ABSTRACT
Human adenoviruses (HAdVs) contain seven species (HAdV-A to -G), each associated with specific disease conditions. Among these, HAdV-D includes those viruses associated with epidemic keratoconjunctivitis (EKC), a severe ocular surface infection. The reasons for corneal tropism for some but not all HAdV-Ds are not known. The fiber protein is a major capsid protein; its C-terminal “knob” mediates binding with host cell receptors to facilitate subsequent viral entry. In a comprehensive phylogenetic analysis of HAdV-D capsid genes, fiber knob gene sequences of HAdV-D types associated with EKC formed a unique clade. By proteotyping analysis, EKC virus-associated fiber knobs were uniquely shared. Comparative structural modeling showed no distinct variations in fiber knobs of EKC types but did show variation among HAdV-Ds in a region overlapping with the known CD46 binding site in HAdV-B. We also found signature amino acid positions that distinguish EKC from non-EKC types, and by in vitro studies we showed that corneal epithelial cell tropism can be predicted by the presence of a lysine or alanine at residue 240. This same amino acid residue in EKC viruses shows evidence for positive selection, suggesting that evolutionary pressure enhances fitness in corneal infection, and may be a molecular determinant in EKC pathogenesis.
IMPORTANCE Viruses adapt various survival strategies to gain entry into target host cells. Human adenovirus (HAdV) types are associated with distinct disease conditions, yet evidence for connections between genotype and cellular tropism is generally lacking. Here, we provide a structural and evolutionary basis for the association between specific genotypes within HAdV species D and epidemic keratoconjunctivitis, a severe ocular surface infection. We find that HAdV-D fiber genes of major EKC pathogens, specifically the fiber knob gene region, share a distinct phylogenetic clade. Deeper analysis of the fiber gene revealed that evolutionary pressure at crucial amino acid sites has a significant impact on its structural conformation, which is likely important in host cell binding and entry. Specific amino acids in hot spot residues provide a link to ocular cell tropism and possibly to corneal pathogenesis.
INTRODUCTION
Human adenoviruses (HAdVs) are major causes of respiratory disease, gastroenteritis, and keratoconjunctivitis (1–4). HAdV infections are also a major concern in immunocompromised hosts, with fatality rates as high as 55% (5). There are 72 HAdVs currently genotyped in GenBank, which are classified into seven species (HAdV-A to -G) as determined by whole-genome analysis. The majority (n = 47) belong to HAdV-D, including types 8, 37, 53, 54, 56, and 64 (6–12). Each of these has been associated with epidemic keratoconjunctivitis (EKC), a highly contagious ocular surface infection (13). HAdV-D evolves by homologous recombination within genes encoding the major HAdV capsid proteins (14). For example, type 64 (previously typed as 19a) is a recombinant with types 19, 22, and 37 in the hexon, penton base, and fiber genes, respectively (12). With no specific antiviral treatment available for adenovirus infections, the evolution of new and potentially more virulent HAdV-Ds represents a threat to public health.
Virus attachment to its host cell receptor is an obligatory step in viral infection. Primary receptors identified for HAdV include the coxsackie adenovirus receptor (CAR), membrane cofactor CD46, and GD1a glycan (15–20), with the last two proposed as receptors for the specific HAdV-D types associated with EKC. The specific ligand on the adenovirus that first engages its host target cell receptor is the fiber knob (16, 21), the most distal component of the viral capsid. The fiber protein is a trimer; each fiber monomer consists of an N-terminal tail, a central shaft, and a C-terminal knob binding domain (22). By crystallography and functional analyses, Nilsson and colleagues showed direct interactions between the HAdV-D37 fiber knob and GD1a glycan (20). However, they found no significant difference in GD1a glycan-fiber knob interactions between EKC-causing and other, non-EKC-causing HAdV-D types. Therefore, GD1a glycan interactions alone do not explain corneal tropism, leaving open the question of why certain HAdV-Ds infect the cornea while others do not.
Amino acid mutations in residues at the molecular interface between a viral ligand and its host cell receptor can facilitate a survival advantage for the virus (23–25). Such positive selection represents an evolutionary tactic for securing and expanding viral tropism. However, evidence for positive selection in the fiber knob leading to corneal tropism is limited (26). In the work described here, we demonstrate that the HAdV-D fiber knobs of viruses associated with EKC form a unique phylogenetic clade, which is not evident for other genomic regions. We also show amino acid variations specific to EKC virus-associated fiber knobs. Finally, we demonstrate how knowledge of specific amino acids in a particular fiber knob can be used to predict corneal epithelial tropism. These results demonstrate for the first time a genomic correlate for corneal tropism in EKC.
MATERIALS AND METHODS
Cell culture and virus purification.
A549 (CCL-185), a human alveolar carcinoma cell line, was obtained from American Type Culture Collection (ATCC), Manassas, VA. Telomerase-immortalized human corneal epithelial (THE) cells were the kind gift of Jerry Shay (University of Texas-Southwestern Medical Center, Dallas, TX) and were maintained in defined keratinocyte serum-free medium with supplements (Gibco, Grand Island, NY). Primary human corneal fibroblasts (HCF) were derived from human donor corneas and processed as previously described (20, 27) and were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. All experiments with primary HCF were performed at third passage.
HAdV-D37 and -D9 (GenBank accession no. DQ900900 and AJ854486) were obtained from the ATCC. HAdV-D56 (accession no. HM770721) was kindly provided by C. Henquell et al. (28). All three virus stocks were grown in A549 cells, purified by CsCl gradient centrifugation, dialyzed, and stored at −80°C. All viruses and cell lines tested negative for endotoxin and mycoplasma contamination prior to use. Virus titration was performed in A549 cells, and the 50% tissue culture infective dose (TCID50) titer was calculated by the formula of Reed and Muench (29).
Proteotype analysis.
The concept of proteotyping was first introduced to study the evolution of avian influenza virus (30) and later applied to HAdV-D recombination (14). A multiple amino acid sequence alignment was performed using CLUSTALW in MEGA6 (31). A phylogenetic tree was constructed using the maximum-likelihood method with a bootstrap test of 1,000 replicates and the JTT matrix-based model algorithm. In the alignment, each amino acid was assigned a unique, arbitrary color. The most commonly represented consensus amino acid was colored white, and gaps in the alignment were colored black. Unique amino acids with a threshold of <10% divergence were used to distinguish individual proteotypes.
Homology modeling and structure validation.
Homology fiber knob models were built in Swiss ExPASy (http://swissmodel.expasy.org) using the crystal structure of HAdV-D37 as the template (Protein Data Bank [PDB] code 1UXA) (32). The structure validation was performed in PROCHECK using the Structure Analysis and Verification Server (http://services.mbi.ucla.edu/SAVES/) for PROCHECK (33) and VERIFY3D (34). The distribution of phi (ϕ)-psi (ψ) torsion angles in Ramachandran plots showed at least 83% residues in most favored core region and fewer than 1.2% residues in disallowed regions for the generated models. The VERIFY3D results showed at least 98.38% of the residues with an averaged 3D-1D score ≥0.2. UCSF Chimera v1.9 (35) was used for visualization and root mean square deviation (RMSD) analysis.
