ABSTRACT
There is accumulating evidence that the viral interleukin-10 (vIL-10) ortholog of both human and rhesus cytomegalovirus (HCMV and RhCMV, respectively) suppresses the functionality of cell types that are critical to contain virus dissemination and help shape long-term immunity during the earliest virus-host interactions. In particular, exposure of macrophages, peripheral blood mononuclear cells, monocyte-derived dendritic cells, and plasmacytoid dendritic cells to vIL-10 suppresses multiple effector functions including, notably, those that link innate and adaptive immune responses. Further, vaccination of RhCMV-uninfected rhesus macaques with nonfunctional forms of RhCMV vIL-10 greatly restricted parameters of RhCMV infection following RhCMV challenge of the vaccinees. Vaccinees exhibited significantly reduced shedding of RhCMV in saliva and urine following RhCMV challenge compared to shedding in unvaccinated controls. Based on the evidence that vIL-10 is critical during acute infection, the role of vIL-10 during persistent infection was analyzed in rhesus macaques infected long term with RhCMV to determine whether postinfection vaccination against vIL-10 could change the virus-host balance. RhCMV-seropositive macaques, which shed RhCMV in saliva, were vaccinated with nonfunctional RhCMV vIL-10, and shedding levels of RhCMV in saliva were evaluated. Following robust increases in vIL-10-binding and vIL-10-neutralizing antibodies, shedding levels of RhCMV modestly declined, consistent with the interpretation that vIL-10 may play a functional role during persistent infection. However, a more significant association was observed between the levels of cellular IL-10 secreted in peripheral blood mononuclear cells exposed to RhCMV antigens and shedding of RhCMV in saliva. This result implies that RhCMV persistence is associated with the induction of cellular IL-10 receptor-mediated signaling pathways.
IMPORTANCE Human health is adversely impacted by viruses that establish lifelong infections that are often accompanied with increased morbidity and mortality (e.g., infections with HIV, hepatitis C virus, or human cytomegalovirus). A longstanding but unfulfilled goal has been to develop postinfection vaccine strategies that could “reboot” the immune system of an infected individual in ways that would enable the infected host to develop immune responses that clear reservoirs of persistent virus infection, effectively curing the host of infection. This concept was evaluated in rhesus macaques infected long term with rhesus cytomegalovirus by repeatedly immunizing infected animals with nonfunctional versions of the rhesus cytomegalovirus-encoded viral interleukin-10 immune-modulating protein. Following vaccine-mediated boosting of antibody titers to viral interleukin-10, there was modest evidence for increased immunological control of the virus following vaccination. More significantly, data were also obtained that indicated that rhesus cytomegalovirus is able to persist due to upregulation of the cellular interleukin-10 signaling pathway.
INTRODUCTION
It is incumbent upon microbes that establish lifelong pathogen-host relationships to modify host immunity in ways that facilitate persistent carriage of the microbe by immunocompetent hosts. Immune-modulating strategies of persistence by “hit-and-stay” pathogens are likely to be fundamentally different from acute, or “hit-and-run,” pathogens, for which viral replication and intra- and interhost dissemination mostly transpire prior to development of de novo adaptive immune responses that could potentially clear the pathogen (1). Cellular interleukin-10 (cIL-10) is an anti-inflammatory cytokine that is considered a master regulator of the immune system due to its positive and negative effects on cells bearing the IL-10 receptor (IL-10R) (2). Manipulation of the cIL-10/IL-10R signaling pathway has been increasingly associated with long-term persistent infections in immunocompetent hosts (3). Multiple evolutionarily diverse microbes (viral, pathogenic and commensal bacterial, fungal, protozoal, and helminthic) activate the IL-10R-mediated signaling pathway as part of their natural histories. Such evolutionary convergence upon a single cytokine signaling pathway (4) suggests cIL-10 is an essential component in creating and maintaining immune niches permissive to long-term infection, particularly in infected hosts with fully functional immune systems. Indeed, murine studies in which IL-10/IL-10R signaling was disrupted, either by neutralizing cIL-10 functionality or through antibody-mediated blockage of IL-10R, have shown this disruption results in significantly decreased pathogen loads (murine cytomegalovirus [MCMV], lymphocytic choriomeningitis virus) (5–7). Based on these findings, blocking pathogen-associated cIL-10 manipulation may be a relevant strategy for blocking either the establishment and/or maintenance of a persistent infection.
Human cytomegalovirus (HCMV) is a ubiquitous persistent pathogen with worldwide seroprevalence rates in adults that range from ∼50 to 100%, and there is accumulating evidence that persistence is mediated through viral modulation of host immunity, including manipulation of the IL-10R pathway (3, 8–10). HCMV is generally considered a virus with low pathogenic potential in immunocompetent hosts, consistent with the interpretation that host immune responses to HCMV infection are protective against HCMV sequelae. In the absence of immune functionality, HCMV can be a significant cause of morbidity and mortality in immunosuppressed transplant recipients, immunodeficient AIDS patients not on highly active antiretroviral therapy, and in congenitally infected infants. Taken together, host immunity mostly protects against HCMV disease during primary infection, but host immune responses are insufficient to clear persistent reservoirs in infected hosts, despite extraordinarily large HCMV-specific T cell responses (11) and the induction of neutralizing antibodies against multiple HCMV glycoproteins (4, 12–14). The viral mechanisms of persistence are incompletely resolved, but there is increasing evidence that cIL-10 signaling through binding to its high-affinity receptor (IL-10R1) is critical for maintaining persistence (2, 3). Studies of MCMV-infected mice revealed that there is a marked increase in the number of CD4+ cells expressing cIL-10 in tissue sites of persistence (salivary glands) but not in tissues where MCMV establishes a latent infection (e.g., spleen) (7). While MCMV does not encode an ortholog of cIL-10, the results of this study imply that MCMV, like many viral and nonviral pathogens (3), has co-opted the cIL-10/IL-10R1 signaling cascade to enable persistence in an immune host.
The complexity of IL-10 signaling induced by HCMV and RhCMV is greater than that observed by MCMV. Unlike MCMV, both HCMV and RhCMV encode a cIL-10 ortholog (cmvIL-10 and rhcmvIL-10, respectively) that binds to IL-10R1, and each viral ortholog exhibits comparable functionality to that of cIL-10 (3, 8, 15, 16). A recent RhCMV study that employed vaccine-mediated disruption of rhcmvIL-10-mediated signaling revealed that vaccine-mediated neutralization of rhcmvIL-10 function significantly restricted long-term parameters of primary RhCMV infection, particularly the frequency and magnitude of RhCMV detection in the saliva and urine of vaccinated/challenged rhesus macaques (17). Similarly, Chang et al. reported that animals infected with a rhcmvIL-10 knockout variant of RhCMV (68-1) had quantitatively enhanced innate immune responses and developed more robust antiviral adaptive immune responses than animals expressing the parental virus, rhcmvIL-10 (18). Taken together, early viral production of rhcmvIL-10 during primary infection appears to be critical in shaping the immune system to facilitate both viral dissemination and persistence in an immune host.
However, little is known about what functional role, if any, rhcmvIL-10 might play in long-term infections. Since the results of previous studies implied that rhcmvIL-10 is important for establishing a persistent infection (17), our study sought to determine whether long-term rhcmvIL-10 expression may be involved in maintaining persistence in immunocompetent animals.
MATERIALS AND METHODS
Study animals and immunization.
Ten RhCMV-seropositive rhesus macaques were immunized with a combination of nickel affinity-purified rhcmvIL-10M1 and rhcmvIL-10M2 (19) proteins mixed (ratio) with Addavax adjuvant (Invivogen). Animals were injected at weeks 0, 6, and 12 (50 μg of each protein intramuscularly [i.m.]). Ten additional animals concurrently received injections of Escherichia coli-derived maltose-binding protein (MBP) in the Addavax adjuvant as a control. Animals were prospectively evaluated to 24 weeks postinfection (p.i.).
Sample collection and DNA extraction.
Blood and saliva were collected from anesthetized animals according to our published protocols (20). RhCMV viral DNA was extracted from plasma and via oral swabs using the QiaSymphony system (Qiagen) according to our published protocols (21). Samples were extracted following the manufacturer's protocols and stored for use at −80°C.
Quantitative real-time PCR.
RhCMV DNA was quantified in plasma and oral swabs by using real-time PCR according to previously published protocols (22). Briefly, primers were designed to amplify the RhCMV gB gene along with a probe that included the reporter dye TET (Roche) on the 5′ end and 6-carboxymethylrhodamine (TAMRA) quencher (Applied Biosystems, Foster City, CA) on the 3′ end. Reaction mixtures included 1× TaqMan universal PCR master mix (Applied Biosystems), 17.5 pmol forward and reverse primers, 2.5 pmol probe, and 5 μl DNA sample in a total volume of 12.5 μl. All samples were run in triplicate using the ABI Prism 7900 sequence detection system (Applied Biosystems). Quantification of DNA was calculated using a 10-fold serial dilution standard curve of RhCMV gB plasmid, purified by two rounds of cesium chloride gradient centrifugation, containing 106 to 100 copies per 5 μl of DNA sample.
RhCMV EGFP neutralization assay.
With a RhCMV-enhanced green fluorescent protein (EGFP) construct, our neutralization assays were performed following previously published protocols (23).
rhcmvIL-10 ELISA.
