ABSTRACT
Despite the advent of combined antiretroviral therapy (cART), the persistence of viral reservoirs remains a major barrier to curing human immunodeficiency virus type 1 (HIV-1) infection. Recently, the shock and kill strategy, by which such reservoirs are eradicated following reactivation of latent HIV-1 by latency-reversing agents (LRAs), has been extensively practiced. It is important to reestablish virus-specific and reliable immune surveillance to eradicate the reactivated virus-harboring cells. In this report, we attempted to reach this goal by using newly developed chimeric antigen receptor (CAR)-T cell technology. To generate anti-HIV-1 CAR-T cells, we connected the single-chain variable fragment of the broadly neutralizing HIV-1-specific antibody VRC01 to a third-generation CAR moiety as the extracellular and intracellular domains and subsequently transduced this into primary CD8+ T lymphocytes. We demonstrated that the resulting VC-CAR-T cells induced T cell-mediated cytolysis of cells expressing HIV-1 Env proteins and significantly inhibited HIV-1 rebound after removal of antiviral inhibitors in a viral infectivity model in cell culture that mimics the termination of the cART in the clinic. Importantly, the VC-CAR-T cells also effectively induced the cytolysis of LRA-reactivated HIV-1-infected CD4+ T lymphocytes isolated from infected individuals receiving suppressive cART. Our data demonstrate that the special features of genetically engineered CAR-T cells make them a particularly suitable candidate for therapeutic application in efforts to reach a functional HIV cure.
IMPORTANCE The presence of latently infected cells remains a key obstacle to the development of a functional HIV-1 cure. Reactivation of dormant viruses is possible with latency-reversing agents, but the effectiveness of these compounds and the subsequent immune response require optimization if the eradication of HIV-1-infected cells is to be achieved. Here, we describe the use of a chimeric antigen receptor, comprised of T cell activation domains and a broadly neutralizing antibody, VRC01, targeting HIV-1 to treat the infected cells. T cells expressing this construct exerted specific cytotoxic activity against wild-type HIV-1-infected cells, resulting in a dramatic reduction in viral rebound in vitro, and showed persistent effectiveness against reactivated latently infected T lymphocytes from HIV-1 patients receiving combined antiretroviral therapy. The methods used in this study constitute an improvement over existing CD4-based CAR-T technology and offer a promising approach to HIV-1 immunotherapy.
INTRODUCTION
HIV-1 replication can be efficiently suppressed with combined antiretroviral therapy (cART). However, treatment must be maintained throughout the lifetime of the patient, as this virus can persist in a stable latent reservoir, constituting a major barrier to the establishment of an HIV-1 cure. To date, many strategies have been proposed for the eradication of HIV-1 reservoirs (1–3). Recent efforts have focused on the reactivation of HIV-1-harboring cells with special latency-reversing agents (LRAs), which expose the virus-infected cells to the immune system without global T cell activation. This modality is also known as the shock and kill strategy (4–9). It is well known that HIV-1 can quickly acquire mutations to evade immune recognition (10–12). Several studies have indicated that CD8+ T lymphocytes in infected patients on cART lack HIV-1-specific or effective immune responses and cannot completely eliminate latently infected cells, even after successful reactivation (13, 14). Therefore, the reestablishment of potent antiviral immunity is required for an ultimate kill strategy to eradicate viral reservoirs (7).
In recent years, chimeric antigen receptor (CAR)-modified immune cells have emerged as a novel tool to kill cancer cells in a high-affinity, T cell receptor-independent, and major histocompatibility complex (MHC)-unrestricted manner (15). Such specific targeting of cancer cells can be achieved through the adoptive transfer of autologous immune cells genetically engineered to express a CAR molecule recognizing the natural antigen on malignant cells (16–19). The clinical use of CARs against leukemia and lymphoma has been shown to be highly effective and successful (20–23).
This strategy has also been proposed for the treatment of viral infection, including HIV-1, hepatitis B virus, and hepatitis C virus (24, 25). Previous reports have described the generation of HIV-1-specific CAR-T cells by connection of an extracellular antigen-binding domain to intracellular T cell activation domains (e.g., CD3ζ and CD28). The former can be a single-chain variable fragment (scFv) (26–30) or a natural molecule, such as CD4 (26, 27, 31–36), capable of directly inducing the death of cells expressing the viral envelope glycoprotein (Env). However, it remains to be determined whether these CAR-T cells can be used to kill the reactivated HIV-1 latently infected cells from the infected individuals receiving suppressive cART.
HIV-1 broadly neutralizing antibodies (bNAbs) have been well characterized in recent years and include molecules such as VRC01-03, 3BNC117, NIH45-46, and PGT125-131, among others. By targeting several conserved sites in the glycoproteins gp120 and gp41, they are capable of neutralizing multiple HIV-1 strains (37). These antibodies can protect against challenge with HIV-1 in humanized mouse models and nonhuman primates (38–40). They have also been shown to exert protection in a recent human HIV-1 vaccine clinical trial (41).
In this report, we developed a novel MHC-independent third-generation anti-HIV-1 CAR molecule (CD3ζ-CD28-CD137). The extracellular domain consisted of an scFv region derived from the bNAb VRC01 capable of redirecting the antigen specificity of primary CD8+ T cell populations against gp120, which comprises the CD4 binding site (CD4bs) of various HIV-1 isolates (42, 43). We have also redesigned the CAR cytoplasmic region, composed of a CD3ζ chain and multiple signaling domains (CD28 and CD137) (44). We found that the newly designed CAR-T cells, here referred to as VC-CAR-T cells, were able to induce T cell-mediated cytolysis after coculture with gp120-expressing cells and wild-type HIV-1-infected CD4+ T cells. We also found that VC-CAR-T cells display superior potency compared to the CD4-CAR described previously (26, 27, 31–36). Importantly, we have also confirmed that they can effectively kill the reactivated HIV-1-infected CD4+ T lymphocytes isolated from HIV-1-infected patients.
MATERIALS AND METHODS
Patient cohort.
This research was approved by the Ethics Review Board of The Eighth People's Hospital at Guangzhou (Guangzhou Infectious Disease Hospital, Guangzhou, China) and the Ethics Review Board of Sun Yat-Sen University. HIV-1-infected patients were recruited at The Eighth People's Hospital at Guangzhou and gave written informed consent with approval of the Ethics Committees. All patients were recruited on the basis of prolonged continuous suppression of plasma HIV-1 viremia on cART to below the limit of detection of standard clinical assays (<50 copies HIV-1 RNA ml−1). Unidentified human peripheral blood mononuclear cells (PBMCs) of healthy blood donors were provided by the Guangzhou Blood Center. We did not have any interaction with these human subjects or protected information; therefore, no informed consent was required.
Cell lines.
HEK293T and HeLa cells were maintained in conditioned Dulbecco's modified Eagle medium (DMEM) (Gibco, Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS) (Gibco, Invitrogen, Carlsbad, CA). Jurkat-internal ribosomal entry site-green fluorescent protein (IRES-GFP) and Jurkat-gp160-IRES-GFP strains, including Jurkat-gp160NL4-3 and Jurkat-gp160BaL, which constitutively express Env from different HIV-1 strains, such as NL4-3 and BaL, were constructed and grown in conditioned RPMI 1640 (Gibco, Invitrogen, Carlsbad, CA) medium plus 10% FBS and 2 mM GlutaMAX (Gibco, Invitrogen, Carlsbad, CA). All cell culture media contained 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Gibco, Invitrogen, Carlsbad, CA). All cell lines were maintained in an environment of 37°C and 5% CO2.
Construction of CAR-encoding lentiviral vector.
