blood-2009-03-213876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi Y.W., Bae S.M., Kim Y.W., Lee H.N., Park T.C., Ro D.Y., Shin J.C., Shin S.J., Seo J.S., Ahn W.S. Gene expression profiles in squamous cell cervical carcinoma using array-based comparative genomic hybridization analysis. Int. J. Gynecol. Cancer. 2007;17:687–696. doi: 10.1111/j.1525-1438.2007.00834.x. [DOI] [PubMed] [Google Scholar]
- 30.Oyama T., Miyoshi Y., Koyama K., Nakagawa H., Yamori T., Ito T., Matsuda H., Arakawa H., Nakamura Y. Isolation of a novel gene on 8p21.3-22 whose expression is reduced significantly in human colorectal cancers with liver metastasis. Genes Chromosomes Cancer. 2000;29:9–15. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1001>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 31.De Vos M.L., Lawrence D.S., Smith C.D. Cellular pharmacology of cerulenin analogs that inhibit protein palmitoylation. Biochem. Pharmacol. 2001;62:985–995. doi: 10.1016/s0006-2952(01)00739-0. [DOI] [PubMed] [Google Scholar]
- 32.Draper J.M., Smith C.D. Palmitoyl acyltransferase assays and inhibitors. Mol. Membr. Biol. 2009;26:5–13. doi: 10.1080/09687680802683839. [Review]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Planey S.L., Zacharias D.A. Identification of targets and inhibitors of protein palmitoylation. Expert Opin. Drug Discov. 2010;5:155–164. doi: 10.1517/17460440903548218. [DOI] [PubMed] [Google Scholar]
- 34.Omura S. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol. Rev. 1976;40:681–697. doi: 10.1128/br.40.3.681-697.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patterson S.I., Skene J.H. Inhibition of dynamic protein palmitoylation in intact cells with tunicamycin. Methods Enzymol. 1995;250:284–300. doi: 10.1016/0076-6879(95)50079-0. [DOI] [PubMed] [Google Scholar]
- 36.Schlag B.D., Lou Z., Fennell M., Dunlop J. Ligand dependency of 5-hydroxytryptamine 2C receptor internalization. J. Pharmacol. Exp. Ther. 2004;310:865–870. doi: 10.1124/jpet.104.067306. [DOI] [PubMed] [Google Scholar]
- 37.Christopher J.A., Brown J., Dore A.S., Errey J.C., Koglin M., Marshall F.H., Myszka D.G., Rich R.L., Tate C.G., Tehan B., Warne T., Congreve M. Biophysical fragment screening of the beta1-adrenergic receptor: identification of high affinity arylpiperazine leads using structure-based drug design. J. Med. Chem. 2013;56:3446–3455. doi: 10.1021/jm400140q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debevec G., Chen W., Yu Y., Houghten R.A., Giulianotti M.A. Libraries from Libraries: A Series of Sulfonamide Linked Heterocycles Derived from the Same Scaffold. Tetrahedron Lett. 2013;54:4296–4299. doi: 10.1016/j.tetlet.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.