For the rhcmvIL-1 enzyme-linked immunosorbent assay (ELISA), anti-rhcmvIL-10 antibodies were quantified from heat-inactivated plasma samples and oral swab specimens by using a previously published protocol with slight variations (19). Briefly, 96-well plates (Immulon 4 HBX; Dynex Technologies Inc.) were coated with 25 ng/well of nickel-purified rhcmvIL-10 protein in coating buffer (phosphate-buffered saline [PBS; Sigma]–0.375% sodium bicarbonate [GIBCO]) overnight at 4°C. Plates were then washed 6 times with PBS–0.05% Tween (PBS/T; Sigma) wash buffer and blocked with 300 μl/well PBS–1% bovine serum albumin (BSA; Sigma) for 2 h at 25°C in a temperature-controlled incubator. Plates were washed 6 times in PBS/T, and 100 μl of sample (1:100 rhesus macaque plasma diluted in PBS/T–1% BSA or 1:6 oral swab diluted in PBS) was added to each well in duplicate and incubated for 2 h at 25°C. Plates were then washed 6 times with PBS/T and loaded with 100 μl/well of peroxidase-conjugated goat anti-monkey IgG (Kirkegaard & Perry Laboratories, Inc. [KPL]) diluted 1:175,000 in PBS/T–1% BSA and incubated at 25°C for 1 h. Plates were washed again (6 times with PBS/T), and 100 μl of tetramethylbenzidine (TMB; Sigma) was added to each well. Plates were incubated in the dark at 25°C for roughly 25 min, and color development was then stopped with the addition of 50 μl/well of 0.5 M sulfuric acid. Color development was then quantified spectrophotometrically using the Bio-Rad model 680 microplate reader at a wavelength of 450 nm. Measurements are reported as relative units (RU), calculated according to a 2-fold dilution standard curve of anti-rhcmvIL-10 hyperimmune serum from a previously vaccinated rhesus macaque.
RhCMV ELISA.
The RhCMV ELISA was done as reported above with slight variations. Ninety-six-well plates were coated overnight at 4°C with 0.25 μg/well of heat-inactivated RhCMV virion diluted in coating buffer. They were subsequently washed with PBS/T and blocked as stated above. A 100-μl aliquot of rhesus macaque plasma diluted 1:100 in PBS/T–1% BSA was then added to wells in duplicate and incubated for 2 h at 25°C. Plates were then washed 6 times with PBS/T, 100 μl of anti-monkey IgG diluted at 1:190,000 with PBS/T–1% BSA was added to each well, and mixtures were incubated at 25°C for 1 h. Plates were then treated exactly as stated for the rhcmvIL-10 ELISA.
cIL-10 ELISA.
cIL-10 secretion by RhCMV-activated peripheral blood mononuclear cells (PBMC) was measured using a rhesus cIL-10 ELISA kit (U-Cytech, Netherlands), according to the manufacturer's protocol with slight variations. Briefly, 96-well microplates (Immulon 4 HBX) were coated with the supplied IL-10 antibody and incubated overnight at 4°C. The plates were washed 6 times with PBS-T and incubated with PBS–1% BSA blocking buffer for 60 min at 37°C. The buffer was removed, 100 μl/well of PBMC supernatant or plasma (1:100 dilution in PBS/T–0.5% BSA) was added, and the plates were incubated at 37° for 2 h. The plates were then washed 6 times with PBS/T wash buffer, 100 μl/well of IL-10 detector antibody was added, and the cells were incubated 1 h at 37°C. After washing, 100 μl/well of streptavidin-horseradish peroxidase (HRP) polymer conjugate (U-Cytech) was added and the mixture was incubated at 37°C for 1 h. After washing, TMB substrate (100 μl/well) was added, and the plates were incubated at 25°C for 10 min. Color development was stopped by the addition of 0.5 M sulfuric acid (50 μl/well). Following a 5-min incubation (25°C), the plates were read at a wavelength of 450 nm on a Bio-Rad model 680 microplate reader. Concentrations of cIL-10 were quantified using a 2-fold serially diluted recombinant cIL-10 standard (U-Cytech) that was included on each plate.
Neutralization of rhcmvIL-10 function in vitro.
Antibodies against rhcmvIL-10 were characterized in a cellular-based protein neutralization assay using a previously published protocol (19) with minor modifications. Briefly, plasma samples were diluted (1:4,000) in RPMI–10% fetal bovine serum with penicillin-streptomycin and l-glutamine (1-ml final volume) in the presence or absence of recombinant rhcmvIL-10 (1.0 ng/ml) for 3 h at 37°C. Mixtures of 200 μl of plasma with or without rhcmvIL-10 were then incubated (each in triplicate) with 4 × 105 Ficoll-purified PBMC/well in a 96-well U-bottom plate (Falcon) for 30 min in a humidified 37°C incubator (5% CO2). Lipopolysaccharide (LPS; from Escherichia coli O127:B8; Sigma) was subsequently added to the cells (5 μg/ml, final concentration) and then incubated for 24 h at 37°C (5% CO2). Cells were spun down, and supernatant was collected the following day and stored at −80°C until assayed for IL-12 production. IL-12 secretion by LPS-activated PBMC was measured in an ELISA (U-Cytech, Netherlands), according to the manufacturer's protocol with slight variations. Briefly, 96-well microplates (Immulon 4 HBX) were coated with the supplied IL-12 antibody pair (p40 plus p70) and incubated overnight at 4°C. The plates were washed 6 times with PBS-T and incubated with PBS–1% BSA blocking buffer for 1 h at 37°C. The buffer was removed, 100 μl/well of PBMC supernatant was added, and the cell mixture was incubated at 4°C overnight. The plates were washed 6 times with PBS-T wash buffer, 100 μl/well of detector antibody was added, and the cells were incubated 1 h at 37°C. After washing, 100 μl/well of HRP conjugate (U-Cytech) was added and incubated at 37°C for 1 h. TMB substrate (100 μl/well) was added after plates were washed, and then the mixtures was incubated at 25°C for 11 min. Color development was stopped by the addition of 0.5 M sulfuric acid (50 μl/well). Following a 5-min incubation (25°C), the plates were read at a wavelength of 450 nm on a model 680 microplate reader (Bio-Rad). Concentrations of IL-12 were quantified using a 2-fold serially diluted recombinant IL-12 standard (U-Cytech) that was included on each plate. Neutralization was calculated as the inverse of the ratio of the IL-12 concentration (rhcmvIL-10 plus plasma) divided by the IL-12 concentration (plasma only), and the results were expressed as the percent rhcmvIL-10 neutralized.
PBMC RhCMV stimulation.
Cryopreserved, Ficoll gradient-purified PBMC were thawed on ice, aliquots of 1 × 106 cells per well were added to a 48-well plate, and the mixtures were rested overnight at 37°C in complete RPMI medium containing 10% endotoxin-free fetal calf serum (FCS), 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 50 μM 2-mercaptoethanol, and 10 mM HEPES. Samples were then incubated with either heat-inactivated RhCMV virion (2.5 μg/ml), nonfunctional rhcmvIL-10 proteins (rhcmvIL-10M1 and rhcmvIL-10M2 [5.0 μg/ml]), or medium in the presence of costimulatory monoclonal antibodies to CD28 (clone 28.2) and CD49d (clone 9F10) (5 μg/ml for each; eBioscience) for 10 h. Cells that were stained for gamma interferon (IFN-γ) and tumor necrosis factor (TNF) were also treated with GolgiPlug (BD Biosciences) after the first 2 h. Supernatants and cells were then centrifuged and used for the cIL-10 ELISA and flow cytometric analyses, respectively.
Intracellular cytokine staining.
For intracellular cytokine staining (ICS), cells were fixed and stained following the published protocol of Chang et al. (18) with slight modifications. Surface staining was done using directly conjugated monoclonal antibodies against human (rhesus-macaque cross-reactive) CD4 (clone L200) and CD8 (clone 3B5) (BD Biosciences). Cells were then fixed and permeabilized using a fixation/permeabilization kit (BD Biosciences), and cells were stained intracellularly for CD3 (clone SP34-2), IFN-γ (clone B27; BD Biosciences), and TNF (clone Mab11). A background baseline value was established for each animal by running a parallel sample without antigen (Ag) stimulation. This value was then subtracted from the corresponding antigen-stimulated sample result to determine the net level of antigen-specific cytokine response.
Flow cytometry.
Five- and seven-color flow cytometric analyses were performed using the Fortessa II system with Diva software (BD Biosciences). Results were analyzed and displayed using FlowJo software (Tree Star).
Statistical analysis.
A linear mixed-effects model was used to estimate the correlation between RhCMV and cIL-10 PBMC and between RhCMV and cIL-10 CD4, with data values log transformed to base 10. RhCMV values were censored at 1,000 (limit of detection), and both cIL-10 in PBMC and cIL-10 in CD4+ cells were censored at 0. For determining whether there was a correlation between RhCMV genome copy numbers in saliva and cIL-10 produced following RhCMV antigen stimulation of PBMC, samples from weeks 21 (10 animals only), 22, 28, and 34 for each animal were incorporated into the analysis (see Fig. 3, below). A mixed-effects model was used to account for the within-animal dependencies of scores due to the repeated measures. The model assumed censored values, using methods described in reference 24.