Cy3-virus labeling.
HAdV-D37, HAdV-D56, and HAdV-D9 were conjugated with Cy3 dye (GE Healthcare, Piscataway, NJ) as previously described (36). Briefly, 1 mg of Cy3 dye was reconstituted in 1 ml of 0.1 M sodium bicarbonate (pH 9.3). Labeling was performed by conjugating the reconstituted Cy3 dye with ∼1012 adenovirus particles/ml. The reconstituted dye was 20% of the final solution, and the mixture was incubated for 30 min in the dark with gentle mixing every 10 min. The Cy3-labeled virus was then dialyzed overnight to remove the excess dye and aliquots stored at 4°C for up to 4 weeks. We have previously established that Cy3 adherence to HAdV capsid and subsequent intensity of fluorescence are independent of the HAdV genotype and that virus labeled with Cy3 grows to a titer equal to that of unlabeled virus (37).
In vitro infection and confocal microscopy.
A549, THE, and HCF cells were grown to ∼80% confluence on slide chambers (Nunc, Rochester, NY), and Cy3-labeled virus was added and kept at 4°C for synchronization of infection. After 30 min, the cells were washed and incubated for 1 h at 37°C. The cells were then fixed in 4% paraformaldehyde for 10 min, washed in phosphate-buffered saline (PBS) containing 2% bovine serum albumin (BSA), permeabilized in a solution containing 0.1% Triton X-100 in 2% BSA for 10 min, blocked in 2% BSA–PBS for 30 min, and treated with 1 μg/ml of Alexa 488-conjugated phalloidin (Thermo Fisher Scientific, Waltham, MA) for 45 min at room temperature. Cells were then washed, and mounting medium containing DAPI (4,6-diamidino-2-phenylindole) (Vectashield; Vector Laboratories, Burlingame, CA) was added. Sealed coverslip slides were then scanned with a confocal laser scanning microscope (TCS SP5; Leica, Heidelberg, MA). Quantification of the Cy3 label was performed using image processing and analysis (ImageJ v1.49, National Institutes of Health, Bethesda, MD). For each virus and cell type, 10 randomly chosen high-power fields (×1,000) with at least 20 cells were quantified and plotted as pixel units per cell. Each experiment was repeated three times.
Quantitative reverse transcription-PCR (qRT-PCR).
Cells were infected with HAdV-D37, -D56, or -D9 at multiplicity of infection (MOI) of 10 for 1 and 2 h, washed with PBS and lysed in TRIzol (Zymo Research, Irvine, CA). RNA was extracted using the Direct-zol RNA kit (Zymo) according to the manufacturer's instructions, and the eluent was treated with Turbo DNase (Ambion, Austin, TX) for 30 min at 37°C. For cDNA synthesis, 500 ng of RNA was reverse transcribed using oligo(dT) and Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI) according to the manufacturer's instructions. Real-time PCR was performed with Fast SYBR green mix (Thermo Fischer Scientific, Waltham, MA) in the QuantStudio 3 system (Thermo Fischer Scientific) with consensus E1A primers for HAdV-D37, -D56, and -D9, designed using NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The forward and reverse primer sequences were 5′-GAA GGT TTT CCT CCC AGC GA and 5′-CTT ATC CCG GTG GTA CTG GCA, respectively. Human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primers were applied as an internal normalization control. Data were analyzed by the comparative threshold cycle (CT) method. Each experimental condition was analyzed in triplicate wells and repeated three times.
Selection pressure analysis.
To identify fiber codons under positive selection, relative rates of synonymous substitutions per site (dS) and nonsynonymous substitutions per site (dN) were calculated using the HyPhy package in Datamonkey (http://www.datamonkey.org/) (38, 39). The sequence alignment was analyzed using genetic algorithm recombination detection (GARD) to account for recombination bias, followed by internal fixed-effects likelihood (IFEL) modeling, which is a more conservative method. A P value of <0.10 was considered significant (40, 41).
Statistical analysis.
The Shapiro-Wilk W test was performed for analysis of normal distribution. Medians with interquartile ranges (IQR) were used to describe variables with skewed distribution and the data analyzed by the Kruskal-Wallis test. Relative gene expression levels were analyzed by two-way analysis of variance (ANOVA), and a P value of <0.05 was considered statistically significant. Except for IFEL modeling, all analyses were performed using STATA 11 (StataCorp. College Station, TX) or GraphPad Prism v6.0 (GraphPad Software, San Diego, CA).
RESULTS
EKC-associated HAdV-D types form a distinct fiber clade and closely adjacent proteotypes.
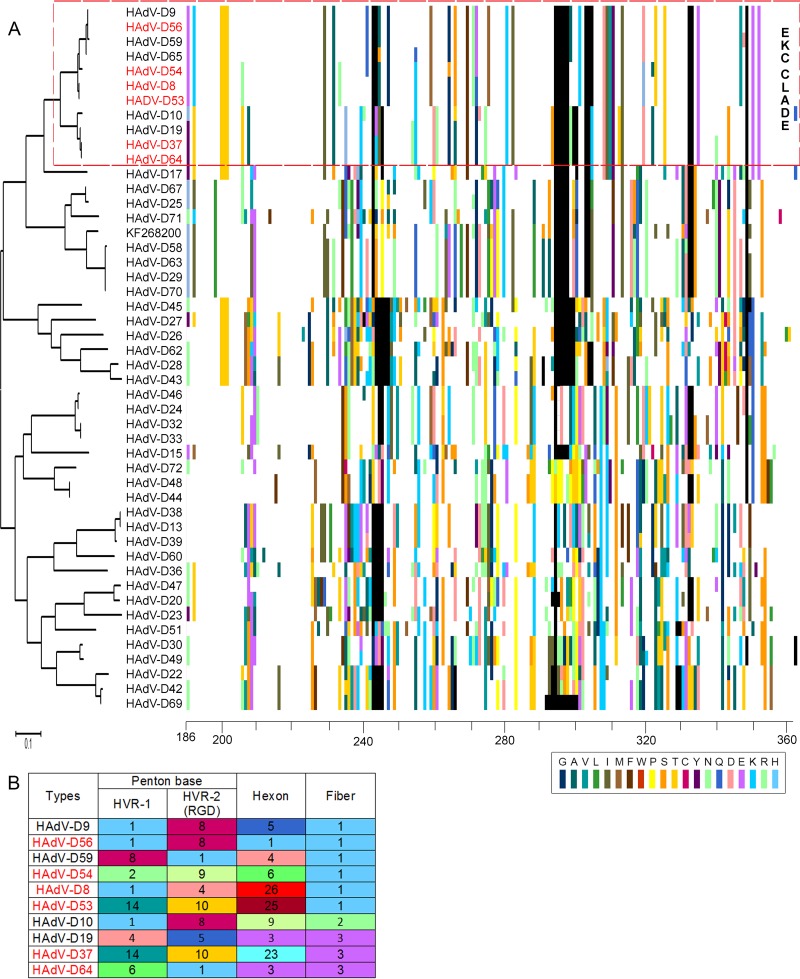
The major proteins of the adenovirus capsid are the hexon, penton base, and fiber. For HAdV-D, the corresponding genes are stereotypically hypervariable in comparison to the rest of the genome (14). We focused our analyses on the fiber knob region, given its known importance to initial binding of HAdV to host cell receptors. In a comprehensive analysis of all 47 HAdV-D types currently in GenBank plus one novel HAdV-D (accession no. KF268200), we applied proteotyping (30) to identify uniquely shared fiber knob protein types. The generated maximum-likelihood tree and the amino acid alignment reveal a pattern where the six EKC-associated viruses, along with five others, form a closely associated “EKC clade” (Fig. 1A). The non-EKC HAdV-Ds also present in the EKC clade were HAdV-D9, -D10, -D19, -D59, and -D65. In total, using an arbitrary threshold of <10% amino acid divergence to distinguish unique proteotypes (14), we found 24 distinct fiber knob proteotypes among 48 HAdV-D viral genotypes (data not shown).
