ABSTRACT
Like almost all of the positive-strand RNA viruses, hepatitis C virus (HCV) induces host intracellular membrane modification to form the membrane-bound viral replication complex (RC), within which viral replicases amplify the viral RNA genome. Despite accumulated information about how HCV co-opts host factors for viral replication, our knowledge of the molecular mechanisms by which viral proteins hijack host factors for replicase assembly has only begun to emerge. Purification of the viral replicase and identification of the replicase-associated host factors to dissect their roles in RC biogenesis will shed light on the molecular mechanisms of RC assembly. To purify the viral replicase in the context of genuine viral replication, we developed an HCV subgenomic replicon system in which two different affinity tags were simultaneously inserted in frame into HCV NS5A and NS5B. After solubilizing the replicon cells, we purified the viral replicase by two-step affinity purification and identified the associated host factors by mass spectrometry. We identified valosin-containing protein (VCP), a member of the ATPases associated with diverse cellular activities (AAA+ATPase) family, as an active viral replication modulator whose ATPase activity is required for viral replication. A transient replication assay indicated that VCP is involved mainly in viral genome amplification. VCP associated with viral replicase and colocalized with a viral RC marker. Further, in an HCV replicase formation surrogate system, abolishing VCP function resulted in aberrant distribution of HCV NS5A. We propose that HCV may co-opt a host AAA+ATPase for its replicase assembly.
IMPORTANCE Almost all of the positive-strand RNA viruses share a replication strategy in which viral proteins modify host membranes to form the membrane-associated viral replicase. Viruses hijack host factors to facilitate this energy-unfavorable process. Understanding of this fundamental process is hampered by the challenges of purifying the replicase because of the technical difficulties involved. In this study, we developed an HCV subgenomic replicon system in which two different affinity tags were simultaneously inserted in frame into two replicase components. Using this dual-affinity-tagged replicon system, we purified the viral replicase and identified valosin-containing protein (VCP) AAA+ATPase as a pivotal viral replicase-associated host factor that is required for viral genome replication. Abolishing VCP function resulted in aberrant viral protein distribution. We propose that HCV hijacks a host AAA+ATPase for its replicase assembly. Understanding the molecular mechanism of VCP regulates viral replicase assembly may lead to novel antiviral strategies targeting the most conserved viral replication step.
INTRODUCTION
The positive-strand RNA viruses are the largest class of viruses and include many medically and economically important pathogens, including hepatitis C virus (HCV); picornaviruses, which cause hand, foot, and mouth disease; and flaviviruses, such as the West Nile virus and the Zika virus. Positive-strand RNA viruses share a conserved replication mechanism in which viral proteins induce host membrane modification to assemble membrane-associated viral replication complexes (RCs) (1). Viruses hijack host factors to facilitate this energy-unfavorable process (2).
HCV, a member of the Flaviviridae family, chronically infects approximately 160 million people worldwide and causes hepatocellular carcinoma in a significant proportion of the chronically infected population (3). Its 9.6-kb positive-sense RNA genome encodes a single polyprotein that is cleaved into at least 10 individual polypeptides by the host and viral proteinase in the following protein order: 5′-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-3′ (reviewed in reference 4). The single open reading frame (ORF) is flanked by the highly conserved 5′ and 3′ untranslated regions (UTRs). The 5′ UTR contains an internal ribosome entry site (IRES) to initiate cap-independent translation. The 3′ UTR is required for RNA replication and is composed of three sequential elements: a nonconserved variable region (30 to 50 nucleotides), a poly(U/C) stretch (20 to 200 nucleotides), and a conserved 98-nucleotide sequence, termed the 3′X region, which contains three stem-loop structures (reviewed in reference 4). Upon polyprotein processing, HCV nonstructural proteins residing in the endoplasmic reticulum (ER) induce the formation of double-membrane vesicles (DMVs), which are protrusions from the ER membranes toward the cytosol (5). Nonstructural proteins NS3, NS4A, NS4B, NS5A, and NS5B constitute the viral replicase. NS3 is a bifunctional enzyme with protease and helicase activities, with NS4A as a protease cofactor. NS4B is a multispanning integral membrane protein. NS5A is a multifunctional viral protein with no enzymatic activity. NS5B is an RNA-dependent RNA polymerase (reviewed in reference 6).