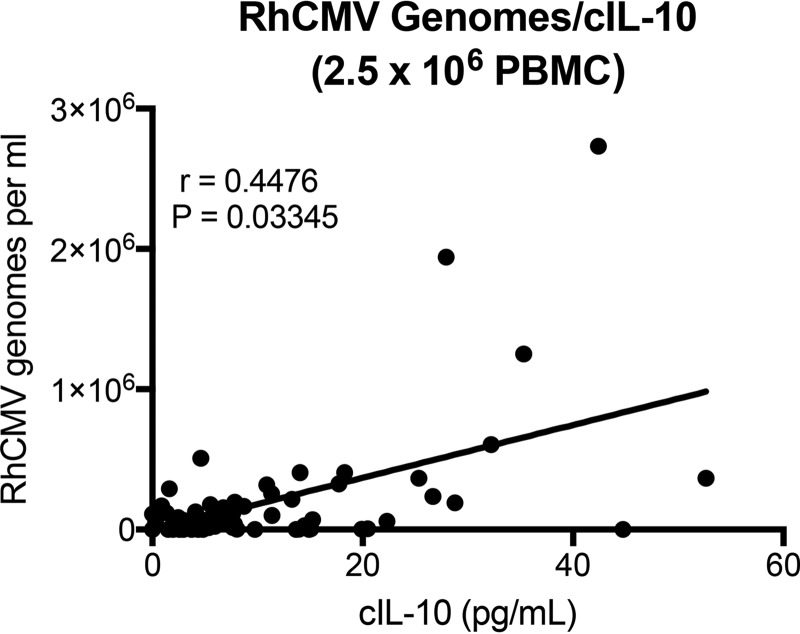
FIG 3.
Antigen-specific cIL-10 levels correlate with chronic viral shedding in saliva. cIL-10 levels secreted by RhCMV antigen-stimulated PBMC (2.5 × 106) isolated from persistently infected macaques were plotted relative to concomitant RhCMV genome copy numbers per milliliter of saliva (r = 0.4476, P = 0.03345; one-tailed t test). Data represent results for samples from weeks 21 (10 animals only), 22, 28, and 34. The limit of detection for RhCMV was 1,000 copies per ml of saliva.
RESULTS
Longitudinal assessment of RhCMV shedding.
RhCMV is endemic in populations of both free-ranging and captive populations of rhesus macaques (Macaca mulatta), and nearly 100% of animals seroconvert to RhCMV infection within the first year of life, well before animals reach sexual maturity (3 to 5 years) (25–29). Once infected with RhCMV, healthy long-term carriers of RhCMV can repeatedly shed virus in bodily fluids (e.g., saliva and urine) for >20 years (20, 30, 31). The cross-sectional frequency of RhCMV in bodily fluids declines as animals age. Whereas ∼75% of adolescent to young adult macaques (3 to 5 years) have detectable RhCMV DNA in saliva and/or urine, 25% of older adults (12 to 20 years) are positive for RhCMV DNA in the same bodily fluids (20).
For this study, a cohort of 20 cohoused RhCMV-infected animals (serologically confirmed [data not shown]) was prospectively evaluated for RhCMV DNA in saliva. Animals ranged in age from 3 years (n = 18) to 6 years (n = 1) to 12 years (n = 1) (Table 1). Based on prior seroepidemiology studies of RhCMV in rhesus macaques at the California National Primate Research Center (CNPRC) (28), the study animals had been infected with RhCMV for >2 to >10 years. Weekly oral swabs (weeks 1 to 13 and weeks 19 to 21) were collected from each animal, and RhCMV genome copy numbers were quantified from DNA purified from each sample. Consistent with previous studies, the preponderance of animals exhibited detectable RhCMV DNA in the majority of samples collected (Table 1; see also Fig. 2, below) (20, 31). Two animals (MMUs 2 and 3) were positive for RhCMV DNA in 14 of the 16 samples (88%), and 15 animals were positive for every oral swab. One animal (MMU 14) was positive for only two time points, and two animals (MMUs 1 and 11) had undetectable RhCMV DNA (<2.3 log10) in any of the 16 saliva samples. Median copy numbers for positive animals ranged from 3.7 to 6.5 log10, and genome copy numbers remained within a relatively tight range during this period of observation. Remarkably, the shedding profile of an individual animal remained relatively constant during this period of observation. The standard deviation (SD) of RhCMV genome copy numbers was ≤0.5 log10 for 17 of the animals, and the remaining 3 animals (MMUs 2, 3, and 12) exhibited a slightly wider range of RhCMV DNA (SD, 0.6 to 0.8 log10).
TABLE 1.
Summary of findings in individual study animals
| Animal ID no. | MMU no.a | Sex | Age (yrs) | No. of RhCMV DNA-positive samplesb | RhCMV copies/ml of saliva (log10)c |
rhcmvIL-10 BAbd | Treatment groupe | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Median | Mean | SD | |||||||
| 32072 | 1 | F | 12 | 0/16 | <2.3 | <2.3 | <2.3 | <2.3 | 0 | 0.75 | V |
| 39547 | 2 | F | 3 | 14/16 | <2.3 | 4.8 | 4.0 | 3.8 | 0.7 | 8.33 | V |
| 39581 | 3 | F | 3 | 14/16 | <2.3 | 5.0 | 3.7 | 3.8 | 0.8 | 8.72 | V |
| 39606 | 4 | M | 3 | 16/16 | 5.3 | 6.2 | 5.8 | 5.7 | 0.3 | 7.30 | V |
| 39629 | 5 | M | 3 | 16/16 | 4.4 | 6.1 | 5.1 | 5.2 | 0.5 | 4.61 | V |
| 39635 | 6 | M | 3 | 16/16 | 4.7 | 5.7 | 5.1 | 5.2 | 0.3 | 11.4 | V |
| 39748 | 7 | F | 3 | 16/16 | 4.9 | 5.8 | 5.4 | 5.4 | 0.3 | 10.00 | V |
| 40013 | 8 | M | 3 | 16/16 | 5.0 | 6.1 | 5.5 | 5.5 | 0.3 | 7.42 | V |
| 40149 | 9 | F | 3 | 16/16 | 3.8 | 5.3 | 4.7 | 4.7 | 0.5 | 3.15 | V |
| 40157 | 10 | M | 3 | 16/16 | 4.8 | 5.7 | 5.2 | 5.3 | 0.3 | 7.72 | V |
| 37429 | 11 | M | 6 | 0/16 | <2.3 | <2.3 | <2.3 | <2.3 | 0 | 4.39 | C |
| 39533 | 12 | F | 3 | 16/16 | 3.4 | 5.3 | 4.5 | 4.4 | 0.6 | 17.33 | C |
| 39544 | 13 | F | 3 | 16/16 | 4.3 | 5.7 | 4.9 | 4.9 | 0.4 | 11.1 | C |
| 39570 | 14 | M | 3 | 2/16 | <2.3 | 3.3 | <2.3 | 2.4 | 0.3 | 0.85 | C |
| 39676 | 15 | M | 3 | 16/16 | 4.2 | 5.7 | 5.2 | 5.1 | 0.4 | 17.55 | C |
| 39692 | 16 | F | 3 | 16/16 | 4.5 | 5.7 | 5.3 | 5.2 | 0.3 | 3.81 | C |
| 39727 | 17 | M | 3 | 16/16 | 4.6 | 5.7 | 5.3 | 5.3 | 0.3 | 13.59 | C |
| 39734 | 18 | M | 3 | 16/16 | 6.1 | 7.1 | 6.5 | 6.5 | 0.3 | 0.54 | C |
| 39735 | 19 | M | 3 | 16/16 | 4.7 | 6.2 | 5.6 | 5.6 | 0.4 | 5.15 | C |
| 40016 | 20 | M | 3 | 16/16 | 4.2 | 6.0 | 4.9 | 5.0 | 0.4 | 7.10 | C |
MMU (an abbreviation used for Macaca mulatta) number for the animal in this study.
Number of saliva samples with detectable RhCMV DNA relative to the total number of samples collected during the 21-week period of observation (n = 16 per group).
The limit of detection of RhCMV DNA was 2.3 log10 (200 copies) per ml.
BAb, rhcmvIL-10-binding antibodies (in relative units [RU]) at week 21.
V, vaccine group; C, control group.
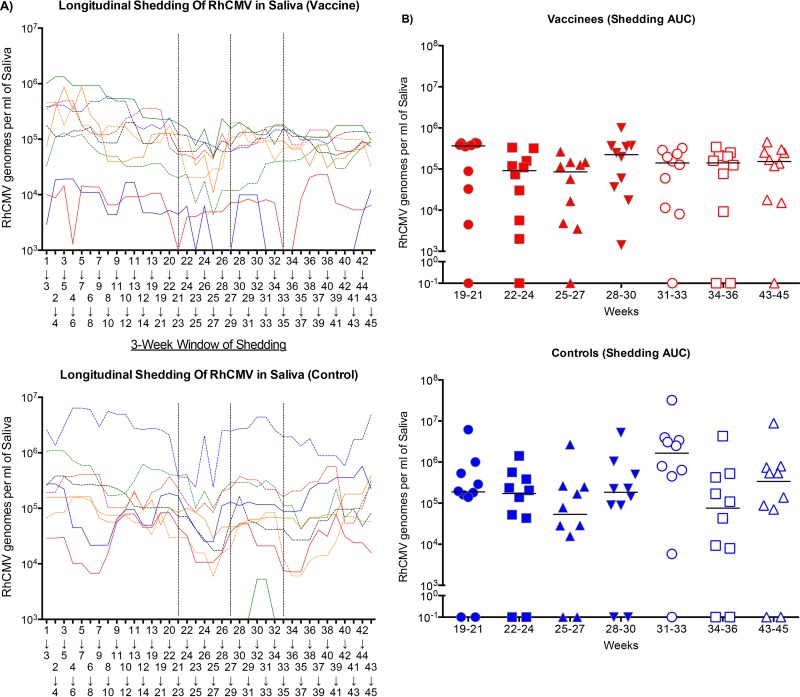
FIG 2.
(A) The 3-week moving average of RhCMV genome copy numbers per milliliter of saliva was plotted relative to the 3-week window of observation for vaccinees (top) and controls (bottom). The time points on the abscissa represent the first week of the 3-week moving average. The dashed lines represent the three immunizations at weeks 21, 27, and 33. (B) The median RhCMV genome copy numbers per milliliter of saliva (solid line) for each treatment group at the time indicated.
