ABSTRACT
Rotaviruses (RVs) of species A (RVA) are a major causative agent of acute gastroenteritis. Recently, histo-blood group antigens (HBGAs) have been reported to interact with human RVA VP8* proteins. Human P[19] is a rare P genotype of porcine origin that infects humans sporadically. The functional and structural characteristics of P[19] VP8* interaction with HBGAs are unknown. In this study, we expressed and purified the VP8* proteins of human and porcine P[19] RVs. In oligosaccharide and saliva binding assays, P[19] VP8* proteins showed obvious binding to A-, B-, and O-type saliva samples irrespective of the secretor status, implying broad binding patterns. However, they did not display specific binding to any of the oligosaccharides tested. In addition, we solved the structure of human P[19] VP8* at 2.4 Å, which revealed a typical galectin-like fold. The structural alignment demonstrated that P[19] VP8* was most similar to that of P[8], which was consistent with the phylogenetic analysis. Structure superimposition revealed the basis for the lack of binding to the oligosaccharides. Our study indicates that P[19] RVs may bind to other oligosaccharides or ligands and may have the potential to spread widely among humans. Thus, it is necessary to place the prevalence and evolution of P[19] RVs under surveillance.
IMPORTANCE Human P[19] is a rare P genotype of porcine origin. Based on phylogenetic analysis of VP8* sequences, P[19] was classified in the P[II] genogroup, together with P[4], P[6], and P[8], which have been reported to interact with HBGAs in a genotype-dependent manner. In this study, we explored the functional and structural characteristics of P[19] VP8* interaction with HBGAs. P[19] VP8* showed binding to A-, B-, and O-type saliva samples, as well as saliva of nonsecretors. This implies that P[19] has the potential to spread among humans with a broad binding range. Careful attention should be paid to the evolution and prevalence of P[19] RVs. Furthermore, we solved the structure of P[19] VP8*. Structure superimposition indicated that P[19] may bind to other oligosaccharides or ligands using potential binding sites, suggesting that further investigation of the specific cell attachment factors is warranted.
INTRODUCTION
Rotaviruses (RVs) are the major causative agents of acute gastroenteritis in young children and animals worldwide (1). The RV genome contains 11 segments of double-stranded RNA encoding 12 proteins: six structural proteins (VP1, VP2, VP3, VP4, VP6, and VP7) and six nonstructural proteins (NSP1 to NSP6) (2). RVs are classified into 8 species (A to H) by the antigenicity of the VP6 protein (3). Group A rotavirus (RVA) is a major cause of human RV-associated gastroenteritis (4). RVA can be classified into various G and P types on the basis of glycoprotein VP7 and protease-sensitive VP4, respectively. To date, at least 27 G and 35 P genotypes have been reported (5). The fact that the segmented genome undergoes point mutations, reassortment, and gene rearrangements accounts for the large genetic diversity of RVs (1).
VP4 can be cleaved by protease to yield N-terminal VP8* and C-terminal VP5* (6). VP8* includes the VP4 spike head and is reported to bind to cell surface glycans essential for cell invasion (7). The ability of a virus to invade host cells is crucial for its replication, host tropism, and pathogenicity. Notably, VP8* is the most variable domain. VP8* proteins of sialidase-sensitive RVs were reported to interact with sialic acids (Sia) (8); however, most animal RVs and almost all human RVs were sialidase insensitive (9). It was noted that many of the sialidase-insensitive RVA strains interact with Sia at subterminal sites of glycoprotein receptors (10). Recently, histo-blood group antigens (HBGAs) have been reported to be attachment factors for human RVs by interacting with VP8* (11, 12). HBGAs are a group of carbohydrates present on the surfaces of red blood cells and mucosal epithelia and as free oligosaccharides in saliva, blood, and milk (13).
Previous studies by oligosaccharide and saliva binding assays showed that P[4] and P[8] bound to Lewis b (Leb) and H1, while P[6] recognized only H1 (11, 14, 15). P[11] showed obvious binding to type 1/type 2 precursors (16, 17). In addition, P[9], P[14], and P[25] all bound to A type HBGA (A-HBGA) (18). Using saturation transfer difference nuclear magnetic resonance (STD-NMR), DS-1 P[4], RV-3 P[6], ST-3 P[6], and HAL1141 P[14] RVs were shown to bind to A-HBGA, whereas Wa P[8] did not (19). In addition, the VP8* protein of Wa also showed no binding to H1 or Leb in the STD-NMR assay. Moreover, epidemiological studies have demonstrated certain correlations between HBGA status and RV infections in different regions and populations (20–22). These studies indicate the complexity of the interactions between RVs and HBGAs. Through phylogenetic analysis of VP8* sequences, RVAs have been classified into five genogroups (P[I] to P[V]) (18). P[I], P[IV], and P[V] mainly infect animals; P[II] infects humans; and P[III] infects both animals and humans. The three major P genotypes in humans (P[4], P[6], and P[8]) are all clustered in P[II].
P[19] is also grouped in P[II]. P[19] is a rare RVA P genotype reported in humans and pigs (4). It was found to be combined with various G genotypes (23). P[19] was first identified in pigs in 1986 (4F) (24). The first human P[19] RV strains were detected in Thailand in 1989 (Mc323 and Mc345) (25). Whole-genome analysis revealed that human P[19] RV Mc323 and Mc345 were of porcine origin (26). It is unknown whether P[19] has HBGA binding patterns similar to those of other members of the P[II] genogroup (P[4]/P[8] or P[6]), which may be essential for understanding P[19] RV infection and transmission in humans and pigs. Therefore, we expressed P[19] VP8* proteins and explored the structural and functional characteristics of P[19] RV VP8* interaction with HBGAs in this study.
MATERIALS AND METHODS
VP8* protein expression and purification.