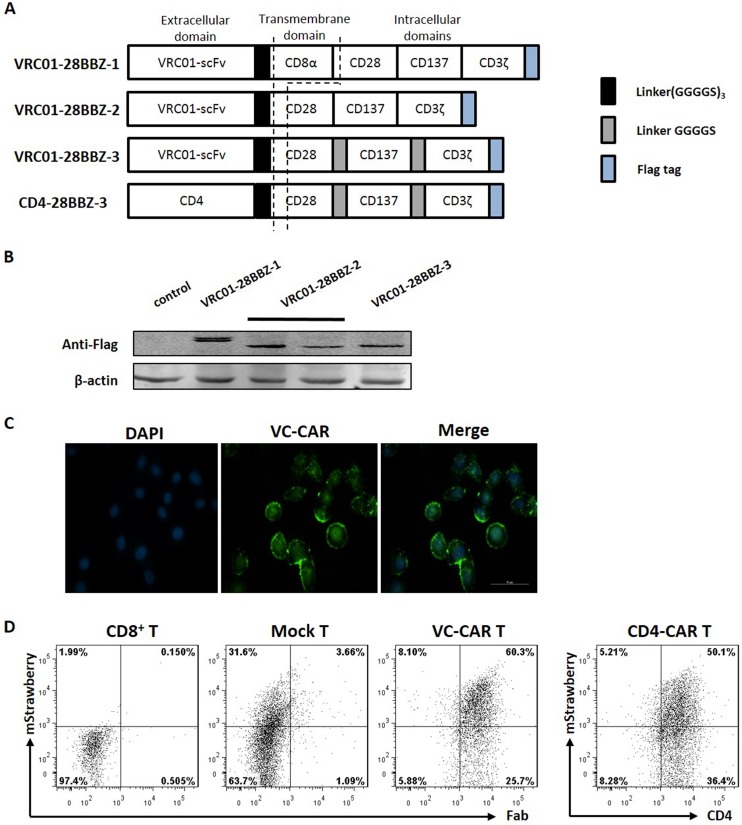
The scFv region derived from HIV-1 broadly neutralizing monoclonal antibody VRC01 was used as the N-terminal region of the CARs directed against the CD4 binding site of envelope glycoprotein gp120. As shown in Fig. 1, the VRC01-28BB CAR was comprised of scFv (VRC01) linked in frame to the hinge domain, transmembrane region, and intracellular signal domains, which contained CD28 (nucleotides 460 to 660; GenBank accession number NM_006139.3), CD137 (nucleotides 640 to 765; NM_001561.5), and CD3ζ (nucleotides 160 to 492; NM_198053.2) in tandem with or without a GGGS sequence inserted between each signaling domain. The transmembrane and intracellular signaling regions of VRC01-28BBZ-1 were from the previous design (44). VRC01-28BBZ-2 contains a modified version of a truncated CD28 transmembrane motif, and VRC01-28BBZ-3 is redesigned with an insert linker (GGGS)3 between each signaling domain (35, 45, 46). The third-generation CAR moieties were inserted into lentiviral vector pCPPT-IRES-mStrawberry. The construction of pCPPT-IRES-mStrawberry is described in Fig. S1 in the supplemental material. Meanwhile, scFv with extracellular domains of the human CD4 molecule was substituted according to a previous description (35). All of the constructs were verified by sequencing, and the complete sequences of all of the constructs are listed in Table S1.
FIG 1.
Characterization of VRC01-28BBZ CAR-T cells. (A) Schematic representation of the VRC01-28BBZ CAR (not to scale). (B) Western blotting was performed to detect the expression of VRC01-28BBZ with anti-Flag tag antibody in HEK293T cells by transfecting three pCPPT-IRES-VRC01-28BBZ-mStrawberry plasmids. The HEK293T cells transfected with pCPPT-IRES-mStrawberry empty vector served as a negative control. (C) Immunofluorescence staining was performed to detect VRC01-28BBZ-3 CAR (VC-CAR) expression in HeLa cells with anti-Flag tag antibody. DAPI staining represents the nuclei. (D) Flow cytometry was performed to detect the transduction efficiencies of VC-CAR and empty vector in human CD8+ T cells by detection of mStrawberry fluorescence. Transduced CD8+ T cells were measured by detecting mStrawberry fluorescence and human Fab stained with goat antibody through flow cytometry. Untransduced CD8+ T cells served as a negative control. The CD4-CAR-transduced CD8+ T cells were detected with anti-CD4 antibody through flow cytometry. CD4-negative gating was set on untransduced control cells (data not shown).
Isolation and culture of primary human T lymphocytes.
The PBMCs derived from healthy donors or HIV-1-infected patients were isolated from buffy coats by Ficoll-Hypaque gradient separation. Excess PBMCs were cryopreserved until ready to use. Primary human CD4+ and CD8+ T cells were obtained from PBMCs by negative magnetic selection through human CD8+ T lymphocyte enrichment set DM (BD-IMag). The isolated T cells were stimulated for 2 days with anti-CD3 antibody at 1 μg ml−1 (R&D Systems), anti-CD28 antibody at 1 μg ml−1 (R&D Systems), and recombinant human interleukin-2 (IL-2) at 10 ng ml−1 (R&D Systems) before wild-type HIV-1 or pseudovirus infection. The transduced T cells were expanded in conditioned medium containing 90% RPMI 1640 (Gibco, Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Gibco, Invitrogen, Carlsbad, CA) and 2 mM GlutaMAX (Gibco, Invitrogen, Carlsbad, CA) at a concentration of 1 × 106 cells ml−1. Cells were fed with IL-2 at 10 ng ml−1 and IL-7 at 10 ng ml−1 (R&D Systems) every other day. The genetically modified T cells were used for functional assay at 1 week after transduction. All cell culture media contained 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Gibco, Invitrogen, Carlsbad, CA), and cell cultures were maintained in an environment of 37°C and 5% CO2.
Pseudovirus production and transduction.
The day before transduction, HEK293T cells were seeded at 8 × 106 cells per 100-mm dish. Twenty-four hours later, the pseudoviruses were generated by cotransfecting HEK293T cells with pCPPT-IRES-mStrawberry plasmids encoding various CAR moieties (13.5 μg), pMD.2G encoding VSV-G envelope (7.5 μg), and a packaging vector, psPAX2 (16.5 μg), using a phosphate transfection system by following the manufacturer's instructions. Supernatants were harvested after 48 h and filtered through a 0.22-μm membrane to remove cell debris. Pseudoviruses were concentrated by centrifuging with a 30% volume of 50% polyethylene glycol 6000 and a 13% volume of 4 M NaCl. For infection, on day 1, 106 purified and anti-CD3− and anti-CD28 antibody-activated T cells were plated onto a 24-well plate, followed by adding 2 ml of pseudovirus supernatant plus Polybrene (Sigma) at 8 μg ml−1. The plates were centrifuged for 90 min at 350 × g and then incubated at 37°C. Twelve hours later, cells were infected for the secondary round with the same procedure. At day 2 postinfection, pseudoviruses were replaced by the fresh culture media as described above.
Real-time qRT-PCR analysis.
Total RNA was isolated with TRIzol reagent (Life Technologies) and then subjected to cDNA synthesis using a PrimeScript reverse transcription (RT) reagent kit (TaKaRa). All primers were annealed at 37°C and RT was processed at 42°C. Quantitative PCR was performed with a SYBR premix Ex Taq II kit (TaKaRa) by following the manufacturer's instructions. The primer sequences are listed in Table S2 in the supplemental material. The expression of viral RNAs was determined by real-time quantitative reverse transcription-PCR (qRT-PCR) with the primer pair SK38 (5′-ATAATCCACCTATCCCAGTAGGAGAAA-3′) and SK39 (5′-TTTGGTCCTTGTCTTATGTCCAGAATGC-3′). An in vitro-synthesized HIV-1 RNA, after quantification, was used as the external control for measuring cell-associated viral RNA (47). Quantification was normalized to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene or β-actin.
Western blotting.
Thirty micrograms of protein was resolved by SDS-PAGE and transferred to nitrocellulose blotting membranes (Bio-Rad, Hercules, CA). The following antibodies and dilutions were used in these experiments: rabbit-anti-Flag (1:1,000 dilution) (MBL) that recognizes the C-terminal region of CARs, mouse anti-human β-actin (1:5,000 dilution) (MBL), goat-anti-rabbit-conjugated IRDye 800CW secondary antibodies (1:5,000 dilution) (LI-COR Biosciences), and goat-anti-mouse-conjugated IRDye 680RD secondary antibodies (1:5,000 dilution) (LI-COR Biosciences). Blots were then detected by a Odyssey two-color infrared scanner (LI-COR Biosciences).
Immunofluorescence.
Cells were fixed with 4% paraformaldehyde. After permeabilization with 0.1% Triton X-100, cells were incubated with 3% bovine serum albumin (BSA) to block nonspecific binding. Cells were then stained with the primary antibody rabbit anti-Flag (MBL) (1:1,000 dilution) at 20°C for 2 h and then were probed with Dylight488 goat anti-mouse secondary antibody (1:5,000 dilution) at 20°C for 2 h (Abcam). Fluorescent signals were detected using a fluorescence microscope (DMI6000B; Leica). 4′,6-Diamidino-2-phenylindole (DAPI) (Life Technologies) was used for nuclear staining.
LDH assay.
The specific killing activity of VRC01-28BBZ CAR-transduced T cells toward Jurkat cells expressing HIV-1 envelope glycoprotein or HIV-1-infected primary CD4+ T lymphocytes at different ratios (from 8:1 to 0.5:1) was measured after coculture for 24 to 48 h in a 96-well U-bottom plate by lactate dehydrogenase (LDH) assay using the CytoTox 96 nonradioactive cytotoxicity kit (G1781; Promega). The manufacturer's instructions were followed. Absorbance values of wells containing effector cells alone and target cells alone were combined and subtracted as the background from the values of the cocultures. Wells containing target cells alone were mixed with a lysis reagent for 30 min at 37°C, and the resulting luminescence was set as 100% lysis.
IFN-γ ELISpot assay.
For enzyme-linked immunosorbent spot (ELISpot) assay, VC-CAR or control effector CD8+ T cells (105 cells) were mixed with target cells at the indicated range of effector-to-target (E:T) ratios and then added to the anti-gamma interferon (IFN-γ) antibody-precoated plates from the human IFN-γ ELISpot assay kit (DKW22-1000-096s; Dakewe), along with a negative control (effector CD8+ T cells alone) or positive control (phytohemagglutinin [PHA] stimulation). Plates were incubated for 16 to 20 h at 37°C and 5% CO2. The ELISpot assays were then performed according to the manufacturer's instructions. The plates were scanned by an S6 ultra immunoscan reader (Cellular Technology Ltd.), and the number of IFN-γ-positive T cells was calculated by ImmunoSpot 5.1.34 software (Cellular Technology Ltd.).