FIG 1.
Fiber knob proteotyping alignments for 48 HAdV-D proteins. (A) Maximum-likelihood phylogenetic tree and amino acid signatures (proteotypes) for fiber knob proteins. For proteotype assignments, each amino acid that varied from the consensus sequence was assigned a unique color (bottom right). Consensus amino acids at each position across all 48 viruses were assigned white, and gaps in the alignment are shown in black. All six EKC-associated viruses form closely associated fiber knob clusters. (B) Proteotypes of aligned vertical colored bars (as shown in panel A) that were <10% different and match the clade-guided sequence alignment are numbered in order for all three major capsid proteins. EKC viruses shared two fiber knob proteotypes (HAdV-D8, -D53, -D54, and -D56 in proteotype 1 and HAdV-D37 and -D64 in proteotype 3). Hexon and penton base proteotypes were considerably different. The HAdV-D65 proteotype was not determined for hexon and penton base and therefore is not included.
A tabular form of capsid protein proteotyping for the EKC clade, when anchored on the fiber knob (Fig. 1B), shows EKC viruses grouped into two closely adjacent proteotypes, suggesting a direct link between the fiber knob and corneal tropism. Other major capsid proteins, for example, the penton base, may also play a role in tropism. The penton base contains two hypervariable loops, the first with unknown function and the second containing an Arg-Gly-Asp (RGD) motif important to viral internalization. After primary binding of the fiber knob to its host cell receptor, the RGD motifs on the five-sided penton base capsomer bind to host cellular integrins to mediate endocytosis of the virus (42–45). We previously showed that the two corresponding hypervariable regions of the penton base open reading frame frequently undergo homologous recombination (14, 46), and we have suggested that penton base hypervariable regions may be important to viral pathogenesis (47). Our proteotyping analysis showed that the two hypervariable loops of the penton base protein of the EKC virus HAdV-D56 and the non-EKC virus HAdV-D9 are shared, as was previously shown (10). The first hypervariable loop of HAdV-D56 is also shared with HAdV-D8 and -D10. Otherwise, no EKC virus falls in the same penton base proteotype as any other EKC virus. The hexon protein is the most abundant protein in the viral capsid, with 240 hexon trimers in every capsid. The hexon protein's two closely adjacent hypervariable loops form the epsilon determinant responsible for humoral immune responses and permit differentiation of serotypes by serum neutralization testing. No EKC virus shared a hexon proteotype with any other EKC virus. Therefore, for the major capsid genes, the fiber knob region was uniquely similar within the six EKC types, consistent with a role in pathogenesis.
EKC-associated viruses do not exhibit a unique fiber knob conformation.
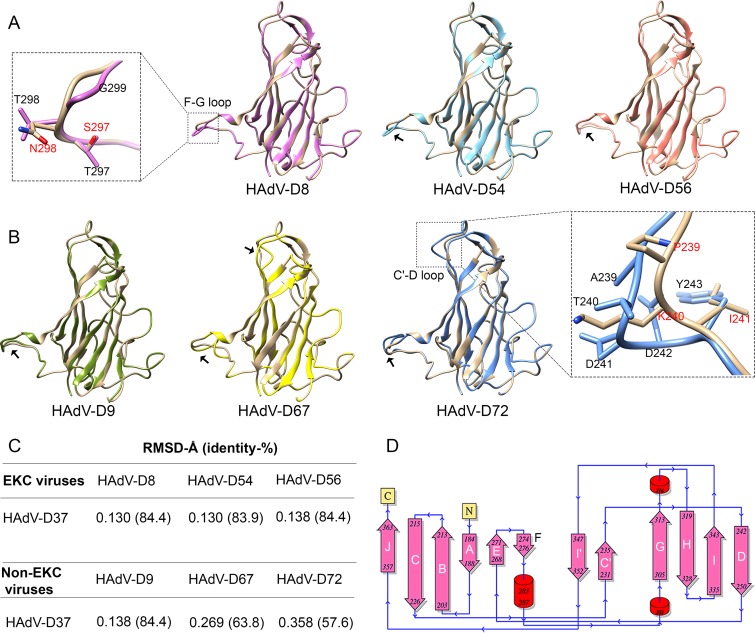
As discussed above, the adenovirus fiber knob mediates primary interaction with the host cell. To determine whether EKC virus fiber knobs share a unique structure, we constructed homology models for structural comparisons of the fiber knobs of EKC (Fig. 2A) and representative non-EKC viruses (Fig. 2B). The models were superimposed on the HAdV-D37 structure for comparison, and the structural divergences were measured by root mean square deviation (RMSD) (Fig. 2C). Each fiber knob monomer had 12 β-strands and 3 α-helices, and observed structural variations were located accordingly (Fig. 2D). Consistent with the phylogenetic data, the RMSD values of superimposed structures showed a definite relationship. Comparison of structural models within EKC viruses showed conformational changes in the F-G connecting loop with RMSD values between 0.130 and 0.138. In addition to T297S and T/A298N substitutions, a glycine residue absent in HAdV-D37 gave an altered conformation at this F-G loop (Fig. 2A). Similar structural divergence was observed within closely identical non-EKC types in the F-G loop. Therefore, although we found structural variations at distinct loci when comparing one HAdV-D virus to another, we could not identify conformational changes specific to EKC viruses. The RMSD values were high for distant clades (representative types 29, 67, and 72), illustrating increased structural divergence, and showed an additional conformational change at C′-D connecting loops (Fig. 2B). Types 10 and 19 had identical residues at these crucial positions and therefore showed no structural variation (data not shown).