The VP8* gene (encoding amino acids 1 to 230) of human P[19] Mc345 (RVA/Human-tc/THA/Mc345/1989/G9P[19]; GenBank accession number D38054) was synthesized by Genewiz Company (Suzhou, China). One porcine G3P[19] RV was identified in our laboratory (unpublished data), and the VP8* sequence (GenBank accession number KX455847) was obtained from a reverse transcription (RT)-PCR replicon. VP8* core fragments (amino acids [aa] 64 to 223) of both human and porcine P[19] RVs were then cloned into the pGEX4T-1 vector with an N-terminal glutathione S-transferase (GST) tag. VP8* core fragments (aa 64 to 223) with a C-terminal hexahistidine (His) tag were cloned into the pET-30a vector. The recombinant proteins were expressed in Escherichia coli strain BL21 induced with isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 0.4 mM at 22°C for 16 h. The GST fusion and His tag proteins were purified as reported previously (27). Briefly, for the GST fusion protein, the supernatant of the bacterial lysate was filtered through a 0.22-μm membrane and bound to glutathione-Sepharose (GE Healthcare Life Sciences). The GST fusion protein of interest was eluted with elution buffer (10 mM reduced glutathione, 50 mM Tris-HCl, pH 8.0) after washing five times with phosphate-buffered saline (PBS). The His-tagged protein was bound to a HisTrap column and eluted with PBS with 300 mM imidazole buffer following washing with PBS and PBS with 20 mM imidazole buffer. Samples of each eluted product were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to check the proteins. VP8* protein was concentrated to ∼5 mg/ml using the 10-kDa concentration tube (Millipore), centrifuged at a speed of 2,000 × g, and then applied to a Superdex 20010/300GL gel filtration column buffered with PBS. The protein from each peak was collected and verified by SDS-PAGE. The protein of interest was concentrated to ∼5 mg/ml.
Saliva binding assay.
A panel of saliva samples with typed A, B, and O and secretor status kept in our laboratory was used for a saliva binding assay as described previously (27, 28). A plate was coated with saliva samples. After blocking with 5% nonfat milk, the VP8*core-GST fusion proteins of both human and porcine P[19] (50 μg/ml) were added at a volume of 100 μl per well. Then, mouse GST antibody (1:1,000; Abcam) was added, followed by horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (1:1,500; Abgent). The reaction was developed using a 3,3′,5,5′-tetramethylbenzidine (TMB) kit, and the absorbance was measured at 450 nm. In each step, the plates were incubated at 37°C for 1 h and washed five times with 0.5% PBS-Tween 20 buffer.
Oligosaccharide binding assay.
A plate was coated with the VP8*core-GST fusion proteins of both human and porcine P[19] at 20 μg per well, as reported previously (11). After blocking with 5% nonfat milk, synthetic-oligosaccharide–polyacrylamide (PAA)–biotin conjugates (Lewis a [Lea], Leb, Lewis x [Lex], Lewis y [Ley], H1, H2, H3, A, B, type 1 precursor, type 2 precursor, Neu5Ac, Neu5Gc, and sialyl-Lex; GlycoTech, Inc., Gaithersburg, MD) were added at 0.2 μg per well. Then, HRP-conjugated streptavidin (Abcam) was added at 0.1 μg per well. The plates were incubated at 37°C for 1 h and washed five times with 0.5% PBS-Tween 20 buffer at each step. The reaction mixture was developed using a TMB kit, and the absorbance was measured at 450 nm.
Protein crystallization.
The VP8*core-His protein was further purified by gel filtration buffered with 20 mM Tris-HCl, 50 mM NaCl, pH 8.0. The protein was concentrated to approximately 10 mg/ml, and crystallization screening was carried out using the sitting-drop vapor diffusion method at 18°C with 1 μl of protein mixed with 1 μl of reservoir solution. Human P[19] VP8* was crystallized under these conditions with 0.2 M ammonium sulfate, 0.1 M MES (morpholineethanesulfonic acid), pH 6.5, and 30% (wt/vol) polyethylene glycol MME 5000.
Data collection and processing.
Crystals were flash-frozen in liquid nitrogen after being dipped briefly in cryoprotectant solutions containing 20% (vol/vol) glycerol. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) BL17U. Original data were processed using HKL2000 software (29). The human P[19] VP8* structure was solved by molecular replacement using Phaser software implemented in the CCP4 program suite (30), with the structure of Wa VP8* (Protein Data Bank [PDB] code 2DWR) as the search model. The initial model was refined by rigid-body refinement using the REFMAC5 program (31), and extensive model building was performed using the COOT program (32). The model was then subjected to iterative cycles of refinement using PHENIX software (33) with energy minimization, isotropic ADP refinement, and bulk solvent modeling. Final statistics for P[19] VP8* are presented in Table 1. The structural analysis was performed using the PyMOL software package. Root mean square deviation (RMSD) values of the matching Cα atoms between P[19] VP8* and other representative VP8*structures, along with the percent sequence identity, were calculated with the align function in PyMOL.
TABLE 1.