ELISA.
VC-CAR or control effector CD8+ T cells (105 cells) were cocultured with Jurkat-based target cells on gradients from 1:1 to 1:8 (E:T ratio) in 96-well round-bottom plates. Supernatants were collected after 20 to 24 h. Cytokine release by effector CD8+ T cells in response to stimulation with target cells was analyzed using granzyme B (Neobioscience) and IL-2 enzyme-linked immunosorbent assay (ELISA) kits (MultiSciences, Lianke BiotechCo., Ltd.) according to the manufacturer's instructions. HIV-1 viral particle production in cell cultures was determined with an HIV-1 p24 ELISA kit by following the manufacturer's protocol (Clontech).
Flow cytometry.
For intracellular HIV-1 Gag (p24) staining, surface staining (CD3, CD8, or CD4) was performed, followed by intracellular staining for HIV-1 p24 (Santa Cruz Biotechnology) with the transcription factor buffer set, including fixation/permeabilization and fixation/wash buffers (BD Biosciences), according to the manufacturer's protocol. The antibody used in surface staining of the VC-CAR moiety was goat F(ab′)2 anti-human IgG-F(ab′)2 (DyLight 488) (catalog number ab98600; Abcam). Other surface antibodies were purchased from BD Biosciences. Data were acquired on a BD FACSAria and were analyzed with FlowJo software (Tree Star, Ashland, OR).
In vitro wild-type HIV-1 infection and drug withdrawal model.
The PBMCs from healthy donors were stimulated by adding 1 mg ml−1 PHA and 10 ng ml−1 IL-2 to the conditioned RPMI 1640 medium with 10% heat-inactivated fetal bovine serum and antibiotics for 2 days before isolation of CD4+ T cells. CD4+ T cells were infected with laboratory virus strain NL4-3 (p24 titer of 1 ng ml−1). Three hours after HIV-1NL4-3 infection, the culture medium was changed by centrifugation. Infected CD4+ T cells were cultured in basal medium plus IL-2 (10 ng ml−1; recombinant human; R&D Systems) and further incubated at 37°C in a humidified incubator with 5% CO2. Six days after HIV-1NL4-3 infection, azidothymidine (Zidovudine; Sigma-Aldrich) and lopinavir (Sigma-Aldrich) were added to the CD4+ T cell culture, both at 50 μM, to inhibit virus production and prevent further infection events. The cells were then cultured in the presence of low-concentration IL-2 (1 ng ml−1). Anti-HIV-1 drugs were withdrawn when the viral production was significantly decreased to the marginal level for p24 detection (about 6 to 8 day after drugs adding), and then 0.5 × 106 CD4+ T cells were mixed with autologous VC-CAR or control CD8+ T cells at 1:2 or 1:4 ratios in the conditioned medium plus IL-2 (10 ng ml−1) at 1 ml in a 24-well plate. Every 2 days the cultures were tested for HIV-1 p24 antigen with the HIV-1 p24 antigen assay kit by following the manufacturer's instructions.
Viral outgrowth assay.
Freshly purified CD4+ T lymphocytes were obtained from a single blood draw from HIV-1-infected patients receiving suppressive cART. Coculture was performed to recover replication-competent viruses as previously described, with some modifications (48). Briefly, at day 1, 1 × 106 CD4+ T lymphocytes from HIV-1-infected patients were stimulated by coculture with 1 × 107 irradiated allogeneic PBMC (5000R, Rs2000; Rad Source) from uninfected donors and 1 μg ml−1 PHA-M (Sigma-Aldrich) or a combination of specific LRAs, including 500 nM suberoylanilide hydroxamic acid (SAHA; Sigma-Aldrich) and 20 nM bryostatin-1 (Sigma-Aldrich), in the conditioned RPMI 1640 medium containing 10% FBS and 10 ng ml−1 IL-2. At day 2, the cell culture was mixed with autologous 1 × 106 VC-CAR or control CD8+ T cells at a 1:1 ratio. At day 3, cell-associated viral RNAs were determined by qRT-PCR, and 4 × 106 activated CD4+ lymphoblasts from healthy donors were added in fresh medium plus IL-2 (10 ng ml−1) to propagate replication-competent viruses in the culture wells. Typically, two additions were made of CD4+ lymphoblasts from uninfected donors as target cells for HIV-1 outgrowth at days 3 and 7. At day 5, owing to the proliferation of both lymphoblasts and patient cells, the culture medium was changed. At day 7, all wells were split in half. Cells and media were gently mixed, and half of each well was discarded. A second addition of lymphoblasts was added to all wells. A total of 4 × 106 lymphoblasts were added to wells containing 1 × 106 patient CD4+ T cells. At day 10, wells again were fed with fresh medium as at day 5. Supernatants from each well were tested for HIV-1 RNA and/or HIV-1 p24 protein at various time points by qRT-PCR and ELISA via an HIV-1 p24 antigen ELISA kit.
RESULTS
Generation of VRC01-28BBZ CAR specifically targeting HIV-1 gp120.
As shown schematically in Fig. 1A, we connected the scFv sequence derived from the VRC01 bNAb active against genetically diverse HIV-1 isolates to three analogous but different third-generation intracellular CAR moieties (42, 43). The transmembrane and intracellular signaling regions of the first of these, VRC01-28BBZ-1, were constructed as described in a previous report (44). VRC01-28BBZ-2 contains a modified version of a truncated CD28 transmembrane motif which is required for CD28 costimulation signal, while VRC01-28BBZ-3 was redesigned with an insert linker between each signaling domain (Fig. 1A; see also Table S1 in the supplemental material) (35, 45, 46). Sequences encoding the three moieties, each with a FLAG tag added to the C terminus, were then inserted into pCPPT-IRES-mStrawberry lentiviral vectors, and the expression of all three CARs was validated by Western blotting and immunofluorescence (Fig. 1B and C). Primary CD8+ T lymphocytes were then transduced with the lentiviral vectors. Flow cytometry further demonstrated the high transduction efficiencies and cell surface expression of the CAR moiety (>60%) using an antibody against human Fab (antigen-binding antibody fragment), as shown in Fig. 1D.
Selection of the most efficient CAR moiety.
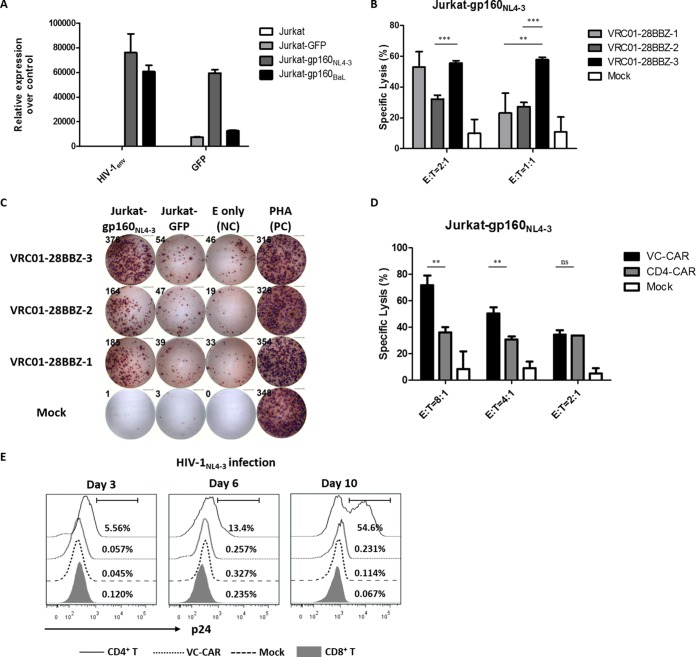
To identify the optimal CAR for use in further experiments, CD8+ T lymphocytes transduced with lentiviral vectors encoding the above-mentioned CAR moieties were mixed with Jurkat-derived target cells expressing HIV-1NL4-3 envelope glycoprotein and enhanced green fluorescent protein (gp160-IRES-eGFP) (Fig. 2A). We compared the cytotoxic potency of each group of CAR-expressing T cells by detecting the release of lactate dehydrogenase (LDH) (49). All three CAR-T cell types displayed specific and potent cytotoxicity against Jurkat-gp160NL4-3 cells. Notably, the VRC01-28BBZ-3 group demonstrated higher cytotoxicity at 2:1 and 1:1 effector-to-target cell ratios than the other two CAR-T cell types tested, while control effector cells showed negligible activity against their targets (Fig. 2B). The results of IFN-γ ELISpot assay also indicated that VRC01-28BBZ-3 exhibited superior efficacy compared to VRC01-28BBZ-1 and VRC01-28BBZ-2 in response to Jurkat-gp160NL4-3 cells (Fig. 2C). Therefore, we chose to use VRC01-28BBZ-3-expressing T cells, here referred to as VC-CAR-T cells, for subsequent experiments. Previous studies have reported that CD4-based CAR-T cells, referred to here as CD4-CAR-T cells, display direct cytotoxic activity against HIV-1 Env-expressing target cells (26, 27, 31–36). To compare the efficiency of CD4-CAR and VC-CAR, we generated the former by replacing the VC-CAR scFv with the human CD4 extracellular domain and transduced this construct into CD8+ T lymphocytes (Fig. 1A and D). Cytolysis experiments indicated that VC-CAR-T cells were much more effective than CD4-CAR-T cells over the indicated range of effector-to-target ratios (Fig. 2D). A potential mechanism to explain this observation is that the HIV-1 bNAb-derived scFv could have a stronger affinity toward the gp120 subunit than the CD4 moiety.