FIG 2.
Comparative structural analysis of EKC and non-EKC fiber knobs. Homology modeling of fiber proteins was performed using an HAdV-D37 template (PDB ID 1UXA). EKC fiber proteins for HAdV-D8 and -D37 are 100% identical to those for HAdV-D53 and -D64, respectively. Therefore, representative types are modeled. (A and B) Superimpositions of HAdV-D37 (tan) with EKC-associated virus types (A) and representative non-EKC types (B). The F-G and C′-D loops with significant conformational changes are indicated by black arrows. The inset shows a close-up view of the conformational changes, with HAdV-D37 amino acids indicated in red. Structural variations observed in the F-G loop were common for both EKC and non-EKC viruses. In addition, the distant non-EKC clade types (HAdV-D67 and -D72) showed structural variations in the C′-D loop. (C) RMSD values of two superimposed structures and corresponding protein sequence identity. (D) The secondary structure of HAdV-D37 fiber knob protein shows α-helices (cylinders), β-strands (arrows), and terminal ends (yellow box). The conformational changes shown in panels A and B indicate the beta strand connecting loops.
Structural variations among fiber knobs overlap the CD46 receptor binding site.
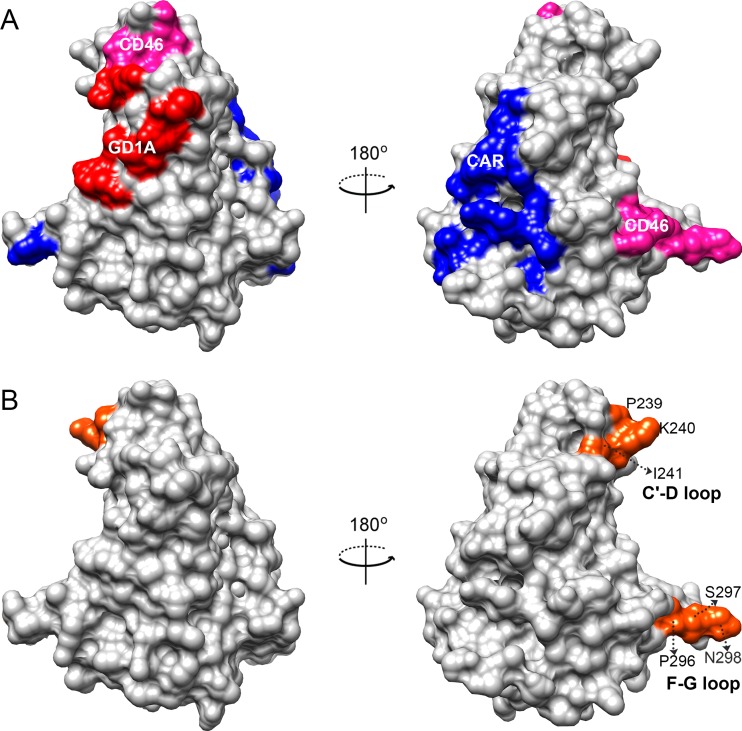
To determine if the observed structural variations overlap cellular receptor binding sites, we mapped the known HAdV-cellular receptor interactions for CAR, CD46, and GD1a glycan on the homology model (16, 20, 32, 44, 48–54). The two structural variations viewed in a surface filled model (Fig. 3) showed partial overlap with the CD46 receptor interaction sites at the F-G loop (Fig. 3B). Further analyses of amino acid differences in the same receptor interaction sites did not identify specific amino acid differences between EKC and non-EKC types within the EKC clade. It is noteworthy that HAdV-D17, chosen for inclusion in the analysis because it was closely related phylogenetically but not in a shared proteotype with the any of the EKC viruses, showed more amino acid variations than the other viruses within the EKC clade (data not shown).
FIG 3.
Receptor binding sites and observed structural variations in fiber knob proteins. (A) Known cellular receptor interactions of CAR, CD46, and GD1a glycan are mapped in the surface-filled representation of the HAdV-D37 structural model (PDB ID 1UXA) and indicated in blue, pink, and red, respectively. (B) Structural variations of EKC and non-EKC viruses identified by comparative modeling were located (orange). The observed conformational changes did not overlap the GD1a glycan or coxsackie adenovirus receptors (CAR), but the conformational change identified in the F-G loop partially overlapped the known CD46 interaction site.
Specific fiber knob amino acids differentiate EKC viruses from non-EKC viruses.
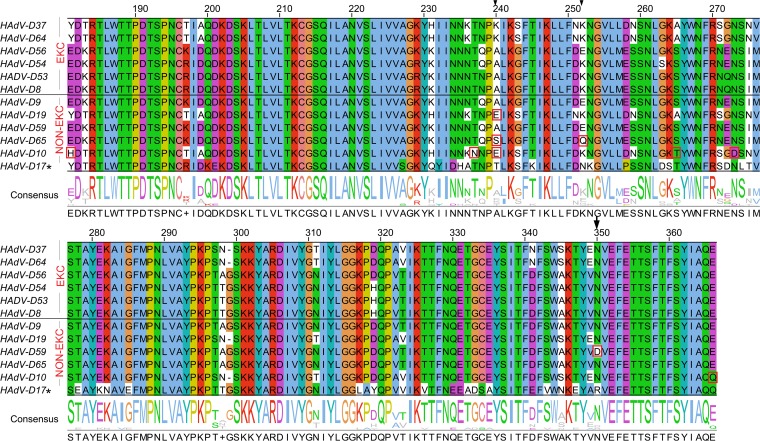
To further probe for characteristics of the fiber knob specific to EKC viruses, we analyzed an alignment of EKC clade sequences (Fig. 1A), including the six viruses associated with EKC, the five other viruses in the EKC clade, and HAdV-D17 (Fig. 4). Upon comparison between EKC and non-EKC viruses in the EKC clade, amino acid residues at three positions showed considerable variation. In particular, residue 240 showed five distinct amino acids. EKC-associated viruses had lysine (K) or alanine (A) at residue 240. Two non-EKC viruses, HAdV-D9 and -D59, also had alanine at residue 240. The other non-EKC types contained glutamic acid (E), serine (S), or threonine (T). Compared to the EKC viruses, the non-EKC type D19 had only a single amino acid substitution at position 240, type 65 had substitutions at positions 240 and 252, and type 59 had an amino acid substitution at position 350. In addition to the amino acid substitution at position 240, HAdV-D10 had amino acid substitutions at five other positions.
FIG 4.
Amino acid differences between EKC and non-EKC viruses in the EKC clade. A multiple-sequence alignment of EKC and non-EKC fiber knobs is shown. Amino acid residues are color coded by residue type. Residues that differ between EKC and non-EKC types are boxed in red and the highly varied positions are indicated by arrows. HAdV-D17, a divergent clade control, showed comparatively high amino acid substitutions. Amino acid start positions are according to the HAdV-D37 sequence (accession no. DQ900900).