Crystallographic X-ray diffraction and refinement statistics
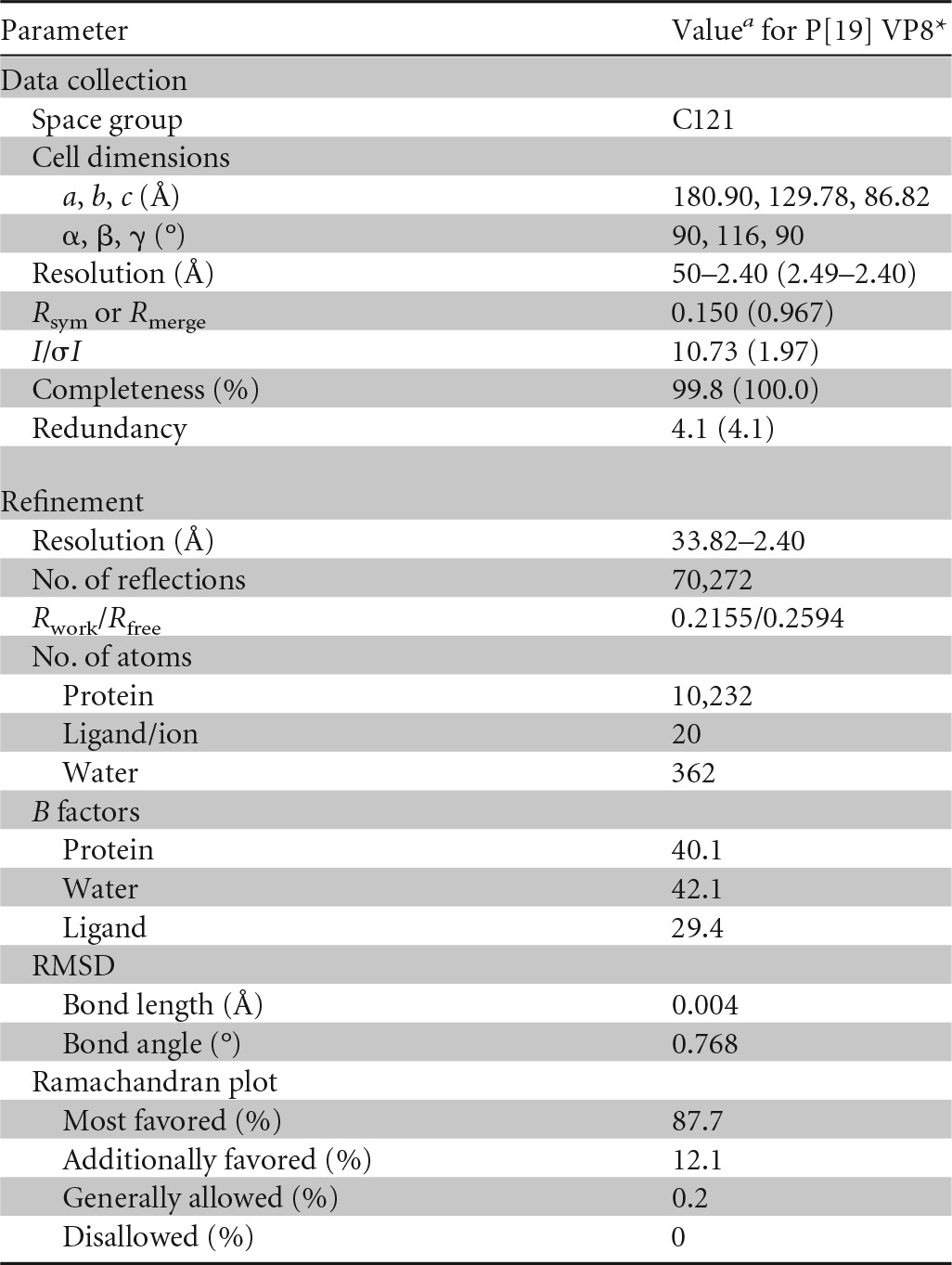
a The values in parentheses are for the highest-resolution shell.
Accession number(s).
The P[19] VP8* structure has been deposited in PDB with the PDB ID 5GJ6. The VP8* sequence has been deposited in GenBank (accession number KX455847).
RESULTS
Expression and purification of P[19] VP8* protein.
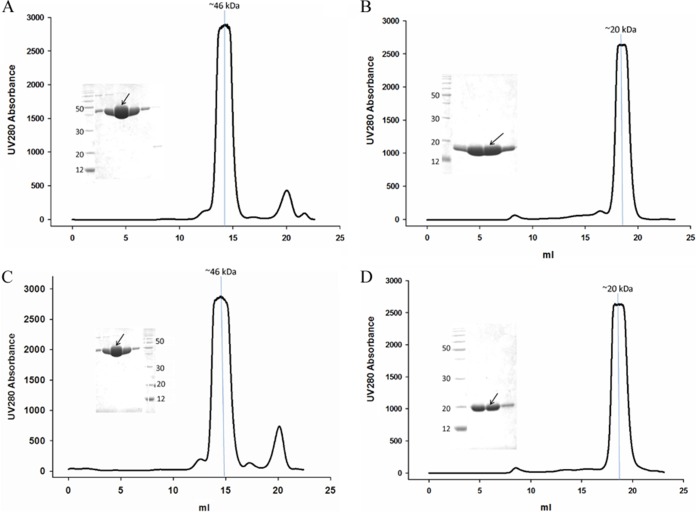
VP8*core proteins of human and porcine P[19] were expressed both in GST fusion form for the functional assay and with a His tag for crystallization. The VP8*core-GST fusion proteins were ∼46 kDa, and the gel filtration peak of the fusion protein was eluted in ∼14.3 ml (Fig. 1A and C). The human and porcine P[19] VP8* core-His proteins were ∼20 kDa, and the gel filtration peak was eluted in ∼18.5 ml (Fig. 1B and D), implying that the VP8* protein existed in the monomer form, in agreement with a previous report on P[4] (34).
FIG 1.
Human and porcine P[19] VP8*core-GST (A and C) and human and porcine P[19] VP8*core-His (B and D) protein purification by gel filtration. The arrows indicate the protein samples at the peak positions in the gel filtration. A molecular mass marker was added to indicate the putative corresponding protein size of the main peak in the gel filtration.
Assay of P[19] VP8* protein binding to saliva samples.
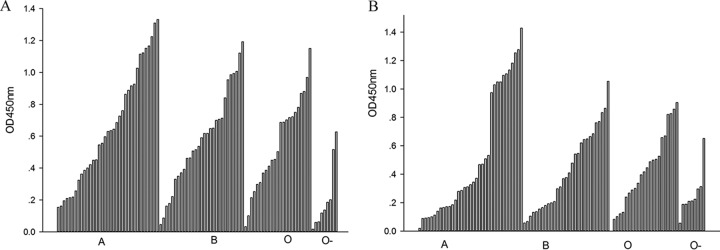
In order to evaluate the binding patterns of P[19] VP8* to A-, B-, and O-type salivas, the purified VP8*-GST proteins were tested in a saliva binding assay using saliva samples with known A, B, and O and Lewis types kept in our laboratory. These saliva samples had been used for the binding assay of Rotateq P[8] and P[14] VP8* proteins. Rotateq P[8] bound to the A, B, and O types, but not to nonsecretors (28), whereas P[14] specifically bound to A-type salivas (35). In this assay of P[19], both human and porcine P[19] VP8* proteins showed obvious binding to A-, B-, and O-type saliva samples (Fig. 2). Notably, P[19] VP8* proteins also bound to salivas of nonsecretors, which did not bind VP8* of P[4] and P[8] RVAs (11, 27).
FIG 2.