FIG 2.
Selection of the most efficient CAR moiety. (A) qRT-PCR was performed to characterize the expression of HIV-1 Env and eGFP in Jurkat-based target cells. Jurkat cells served as a negative control. (B) Cytotoxicity assays were performed using the CytoTox nonradioactive cytotoxicity kit, using as targets (T) Jurkat-gp160NL4-3 cells constitutively expressing Env from HIV-1NL4-3. Cocultures were performed for 24 h with T cells expressing the indicated CARs (E, effector cells); the RFP-CD8+ T cells (Mock) served as a negative control. Shown are the cytotoxic effects of VRC01-28BBZ-transduced T cells on target cells at 2:1 and 1:1 (E:T) ratios for 24 h. (C) Characterization of VRC01-28BBZ-transduced effector CD8+ T cells after coculture with Jurkat-gp160NL4-3 or Jurkat-GFP cells at a 4:1 (E:T) ratio for 20 h by IFN-γ ELISpot assay. Effector cells alone served as the negative control (NC). The PHA-stimulated effector cells served as the positive control (PC). These data represent three independent experiments. (D) Cytotoxic effect of VC-CAR-T cells and CD4-CAR-T cells on Jurkat-gp160NL4-3 at the indicated range of E:T ratios (24 h). (E) The CD8+ T cells expressing VC-CAR were challenged with cell-free HIV-1NL4-3 (p24 titer of 200 ng ml−1). Infection was analyzed at the indicated time points by staining for intracellular p24 after gating the CD3+ CD8+ subpopulation. Untransduced CD8+ T cells and CD8+ T cells transduced with empty vector (mock) served as a negative control, and CD4+ T cells (gate CD3+ CD8−) served as a positive control. The percent value in each histogram (left three panels) indicates the p24-positive population. (A, B, and D) Data reflect means ± standard errors of the means (SEM), and P values were calculated using the two-tailed unpaired Student's t test with equal variances (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data shown in panel E represent two independent experiments.
Susceptibility of VC-CAR-T cells to HIV-1 infection.
It is concerning that the specific interaction between the scFv and HIV-1 gp120 might result in CAR-T cells being susceptible to HIV-1 infection (31, 32, 50). To examine this possibility, unmodified CD8+ T cells and those expressing VC-CAR or an empty vector were challenged with wild-type HIV-1NL4-3 (X4). The cells were analyzed by intracellular p24 staining at days 3, 6, and 10. Percentages calculated from histograms indicated that the population of p24-positive CAR-T cells was not significantly different from those of control CD8+ T cells and much lower than that of the HIV-1 natural target CD4+ T cells in same period, indicating that they are not susceptible to HIV-1 infection (Fig. 2E).
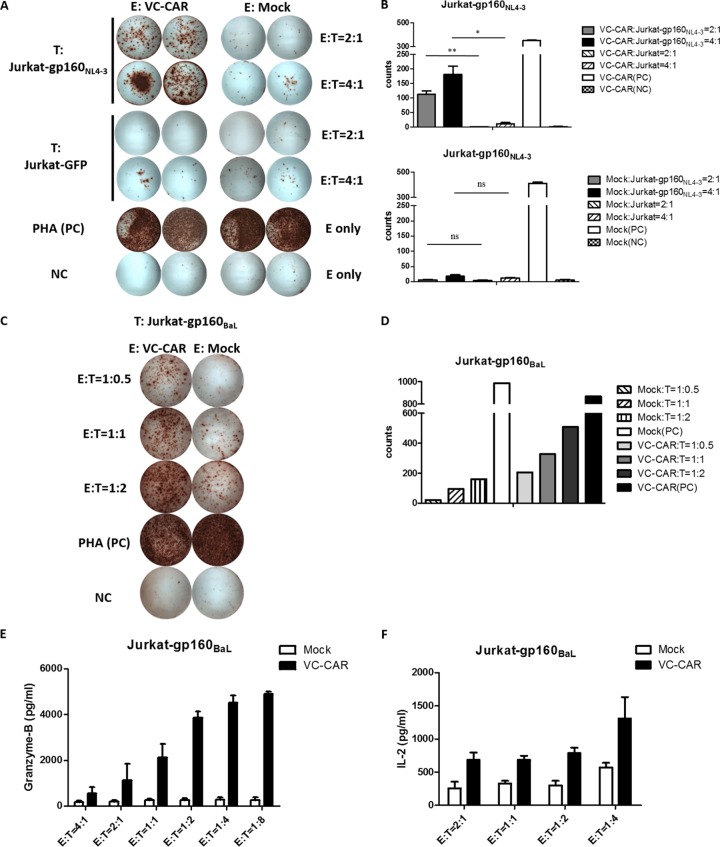
Efficient in vitro cytokine secretion by VC-CAR CD8+ T cells following HIV-1 Env stimulation.
To examine its functionality, the VC-CAR construct was transduced into CD8+ T cells with lentiviral vector. We then cocultured these VC-CAR-T cells with two HIV-1 Env-expressing target cell lines generated by ourselves, named Jurkat-gp160NL4-3 and Jurkat-gp160BaL. We found that secretion of IFN-γ was robustly increased compared to that observed from cocultures with HIV-1 Env-negative cells (Jurkat-GFP) (Fig. 3A to D). The specificity of these cells was also demonstrated by the absence of IFN-γ secretion when HIV-1 Env-expressing target cell lines were cocultured with control effector cells (Fig. 3A and B). The functional potency of effector VC-CAR-T cells in response to Jurkat-gp160NL4-3 cells was further validated by the specific secretion of IL-2 and granzyme B in a dose-dependent manner (Fig. 3E and F).
FIG 3.
Cytokine release of VC-CAR-T cells after coculture with Env-expressing Jurkat target cells. (A) Characterization of VC-CAR or control transduced effector CD8+ T cells cocultured with Jurkat-gp160NL4-3 or Jurkat-GFP cells at 2:1 and 4:1 (E:T) ratios for 20 h by IFN-γ ELISpot assay. The PHA-stimulated effector cells served as the positive control (PC). Effector cells alone served as the negative control (NC). (B) Summary of IFN-γ ELISpot assays described for panel A. The count number of the y axis was measured as spot-forming cells (SFCs)/104 CD8+ T cells. (C) Characterization of VC-CAR or control transduced effector CD8+ T cells cocultured with Jurkat-gp160BaL cells. (D) Summary of IFN-γ ELISpot assay described for panel C. The count number of the y axis was measured as SFCs/104 CD8+ T cells. (E) Granzyme B production in cocultures of Jurkat-gp160NL4-3 target cells with VC-CAR-transduced effector CD8+ T cells at the indicated E:T ratios for 20 h. The effector cells expressing red fluorescent protein served as negative controls. (F) IL-2 production from triple VC-CAR or control effector CD8+ T cells cocultured with Jurkat-gp160NL4-3 cells for 18 h. (B, E, and F) Data reflects means ± SEM. P values were calculated using the two-tailed unpaired Student's t test with equal variances (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
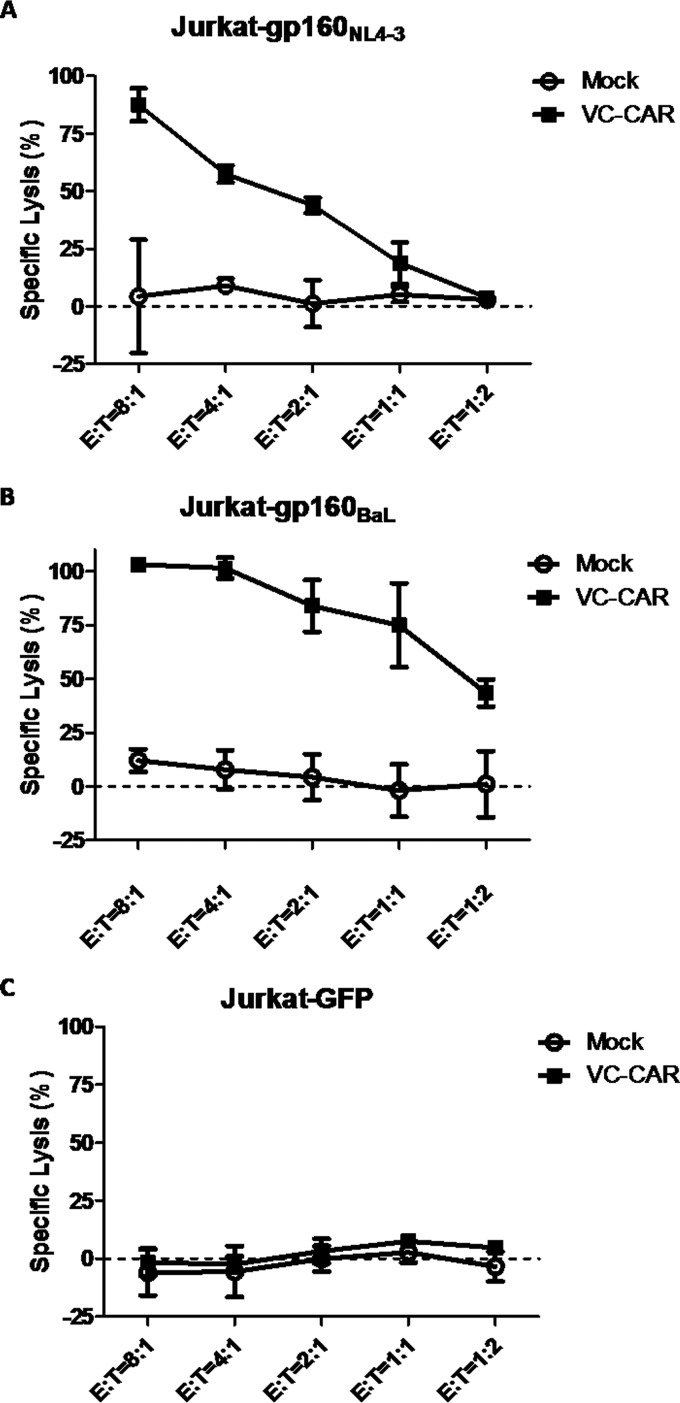
Specific cytotoxicity of HIV-1 Env-expressing Jurkat cells induced by VC-CAR-T cells.