Based on patterns of RhCMV DNA detection in saliva, study animals were assigned to either of two groups, vaccine (V) or control (C) (Table 1). The vaccine group comprised 5 females and 5 males (9 of which were persistently positive for RhCMV DNA in saliva), and the control group comprised 3 females and 7 males (8 of which were persistently RhCMV DNA positive in saliva). The frequency and magnitude of RhCMV in the vaccine and control groups were comparable (median RhCMV copy number/ml of saliva, 5.1 and 5.0 log10, respectively). All animals had detectable binding antibodies to rhcmvIL-10 (Table 1), and both the vaccine and control groups exhibited comparable ranges and medians of rhcmvIL-10 binding antibodies (V range, 0.75 to 11.41; V median, 7.57; C range, 0.54 to 17.55; C median, 6.13).
rhcmvIL-10M1/M2 postinfection vaccination boosts memory antibody responses.
It is well established that rhcmvIL-10 M1/M2 vaccination is capable of stimulating robust humoral immune responses in both naive and RhCMV-infected animals and that preexposure vaccination alters the course of the RhCMV challenge infection, significantly lowering shedding titers and frequency (17, 19). The fundamental goal of this study was to determine if postinfection vaccination was able to modify one phenotype of persistent infection, namely, chronic detection of RhCMV DNA in the saliva of long-term-infected animals. The 10 animals in the vaccine group were immunized intramuscularly three times (weeks 21, 27, and 33) with two nonfunctional forms of rhcmvIL-10 (M1 and M2, described previously [19]) adjuvanted in a squalene-based, oil-in-water nanoemulsion. The 10 animals in the control group were immunized three times MBP of E. coli. Animals were sampled weekly (saliva, blood) through week 45 to determine whether there were changes in the frequency and/or magnitude of RhCMV DNA in either bodily fluid.
Consistent with previous findings, rhcmvIL-10 M1/M2 vaccination stimulated robust memory binding and neutralizing antibody responses postvaccination in immune animals (19). Eight of the 10 vaccinated animals exhibited large increases in rhcmvIL-10-binding antibodies after the first immunization (Fig. 1, left graph), and all demonstrated still higher responses after the second and third immunizations. Significant increases were observed after the second immunization, compared to the peak responses after the first immunization, but the peak responses after the second and third immunizations were indistinguishable (Fig. 1, middle). Antibody responses declined after each immunization, although the binding antibody levels 12 weeks after the third immunization were significantly elevated relative to the baseline responses at week 21. Control animals, in contrast, maintained relatively consistent antibody titers following the three immunizations with MBP.
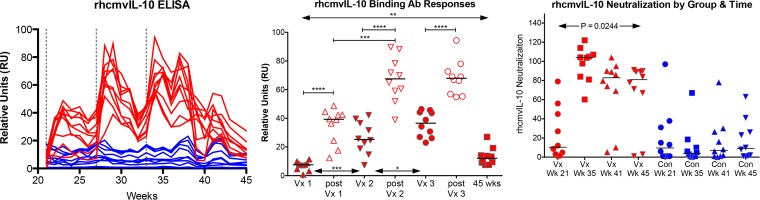
FIG 1.
rhcmvIL-10 antibody responses postimmunization. (Left) Longitudinal rhcmvIL-10 binding antibody responses following immunizations at weeks 21, 27, and 33 (dotted lines) for vaccinees (red) and controls (blue). (Middle) Peak rhcmvIL-10 binding antibody responses at the times of immunization (Vx 1 to 3), within 3 weeks following each immunization (post Vx 1 to 3), or 12 weeks post Vx3 (45 weeks). ****, P < 0.0001; ***, P = 0.0003; **, P = 0.0068; *, P = 0.0431. (Right) vIL-10 neutralizing antibody responses for vaccinees (red) and controls (blue) at weeks 21, and 2, 8, and 12 weeks after the third immunization at week 33 (weeks 35, 41, and 45, respectively).
The three-dose vaccination regimen similarly induced a significant increase in antibodies that neutralized wild-type rhcmvIL-10 function in a bioassay that quantified the ability of antibodies to neutralize rhcmvIL-10-mediated suppression of IL-12 production in LPS-stimulated PBMC (Fig. 1, right graph). Notably, rhcmvIL-10-neutralizing responses remained significantly elevated during the 12-week interval after the third immunization for 8 of the 10 vaccinated animals. During the same period of observation, binding antibodies to total RhCMV antigens and fibroblast neutralization titers remained unchanged (data not shown), consistent with the interpretation that vaccination against rhcmvIL-10 in infected animals does not alter host antibody responses to total RhCMV antigens.
Postinfection rhcmvIL-10 vaccination effects on established viral shedding patterns.
The therapeutic efficacy of rhcmvIL-10 postinfection vaccination was assessed by evaluating changes in chronic viral shedding patterns postvaccination. RhCMV genome copy numbers in saliva were quantified from oral swab samples for the two 6-week intervals between the three immunizations (weeks 21, 27, and 33) and for 12 weeks after the third immunization. The results were compared to the shedding profiles determined during the prevaccination phase (Table 1). The longitudinal pattern of shedding was determined for each animal by using a 3-week moving average of RhCMV genome copy number per ml of saliva (Fig. 2A). For simplicity, the presence of RhCMV genomes in saliva is termed “shedding,” although the quantification of infectious virions in saliva was not performed. When the 3-week shedding profiles at the time of the first immunization (weeks 19 to 21) were compared to those for the final 3 weeks of observation (weeks 43 to 45), there was a decline in the mean shedding loads in the vaccinated animals compared to the controls that approached but did not quite reach statistical significance (P = 0.0547 versus P = 0.6406 for vaccinated and control animals, respectively) (Fig. 2B).
An alternative approach was used to analyze pre- and postvaccination shedding profiles. For this, the area under the curve was also calculated to represent changes in the total infectious burden of RhCMV in saliva. The results were comparable in that there was a trend toward reduced shedding in the vaccinated animals compared to the controls (P = 0.0977 and P = 0.7422, respectively [data not shown]). The modest reduction in RhCMV DNA in saliva after three vaccinations with nonfunctional rhcmvIL-10 suggests that it may be possible to disrupt the virus-host détente in long-term RhCMV infection through optimization of postinfection immunization strategies. While the results further suggested that rhcmvIL-10 might play a role in maintaining viral persistence in an immune host, an alternative approach indicated that activation of the IL-10R1-mediated signaling pathway through activation of cIL-10 plays a more prominent role.
RhCMV-specific cIL-10 responses in PBMC correlate with RhCMV shedding.
Previous murine CMV studies have reported a link between viral persistence and T cell-derived cIL-10 (7, 32). Due to the uncertainty of a definitive impact on viral shedding by the postexposure vaccine with rhcmvIL-10, cIL-10 levels were quantified in PBMC exposed to purified heat-inactivated RhCMV virions (RhCMV antigens) and evaluated in the context of RhCMV shedding. PBMC harvested at 3 to 4 time points from each animal (weeks 21 [10 animals only], 22, 28, and 34) were stimulated with RhCMV antigens and subsequently analyzed for changes in secreted cIL-10 compared to matched unstimulated PBMC. PBMC that were stimulated by RhCMV antigens exhibited variable levels of detectable cIL-10, ranging from 0 to >50 pg/ml (Fig. 3A). When analyzed in relation to detectable RhCMV genomes per ml of saliva at the same time point for which the PBMC were obtained, cIL-10 levels in RhCMV-stimulated PBMC significantly correlated with viral shedding titers (r = 0. 4476, P = 0.03345).
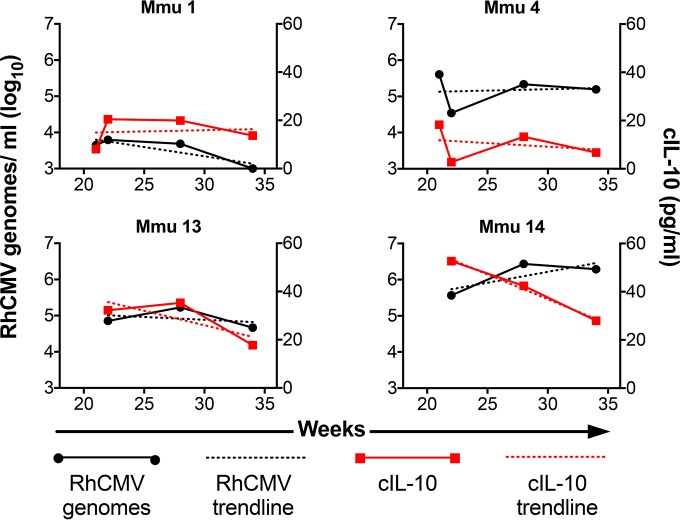
The relation of cIL-10 production and viral shedding was also observed within individuals over time. Longitudinal levels of cIL-10 produced by RhCMV antigen-stimulated PBMC were plotted along with concurrent RhCMV genome copy numbers in saliva to evaluate the patterns of production within the context of a single animal (3 to 4 time points per animal) (Fig. 4; see also Fig. S1 in the supplemental material). Forty-one percent (7 out of 17) displayed a direct mirroring of cIL-10 output patterns and viral shedding patterns in individual animals, while 70% (12 out of 17) displayed similar trends (determined by the slope value of the trend line) of increased or decrease PBMC-derived cIL-10 and viral genome copies in saliva (3 animals were removed from analysis due to a complete absence of detectable viral shedding) (see Fig. S1 in the supplemental material). Although ∼30% of animals showed differing trends of viral shedding and cIL-10 production, the overall results were congruent with the interpretation drawn above that there is a direct relation between the level of cIL-10 produced following exposure of PBMC to RhCMV antigen and the level of RhCMV detected in saliva.