Saliva-based binding assay of human (A) and porcine (B) P[19] VP8*core proteins. VP8* proteins bound to saliva samples of A, B, and O types and saliva of nonsecretors (O−). The cutoff value was the value of a negative-control well with PBS instead of saliva samples. In total, 93 saliva samples were tested in the assay. OD450nm, optical density at 450 nm.
Detection of P[19] VP8* attachment to synthetic oligosaccharides.
To investigate the specific binding ligand of P[19] VP8*, we conducted a synthetic-oligosaccharide binding assay. Human and porcine P[19] VP8* proteins displayed no detectable binding to the synthetic oligosaccharides (Fig. 3), including Lea, Leb, Lex, Ley, H1, H2, H3, A, B, type I precursor, type II precursor, Neu5Ac, Neu5Gc, and sialyl-Lex. P[14] VP8* bound to A-HBGAs was used as a positive control, and GST protein was a negative control.
FIG 3.
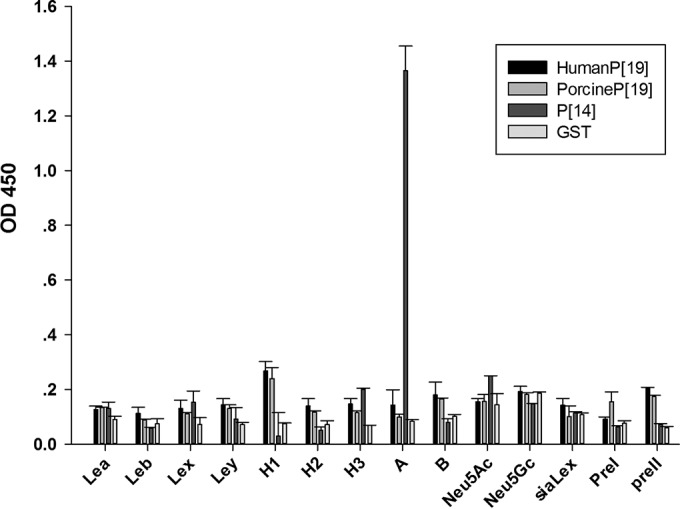
Oligosaccharide binding assay of human and porcine P[19] VP8*core proteins. The VP8* proteins did not bind to any HBGA tested in the study. P[14] VP8*, which binds to A-HBGA, was used as a positive control. GST protein was used as a negative control. The error bars indicate standard deviations.
Structure of human P[19] VP8*.
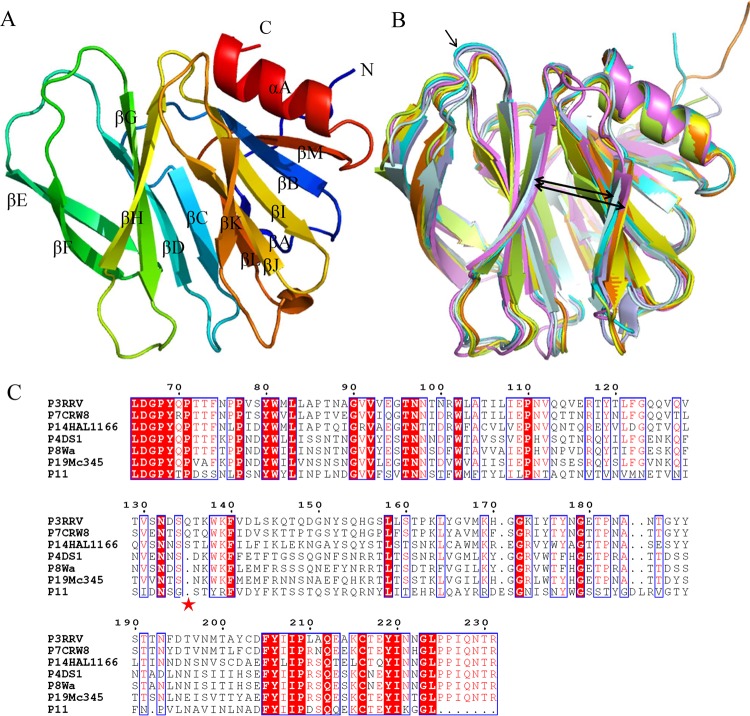
We solved the P[19] VP8* structure at 2.4 Å (Table 1) (PDB ID 5GJ6). Similarly to other solved VP8* structures, it shows a typical galectin-like fold (36) with two twisted antiparallel β-sheets consisting of strands A, L, C, D, G, and H and M, B, I, J, and K, respectively (Fig. 4A). The two β-sheets are separated by a shallow cleft, which forms the main ligand binding sites. Structure alignment revealed that P[19] VP8* was most similar to Wa P[8] VP8* (PDB code 2DWR) with an RMSD of 0.422. P[19] VP8* has a cleft that separates the two β-barrel sheets (Fig. 4B, thick arrows) wider than those of P[14] (PDB accession no. 2DWR) and animal P[3] (PDB accession no. 1KQR) and similar to those of P[4] (PDB accession no. 2AEN) and Wa P[8] (Fig. 4B). The ligand binding clefts in P[4], P[8], and P[19] VP8* are ∼0.5 Å wider than those in P[3] and P[14] VP8* proteins. The distance between the Cα atoms of Pro157 and Thr186 in P[3] is ∼10.7 Å, and that of Thr157 and Thr186 in P[19] VP8* is ∼11.2 Å. Residue 135 of P[3] and P[14] (Fig. 4B, thin arrow), which was deleted in P[4], P[8], and P[19] (Fig. 4C), was reported to influence the width of the binding cleft (37). From the sequence alignment, amino acids 101, 155, and 187 to 190 of the putative ligand binding site are quite varied among P genotypes (Fig. 4C).
FIG 4.