We then tested VC-CAR-T cells for specific killing activity against Env-expressing Jurkat cell lines by detecting LDH release. VC-CAR-T cells displayed potent cytotoxic activity against two cell lines, Jurkat-gp160NL4-3 and Jurkat-gp160BaL, which constitutively express the Env protein of different HIV-1 isolates (Fig. 4A and B). In addition, the specificity of their function was confirmed by the lack of killing activity exhibited by control effector CD8+ T cells and by the absence of Env-negative Jurkat-GFP cell cytotoxicity (Fig. 4C).
FIG 4.
Specific killing of Env-expressing cells by VC-CAR-transduced effector CD8+ T cells. Direct killing of target cell lines was performed using the CytoTox nonradioactive cytotoxicity kit with Jurkat-gp160NL4-3 (A), Jurkat-gp160BaL (B), and Jurkat-GFP (C) cells as targets. Cocultures were performed for 24 h with CD8+ T cells expressing the VC-CAR (E, effector cells); the red fluorescent protein-transduced CD8+ T cells served as a negative control. Data reflect means ± SEM.
Specific cytotoxicity of wild-type HIV-1-infected cells induced by VC-CAR-T cells.
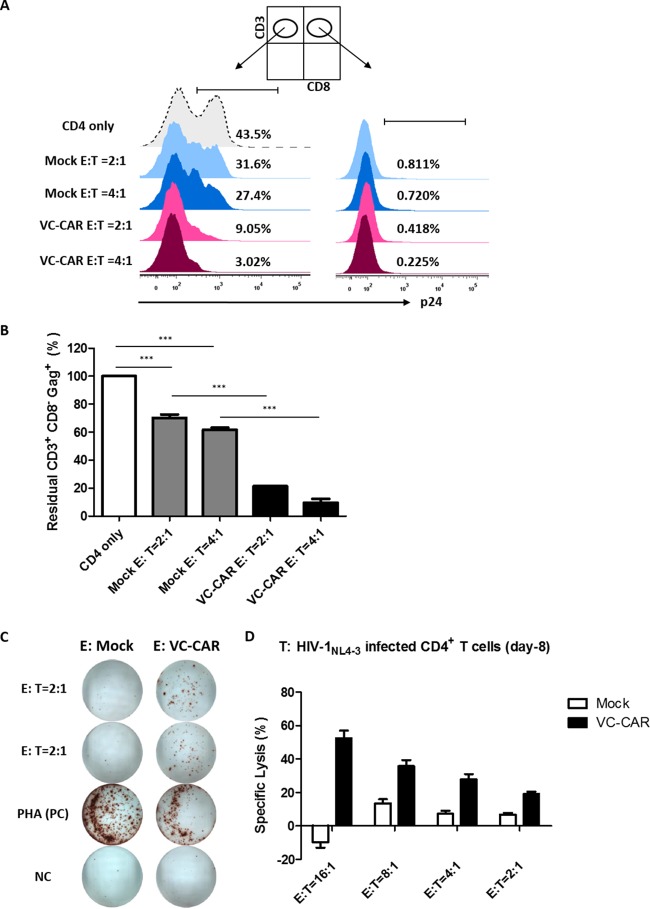
To further characterize the effectiveness of VC-CAR-T cells in eliminating cells infected by wild-type HIV-1, activated CD4+ T cells isolated from healthy donors were infected with wild-type HIV-1NL4-3. Eight days after infection, the cells were cocultured with autologous VC-CAR-T cells at ratios of 1:2 and 1:4 for 72 h, and cytotoxicity was analyzed using residual CD3+ CD8− Gag+ T cells. VC-CAR-T cells efficiently killed infected CD4+ T cells and achieved an elimination rate of more than 92% at an E:T ratio of 4:1, while for control effector CD8+ T cells this figure was less than 38%, representing a significant difference (Fig. 5A, left, and B). Although the close intercellular contacts between the effector CD8+ and infected CD4+ T cells at the cytotoxic T-lymphocyte immunological synapse would likely foster highly efficient cell-to-cell infection of effector cells, VC-CAR-T cells remained HIV-1 Gag negative as control effector CD8+ T cells did, further confirming that they are not susceptible to HIV-1 infection (Fig. 5A, right). The functional potency of the specific effector response of VC-CAR-T cells on wild-type HIV-1-infected CD4+ T cells was further validated by the secretion of IFN-γ and release of LDH (Fig. 5C and D).
FIG 5.
Specific killing of HIV-1NL4-3-infected primary CD4+ T cells by VC-CAR-transduced effector CD8+ T cells. (A) Primary CD4+ T cells were infected with HIV-1NL4-3 (p24 titer of 200 ng ml−1). At day 8 postinfection, CD4+ T cells were mixed with autologous VC-CAR or control transduced effector CD8+ T cells at a 1:2 or 1:4 ratio. Seventy-two hours after coculture, specific cytotoxicity was analyzed by flow cytometry for the intracellular staining of Gag+ T cells gated on CD3+ CD8− or CD3+ CD8+ subpopulations, respectively, as indicated at the top. (B) Summary of residual CD3+ CD8− Gag+ T cells from the flow cytometry analysis. (C) VC-CAR or control transduced effector CD8+ T cells were cocultured with HIV-1NL4-3-infected primary CD4+ T cells at a 2:1 (E:T) ratio for 20 h. IFN-γ secretion was analyzed by ELISpot assay. The PHA-stimulated effector cells served as the positive control (PC). Effector cells alone served as the negative control (NC). (D) Specific killing of HIV-1NL4-3-infected primary CD4+ T cells by VC-CAR-transduced effector CD8+ T cells. Cocultures were performed for 24 h. (B and D) Data reflect means ± SEM from triplicates. P values were calculated using the two-tailed unpaired Student's t test with equal variances (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Effective suppression of HIV-1 rebound after withdrawal of antiviral treatment in vitro.