FIG 4.
Longitudinal cIL-10 production mirrors viral shedding patterns. Production of cIL-10 by RhCMV antigen-stimulated PBMC (red; right ordinate) in individual animals over time was graphed alongside RhCMV genome copy number per milliliter (log10) of saliva (black; left ordinate) at concurrent time points (3 to 4 per animal). The dashed lines represent the trend line determined by linear regression. This is a representative sampling of the 20 study animals (refer to Fig. S1 in the supplemental material for results of the total analysis).
Peripheral activation of cIL-10 following primary RhCMV infection.
The apparent relation between the levels of cIL-10 secreted from RhCMV-specific T cells and shedding of RhCMV in saliva led to the investigation of the temporal kinetics of the development of this relationship following primary RhCMV infection. To accomplish this, PBMC samples from a separate study, in which naive macaques had been experimentally inoculated by a subcutaneous route with an epitheliotropic strain of RhCMV (UCD59), were longitudinally analyzed for levels of cIL-10 secreted from unstimulated cells, in addition to cIL-10 produced following RhCMV antigen stimulation. The results suggested that primary RhCMV infection is associated with a systemic, transient, nonspecific activation of cIL-10 in PBMC.
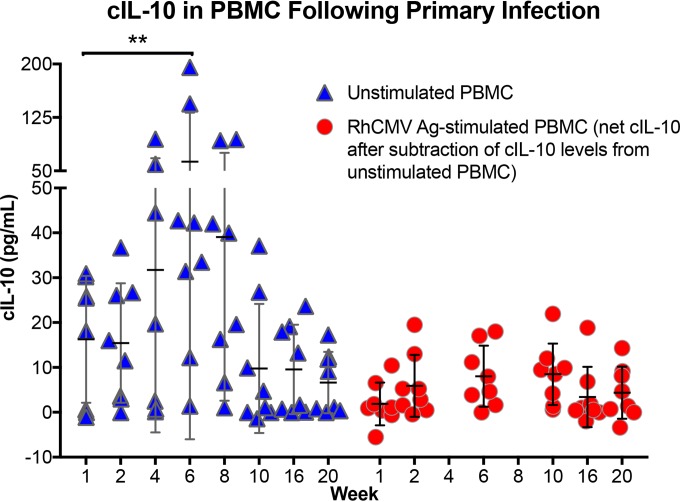
For this experiment, cIL-10 levels from unstimulated and RhCMV antigen-stimulated PBMC were longitudinally compared to 1-week post-RhCMV inoculation samples, since samples from the time of inoculation (week 0) were no longer available from this other study (Fig. 5, unstimulated versus RhCMV Ag stimulated). For the PBMC samples stimulated with RhCMV antigen, only the net cIL-10 levels are presented, because background concentrations of cIL-10 from unstimulated PBMC were subtracted from the RhCMV antigen-stimulated cIL-10 levels. Notably, there was a significant increase in basal cIL-10 in week 6 samples from unstimulated PBMC, and there was also an increase with week 4 and 8 samples, although the differences for these two intervals did not reach statistical significance. For all eight animals, the time point for peak cIL-10 detection in unstimulated cells was either week 4, 6, or 8, with peak concentrations of cIL-10 ranging from 19.6 to 195.6 pg/ml. The one remaining animal exhibited peak basal cIL-10 expression at week 1. By week 20, unstimulated PBMC from four animals had barely detectable levels of cIL-10 (0 to 1.1 pg/ml), and in the other four concentrations ranged from 9.2 to 17.3 pg/ml.
FIG 5.
cIL-10 levels produced by PBMC following primary UCD59 inoculation. RhCMV-uninfected macaques were inoculated with 105 PFU of the epitheliotropic UCD59 strain of RhCMV (83) by a subcutaneous route. PBMC were isolated at the indicated times postinoculation (abscissa), and basal levels of cIL-10 produced from unstimulated (blue triangles) or RhCMV antigen-stimulated (red circles) PBMC were determined. For RhCMV antigen-stimulated PBMC, the values represent the net values of cIL-10 produced after subtraction of the cIL-10 level produced from unstimulated cells. **, P < 0.05.
A quite distinct pattern was observed for cIL-10 production in RhCMV Ag-stimulated PBMC. While there was a slight trend of increased cIL-10 production over the same time frame, the magnitude of cIL-10 produced following antigen stimulation (minus the cIL-10 from unstimulated PBMC) was far lower than the basal level of cIL-10 production from unstimulated cells, ranging from 0 to 18.8 pg/ml (Fig. 5, red circles). The results suggested that primary RhCMV infection is associated with a nonspecific systemic increase in cIL-10 from mononuclear cells in the peripheral circulation. It is important to note that the period during which cIL-10 expression is increased from unstimulated cells (4 to 8 weeks) is contemporaneous with the first detection of RhCMV DNA in saliva and urine and also the first detection of antibodies that neutralize rhcmvIL-10 function (17). Put another way, distal sites, from which RhCMV is persistently shed following primary infection, are seeded with progeny virions during the period of time when there appears to be an immunosuppressive milieu for innate and adaptive effector cells. Moreover, the decline in basal cIL-10 production in unstimulated cells is just prior to the appearance of de novo antibodies that neutralize rhcmvIL-10 function (17), suggesting that early expression of rhcmvIL-10 may drive the peripheral production of cIL-10.
DISCUSSION
Together with previous in vitro and in vivo studies investigating the functionality of HCMV- and RhCMV-encoded viral IL-10 orthologs (cmvIL-10 and rhcmvIL-10, respectively), the results of this study lead to a model in which the virus manipulates the IL-10R1 signaling pathway throughout its infectious cycle within an immunocompetent host. Multiple studies have demonstrated that viral IL-10 alters the functionalities of key innate cell types that would be particularly relevant during acute infection, including macrophages, monocyte-derived dendritic cells (DC), and plasmacytoid DC (PDC), and in vitro studies emphasize that manipulation of these cell types would be critical for engendering adaptive immune responses that facilitate lifelong persistence in immune hosts (15, 18, 19, 33–40). In support of the conclusions drawn from these in vitro experiments, inoculation of macaques with a variant of RhCMV strain 68-1, in which the rhcmvIL-10 open reading frame (ORF; UL111A) had been deleted (RhCMVΔUL111A), resulted in dampened innate and adaptive immune responses to primary RhCMV infection (18). The central importance of early viral manipulation of host immunity was bolstered by a study demonstrating that vaccinating naive macaques with nonfunctional forms of rhcmvIL-10 significantly reduces long-term parameters of RhCMV challenge, presumably by blunting the immediate effects of rhcmvIL-10 on local immune cells at the subcutaneous site of RhCMV inoculation (17). As such, rhcmvIL-10 can be viewed as a virulence factor for RhCMV's natural history, since the absence of full functionality immediately after infection reduces the ability to colonize select niches (e.g., salivary glands, genitourinary tract) within the infected host.
The current study was initiated to determine whether rhcmvIL-10 can be considered a virulence factor for maintenance of a persistent infection. While postinfection vaccination with nonfunctional forms of rhcmvIL-10 stimulated large and durable increases in antibodies that neutralized wild-type rhcmvIL-10 function in an ex vivo bioassay, there was no significant impact on RhCMV genomes detected in saliva. Assuming that there would be equal penetrance of rhcmvIL-10-neutralizing antibodies in all tissues, including the salivary glands, it must be concluded that rhcmvIL-10 is not essential for maintenance of persistent reservoirs of infected cells that reactivate to excrete virions in bodily fluids such as saliva. There were two notable findings that indicated that RhCMV takes advantage of cIL-10-mediated activation of IL-10R1 signaling pathways. The first was the observation that there is a transient increase in basal levels of cIL-10 production in unstimulated PBMC 4 to 8 weeks post-RhCMV infection (Fig. 5). This increase in cIL-10, which was not observed in RhCMV antigen-stimulated PBMC, was contemporaneous with the first detection of RhCMV DNA in urine and saliva. This suggests that primary RhCMV infection is associated with a generalized, transient immune suppression of the infected host during the point in the RhCMV infectious cycle when progeny virions disseminate from the primary site of inoculation to distal sites. In this regard, the phenotype of elevated cIL-10 levels is common during the early phases of bacterial and viral infections in multiple species. As examples, early infections of cattle, swine, chickens, mice, and humans with foot-and-mouth disease virus, classical swine fever virus, Salmonella enterica, MCMV and Ehrlichia muris, and Helicobacter pylori and hepatitis C virus, respectively, are also associated with elevated cIL-10 expression during the early stage of infection (41–47). In addition, there is increasing evidence that HCMV infection, particularly within CD34+ myeloid progenitor cells, is associated with HCMV-induced upregulation of cIL-10 expression (48–52). It has been noted that cmvIL-10 expressed in CD34+ cells appears to attenuate recognition of virally infected cells from HCMV-specific CD4+T cells through downregulation of major histocompatibility class II (MHC-II) expression (53). However, expression of cmvIL-10 does not appear to affect MHC-I antigen presentation in neighboring antigen-presenting cells (54). Together with other examples cited in a recent review (3), one unifying theme for the evolutionarily diverse organisms that co-opt IL-10 is that early induction of IL-10R1-mediated signaling pathways is essential for establishment and maintenance of persistence. In the case of RhCMV, this appears to be a systemic effect early in the infection process and not one driven by antigen-specific T cells, as the net increases postinfection were observed only in unstimulated PBMC.