Structural analysis of human P[19] Mc345 VP8*. (A) Overall structure of human P[19] VP8* with two twisted antiparallel sheets consisting of strands A, L, D, G, and H and M, B, I, J, and K. (B) Superimposition of Mc345 human P[19] VP8* structure (5GJ6) (lemon) on those of rhesus rotavirus (RRV) P[3] (1KQR) (pale cyan), HAL1166 P[14] (4DRV) (cyan), CRW-7 P[7] (2I2S) (light blue), DS-1 P[4] (2AEN) (orange), Wa P[8] (2DWR) (yellow), and human rotavirus (HRV) P[11] (4YG0) (violet). The width of the cleft that separates the two β-barrel sheets is shown by the thick arrows. Residue 135 of P[3] and P[14], which was deleted in P[4], P[8], P[11], and P[19], is indicated by the thin arrow. (C) Sequence alignment of VP8* proteins of RRV P[3], HAL1166 P[14], CRW-7 P[7], DS-1 P[4], Wa P[8], P[11], and Mc345 P[19]. The position of residue 135 is labeled with an asterisk. The alignment was done with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/), and the colors and box labels were generated with ESPript (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Red shading indicates residues that are the same in all the aligned sequences, and red letters indicate the residues with high conservation.
Structural comparison of RV VP8* receptor binding sites.
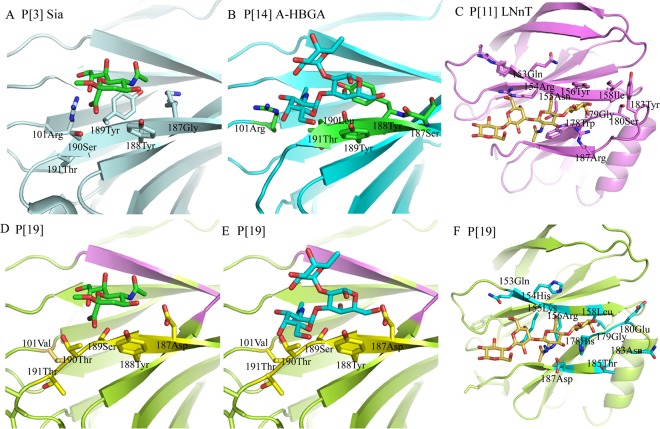
Superimposition of the complex structures of P[3] and Sia (36) (Fig. 5A), P[14] and A-HBGA (12) (Fig. 5B), and P[11] and LNnT (37) (Fig. 5C) on P[19] VP8* demonstrated that the residues that interact with these ligands differ. The residues Arg101, Tyr189, and Leu190 participating in the interaction in P[3] and P[14] were replaced by Val101, Ser189, and Thr190 in P[19] (Fig. 5D and E). The putative binding site of P[19] VP8* showed no specific interaction with Sia. Steric hindrance was observed for A-HBGA with residue 190Thr (Fig. 5E). For LNnT, the interaction residues were quite different between P[11] and P[19]. In P[11] and LNnT, Asn153 and Arg154 formed an H bond; Asn155, Tyr156, Asp185, and Arg187 involved hydrophobic and H-bond interactions; and the corresponding residues in P[19] were Gln153, His154, Lys155, Arg156, Thr185, and Asp187. The overall conformation changed enormously, and no specific binding was seen (Fig. 5F).
FIG 5.
Detailed analysis of the interactions between VP8* and various ligands. (A to C) The residues involved in the interactions between RRV P[3] VP8* and Sia (1KQR) (A), HAL1166 P[14] VP8* and A-HBGA (4DRV) (B), and HRV P[11] VP8* and LNnT (4YG0) (C) are labeled. (D to F) Superimposition of Sia, A-HBGA, and LNnT on the Mc345 P[19] VP8* (5GJ6) structure. The residues in P[19] corresponding to those in P[3], P[14], and P[11] are also shown.
Electrostatic potential surfaces of VP8* structures.
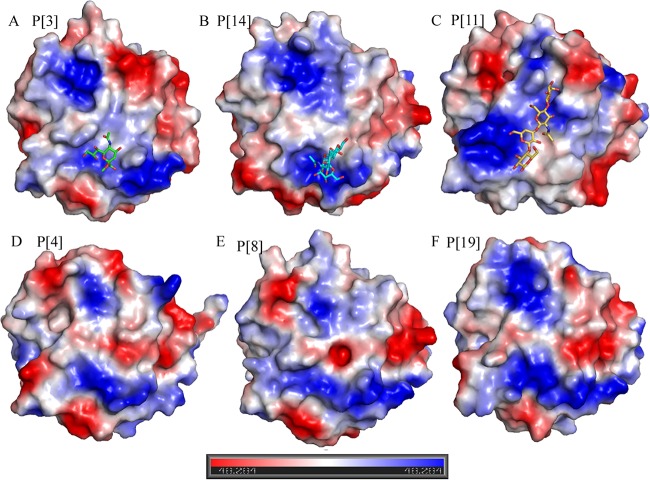
To further analyze the VP8* structures of different genotypes, the electrostatic potential surfaces of P[3], P[14], P[11], P[4], P[8], and P[19] VP8* proteins were analyzed, showing similarities and differences among the putative ligand binding sites (Fig. 6). The structure of the P[3] Sia binding site was slightly positive (Fig. 6A), whereas in P[14], the A-HBGA binding site was positively charged (Fig. 6B). In P[11], the binding site of LNnT presented a positive charge on the top and was both neutral and positive on the bottom (Fig. 6C). In P[4], P[8], and P[19], the putative Sia binding site showed a positive charge, whereas the putative LNnT binding site displayed a somewhat negative charge in the middle region (Fig. 6D, E, and F), indicating a variation in potential ligand binding for different P genotypes.
FIG 6.
Electrostatic-potential presentation of VP8* structures. Shown are RRV P[3] with sialic acid (A), HAL1166 P[14] with A-HBGAs (B), HRV P[11] with LNnT (C), DS-1 P[4] (D), Wa P[8] (E), and Mc345 P[19] (F) VP8*. The red and blue bar indicates the negative and positive charges; the values refer to the relative degrees of electrostatic potential.
DISCUSSION
P[19] RVs have been reported sporadically in humans. Following the report of Mc323 and Mc345, a human rotavirus G9P[19] isolate (RMC321) with porcine rotavirus characteristics was reported following an outbreak of infantile gastroenteritis in India (38). In addition, P[19] RVs with novel lineages of G3, G5, G9, and G26 were detected in children with acute diarrhea in Asia (23, 39). Furthermore, a rare G3P[19] RV infection in humans that showed high sequence identity with G3P[19] from Asia (40) was identified in Italy. In pigs, G3P[19], G4P[19], and G9P[19] were also detected in Thailand (4, 41), and G3P[19] was also found in China (unpublished data from our laboratory). Sequence analysis has indicated that all human P[19] RVs are of porcine origin. In this study, human P[19] Mc345 showed binding to the saliva samples similar to that of porcine P[19], which was consistent with a previous report that Mc345 was of porcine origin (26). From the sequence alignment of VP8* core fragments, the two sequences showed ∼95.6% identity, and only 7 amino acids were different, as revealed in the Mc345 VP8* structure (data not shown). These amino acids were not located in the putative ligand binding sites, also consistent with the saliva binding assay.