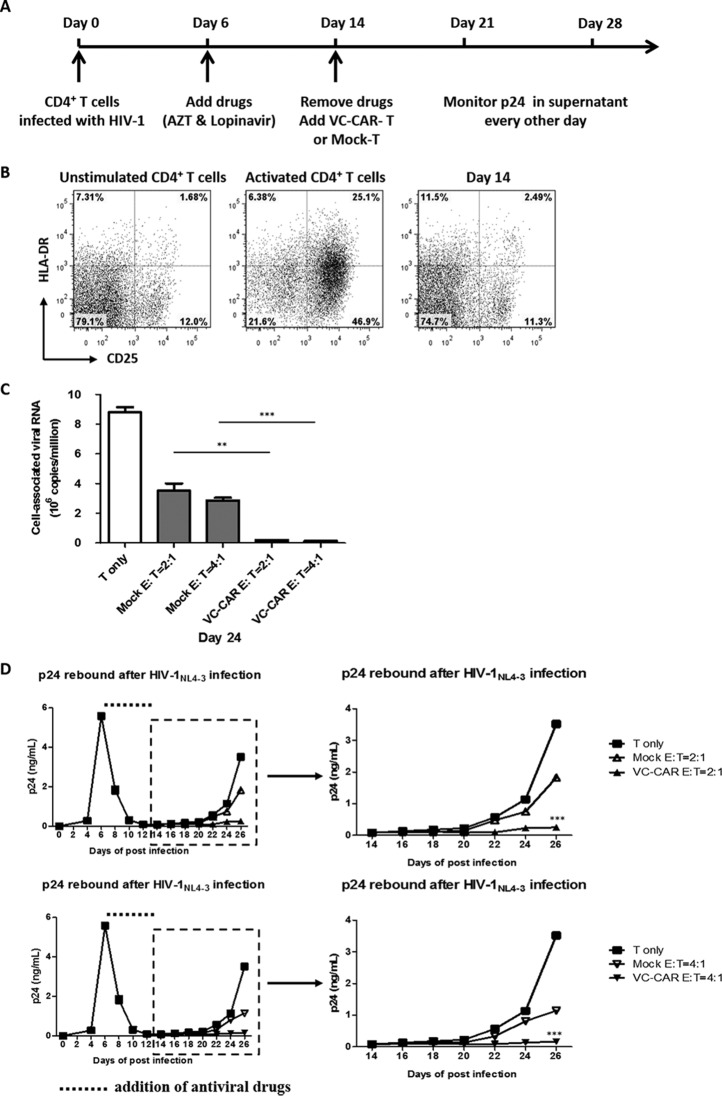
We withdrew antiviral treatment for viral infection in vitro in an attempt to mimic the in vivo viral rebound process. Peripheral blood mononuclear cells were isolated from healthy donors and divided into two populations. CD4+ T lymphocytes were used as target cells for HIV-1 infection, while CD8+ T lymphocytes were used to generate VC-CAR-T cells. Six days after wild-type HIV-1NL4-3 infection, antiretroviral compounds (azidothymidine and lopinavir) were added to the CD4+ T cell culture to inhibit virus production and prevent further infection events (Fig. 6A). Meanwhile, the cells were maintained in the presence of a very low concentration of IL-2. After approximately 8 to 10 days, virus production had significantly decreased to the lower limit for p24 detection, and infected CD4+ T cells were close to quiescence (Fig. 6B). We then withdrew anti-HIV-1 drugs and added autologous VC-CAR or control CD8+ T cells concurrently (Fig. 6A). HIV-1 was seen to rebound very quickly in CD4+ T lymphocytes in the absence of coculture, and control CD8+ T cells exhibited only limited inhibitory action on viral propagation (Fig. 6D). However, viral production was significantly and persistently suppressed following coculture with VC-CAR-T cells (Fig. 6D). The level of cell-associated viral RNA in infected CD4+ T lymphocytes also substantially decreased by nearly 50-fold (Fig. 6C), indicating effective elimination of virus-producing cells. Furthermore, this inhibitory effect on viral rebound was enhanced by increasing the effector-to-target ratio to 4:1 (Fig. 6C and D, lower).
FIG 6.
Effective suppression of HIV-1 rebound after the withdrawal of antiviral treatment in vitro. The primary CD4+ T cells were infected with HIV-1NL4-3 (1 ng ml−1 p24). (A) Experimental design. (B) The expression of the activation markers CD25 and HLA-DR on unstimulated and activated CD4+ T cells and CD4+ T cells at day 14 postinfection. The percentage of cells in each quadrant is indicated. (C) After the withdrawal of antiviral drugs, 0.5 × 106 CD4+ T cells were mixed with autologous VC-CAR engineered or control CD8+ T cells at a 1:2 or 1:4 ratio. On day 24, cells were collected and viral RNAs were isolated and amplified by real-time RT-qPCR with primer SK38 and SK39. The cutoff for cell-associated viral RNA is 800 copies ml−1. (D) Every 2 days the cultures were tested for the presence of p24 in the supernatant by ELISA. The dotted line represents the addition of antiviral drugs. (C and D) Data reflect means ± SEM. P values were calculated using the two-tailed unpaired Student's t test with equal variances (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Elimination of reactivated HIV-1 latently infected CD4+ T lymphocytes from virus-infected individuals receiving cART.
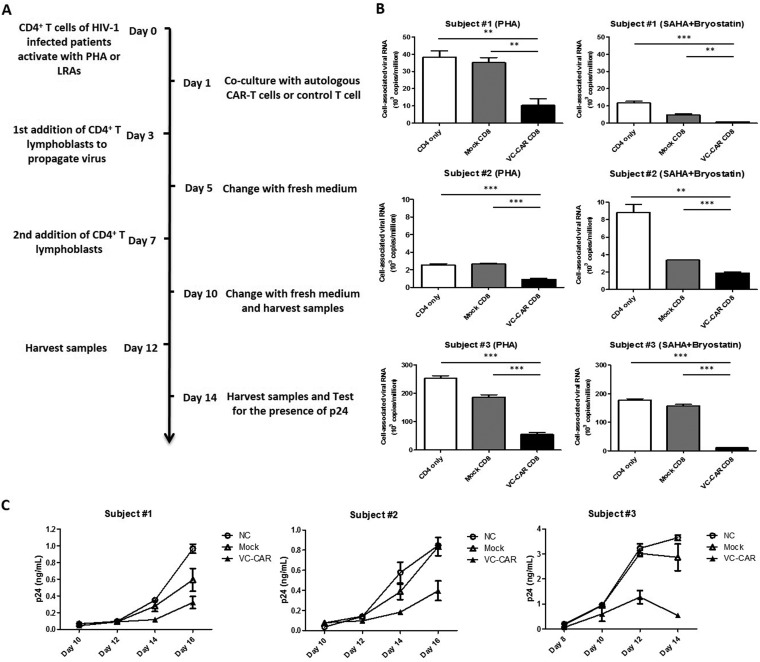
In order to determine whether VC-CAR-T cells are able to recognize and kill activated infected CD4+ T lymphocytes isolated from HIV-1-infected patients receiving suppressive cART, we performed a viral outgrowth assay in the presence of VC-CAR-T cells (48). Figure 7A shows a schematic of this procedure. We first activated the CD4+ T cells isolated from HIV-1-infected patients with PHA or a combination of specific LRAs, including a vorinostat (SAHA), a histone deacetylase inhibitor (HDACi), and bryostatin 1, a protein kinase C/nuclear factor-κB pathway activator. One day following treatment, the activated CD4+ T cells were cocultured with VC-CAR-T or control effector cells generated from autologous CD8+ T lymphocytes. We found cell-associated viral RNA levels to be significantly decreased by VC-CAR-T cells in both PHA- and LRA mixture-activated CD4+ T lymphocytes (Fig. 7B), while control CD8+ T cells from the same subjects showed only minimal or significantly diminished effects (Fig. 7B). When cocultures were maintained over a length of time, viral replication was seen to be persistently and effectively inhibited by VC-CAR-T cells (Fig. 7C). These results demonstrate that VC-CAR-T cells can recognize and effectively eliminate autologous reactivated HIV-1-infected CD4+ T cells from patients receiving suppressive cART.
FIG 7.
Elimination of reactivated HIV-1 latently infected CD4+ T lymphocytes from HIV-1-infected individuals receiving suppressive cART. (A) Experimental design. (B) On day 0, CD4+ T cells from HIV-1-infected patients were stimulated by PHA-M or by a combination of specific LRAs (SAHA plus bryostatin-1). After 1 day, 1 × 106 CD4+ T cells were mixed with autologous VC-CAR-engineered or control CD8+ T cells at a 1:1 ratio. On day 3, cells were collected and viral RNAs were isolated and amplified by real-time qRT-PCR with primers SK38 and SK39. The cutoff for cell-associated viral RNA is 800 copies ml−1. (C) The cultures were further tested for the presence of p24 with viral outgrowth by ELISA at the indicated time points. (B and C) Data reflect means ± SEM. P values were calculated using the two-tailed unpaired Student's t test with equal variances (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Despite decades of effort, clinical progress in the treatment of HIV-1 infection remains limited principally by high viral mutation rates and the persistence of latently infected cells (51–53). In the present study, we combined an extracellular domain derived from the scFv of the HIV-1 bNAb VRC01 and a third-generation CAR moiety to develop novel MHC-independent, anti-HIV-1 VC-CAR-T cells. These cells exhibited specific antiviral activity and induced broad T cell-mediated cytotoxicity against diverse HIV-1 isolates. Importantly, the VC-CAR described here displays augmented potency compared to previously described CD4-CAR (Fig. 2D). This may be explained by differences in affinity toward the gp120 subunit between the HIV-1 bNAb-derived scFv and the CD4 moiety (42, 43). The third-generation CAR moiety, combining multiple intracellular signaling domains (CD3ζ-CD28-41BB), offers improved amplification of activation signals and enhanced T cell-mediated cytolysis of HIV-1-infected cells (17). To enhance immune potency and thereby prevent viral entry, two groups have used the previously described CD4-CAR system to express an scFv of 17b, a monoclonal antibody targeting the coreceptor binding region of gp120, and short hairpin RNAs specific to the coreceptor chemokine (C-C motif) receptor 5 and the HIV-1 long terminal repeat (35, 36). In the present work, our VC-CAR-T cells were not susceptible to HIV-1 infection, perhaps because the interaction between the VRC01 scFv and viral envelope gp120 did not lead to the exposure of gp41 to the cell membrane, thus not fully permitting viral entry into CD8+ T lymphocytes.