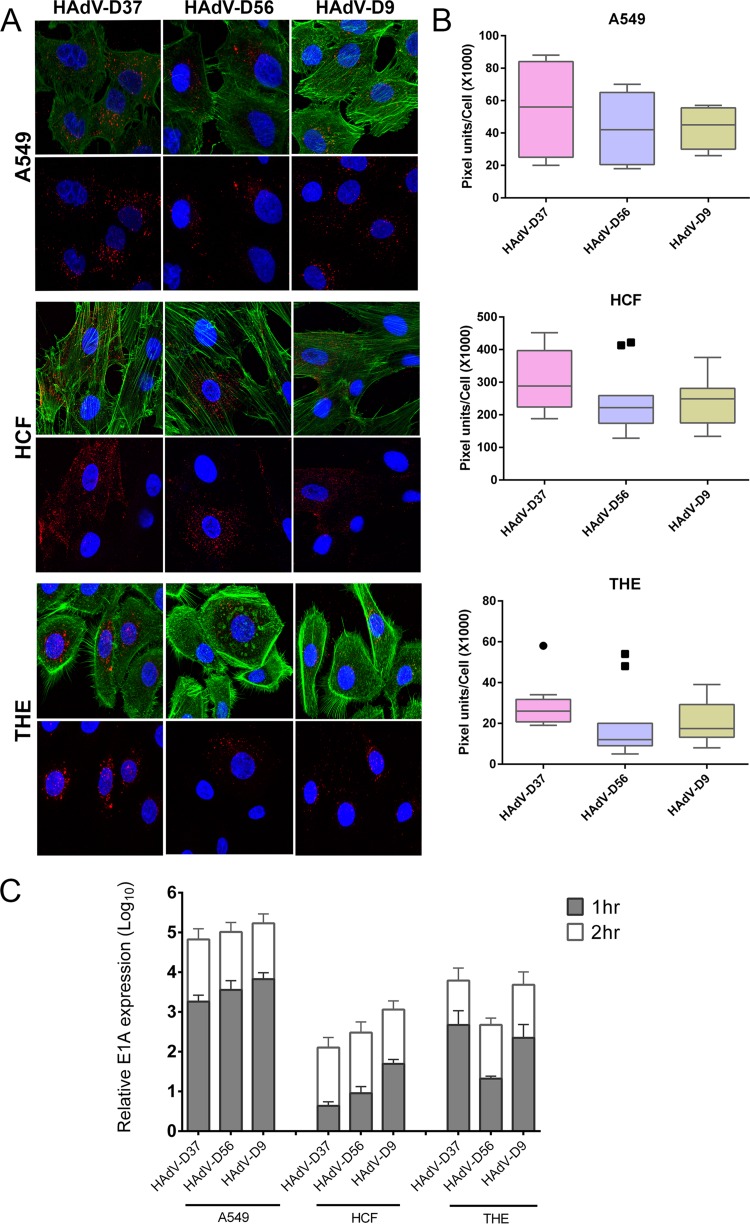
In natural infection of the human cornea during EKC, adenovirus enters and replicates first in the corneal epithelium (55). We have previously shown that a telomerase-immortalized human corneal epithelial cell line (THE) can recapitulate corneal tropism; EKC viruses readily enter and traffic in THE cells, while non-EKC viruses do not (12, 37). EKC is defined in part by corneal infection, and adenovirus keratitis occurs only in the context of EKC. Therefore, corneal tropism appears to be a requisite component of EKC. The non-EKC virus HAdV-D9 contained an alanine at residue 240 and closely resembled the EKC virus type 56 in the fiber knob and in the consensus EKC fiber shaft (reference 10 and data not shown). Therefore, we predicted that type 9 should also enter THE cells. We tested this hypothesis by comparing cellular entry of Cy3-labeled HAdV-D9 virus with that of two EKC virus types, HAdV-D56 and HAdV-D37, in both THE cells and primary HCF, with A549 cells as a control. At 1 h postinfection (hpi), a time at which we previously demonstrated differential entry of HAdV-D19 and -D64 into the same two corneal cell types (12), HAdV-D9 appeared to enter both (Fig. 5A). Quantification of red signal with ImageJ that showed entry of HAdV-D9 into THE cells at 1 hpi was greater than that of the EKC virus HAdV-D56 (P < 0.05) (Fig. 5B) although less than HAdV-D37. By real-time RT-PCR, HAdV-D9 E1A gene expression in THE cells was greater than that of HAdV-D56 at 1 hpi (P < 0.05) (Fig. 5C) and was essentially equal to that of HAdV-D37 (the latter is known to cause particularly severe EKC infections). By 2 hpi, there was no significant difference in viral gene expression between viruses (P > 0.05).
FIG 5.
Cellular entry and virus gene expression. (A) Comparison of infection by Cy3-labeled HAdV-D37, -D56, and -D9 in A549 cells, primary human corneal fibroblasts (HCF), and telomerase-immortalized human corneal epithelial (THE) cells by confocal microscopy at 1 h postinfection. Cy3 labels virus red, DAPI stains nuclei blue, and phalloidin stains cellular actin green. (B) Quantification of Cy3-labeled virus was performed using ImageJ software and graphed in box plots. The circle and squares indicate statistical outliers. (C) Early viral gene expression (E1A) as an indirect measure of viral nuclear entry at 1 and 2 hpi. Values represent the mean and standard deviation.
Fiber knob residues under positive selection determine the cellular specificity of EKC viruses.
Selection pressure on a gene can be estimated by the ratio of nonsynonymous substitutions (dN) to synonymous substitutions (dS). A finding of a dN/dS ratio of >1 for a given amino acid position indicates positive selection, in which individual changes in hot spot residues are believed to increase viral fitness. In order to determine if certain virus types have an evolutionary advantage in EKC, we performed selection pressure analysis of the complete fiber genes of all 48 HAdV-Ds analyzed above. By codon-based selection pressure analysis using IFEL models, seven positively selected sites were identified (Table 1). Interestingly, the amino acid at position 240, shown above to be involved in an altered structural conformation at the C′-D loop and which was also consistently identified as an amino acid that differentiates EKC and non-EKC types, appeared to be under positive selection pressure (P = 0.01). Three other positively selected sites were identified in the N-terminal tail region and another three codons in the fiber knob at positions 306, 321, and 324, all in close proximity to the GD1a glycan binding site (data not shown). These data suggest that these latter positively selected sites may enhance receptor binding interactions for other tissue sites but that amino acid 240 is a crucial residue for corneal tropism.
TABLE 1.