P[19] RV is classified in P[II], along with P[4], P[6], and P[8] RVs, which are prevalent in humans and reported to interact with HBGAs (13). P[4] and P[8] RV VP8* bound to saliva types A, B, and O of secretors and did not bind to saliva of nonsecretors. Meanwhile, P[6] also recognized A, B, and O saliva types of secretors with relatively low binding affinity. In this study, P[19] RV VP8* bound to A-, B-, and O-type saliva samples irrespective of the secretor status, implying that P[19] may have a broad binding range. Since all of the reported human P[19] RVs have been shown to be of porcine origin, it is suggested that P[19] may infect humans sporadically and may still not have the capacity to spread among humans. However, based on saliva binding patterns broader than those of other P genotypes (11, 18), if the P[19] RVs acquired the ability to cross the species barrier and infect humans fully, it might cause widespread prevalence, as P[8] RVs did. Recently, a new GII.17 norovirus (NoV) variant caused significantly increased acute gastroenteritis outbreaks in China and other regions of Asia during 2014 and 2015 (42), which highlights the notion that a rare human NoV genotype has the potential to emerge as an epidemiologically important pathogen. The rare GII.17 NoVs also showed good binding in the saliva binding assay but did not bind to the oligosaccharides tested (42, 43). This underscores the need for surveillance of the prevalence and evolution of P[19] RVs in humans and pigs.
In our analysis of specific HBGA ligands of P[19] RV, the P[19] VP8* proteins showed no obvious binding to any synthetic oligosaccharides in the oligosaccharide binding assay, whereas P[4] and P[8] VP8* showed binding to H1 and Leb, and P[6] bound to only H1. More kinds of oligosaccharides or other ligands still need to be explored to fully characterize the receptor binding specificity of P[19] RV.
P[19] VP8* showed a typical galectin-like fold (36) that was most similar to that of P[8]. The structure alignment of P[19] VP8* with other solved VP8* structures is presented in Table 2 with RMSD values. The values reflect the relative structural similarity, with a smaller value indicating higher similarity. P[19] VP8* shares the greatest structural similarity with P[8] and P[4], with RMSD values of 0.422 and 0.520, respectively. This is consistent with the phylogenetic analysis based on the VP8* sequence, which classified P[19] with P[4], P[8], and P[6] in genogroup II (18). The VP8* structures of P[4] and P[8] have been solved, albeit with no ligand bound, so details of the ligand binding sites remain unknown. Superimposition of the P[19] VP8* structure on those of P[3], P[11], and P[14] with ligands showed a lack of interactions or steric hindrance with the ligands, which may provide a structural basis for the lack of binding to the oligosaccharides in the enzyme immunoassay (EIA).
TABLE 2.
Structural alignment of VP8* of different P genotypesa
| P genotype | RMSD (Å)b for: |
|||||
|---|---|---|---|---|---|---|
| P[3] | P[7] | P[14] | P[4] | P[8] | P[11] | |
| P[7] | 0.418 | |||||
| P[14] | 0.623 | 0.611 | ||||
| P[4] | 0.845 | 0.883 | 0.703 | |||
| P[8] | 0.894 | 0.879 | 0.784 | 0.386 | ||
| P[11] | 2.019 | 1.137 | 1.068 | 1.050 | 1.018 | |
| P[19] | 0.760 | 0.724 | 0.631 | 0.520 | 0.422 | 1.750 |
P[3] (PDB accession no. 1KQR), CRW-7 P[7] (PDB accession no. 2I2S), HAL1166 P[14] (PDB accession no. 4DRV), DS-1 P[4] (PDB accession no. 2AEN), Wa P[8] (PDB accession no. 2DWR), HRV P[11] (PDB accession no. 4YG0), and Mc345 P[19] (PDB accession no. 5GJ6).
The values represent RMSDs of the Cα atoms of one VP8 monomer.
Furthermore, the electrostatic potential surface comparison revealed the similarities and differences among different P genotypes, which may explain to some extent the various binding patterns observed in the functional assays (11, 12, 16–18, 36). However, for the potential binding site of P[19], the surface presentation of P[3] and Sia, P[14] and A, P[11] and LNnT, and P[19] showed that there was indeed a cavity in the potential binding site in P[19] VP8* (Fig. 7), suggesting that P[19] VP8* may bind to some other oligosaccharides or ligands at the site. Further studies are needed to explore the specific cell attachment factors for P[19] RVs.
FIG 7.
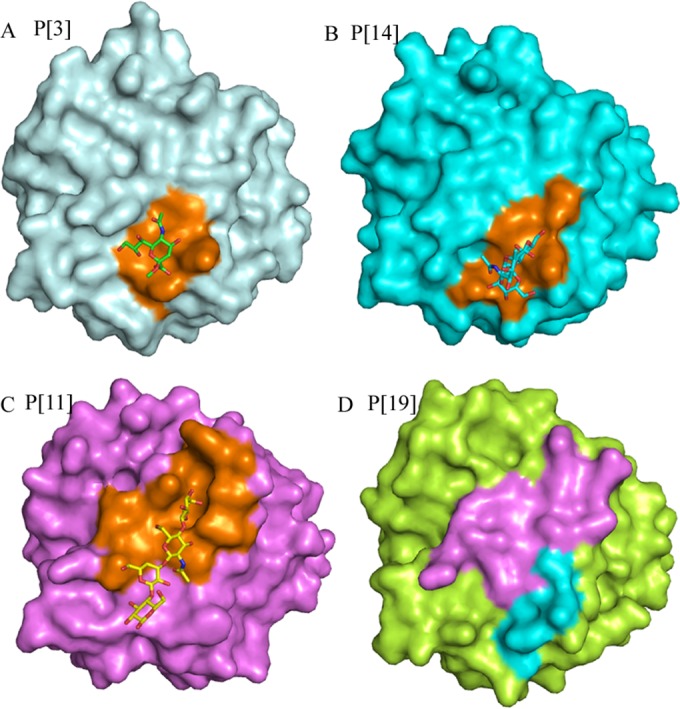
Surface presentation of VP8* structures. Shown are RRV P[3] with sialic acid (A), HAL1166 P[14] with A-HBGAs (B), HRV P[11] with LNnT (C), and Mc345 P[19] (D) VP8*. The binding sites of Sia, A-HBGA, and LNnT are shown in orange. The putative binding regions of LNnT and sialic acid/A-HBGAs of Mc345 P[19] are violet and cyan, respectively.