The special features of the VC-CAR approach to genetically engineering CD8+ T cells make it a particularly suitable candidate for the enhancement of HIV-1-specific immune surveillance. Since the safety and feasibility of CAR-T cell therapy have been established in several clinical trials (54–56), it seems reasonable to develop therapeutic applications of this technique in the pursuit of a functional HIV-1 cure (7). Our data indicate that VC-CAR-T cells can significantly delay HIV-1 rebound in an in vitro system. Importantly, the VC-CAR-T cells also effectively eradicated the reactivated HIV-1-infected CD4+ T lymphocytes isolated from infected individuals receiving suppressive cART. Accordingly, the next steps will focus on maintaining the self-renewal of reconstituted CAR-T cells to prolong their persistence in vivo after adoptive transfer (57, 58). In addition, a combination of CAR-T cells recognizing several independent sites on the HIV-1 envelope might further potentiate antiviral activity. Furthermore, based upon our current work, the CAR-T cell therapy in combination with highly efficient LRAs merits being evaluated in future clinical trials (7).
Latent infection remains a major barrier to the realization of an HIV-1 cure, although the shock and kill (also known as kick and kill) strategy has been extensively discussed (59–63). However, recent studies suggest that the latent reservoir size is 60 times larger than previously estimated from patients on cART, and only a small fraction of replication-competent viruses can be recovered from this reservoir by LRAs (64). For this reason, the viral reservoir can be categorized into shallow and deep tiers, as we proposed previously (7). The former consists of infected cells harboring inducible proviruses (7). According to the shock and kill strategy, when latent viruses are reactivated, potent reconstituted immune surveillance, comprised of CAR-T cells, natural cytotoxic T lymphocytes, and bNAbs, could eradicate these HIV-1-producing cells and persistently control viral replication without the continuation of cART (7, 13). As for the deep reservoir that cannot be activated by current methods, highly effective LRAs are urgently needed to reduce its size to the greatest extent possible. Thereafter, strategies such as the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system, RNA interference, and small-molecule approaches might be utilized to permanently extirpate these deeply silenced viruses (65–71). In any case, the CAR-T cell method could be used to enhance and maintain immune surveillance (7). As such, a long-term, even lifelong functional HIV-1 cure may be achievable.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Special Research Program for Important Infectious Diseases (2013ZX10001004) and (2012ZX10001003-004-002), the Introduction of Innovative R&D Team Program of Guangdong Province (2009010058), the International Collaboration Program of Natural Science Foundation of China (81561128007), the Important Key Program of Natural Science Foundation of China (81590765), and the Joint-Innovation Program in Healthcare for Special Scientific Research Projects of Guangzhou (201508020256).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00852-16.
REFERENCES
- 1.Trono D, Van Lint C, Rouzioux C, Verdin E, Barre-Sinoussi F, Chun TW, Chomont N. 2010. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science (New York, NY) 329:174–180. doi: 10.1126/science.1191047. [DOI] [PubMed] [Google Scholar]
- 2.Strain MC, Gunthard HF, Havlir DV, Ignacio CC, Smith DM, Leigh-Brown AJ, Macaranas TR, Lam RY, Daly OA, Fischer M, Opravil M, Levine H, Bacheler L, Spina CA, Richman DD, Wong JK. 2003. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci U S A 100:4819–4824. doi: 10.1073/pnas.0736332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 4.Shan L, Xing S, Yang HC, Zhang H, Margolick JB, Siliciano RF. 2014. Unique characteristics of histone deacetylase inhibitors in reactivation of latent HIV-1 in Bcl-2-transduced primary resting CD4+ T cells. J Antimicrob Chemother 69:28–33. doi: 10.1093/jac/dkt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. 2009. The challenge of finding a cure for HIV infection. Science (New York, NY) 323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 6.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol 10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Ma X, Liu B, Chen C, Zhang H. 2015. HIV-1 functional cure: will the dream come true? BMC Med 13:284. doi: 10.1186/s12916-015-0517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, Coffin JM, Bosch RJ, Margolis DM. 2005. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet (London, England) 366:549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulkosky J, Culnan DM, Roman J, Dornadula G, Schnell M, Boyd MR, Pomerantz RJ. 2001. Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98:3006–3015. doi: 10.1182/blood.V98.10.3006. [DOI] [PubMed] [Google Scholar]
- 10.Goulder PJ, Watkins DI. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol 4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 11.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Perelson AS, Hahn BH, Shaw GM, Korber BT, McMichael AJ. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, Zhang H, Margolick JB, Blankson JN, Siliciano RF. 2012. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36:491–501. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, Lai J, McHugh HL, Hao H, Zhang H, Margolick JB, Gurer C, Murphy AJ, Valenzuela DM, Yancopoulos GD, Deeks SG, Strowig T, Kumar P, Siliciano JD, Salzberg SL, Flavell RA, Shan L, Siliciano RF. 2015. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dotti G, Gottschalk S, Savoldo B, Brenner MK. 2014. Design and development of therapies using chimeric antigen receptor-expressing T cells. Immunol Rev 257:107–126. doi: 10.1111/imr.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kershaw MH, Westwood JA, Darcy PK. 2013. Gene-engineered T cells for cancer therapy. Nat Rev Cancer 13:525–541. doi: 10.1038/nrc3565. [DOI] [PubMed] [Google Scholar]
- 17.Maus MV, Grupp SA, Porter DL, June CH. 2014. Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood 123:2625–2635. doi: 10.1182/blood-2013-11-492231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinrichs CS, Rosenberg SA. 2014. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol Rev 257:56–71. doi: 10.1111/imr.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg SA, Restifo NP. 2015. Adoptive cell transfer as personalized immunotherapy for human cancer. Science (New York, NY) 348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, Wright JF, Milone MC, Levine BL, June CH. 2013. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med 368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kochenderfer JN, Dudley ME, Feldman SA, Wilson WH, Spaner DE, Maric I, Stetler-Stevenson M, Phan GQ, Hughes MS, Sherry RM, Yang JC, Kammula US, Devillier L, Carpenter R, Nathan DA, Morgan RA, Laurencot C, Rosenberg SA. 2012. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood 119:2709–2720. doi: 10.1182/blood-2011-10-384388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M, Shah NN, Steinberg SM, Stroncek D, Tschernia N, Yuan C, Zhang H, Zhang L, Rosenberg SA, Wayne AS, Mackall CL. 2015. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet (London, England) 385:517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter DL, Levine BL, Kalos M, Bagg A, June CH. 2011. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med 365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krebs K, Bottinger N, Huang LR, Chmielewski M, Arzberger S, Gasteiger G, Jager C, Schmitt E, Bohne F, Aichler M, Uckert W, Abken H, Heikenwalder M, Knolle P, Protzer U. 2013. T cells expressing a chimeric antigen receptor that binds hepatitis B virus envelope proteins control virus replication in mice. Gastroenterology 145:456–465. doi: 10.1053/j.gastro.2013.04.047. [DOI] [PubMed] [Google Scholar]
- 25.Sautto GA, Wisskirchen K, Clementi N, Castelli M, Diotti RA, Graf J, Clementi M, Burioni R, Protzer U, Mancini N. 2016. Chimeric antigen receptor (CAR)-engineered T cells redirected against hepatitis C virus (HCV) E2 glycoprotein. Gut 65:512–523. doi: 10.1136/gutjnl-2014-308316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts MR, Qin L, Zhang D, Smith DH, Tran AC, Dull TJ, Groopman JE, Capon DJ, Byrn RA, Finer MH. 1994. Targeting of human immunodeficiency virus-infected cells by CD8+ T lymphocytes armed with universal T-cell receptors. Blood 84:2878–2889. [PubMed] [Google Scholar]
- 27.Yang OO, Tran AC, Kalams SA, Johnson RP, Roberts MR, Walker BD. 1997. Lysis of HIV-1-infected cells and inhibition of viral replication by universal receptor T cells. Proc Natl Acad Sci U S A 94:11478–11483. doi: 10.1073/pnas.94.21.11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bitton N, Verrier F, Debre P, Gorochov G. 1998. Characterization of T cell-expressed chimeric receptors with antibody-type specificity for the CD4 binding site of HIV-1 gp120. Eur J Immunol 28:4177–4187. doi:. [DOI] [PubMed] [Google Scholar]
- 29.Masiero S, Del Vecchio C, Gavioli R, Mattiuzzo G, Cusi MG, Micheli L, Gennari F, Siccardi A, Marasco WA, Palu G, Parolin C. 2005. T-cell engineering by a chimeric T-cell receptor with antibody-type specificity for the HIV-1 gp120. Gene Ther 12:299–310. doi: 10.1038/sj.gt.3302413. [DOI] [PubMed] [Google Scholar]