At some point after 20 weeks post-RhCMV inoculation (Fig. 5) but before the approximate 2 years post-natural RhCMV exposure in the animals in this study, a shift occurs such that PBMC production of cIL-10 is only detected after stimulation with RhCMV virion antigen. One striking finding of this study is that there are significant correlations between the levels of antigen-stimulated cIL-10 production in PBMC and RhCMV genome copy numbers detected in saliva. This is not unique to RhCMV, as cIL-10 levels have been positively associated, either alone or with other biomarkers, with increased burdens of other pathogens, including human papillomavirus, HIV, hepatitis B virus, Epstein-Barr virus (EBV), Mycobacterium tuberculosis, Leishmania spp., dengue virus, and murine and human CMV (45, 55–72). While the specific cell type(s) responsible for elevated cIL-10 expression in response to RhCMV antigen exposure was not explored, multiple cell types have been implicated for other pathogens, including regulatory B and T cells. Taken together, the results emphasize that both early and sustained activation of IL-10 signaling is a common strategy for both intra- and extracellular pathogens and microbes (3). It is worth noting that the original 1990 description of what was then referred to as cytokine synthesis inhibitory factor (now cIL-10) noted the sequence homology with an uncharacterized ORF in EBV (73). The authors of that initial publication on IL-10 speculated that, “an intriguing possibility is that EBV has exploited the biological activity of the product of a captured cytokine gene to manipulate the immune response against virally infected cells, thereby promoting survival of the virus.” The apparent convergent evolution of extensive microbial/viral targeting of the cIL-10 pathway certainly reinforces this prescient description of infectious agent-host interactions. This commonality of pathogen natural histories further suggests that prophylactic and/or postinfection intervention strategies specifically disrupting, or possibly augmenting, cIL-10 signaling may be useful for preventing and/or treating infection and disease.
Of all the identified pathogens or commensals whose natural histories are implicated with alterations in cIL-10R1-mediated signaling (3), all but select members of the Herpesviridae and Poxviridae use cIL-10 as the ligand for activating the high-affinity receptor. Apart from members of the Herpesviridae and Poxviridae, microbe-encoded products, when they have been identified, lead to upregulation of cIL-10 expression, which leads to increased activation of the IL-10 signaling pathway. In contrast, some members of the Herpesviridae and Poxviridae, including HCMV and RhCMV, and also EBV, transduced cIL-10 genes during viral evolution and express the viral orthologs in the context of the viral genome. Although examples are limited, vaccination against either the viral cIL-10 ortholog or the microbial inducer of cIL-10 expression has conferred protective efficacy against challenge. As noted, vaccination of naive macaques with nonfunctional forms of rhcmvIL-10 reduced long-term shedding of RhCMV in saliva and urine following RhCMV challenge (17). Similarly, vaccination of mice with bacterially encoded glyceraldehyde phosphate dehydrogenase protein of group B streptococcus (74), the CyaA toxin of Bordetella pertussis (75), or the BopN virulence factor of Bordetella bronchiseptica (76), all of which have been identified as stimulators of expression of cIL-10, confers significant measures of protection against bacterial challenge. By extension, interruption of IL-10R1 signaling through antibody-mediated blockade of cIL-10R1 significantly reduced virus production and in some cases cleared the infection from sites of persistence following MCMV or lymphocytic choriomeningitis virus infections (5, 7, 77–79). Collectively, all of these studies open the possibility of targeting the IL-10R1 signaling pathway to prevent or treat ongoing pathogenic infections.
Renowned vaccinologist Maurice Hilleman wrote that, “Persistent [infections] causing serious diseases derive primarily from altered function of the immune system” (1). One unifying theme for persistent pathogens is that primary infections can stimulate host pathogen-specific immune responses that are insufficient to effectively restrict ongoing pathogen replication and/or antigen expression, which can result in direct pathogen sequelae and/or indirect immunopathologies. In effect, pathogens skew immune responses in ways that favor pathogen persistence at the expense of host-mediated immune clearance. This is the antithesis of adjuvant-stimulated, vaccine-mediated immune responses, which favor the host. One implication is that vaccination strategies prior to infection should induce a distinctly different vaccine-mediated immune state from that observed following natural infection. It might be possible to counteract the immune environment during an ongoing infection, such as neutralizing IL-10R1 signaling, and concurrently inducing a novel de novo immune response to HCMV antigens to shift the pathogen-host balance to one favoring the host (5–7, 80–82). The RhCMV model of persistence provides an experimental framework to investigate strategies that prevent and/or abrogate the privileged state of infected cells within an immunocompetent host, particularly those that focus on IL-10 signaling pathways.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 AI49342 (P.A.B. and M.R.W.), R01 AI047300 and AI047300-S1 (M.R.W.), P51 OD011107* (California National Primate Research Center), and the Margaret Deterding Infectious Disease Research Support Fund (P.A.B.).
Funding Statement
The Margaret Deterding Infectious Disease Research Support Fund is a private research fund for Peter A. Barry. The California National Primate Research Center is supported by NIH grant P51 OD011107*.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00635-16.
REFERENCES
- 1.Hilleman MR. 2004. Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections. Proc Natl Acad Sci U S A 101(Suppl 2):S14560–S14566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walter MR. 2014. The molecular basis of IL-10 function: from receptor structure to the onset of signaling. Curr Top Microbiol Immunol 380:191–212. doi: 10.1007/978-3-662-43492-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eberhardt MK, Barry PA. 2014. Pathogen manipulation of cIL-10 signaling pathways: opportunities for vaccine development? Curr Top Microbiol Immunol 380:93–128. doi: 10.1007/978-3-662-43492-5_5. [DOI] [PubMed] [Google Scholar]
- 4.Chiuppesi F, Wussow F, Johnson E, Bian C, Zhuo M, Rajakumar A, Barry PA, Britt WJ, Chakraborty R, Diamond DJ. 2015. Vaccine-derived neutralizing antibodies to the human cytomegalovirus gH/gL pentamer potently block primary cytotrophoblast infection. J Virol 89:11884–11898. doi: 10.1128/JVI.01701-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks DG, Lee AM, Elsaesser H, McGavern DB, Oldstone MB. 2008. IL-10 blockade facilitates DNA vaccine-induced T cell responses and enhances clearance of persistent virus infection. J Exp Med 205:533–541. doi: 10.1084/jem.20071948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks DG, Walsh KB, Elsaesser H, Oldstone MB. 2010. IL-10 directly suppresses CD4 but not CD8 T cell effector and memory responses following acute viral infection. Proc Natl Acad Sci U S A 107:3018–3023. doi: 10.1073/pnas.0914500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphreys IR, de Trez C, Kinkade A, Benedict CA, Croft M, Ware CF. 2007. Cytomegalovirus exploits IL-10-mediated immune regulation in the salivary glands. J Exp Med 204:1217–1225. doi: 10.1084/jem.20062424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slobedman B, Barry PA, Spencer JV, Avdic S, Abendroth A. 2009. Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J Virol 83:9618–9629. doi: 10.1128/JVI.01098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navarro D. 2016. Expanding role of cytomegalovirus as a human pathogen. J Med Virol 88:1103–1112. doi: 10.1002/jmv.24450. [DOI] [PubMed] [Google Scholar]
- 10.Britt W. 2015. Controversies in the natural history of congenital human cytomegalovirus infection: the paradox of infection and disease in offspring of women with immunity prior to pregnancy. Med Microbiol Immunol 204:263–271. doi: 10.1007/s00430-015-0399-9. [DOI] [PubMed] [Google Scholar]
- 11.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britt WJ, Mach M. 1996. Human cytomegalovirus glycoproteins. Intervirology 39:401–412. [DOI] [PubMed] [Google Scholar]
- 13.Shimamura M, Mach M, Britt WJ. 2006. Human cytomegalovirus infection elicits a glycoprotein M (gM)/gN-specific virus-neutralizing antibody response. J Virol 80:4591–4600. doi: 10.1128/JVI.80.9.4591-4600.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lilleri D, Kabanova A, Lanzavecchia A, Gerna G. 2012. Antibodies against neutralization epitopes of human cytomegalovirus gH/gL/pUL128-130-131 complex and virus spreading may correlate with virus control in vivo. J Clin Immunol 32:1324–1331. doi: 10.1007/s10875-012-9739-3. [DOI] [PubMed] [Google Scholar]
- 15.Lockridge KM, Zhou SS, Kravitz RH, Johnson JL, Sawai ET, Blewett EL, Barry PA. 2000. Primate cytomegaloviruses encode and express an IL-10-like protein. Virology 268:272–280. doi: 10.1006/viro.2000.0195. [DOI] [PubMed] [Google Scholar]