ACKNOWLEDGMENTS
We are grateful to George F. Gao at the Institute of Microbiology, Chinese Academy of Sciences, for help with X-ray data collection and data processing. We also thank Hong Wang, Huiying Li, and Xiangyu Kong from our laboratory for help with the experiments. Assistance by the staff at the Shanghai Synchrotron Radiation Facility (SSRF-beamline 17U) is acknowledged.
We declare no potential conflict of interest.
Z.-J.D. and X.S. designed the experiments; X.S. and N.G. performed the experiments; X.S., D.L., M.J., Y.Z., G.X., L.P., and Q.Z. analyzed the data; X.S., R.P., and J.Q. solved the structure. X.S. wrote the draft manuscript. Z.-J.D. revised the manuscript. We all reviewed the manuscript.
The research was supported by grants from the National Natural Science Foundation of China (NSFC) (no. 81472003 and no. 31500139).
REFERENCES
- 1.Greenberg HB, Estes MK. 2009. Rotaviruses: from pathogenesis to vaccination. Gastroenterology 136:1939–1951. doi: 10.1053/j.gastro.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed). 2013. Fields virology, 6th ed, p 1347–1401. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Van Ranst M, Johne R. 2012. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol 157:1177–1182. doi: 10.1007/s00705-012-1273-3. [DOI] [PubMed] [Google Scholar]
- 4.Saikruang W, Khamrin P, Chaimongkol N, Suantai B, Kongkaew A, Kongkaew S, Ushijima H, Maneekarn N. 2013. Genetic diversity and novel combinations of G4P[19] and G9P[19] porcine rotavirus strains in Thailand. Vet Microbiol 161:255–262. doi: 10.1016/j.vetmic.2012.07.036. [DOI] [PubMed] [Google Scholar]
- 5.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padilla-Noriega L, Dunn SJ, Lopez S, Greenberg HB, Arias CF. 1995. Identification of two independent neutralization domains on the VP4 trypsin cleavage products VP5* and VP8* of human rotavirus ST3. Virology 206:148–154. doi: 10.1016/S0042-6822(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 7.Fiore L, Greenberg HB, Mackow ER. 1991. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology 181:553–563. doi: 10.1016/0042-6822(91)90888-I. [DOI] [PubMed] [Google Scholar]
- 8.Dormitzer PR, Sun ZY, Blixt O, Paulson JC, Wagner G, Harrison SC. 2002. Specificity and affinity of sialic acid binding by the rhesus rotavirus VP8* core. J Virol 76:10512–10517. doi: 10.1128/JVI.76.20.10512-10517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciarlet M, Estes MK. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J Gen Virol 80:943–948. doi: 10.1099/0022-1317-80-4-943. [DOI] [PubMed] [Google Scholar]
- 10.Haselhorst T, Fleming FE, Dyason JC, Hartnell RD, Yu X, Holloway G, Santegoets K, Kiefel MJ, Blanchard H, Coulson BS, von Itzstein M. 2009. Sialic acid dependence in rotavirus host cell invasion. Nat Chem Biol 5:91–93. doi: 10.1038/nchembio.134. [DOI] [PubMed] [Google Scholar]
- 11.Huang P, Xia M, Tan M, Zhong W, Wei C, Wang L, Morrow A, Jiang X. 2012. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J Virol 86:4833–4843. doi: 10.1128/JVI.05507-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. 2012. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 485:256–259. doi: 10.1038/nature10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto FI. 1994. Recent progress in the molecular genetic study of the histo-blood group ABO system. Immunohematology 10:1–7. [PubMed] [Google Scholar]
- 14.Imbert-Marcille BM, Barbe L, Dupe M, Le Moullac-Vaidye B, Besse B, Peltier C, Ruvoen-Clouet N, Le Pendu J. 2014. A FUT2 gene common polymorphism determines resistance to rotavirus A of the P[8] genotype. J Infect Dis 209:1227–1230. doi: 10.1093/infdis/jit655. [DOI] [PubMed] [Google Scholar]
- 15.Le Pendu J, Nystrom K, Ruvoen-Clouet N. 2014. Host-pathogen co-evolution and glycan interactions. Curr Opin Virol 7:88–94. doi: 10.1016/j.coviro.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Ramani S, Cortes-Penfield NW, Hu L, Crawford SE, Czako R, Smith DF, Kang G, Ramig RF, Le Pendu J, Prasad BV, Estes MK. 2013. The VP8* domain of neonatal rotavirus strain G10P[11] binds to type II precursor glycans. J Virol 87:7255–7264. doi: 10.1128/JVI.03518-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Huang P, Jiang B, Tan M, Morrow AL, Jiang X. 2013. Poly-LacNAc as an age-specific ligand for rotavirus P[11] in neonates and infants. PLoS One 8:e78113. doi: 10.1371/journal.pone.0078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Huang P, Tan M, Liu Y, Biesiada J, Meller J, Castello AA, Jiang B, Jiang X. 2012. Rotavirus VP8*: phylogeny, host range, and interaction with histo-blood group antigens. J Virol 86:9899–9910. doi: 10.1128/JVI.00979-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohm R, Fleming FE, Maggioni A, Dang VT, Holloway G, Coulson BS, von Itzstein M, Haselhorst T. 2015. Revisiting the role of histo-blood group antigens in rotavirus host-cell invasion. Nat Commun 6:5907. doi: 10.1038/ncomms6907. [DOI] [PubMed] [Google Scholar]