- 30.Ali A, Kitchen SG, Chen IS, Ng HL, Zack JA, Yang OO. 2016. HIV-1-specific chimeric antigen receptors based on broadly neutralizing antibodies. J Virol 90:6999–7006. doi: 10.1128/JVI.00805-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romeo C, Seed B. 1991. Cellular immunity to HIV activated by CD4 fused to T cell or Fc receptor polypeptides. Cell 64:1037–1046. doi: 10.1016/0092-8674(91)90327-U. [DOI] [PubMed] [Google Scholar]
- 32.Sahu GK, Sango K, Selliah N, Ma Q, Skowron G, Junghans RP. 2013. Anti-HIV designer T cells progressively eradicate a latently infected cell line by sequentially inducing HIV reactivation then killing the newly gp120-positive cells. Virology 446:268–275. doi: 10.1016/j.virol.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni Z, Knorr DA, Bendzick L, Allred J, Kaufman DS. 2014. Expression of chimeric receptor CD4zeta by natural killer cells derived from human pluripotent stem cells improves in vitro activity but does not enhance suppression of HIV infection in vivo. Stem Cells (Dayton, OH) 32:1021–1031. doi: 10.1002/stem.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacLean AG, Walker E, Sahu GK, Skowron G, Marx P, von Laer D, Junghans RP, Braun SE. 2014. A novel real-time CTL assay to measure designer T-cell function against HIV Env(+) cells. J Med Primatol 43:341–348. doi: 10.1111/jmp.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Patel B, Ghanem MH, Bundoc V, Zheng Z, Morgan RA, Rosenberg SA, Dey B, Berger EA. 2015. Novel CD4-based bispecific chimeric antigen receptor designed for enhanced anti-HIV potency and absence of HIV entry receptor activity. J Virol 89:6685–6694. doi: 10.1128/JVI.00474-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhen A, Kamata M, Rezek V, Rick J, Levin B, Kasparian S, Chen IS, Yang OO, Zack JA, Kitchen SG. 2015. HIV-specific immunity derived from chimeric antigen receptor-engineered stem cells. Mol Ther 23:1358–1367. doi: 10.1038/mt.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy LE, Weiss RA. 2013. Neutralizing antibodies to HIV-1 induced by immunization. J Exp Med 210:209–223. doi: 10.1084/jem.20121827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. 2012. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halper-Stromberg A, Lu CL, Klein F, Horwitz JA, Bournazos S, Nogueira L, Eisenreich TR, Liu C, Gazumyan A, Schaefer U, Furze RC, Seaman MS, Prinjha R, Tarakhovsky A, Ravetch JV, Nussenzweig MC. 2014. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 158:989–999. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, Nason MC, Montefiori D, Moldt B, Poignard P, Diskin R, Bjorkman PJ, Eckhaus MA, Klein F, Mouquet H, Cetrulo Lorenzi JC, Gazumyan A, Burton DR, Nussenzweig MC, Martin MA, Nishimura Y. 2014. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med 211:2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caskey M, Klein F, Lorenzi JC, Seaman MS, West AP Jr, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fatkenheuer G, Schlesinger SJ, Nussenzweig MC. 2015. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science (New York, NY) 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (New York, NY) 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA. 2010. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther 18:843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esensten JH, Helou YA, Chopra G, Weiss A, Bluestone JA. 2016. CD28 costimulation: from mechanism to therapy. Immunity 44:973–988. doi: 10.1016/j.immuni.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kochenderfer JN, Feldman SA, Zhao Y, Xu H, Black MA, Morgan RA, Wilson WH, Rosenberg SA. 2009. Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. J Immunother (Hagerstown, MD) 32:689–702. doi: 10.1097/CJI.0b013e3181ac6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Fan M, Geng G, Liu B, Huang Z, Luo H, Zhou J, Guo X, Cai W, Zhang H. 2014. A novel HIV-1-encoded microRNA enhances its viral replication by targeting the TATA box region. Retrovirology 11:23. doi: 10.1186/1742-4690-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, Siliciano JD, Siliciano RF. 2013. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog 9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hernandez JM, Bui MH, Han KR, Mukouyama H, Freitas DG, Nguyen D, Caliliw R, Shintaku PI, Paik SH, Tso CL, Figlin RA, Belldegrun AS. 2003. Novel kidney cancer immunotherapy based on the granulocyte-macrophage colony-stimulating factor and carbonic anhydrase IX fusion gene. Clin Cancer Res 9:1906–1916. [PubMed] [Google Scholar]
- 50.Schiffner T, Sattentau QJ, Duncan CJ. 2013. Cell-to-cell spread of HIV-1 and evasion of neutralizing antibodies. Vaccine 31:5789–5797. doi: 10.1016/j.vaccine.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 51.Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med 3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 52.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med 3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 53.Chapuis AG, Casper C, Kuntz S, Zhu J, Tjernlund A, Diem K, Turtle CJ, Cigal ML, Velez R, Riddell S, Corey L, Greenberg PD. 2011. HIV-specific CD8+ T cells from HIV+ individuals receiving HAART can be expanded ex vivo to augment systemic and mucosal immunity in vivo. Blood 117:5391–5402. doi: 10.1182/blood-2010-11-320226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitsuyasu RT, Anton PA, Deeks SG, Scadden DT, Connick E, Downs MT, Bakker A, Roberts MR, June CH, Jalali S, Lin AA, Pennathur-Das R, Hege KM. 2000. Prolonged survival and tissue trafficking following adoptive transfer of CD4zeta gene-modified autologous CD4(+) and CD8(+) T cells in human immunodeficiency virus-infected subjects. Blood 96:785–793. [PubMed] [Google Scholar]
- 55.Walker RE, Bechtel CM, Natarajan V, Baseler M, Hege KM, Metcalf JA, Stevens R, Hazen A, Blaese RM, Chen CC, Leitman SF, Palensky J, Wittes J, Davey RT Jr, Falloon J, Polis MA, Kovacs JA, Broad DF, Levine BL, Roberts MR, Masur H, Lane HC. 2000. Long-term in vivo survival of receptor-modified syngeneic T cells in patients with human immunodeficiency virus infection. Blood 96:467–474. [PubMed] [Google Scholar]
- 56.Deeks SG, Wagner B, Anton PA, Mitsuyasu RT, Scadden DT, Huang C, Macken C, Richman DD, Christopherson C, June CH, Lazar R, Broad DF, Jalali S, Hege KM. 2002. A phase II randomized study of HIV-specific T-cell gene therapy in subjects with undetectable plasma viremia on combination antiretroviral therapy. Mol Ther 5:788–797. doi: 10.1006/mthe.2002.0611. [DOI] [PubMed] [Google Scholar]
- 57.Till BG, Jensen MC, Wang J, Qian X, Gopal AK, Maloney DG, Lindgren CG, Lin Y, Pagel JM, Budde LE, Raubitschek A, Forman SJ, Greenberg PD, Riddell SR, Press OW. 2012. CD20-specific adoptive immunotherapy for lymphoma using a chimeric antigen receptor with both CD28 and 4-1BB domains: pilot clinical trial results. Blood 119:3940–3950. doi: 10.1182/blood-2011-10-387969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gattinoni L, Klebanoff CA, Restifo NP. 2012. Paths to stemness: building the ultimate antitumour T cell. Nat Rev Cancer 12:671–684. doi: 10.1038/nrc3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dahabieh MS, Battivelli E, Verdin E. 2015. Understanding HIV latency: the road to an HIV cure. Annu Rev Med 66:407–421. doi: 10.1146/annurev-med-092112-152941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. 2014. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science (New York, NY) 345:179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. 2014. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science (New York, NY) 345:570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, McElrath MJ, Lehmann C, Klein F, Caskey M, Walker BD, Siliciano JD, Siliciano RF, Jankovic M, Nussenzweig MC. 2015. HIV-1 integration landscape during latent and active infection. Cell 160:420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Razooky BS, Pai A, Aull K, Rouzine IM, Weinberger LS. 2015. A hardwired HIV latency program. Cell 160:990–1001. doi: 10.1016/j.cell.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liao HK, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang CJ, Esteban CR, Young J, Izpisua Belmonte JC. 2015. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 66.Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J, Mo X, Khalili K. 2014. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A 111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson J, Banerjea A, Akkina R. 2003. Bispecific short hairpin siRNA constructs targeted to CD4, CXCR4, and CCR5 confer HIV-1 resistance. Oligonucleotides 13:303–312. doi: 10.1089/154545703322616989. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, Kelleher A. 2005. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J RNAi Gene Silencing 1:66–78. [PMC free article] [PubMed] [Google Scholar]
- 69.Yamagishi M, Ishida T, Miyake A, Cooper DA, Kelleher AD, Suzuki K, Watanabe T. 2009. Retroviral delivery of promoter-targeted shRNA induces long-term silencing of HIV-1 transcription. Microbes Infect 11:500–508. doi: 10.1016/j.micinf.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 70.Mousseau G, Kessing CF, Fromentin R, Trautmann L, Chomont N, Valente ST. 2015. The Tat inhibitor didehydro-cortistatin A prevents HIV-1 reactivation from latency. mBio 6:e00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heredia A, Le N, Gartenhaus RB, Sausville E, Medina-Moreno S, Zapata JC, Davis C, Gallo RC, Redfield RR. 2015. Targeting of mTOR catalytic site inhibits multiple steps of the HIV-1 lifecycle and suppresses HIV-1 viremia in humanized mice. Proc Natl Acad Sci U S A 112:9412–9417. doi: 10.1073/pnas.1511144112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.