- 16.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc Natl Acad Sci U S A 97:1695–1700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberhardt M, Deshpande A, Chang W-L, Barthold S, Walter M, Barry P. 2013. Vaccination against a viral encoded cytokine significantly restricts viral challenge. J Virol 87:11323–11331. doi: 10.1128/JVI.01925-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang WL, Barry PA. 2010. Attenuation of innate immunity by cytomegalovirus IL-10 establishes a long-term deficit of adaptive antiviral immunity. Proc Natl Acad Sci U S A 107:22647–22652. doi: 10.1073/pnas.1013794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logsdon NJ, Eberhardt MK, Allen CE, Barry PA, Walter MR. 2011. Design and analysis of rhesus cytomegalovirus IL-10 mutants as a model for novel vaccines against human cytomegalovirus. PLoS One 6:e28127. doi: 10.1371/journal.pone.0028127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oxford K, DeLa Pena-ponce M, Jensen K, Eberhardt M, Spinner A, Van Rompay K, Rigdon J, Mollan K, Krishnan V, Hudgens M, Barry P, De Paris K. 2015. The interplay between immune maturation, age, chronic viral infection and environment. Immun Ageing 12:3. doi: 10.1186/s12979-015-0030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oxford KL, Strelow L, Yue Y, Chang WL, Schmidt KA, Diamond DJ, Barry PA. 2011. Open reading frames carried on UL/b′ are implicated in shedding and horizontal transmission of rhesus cytomegalovirus in rhesus monkeys. J Virol 85:5105–5114. doi: 10.1128/JVI.02631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sequar G, Britt WJ, Lakeman FD, Lockridge KM, Tarara RP, Canfield DR, Zhou SS, Gardner MB, Barry PA. 2002. Experimental coinfection of rhesus macaques with rhesus cytomegalovirus and simian immunodeficiency virus: pathogenesis. J Virol 76:7661–7671. doi: 10.1128/JVI.76.15.7661-7671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abel K, Martinez J, Yue Y, Lacey SF, Wang Z, Strelow L, Dasgupta A, Li Z, Schmidt KA, Oxford KL, Assaf B, Longmate JA, Diamond DJ, Barry PA. 2011. Vaccine-induced control of viral shedding following rhesus cytomegalovirus challenge in rhesus macaques. J Virol 85:2878–2890. doi: 10.1128/JVI.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiebaut R, Jacqmin-Gadda H. 2004. Mixed models for longitudinal left-censored repeated measures. Comput Methods Programs Biomed 74:255–260. doi: 10.1016/j.cmpb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Andrade MR, Yee J, Barry P, Spinner A, Roberts JA, Cabello PH, Leite JP, Lerche NW. 2003. Prevalence of antibodies to selected viruses in a long-term closed breeding colony of rhesus macaques (Macaca mulatta) in Brazil. Am J Primatol 59:123–128. doi: 10.1002/ajp.10069. [DOI] [PubMed] [Google Scholar]
- 26.Barry P, Chang W-L. 2007. Primate betaherpesviruses, p 1051–1075. In Arvin A, Campadielli G, Moore P, Mocarski E, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 27.Früh K, Malouli D, Oxford K, Barry P. 2013. Non-human-primate models of cytomegalovirus infection, prevention, and therapy, p 463–493. In Reddehase M. (ed), Cytomegaloviruses: from molecular pathogenesis to therapy, vol II Caister Academic Press/Horizon, Norfolk, United Kingdom. [Google Scholar]
- 28.Vogel P, Weigler BJ, Kerr H, Hendrickx A, Barry PA. 1994. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab Anim Sci 44:25–30. [PubMed] [Google Scholar]
- 29.Jones-Engel L, Engel GA, Heidrich J, Chalise M, Poudel N, Viscidi R, Barry PA, Allan JS, Grant R, Kyes R. 2006. Temple monkeys and health implications of commensalism, Kathmandu, Nepal. Emerg Infect Dis 12:900–906. doi: 10.3201/eid1206.060030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asher DM, Gibbs CJ Jr, Lang DJ, Gajdusek DC, Chanock RM. 1974. Persistent shedding of cytomegalovirus in the urine of healthy rhesus monkeys. Proc Soc Exp Biol Med 145:794–801. doi: 10.3181/00379727-145-37897. [DOI] [PubMed] [Google Scholar]
- 31.Huff JL, Eberle R, Capitanio J, Zhou SS, Barry PA. 2003. Differential detection of B virus and rhesus cytomegalovirus in rhesus macaques. J Gen Virol 84:83–92. doi: 10.1099/vir.0.18808-0. [DOI] [PubMed] [Google Scholar]
- 32.Stacey MA, Marsden M, Wang EC, Wilkinson GW, Humphreys IR. 2011. IL-10 restricts activation-induced death of NK cells during acute murine cytomegalovirus infection. J Immunol 187:2944–2952. doi: 10.4049/jimmunol.1101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang WL, Barry PA, Szubin R, Wang D, Baumgarth N. 2009. Human cytomegalovirus suppresses type I interferon secretion by plasmacytoid dendritic cells through its interleukin 10 homolog. Virology 390:330–337. doi: 10.1016/j.virol.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang WL, Baumgarth N, Eberhardt MK, Lee CY, Baron CA, Gregg JP, Barry PA. 2007. Exposure of myeloid dendritic cells to exogenous or endogenous IL-10 during maturation determines their longevity. J Immunol 178:7794–7804. doi: 10.4049/jimmunol.178.12.7794. [DOI] [PubMed] [Google Scholar]
- 35.Chang WL, Baumgarth N, Yu D, Barry PA. 2004. Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J Virol 78:8720–8731. doi: 10.1128/JVI.78.16.8720-8731.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spencer JV, Lockridge KM, Barry PA, Lin G, Tsang M, Penfold ME, Schall TJ. 2002. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J Virol 76:1285–1292. doi: 10.1128/JVI.76.3.1285-1292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avdic S, McSharry BP, Steain M, Poole E, Sinclair J, Abendroth A, Slobedman B. 2016. Human cytomegalovirus encoded cmvIL-10 amplifies its immunomodulatory potential by upregulating human IL-10 in monocytes. J Virol 90:3819–3827. doi: 10.1128/JVI.03066-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avdic S, Cao JZ, Cheung AK, Abendroth A, Slobedman B. 2011. Viral interleukin-10 expressed by human cytomegalovirus during the latent phase of infection modulates latently infected myeloid cell differentiation. J Virol 85:7465–7471. doi: 10.1128/JVI.00088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avdic S, McSharry BP, Slobedman B. 2014. Modulation of dendritic cell functions by viral IL-10 encoded by human cytomegalovirus. Front Microbiol 5:337. doi: 10.3389/fmicb.2014.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Avdic S, Cao JZ, McSharry BP, Clancy LE, Brown R, Steain M, Gottlieb DJ, Abendroth A, Slobedman B. 2013. Human cytomegalovirus interleukin-10 polarizes monocytes toward a deactivated M2c phenotype to repress host immune responses. J Virol 87:10273–10282. doi: 10.1128/JVI.00912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Doel C, Bashiruddin JB. 2015. Interleukin-10 production at the early stage of infection with foot-and-mouth disease virus related to the likelihood of persistent infection in cattle. Vet Res 46:132. doi: 10.1186/s13567-015-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kogut MH, Arsenault RJ. 2015. A role for the non-canonical Wnt-beta-catenin and TGF-beta signaling pathways in the induction of tolerance during the establishment of a Salmonella enterica serovar Enteritidis persistent cecal infection in chickens. Front Vet Sci 2:33. doi: 10.3389/fvets.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Razavi A, Bagheri N, Azadegan-Dehkordi F, Shirzad M, Rahimian G, Rafieian-Kopaei M, Shirzad H. 2015. Comparative immune response in children and adults with H. pylori infection. J Immunol Res 2015:315957. doi: 10.1155/2015/315957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niesen E, Schmidt J, Flecken T, Thimme R. 2015. Suppressive effect of interleukin 10 on priming of naive hepatitis C virus-specific CD8+ T cells. J Infect Dis 211:821–826. doi: 10.1093/infdis/jiu541. [DOI] [PubMed] [Google Scholar]
- 45.Jost NH, Abel S, Hutzler M, Sparwasser T, Zimmermann A, Roers A, Muller W, Klopfleisch R, Hengel H, Westendorf AM, Buer J, Hansen W. 2014. Regulatory T cells and T-cell-derived IL-10 interfere with effective anti-cytomegalovirus immune response. Immunol Cell Biol 92:860–871. doi: 10.1038/icb.2014.62. [DOI] [PubMed] [Google Scholar]
- 46.Yue F, Zhu YP, Zhang YF, Sun GP, Yang Y, Guo DG, Wang AG, Li BW, Yin M, Cheng AC, Wang MS, Wang XN. 2014. Up-regulated expression of PD-1 and its ligands during acute classical swine fever virus infection in swine. Res Vet Sci 97:251–256. doi: 10.1016/j.rvsc.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 47.Singh AK, Thirumalapura NR. 2014. Early induction of interleukin-10 limits antigen-specific CD4(+) T cell expansion, function, and secondary recall responses during persistent phagosomal infection. Infect Immun 82:4092–4103. doi: 10.1128/IAI.02101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mason GM, Jackson S, Okecha G, Poole E, Sissons JG, Sinclair J, Wills MR. 2013. Human cytomegalovirus latency-associated proteins elicit immune-suppressive IL-10 producing CD4(+) T cells. PLoS Pathog 9:e1003635. doi: 10.1371/journal.ppat.1003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poole E, Avdic S, Hodkinson J, Jackson S, Wills M, Slobedman B, Sinclair J. 2014. Latency-associated viral interleukin-10 (IL-10) encoded by human cytomegalovirus modulates cellular IL-10 and CCL8 secretion during latent infection through changes in the cellular microRNA hsa-miR-92a. J Virol 88:13947–13955. doi: 10.1128/JVI.02424-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poole E, Lau JC, Sinclair J. 2015. Latent infection of myeloid progenitors by human cytomegalovirus protects cells from FAS-mediated apoptosis through the cellular IL-10/PEA-15 pathway. J Gen Virol 96:2355–2359. doi: 10.1099/vir.0.000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poole E, Sinclair J. 2015. Sleepless latency of human cytomegalovirus. Med Microbiol Immunol 204:421–429. doi: 10.1007/s00430-015-0401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poole E, McGregor Dallas SR, Colston J, Joseph RS, Sinclair J. 2011. Virally induced changes in cellular microRNAs maintain latency of human cytomegalovirus in CD34(+) progenitors. J Gen Virol 92:1539–1549. doi: 10.1099/vir.0.031377-0. [DOI] [PubMed] [Google Scholar]