Selection pressure analysis of human adenovirus D fiber codons for the total 48 reference sequences
| Codona | dS | dN | dN/dS | Normalized dN − dS | P valueb |
|---|---|---|---|---|---|
| 2 | 0 | 0.324 | Infinite | 0.030 | 0.016 |
| 73 | 0 | 0.515 | Infinite | 0.048 | 0.08 |
| 87 | 0 | 0.343 | Infinite | 0.032 | 0.06 |
| 240 | 0 | 10.111 | Infinite | 0.94 | 0.01 |
| 306 | 1.538 | 6.203 | 4.032 | 0.433 | 0.07 |
| 321 | 0.235 | 1.659 | 7.067 | 0.132 | 0.06 |
| 324 | 0.534 | 3.749 | 7.023 | 0.29 | 0.08 |
Codon positions are according to the HAdV-D37 sequence.
P value with default GARD and IFEL analysis models (significance levels of <0.1).
DISCUSSION
Our analyses provide a structural basis for understanding HAdV-D fiber knob evolution and tropism. By phylogenetic and proteotyping analysis of the major capsid proteins of characterized HAdV-D types, we demonstrated a unique relatedness among all six EKC-associated viruses in the whole fiber protein and also in the fiber knob alone, but not in the penton base or hexon. We next aimed to determine potential structural relatedness within EKC viruses that might influence cellular binding and contribute to disease. Comparisons of fiber knob protein models showed substantial structural variations at two positions, the F-G and C′-D connecting loops, but conformational changes in these regions were observed in both EKC and non-EKC types and did not distinguish them.
The fiber protein is one of three major capsid proteins and is responsible for initial attachment of adenovirus to host cell receptors. Despite an absence of distinct structural features at the fiber knob that might distinguish EKC types, we sought to determine if the observed conformational changes overlap the known binding sites for the known HAdV host cell receptors CAR, CD46, and GD1a glycan. Indeed, the conformational variation at the HAdV-D fiber knob F-G loop partially overlapped with the CD46 binding site established for HAdV-B (50). However, our data showed that the conformational change at this position is not specifically related to tropism specificity in EKC. Structural variations in the F-G loop were common for both EKC and non-EKC types, and no defining amino acid differences were identified in the recognized interacting sites. We also mapped the interacting positions previously established for HAdV-B11 and HAdV-A12 (17, 50) and found a similar knob F-G loop (data not shown).
Wu and coworkers previously showed that the HAdV-D37 fiber knob directly binds the extracellular domain of CD46, but the interacting amino acid determinants were not studied (19). In other work, the rigidity of the fiber shaft was shown to be a possible factor in tropism (56). However, using a human conjunctival cell line also shown to contain HeLa markers (57) and recombinant wild-type and mutant fiber proteins, Huang and colleagues demonstrated that a single amino acid substitution in the non-EKC-causing Ad19 fiber knob, Glu240 to Lys, conferred binding, while the reverse substitution in the Ad37 fiber knob abrogated binding (26). Amino acid 240 is not in a known cellular interaction site. In our analysis of amino acids at residue 240, we noted that EKC-associated types had lysine or alanine. Two non-EKC viruses, HAdV-D9 and -D59, also had alanine substitutions at amino acid position 240, but HAdV-D59 had an additional N350D substitution. Therefore, we tested whether HAdV-D9, with an alanine at fiber knob amino acid 240 similar to the case for EKC viruses, would confer corneal epithelial cell tropism. To perform these studies, we used THE cells, which were previously shown to accurately recapitulate corneal tropism for EKC-associated viruses within HAdV-D (12, 37, 47). We compared HAdV-D9 entry and early gene expression with those of the EKC viruses HAdV-D37 and -D56. HAdV-D9 infection of THE cells was as robust as that of HAdV-D37 and greater than that of HAdV-D56. Interestingly, the ATCC reported years previously that their HAdV-D8 isolate was contaminated with HAdV-D9 and -D10 (58), raising the speculation that HAdV-D9 might also be a cause of EKC, albeit rarely isolated. We previously showed that HAdV-D19 demonstrates reduced viral entry into THE cells in comparison with HAdV-D64 (12). The fiber knobs of these viruses differ in only two amino acids in total, at positions 240 and 340. In the latter, the asparagine (N) in HAdV-D19 is also found in other EKC viruses, not including HAdV-D37 and -D64. These data add further evidence for the possible importance of amino acid 240 to corneal tropism and, by association, to EKC.
Lysine is a positively charged amino acid, and its polarity can enhance electrostatic interactions between a ligand and its receptor. Lys345 was suggested to play a crucial role in HAdV-D37 binding to sialic acid (59). Similarly, lysine residues are involved in HAdV-CAR interactions (4, 24). It is possible that three adjacent lysine residues, at positions 236, 242, and 247, stabilize the interactions to result in increased virulence. Similar to the crucial lysine fence in the globular head of hemagglutinin, and associated with pandemic influenza A/H1N1 (2009) virus (60, 61), our proposed crucial lysine residues are structurally exposed on the outer surface of the fiber knob (data not shown). We therefore speculate that position 240 is a hot spot and a novel interaction site for virus attachment and cell entry.
In conclusion, protein structure and evolutionary fitness are closely linked (62). Selection pressure leads to evolutionary consequences, wherein individual changes at hot spot residues can select for a survival benefit or disadvantage. A ratio of nonsynonymous to synonymous mutations of greater than 1 at a particular codon indicates positive selection and is consistent with increased fitness. Our analysis revealed positive selection at seven positions in the fiber domain, including codon 240. Positive selection at this codon correlated with structural variation in the fiber knob C′-D loop and paralleled the finding of amino acids specific to EKC viruses. These data provide circumstantial evidence for the possible role of amino acid 240 as a molecular determinant for corneal tropism and EKC pathogenesis.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) (grants EY013124, EY021558, and P30), a Senior Scientific Investigator Award grant (to J.C.) from Research to Prevent Blindness, Inc., New York, NY, The Falk Foundation, and the Massachusetts Lions Eye Research Fund. This project was additionally supported by a summer research grant (to D.S.) from the Office of the Vice President for Research at George Mason University.