- 20.Nordgren J, Sharma S, Bucardo F, Nasir W, Gunaydin G, Ouermi D, Nitiema LW, Becker-Dreps S, Simpore J, Hammarstrom L, Larson G, Svensson L. 2014. Both Lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype-dependent manner. Clin Infect Dis 59:1567–1573. doi: 10.1093/cid/ciu633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Trang N, Vu HT, Le NT, Huang P, Jiang X, Anh DD. 2014. Association between norovirus and rotavirus infection and histo-blood group antigen types in Vietnamese children. J Clin Microbiol 52:1366–1374. doi: 10.1128/JCM.02927-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayouni S, Sdiri-Loulizi K, de Rougemont A, Estienney M, Ambert-Balay K, Aho S, Hamami S, Aouni M, Neji-Guediche M, Pothier P, Belliot G. 2015. Rotavirus P[8] infections in persons with secretor and nonsecretor phenotypes, Tunisia. Emerg Infect Dis 21:2055–2058. doi: 10.3201/eid2111.141901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu FT, Banyai K, Huang JC, Wu HS, Chang FY, Yang JY, Hsiung CA, Huang YC, Lin JS, Hwang KP, Jiang B, Gentsch JR. 2011. Diverse origin of P[19] rotaviruses in children with acute diarrhea in Taiwan: detection of novel lineages of the G3, G5, and G9 VP7 genes. J Med Virol 83:1279–1287. doi: 10.1002/jmv.22052. [DOI] [PubMed] [Google Scholar]
- 24.Burke B, McCrae MA, Desselberger U. 1994. Sequence analysis of two porcine rotaviruses differing in growth in vitro and in pathogenicity: distinct VP4 sequences and conservation of NS53, VP6 and VP7 genes. J Gen Virol 75:2205–2212. doi: 10.1099/0022-1317-75-9-2205. [DOI] [PubMed] [Google Scholar]
- 25.Okada J, Urasawa T, Kobayashi N, Taniguchi K, Hasegawa A, Mise K, Urasawa S. 2000. New P serotype of group A human rotavirus closely related to that of a porcine rotavirus. J Med Virol 60:63–69. [PubMed] [Google Scholar]
- 26.Ghosh S, Urushibara N, Taniguchi K, Kobayashi N. 2012. Whole genomic analysis reveals the porcine origin of human G9P[19] rotavirus strains Mc323 and Mc345. Infect Genet Evol 12:471–477. doi: 10.1016/j.meegid.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Ma X, Li DD, Sun XM, Guo YQ, Xiang JY, Wang WH, Zhang LX, Gu QJ, Duan ZJ. 2015. Binding patterns of rotavirus genotypes P[4], P[6], and P[8] in China with histo-blood group antigens. PLoS One 10:e0134584. doi: 10.1371/journal.pone.0134584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Guo N, Li D, Jin M, Zhou Y, Xie G, Pang L, Zhang Q, Cao Y, Duan ZJ. 2016. Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens. Virology 495:129–135. doi: 10.1016/j.virol.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Read RJ. 2001. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr 57:1373–1382. doi: 10.1107/S0907444901012471. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monnier N, Higo-Moriguchi K, Sun ZY, Prasad BV, Taniguchi K, Dormitzer PR. 2006. High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain. J Virol 80:1513–1523. doi: 10.1128/JVI.80.3.1513-1523.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun XM GN, Xu ZQ, Li DD, Duan ZJ. 2016. P[14] Rotavirus VP8* protein specifically binds to A type histo-blood group antigen. Chinese J Exp Clin Virol 30:315–318. [Google Scholar]
- 36.Dormitzer PR, Sun ZY, Wagner G, Harrison SC. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J 21:885–897. doi: 10.1093/emboj/21.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu L, Ramani S, Czako R, Sankaran B, Yu Y, Smith DF, Cummings RD, Estes MK, Venkataram Prasad BV. 2015. Structural basis of glycan specificity in neonate-specific bovine-human reassortant rotavirus. Nat Commun 6:8346. doi: 10.1038/ncomms9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varghese V, Das S, Singh NB, Kojima K, Bhattacharya SK, Krishnan T, Kobayashi N, Naik TN. 2004. Molecular characterization of a human rotavirus reveals porcine characteristics in most of the genes including VP6 and NSP4. Arch Virol 149:155–172. [DOI] [PubMed] [Google Scholar]
- 39.My PV, Rabaa MA, Donato C, Cowley D, Phat VV, Dung TT, Anh PH, Vinh H, Bryant JE, Kellam P, Thwaites G, Woolhouse ME, Kirkwood CD, Baker S. 2014. Novel porcine-like human G26P[19] rotavirus identified in hospitalized paediatric diarrhoea patients in Ho Chi Minh City, Vietnam. J Gen Virol 95:2727–2733. doi: 10.1099/vir.0.068403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ianiro G, Delogu R, Graffeo R, Sanguinetti M, Fiore L, Ruggeri FM. 2014. Detection of rare G3P[19] group A rotavirus in human patient, Italy. Emerg Infect Dis 20:1906–1910. doi: 10.3201/2011.131699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maneekarn N, Khamrin P, Chan-It W, Peerakome S, Sukchai S, Pringprao K, Ushijima H. 2006. Detection of rare G3P[19] porcine rotavirus strains in Chiang Mai, Thailand, provides evidence for origin of the VP4 genes of Mc323 and Mc345 human rotaviruses. J Clin Microbiol 44:4113–4119. doi: 10.1128/JCM.00954-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan MC, Lee N, Hung TN, Kwok K, Cheung K, Tin EK, Lai RW, Nelson EA, Leung TF, Chan PK. 2015. Rapid emergence and predominance of a broadly recognizing and fast-evolving norovirus GII.17 variant in late 2014. Nat Commun 6:10061. doi: 10.1038/ncomms10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh BK, Koromyslova A, Hefele L, Gurth C, Hansman GS. 2016. Structural evolution of the emerging 2014-2015 GII.17 noroviruses. J Virol 90:2710–2715. doi: 10.1128/JVI.03119-15. [DOI] [PMC free article] [PubMed] [Google Scholar]