- 53.Cheung AK, Gottlieb DJ, Plachter B, Pepperl-Klindworth S, Avdic S, Cunningham AL, Abendroth A, Slobedman B. 2009. The role of the human cytomegalovirus UL111A gene in down-regulating CD4+ T-cell recognition of latently infected cells: implications for virus elimination during latency. Blood 114:4128–4137. doi: 10.1182/blood-2008-12-197111. [DOI] [PubMed] [Google Scholar]
- 54.Pepperl-Klindworth S, Besold K, Frankenberg N, Farkas M, Kuball J, Theobald M, Plachter B. 2006. Cytomegalovirus interleukin-10 expression in infected cells does not impair MHC class I restricted peptide presentation on bystanding antigen-presenting cells. Viral Immunol 19:92–101. doi: 10.1089/vim.2006.19.92. [DOI] [PubMed] [Google Scholar]
- 55.Huang A, Zhang B, Yan W, Wang B, Wei H, Zhang F, Wu L, Fan K, Guo Y. 2014. Myeloid-derived suppressor cells regulate immune response in patients with chronic hepatitis B virus infection through PD-1-induced IL-10. J Immunol 193:5461–5469. doi: 10.4049/jimmunol.1400849. [DOI] [PubMed] [Google Scholar]
- 56.Prata TT, Bonin CM, Ferreira AM, Padovani CT, Fernandes CE, Machado AP, Tozetti IA. 2015. Local immunosuppression induced by high viral load of human papillomavirus: characterization of cellular phenotypes producing interleukin-10 in cervical neoplastic lesions. Immunology 146:113–121. doi: 10.1111/imm.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chetty S, Porichis F, Govender P, Zupkosky J, Ghebremichael M, Pillay M, Walker BD, Ndung'u T, Kaufmann DE, Kasprowicz VO. 2014. Tuberculosis distorts the inhibitory impact of interleukin-10 in HIV infection. AIDS 28:2671–2676. doi: 10.1097/QAD.0000000000000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michelin ADEF, Perri SH, De Lima VM. 2011. Evaluation of TNF-alpha, IL-4, and IL-10 and parasite density in spleen and liver of L. (L) chagasi naturally infected dogs. Ann Trop Med Parasitol 105:373–383. doi: 10.1179/1364859411Y.0000000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blanquer J, Chilet M, Benet I, Aguilar G, Munoz-Cobo B, Tellez A, Costa E, Bravo D, Navarro D. 2011. Immunological insights into the pathogenesis of active CMV infection in non-immunosuppressed critically ill patients. J Med Virol 83:1966–1971. doi: 10.1002/jmv.22202. [DOI] [PubMed] [Google Scholar]
- 60.Das A, Ellis G, Pallant C, Lopes AR, Khanna P, Peppa D, Chen A, Blair P, Dusheiko G, Gill U, Kennedy PT, Brunetto M, Lampertico P, Mauri C, Maini MK. 2012. IL-10-producing regulatory B cells in the pathogenesis of chronic hepatitis B virus infection. J Immunol 189:3925–3935. doi: 10.4049/jimmunol.1103139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.do Nascimento PR, Martins DR, Monteiro GR, Queiroz PV, Freire-Neto FP, Queiroz JW, Morais Lima AL, Jeronimo SM. 2013. Association of pro-inflammatory cytokines and iron regulatory protein 2 (IRP2) with Leishmania burden in canine visceral leishmaniasis. PLoS One 8:e73873. doi: 10.1371/journal.pone.0073873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katara GK, Ansari NA, Verma S, Ramesh V, Salotra P. 2011. Foxp3 and IL-10 expression correlates with parasite burden in lesional tissues of post kala azar dermal leishmaniasis (PKDL) patients. PLoS Negl Trop Dis 5:e1171. doi: 10.1371/journal.pntd.0001171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu J, Zhan W, Kim CJ, Clayton K, Zhao H, Lee E, Cao JC, Ziegler B, Gregor A, Yue FY, Huibner S, MacParland S, Schwartz J, Song HH, Benko E, Gyenes G, Kovacs C, Kaul R, Ostrowski M. 2014. IL-10-producing B cells are induced early in HIV-1 infection and suppress HIV-1-specific T cell responses. PLoS One 9:e89236. doi: 10.1371/journal.pone.0089236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petrara MR, Cattelan AM, Zanchetta M, Sasset L, Freguja R, Gianesin K, Cecchetto MG, Carmona F, De Rossi A. 2012. Epstein-Barr virus load and immune activation in human immunodeficiency virus type 1-infected patients. J Clin Virol 53:195–200. doi: 10.1016/j.jcv.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 65.Ritter JT, Tang-Feldman YJ, Lochhead GR, Estrada M, Lochhead S, Yu C, Ashton-Sager A, Tuteja D, Leutenegger C, Pomeroy C. 2010. In vivo characterization of cytokine profiles and viral load during murine cytomegalovirus-induced acute myocarditis. Cardiovasc Pathol 19:83–93. doi: 10.1016/j.carpath.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Schaffer K, Moran J, Duffy M, McCormick AP, Hall WW, Hassan J. 2009. Kinetics of host immune responses and cytomegalovirus resistance in a liver transplant patient. Liver Transpl 15:1199–1203. doi: 10.1002/lt.21832. [DOI] [PubMed] [Google Scholar]
- 67.Siewe B, Stapleton JT, Martinson J, Keshavarzian A, Kazmi N, Demarais PM, French AL, Landay A. 2013. Regulatory B cell frequency correlates with markers of HIV disease progression and attenuates anti-HIV CD8(+) T cell function in vitro. J Leukoc Biol 93:811–818. doi: 10.1189/jlb.0912436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Silva BD, Trentini MM, da Costa AC, Kipnis A, Junqueira-Kipnis AP. 2014. Different phenotypes of CD8+ T cells associated with bacterial load in active tuberculosis. Immunol Lett 160:23–32. doi: 10.1016/j.imlet.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 69.Tsai TT, Chuang YJ, Lin YS, Wan SW, Chen CL, Lin CF. 2013. An emerging role for the anti-inflammatory cytokine interleukin-10 in dengue virus infection. J Biomed Sci 20:40. doi: 10.1186/1423-0127-20-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verma S, Kumar R, Katara GK, Singh LC, Negi NS, Ramesh V, Salotra P. 2010. Quantification of parasite load in clinical samples of leishmaniasis patients: IL-10 level correlates with parasite load in visceral leishmaniasis. PLoS One 5:e10107. doi: 10.1371/journal.pone.0010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Howard AD, Zwilling BS. 1998. Cytokine production by CD4 and CD8 T cells during the growth of Mycobacterium tuberculosis in mice. Clin Exp Immunol 113:443–449. doi: 10.1046/j.1365-2249.1998.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stager S, Maroof A, Zubairi S, Sanos SL, Kopf M, Kaye PM. 2006. Distinct roles for IL-6 and IL-12p40 in mediating protection against Leishmania donovani and the expansion of IL-10+ CD4+ T cells. Eur J Immunol 36:1764–1771. doi: 10.1002/eji.200635937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore KW, Vieira P, Fiorentino DF, Trounstine ML, Khan TA, Mosmann TR. 1990. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science 248:1230–1234. doi: 10.1126/science.2161559. [DOI] [PubMed] [Google Scholar]
- 74.Alves J, Madureira P, Baltazar MT, Barros L, Oliveira L, Dinis-Oliveira RJ, Andrade EB, Ribeiro A, Vieira LM, Trieu-Cuot P, Duarte JA, Carvalho F, Ferreira P. 2015. A safe and stable neonatal vaccine targeting GAPDH confers protection against group B Streptococcus infections in adult susceptible mice. PLoS One 10:e0144196. doi: 10.1371/journal.pone.0144196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hormozi K, Parton R, Coote J. 1999. Adjuvant and protective properties of native and recombinant Bordetella pertussis adenylate cyclase toxin preparations in mice. FEMS Immunol Med Microbiol 23:273–282. doi: 10.1111/j.1574-695X.1999.tb01248.x. [DOI] [PubMed] [Google Scholar]
- 76.Nagamatsu K, Kuwae A, Konaka T, Nagai S, Yoshida S, Eguchi M, Watanabe M, Mimuro H, Koyasu S, Abe A. 2009. Bordetella evades the host immune system by inducing IL-10 through a type III effector, BopN. J Exp Med 206:3073–3088. doi: 10.1084/jem.20090494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ejrnaes M, Filippi CM, Martinic MM, Ling EM, Togher LM, Crotty S, von Herrath MG. 2006. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med 203:2461–2472. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Richter K, Perriard G, Behrendt R, Schwendener RA, Sexl V, Dunn R, Kamanaka M, Flavell RA, Roers A, Oxenius A. 2013. Macrophage and T cell produced IL-10 promotes viral chronicity. PLoS Pathog 9:e1003735. doi: 10.1371/journal.ppat.1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richter K, Perriard G, Oxenius A. 2013. Reversal of chronic to resolved infection by IL-10 blockade is LCMV strain dependent. Eur J Immunol 43:649–654. doi: 10.1002/eji.201242887. [DOI] [PubMed] [Google Scholar]
- 80.Brooks DG, McGavern DB, Oldstone MB. 2006. Reprogramming of antiviral T cells prevents inactivation and restores T cell activity during persistent viral infection. J Clin Invest 116:1675–1685. doi: 10.1172/JCI26856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. 2006. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med 12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Campbell AE, Cavanaugh VJ, Slater JS. 2008. The salivary glands as a privileged site of cytomegalovirus immune evasion and persistence. Med Microbiol Immunol 197:205–213. doi: 10.1007/s00430-008-0077-2. [DOI] [PubMed] [Google Scholar]
- 83.Yue Y, Kaur A, Lilja A, Diamond DJ, Walter MR, Barry PA. 2016. The susceptibility of primary cultured rhesus Macaque kidney epithelial cells to rhesus cytomegalovirus strains. J Gen Virol 97:1426–1438. doi: 10.1099/jgv.0.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.