REFERENCES
- 1.Wallot MA, Dohna-Schwake C, Auth M, Nadalin S, Fiedler M, Malago M, Broelsch C, Voit T. 2006. Disseminated adenovirus infection with respiratory failure in pediatric liver transplant recipients: impact of intravenous cidofovir and inhaled nitric oxide. Pediatr Transplant 10:121–127. doi: 10.1111/j.1399-3046.2005.00411.x. [DOI] [PubMed] [Google Scholar]
- 2.Fletcher SM, McLaws ML, Ellis JT. 2013. Prevalence of gastrointestinal pathogens in developed and developing countries: systematic review and meta-analysis. J Public Health Res 2:42–53. doi: 10.4081/jphr.2013.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jhanji V, Chan TC, Li EY, Agarwal K, Vajpayee RB. 2015. Adenoviral keratoconjunctivitis. Surv Ophthalmol 60:435–443. doi: 10.1016/j.survophthal.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Kajon AE, Hang J, Hawksworth A, Metzgar D, Hage E, Hansen CJ, Kuschner RA, Blair P, Russell KL, Jarman RG. 2015. Molecular epidemiology of adenovirus type 21 respiratory strains isolated from US military trainees (1996-2014). J Infect Dis 212:871–880. doi: 10.1093/infdis/jiv141. [DOI] [PubMed] [Google Scholar]
- 5.Hierholzer JC. 1992. Adenoviruses in the immunocompromised host. Clin Microbiol Rev 5:262–274. doi: 10.1128/CMR.5.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujii SI, Nakazono N, Sawada H, Ishii K, Kato M, Aoki K, Ohtsuka H, Fujinaga K. 1983. Restriction endonuclease cleavage analysis of adenovirus type 8: two new subtypes from patients with epidemic keratoconjunctivitis in Sapporo, Japan. Jpn J Med Sci Biol 36:307–313. doi: 10.7883/yoken1952.36.307. [DOI] [PubMed] [Google Scholar]
- 7.Warren D, Nelson KE, Farrar JA, Hurwitz E, Hierholzer J, Ford E, Anderson LJ. 1989. A large outbreak of epidemic keratoconjunctivitis: problems in controlling nosocomial spread. J Infect Dis 160:938–943. doi: 10.1093/infdis/160.6.938. [DOI] [PubMed] [Google Scholar]
- 8.Engelmann I, Madisch I, Pommer H, Heim A. 2006. An outbreak of epidemic keratoconjunctivitis caused by a new intermediate adenovirus 22/H8 identified by molecular typing. Clin Infect Dis 43:e64–66. doi: 10.1086/507533. [DOI] [PubMed] [Google Scholar]
- 9.Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS. 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4:e5635. doi: 10.1371/journal.pone.0005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson CM, Singh G, Henquell C, Walsh MP, Peigue-Lafeuille H, Seto D, Jones MS, Dyer DW, Chodosh J. 2011. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 409:141–147. doi: 10.1016/j.virol.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiroi S, Koike N, Nishimura T, Takahashi K, Morikawa S, Kase T. 2011. Genetic analysis of human adenovirus type 54 detected in Osaka, Japan. Jpn J Infect Dis 64:535–537. [PubMed] [Google Scholar]
- 12.Zhou X, Robinson CM, Rajaiya J, Dehghan S, Seto D, Jones MS, Dyer DW, Chodosh J. 2012. Analysis of human adenovirus type 19 associated with epidemic keratoconjunctivitis and its reclassification as adenovirus type 64. Invest Ophthalmol Vis Sci 53:2804–2811. doi: 10.1167/iovs.12-9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butt AL, Chodosh J. 2006. Adenoviral keratoconjunctivitis in a tertiary care eye clinic. Cornea 25:199–202. doi: 10.1097/01.ico.0000170693.13326.fb. [DOI] [PubMed] [Google Scholar]
- 14.Robinson CM, Singh G, Lee JY, Dehghan S, Rajaiya J, Liu EB, Yousuf MA, Betensky RA, Jones MS, Dyer DW, Seto D, Chodosh J. 2013. Molecular evolution of human adenoviruses. Sci Rep 3:1812. doi: 10.1038/srep01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louis N, Fender P, Barge A, Kitts P, Chroboczek J. 1994. Cell-binding domain of adenovirus serotype 2 fiber. J Virol 68:4104–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 17.Bewley MC, Springer K, Zhang YB, Freimuth P, Flanagan JM. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 18.Gaggar A, Shayakhmetov DM, Lieber A. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat Med 9:1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 19.Wu E, Trauger SA, Pache L, Mullen TM, von Seggern DJ, Siuzdak G, Nemerow GR. 2004. Membrane cofactor protein is a receptor for adenoviruses associated with epidemic keratoconjunctivitis. J Virol 78:3897–3905. doi: 10.1128/JVI.78.8.3897-3905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsson EC, Storm RJ, Bauer J, Johansson SM, Lookene A, Angstrom J, Hedenstrom M, Eriksson TL, Frangsmyr L, Rinaldi S, Willison HJ, Pedrosa Domellof F, Stehle T, Arnberg N. 2011. The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis. Nat Med 17:105–109. doi: 10.1038/nm.2267. [DOI] [PubMed] [Google Scholar]
- 21.van Raaij MJ, Mitraki A, Lavigne G, Cusack S. 1999. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401:935–938. doi: 10.1038/44880. [DOI] [PubMed] [Google Scholar]
- 22.Green NM, Wrigley NG, Russell WC, Martin SR, McLachlan AD. 1983. Evidence for a repeating cross-beta sheet structure in the adenovirus fibre. EMBO J 2:1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawyer SL, Wu LI, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A 102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shih AC, Hsiao TC, Ho MS, Li WH. 2007. Simultaneous amino acid substitutions at antigenic sites drive influenza A hemagglutinin evolution. Proc Natl Acad Sci U S A 104:6283–6288. doi: 10.1073/pnas.0701396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demogines A, Abraham J, Choe H, Farzan M, Sawyer SL. 2013. Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol 11:e1001571. doi: 10.1371/journal.pbio.1001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S, Reddy V, Dasgupta N, Nemerow GR. 1999. A single amino acid in the adenovirus type 37 fiber confers binding to human conjunctival cells. J Virol 73:2798–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Natarajan K, Rajala MS, Chodosh J. 2003. Corneal IL-8 expression following adenovirus infection is mediated by c-Src activation in human corneal fibroblasts. J Immunol 170:6234–6243. doi: 10.4049/jimmunol.170.12.6234. [DOI] [PubMed] [Google Scholar]
- 28.Henquell C, Boeuf B, Mirand A, Bacher C, Traore O, Dechelotte P, Labbe A, Bailly JL, Peigue-Lafeuille H. 2009. Fatal adenovirus infection in a neonate and transmission to health-care workers. J Clin Virol 45:345–348. doi: 10.1016/j.jcv.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 29.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg (Lond) 27:493–497. [Google Scholar]
- 30.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burmeister WP, Guilligay D, Cusack S, Wadell G, Arnberg N. 2004. Crystal structure of species D adenovirus fiber knobs and their sialic acid binding sites. J Virol 78:7727–7736. doi: 10.1128/JVI.78.14.7727-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486. [DOI] [PubMed] [Google Scholar]
- 34.Eisenberg D, Luthy R, Bowie JU. 1997. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404. doi: 10.1016/S0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Leopold PL, Ferris B, Grinberg I, Worgall S, Hackett NR, Crystal RG. 1998. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum Gene Ther 9:367–378. doi: 10.1089/hum.1998.9.3-367. [DOI] [PubMed] [Google Scholar]
- 37.Singh G, Zhou X, Lee JY, Yousuf MA, Ramke M, Ismail AM, Lee JS, Robinson CM, Seto D, Dyer DW, Jones MS, Rajaiya J, Chodosh J. 2015. Recombination of the epsilon determinant and corneal tropism: human adenovirus species D types 15, 29, 56, and 69. Virology 485:452–459. doi: 10.1016/j.virol.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pond SL, Frost SD. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 39.Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 40.Kosakovsky Pond SL, Frost SD. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 41.Pond SL, Frost SD, Grossman Z, Gravenor MB, Richman DD, Brown AJ. 2006. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput Biol 2:e62. doi: 10.1371/journal.pcbi.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li E, Stupack D, Klemke R, Cheresh DA, Nemerow GR. 1998. Adenovirus endocytosis via alpha(v) integrins requires phosphoinositide-3-OH kinase. J Virol 72:2055–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiu CY, Mathias P, Nemerow GR, Stewart PL. 1999. Structure of adenovirus complexed with its internalization receptor, alphavbeta5 integrin. J Virol 73:6759–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lord R, Parsons M, Kirby I, Beavil A, Hunt J, Sutton B, Santis G. 2006. Analysis of the interaction between RGD-expressing adenovirus type 5 fiber knob domains and alphavbeta3 integrin reveals distinct binding profiles and intracellular trafficking. J Gen Virol 87:2497–2505. doi: 10.1099/vir.0.81620-0. [DOI] [PubMed] [Google Scholar]
- 45.Zubieta C, Blanchoin L, Cusack S. 2006. Structural and biochemical characterization of a human adenovirus 2/12 penton base chimera. FEBS J 273:4336–4345. doi: 10.1111/j.1742-4658.2006.05430.x. [DOI] [PubMed] [Google Scholar]
- 46.Robinson CM, Rajaiya J, Walsh MP, Seto D, Dyer DW, Jones MS, Chodosh J. 2009. Computational analysis of human adenovirus type 22 provides evidence for recombination among species D human adenoviruses in the penton base gene. J Virol 83:8980–8985. doi: 10.1128/JVI.00786-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson CM, Zhou X, Rajaiya J, Yousuf MA, Singh G, DeSerres JJ, Walsh MP, Wong S, Seto D, Dyer DW, Chodosh J, Jones MS. 2013. Predicting the next eye pathogen: analysis of a novel adenovirus. mBio 4:e00595-12. doi: 10.1128/mBio.00595-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirby I, Davison E, Beavil AJ, Soh CP, Wickham TJ, Roelvink PW, Kovesdi I, Sutton BJ, Santis G. 2000. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J Virol 74:2804–2813. doi: 10.1128/JVI.74.6.2804-2813.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirby I, Lord R, Davison E, Wickham TJ, Roelvink PW, Kovesdi I, Sutton BJ, Santis G. 2001. Adenovirus type 9 fiber knob binds to the coxsackie B virus-adenovirus receptor (CAR) with lower affinity than fiber knobs of other CAR-binding adenovirus serotypes. J Virol 75:7210–7214. doi: 10.1128/JVI.75.15.7210-7214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Persson BD, Reiter DM, Marttila M, Mei YF, Casasnovas JM, Arnberg N, Stehle T. 2007. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat Struct Mol Biol 14:164–166. doi: 10.1038/nsmb1190. [DOI] [PubMed] [Google Scholar]
- 51.Gustafsson DJ, Segerman A, Lindman K, Mei YF, Wadell G. 2006. The Arg279Gln [corrected] substitution in the adenovirus type 11p (Ad11p) fiber knob abolishes EDTA-resistant binding to A549 and CHO-CD46 cells, converting the phenotype to that of Ad7p. J Virol 80:1897–1905. doi: 10.1128/JVI.80.4.1897-1905.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pache L, Venkataraman S, Reddy VS, Nemerow GR. 2008. Structural variations in species B adenovirus fibers impact CD46 association. J Virol 82:7923–7931. doi: 10.1128/JVI.00754-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, Liaw YC, Stone D, Kalyuzhniy O, Amiraslanov I, Tuve S, Verlinde CL, Shayakhmetov D, Stehle T, Roffler S, Lieber A. 2007. Identification of CD46 binding sites within the adenovirus serotype 35 fiber knob. J Virol 81:12785–12792. doi: 10.1128/JVI.01732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pache L, Venkataraman S, Nemerow GR, Reddy VS. 2008. Conservation of fiber structure and CD46 usage by subgroup B2 adenoviruses. Virology 375:573–579. doi: 10.1016/j.virol.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 55.Chodosh J, Miller D, Stroop WG, Pflugfelder SC. 1995. Adenovirus epithelial keratitis. Cornea 14:167–174. [PubMed] [Google Scholar]
- 56.Chiu CY, Wu E, Brown SL, Von Seggern DJ, Nemerow GR, Stewart PL. 2001. Structural analysis of a fiber-pseudotyped adenovirus with ocular tropism suggests differential modes of cell receptor interactions. J Virol 75:5375–5380. doi: 10.1128/JVI.75.11.5375-5380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lavappa KS, Macy ML, Shannon JE. 1976. Examination of ATCC stocks for HeLa marker chromosomes in human cell lines. Nature 259:211–213. doi: 10.1038/259211a0. [DOI] [PubMed] [Google Scholar]
- 58.Fahnestock PKS, Yadav D, Benson J, Cooper J, Ikonomi P. 2008. Molecular authentication as a quality control tool in a biological repository, abstr P4-14, p 225 Abstr Am Soc Virol 27th Annu Meet, Cornell University, Ithaca, NY. [Google Scholar]
- 59.Singh G, Robinson CM, Dehghan S, Schmidt T, Seto D, Jones MS, Dyer DW, Chodosh J. 2012. Overreliance on the hexon gene, leading to misclassification of human adenoviruses. J Virol 86:4693–4695. doi: 10.1128/JVI.06969-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim JI, Lee I, Park S, Hwang MW, Bae JY, Lee S, Heo J, Park MS, Garcia-Sastre A. 2013. Genetic requirement for hemagglutinin glycosylation and its implications for influenza A H1N1 virus evolution. J Virol 87:7539–7549. doi: 10.1128/JVI.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tse H, Kao RY, Wu WL, Lim WW, Chen H, Yeung MY, Woo PC, Sze KH, Yuen KY. 2011. Structural basis and sequence co-evolution analysis of the hemagglutinin protein of pandemic influenza A/H1N1 (2009) virus. Exp Biol Med 236:915–925. doi: 10.1258/ebm.2011.010264. [DOI] [PubMed] [Google Scholar]
- 62.Parisi G, Zea DJ, Monzon AM, Marino-Buslje C. 2015. Conformational diversity and the emergence of sequence signatures during evolution. Curr Opin Struct Biol 32:58–65. doi: 10.1016/j.sbi.2015.02.005. [DOI] [PubMed] [Google Scholar]