Overexpression of the HCV polyprotein encompassing NS3 to NS5B can induce DMVs, as observed in virus-infected cells. Overexpression of NS5A alone, although it is less efficient, can also induce DMVs, which argues for the central role of NS5A in RC assembly (5). HCV NS5A recruits host factors and, combined with other viral proteins, locally modifies the membrane lipid composition to facilitate RC assembly (7). Despite great advances in the studies of HCV RC biogenesis, insights into the underlying molecular mechanism of how viral proteins hijack host factors for replicase assembly have only begun to emerge.
Identification of replicase-associated host factors and dissection of their roles in replicase assembly will shed light on the molecular mechanisms of RC biogenesis. Purification of the viral replicase during authentic viral replication is hampered by the membrane association of the replicase, the low expression levels of the replicase components, and the complexity of the replicase modules. The replicase-enriched intracellular membrane fractions and the associated host factors have been studied by selective fractionation of the RC-associated detergent-resistant membranes (8). Anti-NS4B antibody (9) and tagged-NS4B replicons (10) were used to affinity capture the replicase-containing membrane fractions. Chung et al. reported purification of the tagged-NS5A-interacting proteins during viral infection (11). There is no report regarding purification of the solubilized HCV replicase as a protein complex.
In this study, we developed an HCV subgenomic replicon system in which two different affinity tags were simultaneously inserted in frame into HCV NS5A and NS5B without obviously attenuating viral replication, which allows us to purify the NS5A- and NS5B-containing replicase by sequential affinity purification. Affinity purification of the viral replicase and identification of the associated host factors uncovered valosin-containing protein (VCP), a member of the ATPases associated with diverse cellular activities (AAA+ATPase) family, as a pivotal host factor required for viral replication.
MATERIALS AND METHODS
Plasmids.
The plasmid encoding HCV Jc1FLAG2 (p7-nsGluc2A, named Jc1G here) was reported previously (12). To generate Jc1G with the duplicated NS5B stem-loop SL3, first a PacI site was introduced into the Jc1G downstream stop codon of NS5B and then a PCR-amplified NS5B SL3 fragment flanked by PacI recognition sites was ligated into the PacI site in Jc1G to get Jc1G-DU. To generate Jc1G-NS5B.HA, a nucleotide sequence encoding a hemagglutinin (HA) peptide (TAC CCA TAC GAT GTT CCA GAT TAC GCT) was inserted into the NS5B region by fusion PCR. HCV subgenomic replicon construct sgJFH1 was generated by replacing the fragment of Jc1G that encompasses the sequence extending from the 5′ UTR to the structural protein region with a fragment from the BB7 subgenomic replicon encompassing the 5′ UTR-BSD-EMCV IRES region (13). To generate sgJFH1-DU-NS5B.HA1, sgJFH1-DU-NS5B.HA2, and sgJFH1-DU-NS5B.His constructs, first an RsrII/XbaI fragment was transferred from Jc1G-DU into sgJFH1 to get the sgJFH1-DU construct and then the nucleotides encoding the HA peptide, the HA peptide with flanking amino acids (GAC GCT TAC CCA TAC GAT GTT CCA GAT TAC GCT AGC CTA), and the His6 (CAT CAT CAT CAT CAT CAC) peptide were inserted into the NS5B region by fusion PCR to get sgJFH1-DU-NS5B.HA1, sgJFH1-DU-NS5B.HA2, and sgJFH1-DU-NS5B.His, respectively. To generate the sgJFH1-NS5A.HA construct, the nucleotide encoding the HA peptide (TAC CCA TAC GAT GTT CCA GAT TAC GCT) was inserted into the NS5A region of sgJFH1 by fusion PCR as previously reported (14). The sgJFH1-NS4B.HA construct was generated essentially as previously reported, with the additional adaptive mutations (10). To generate the sgJFH1-NS5A.HA.NS5B.His construct, a BsrG1/SanD1 fragment was transferred from the sgJFH1-NS5A.HA construct into the sgJFH1-DU-NS5B.His construct. Plasmid sgJFH1-NS5A.ypet was generated by transferring an SpeI/RsrII fragment from the Jc1G-378-1 construct that includes an in-frame fusion of NS5A.Ypet (Charles Rice, unpublished data) into the sgJFH1 plasmid. To generate the phCMV-3-5B.ypet construct, first plasmid phCMV-3-5B was generated by inserting the PCR-amplified HCV NS3-5B region from Jc1G into the BglII/EcoRI sites in the phCMV vector (Genlantis) and then a SanDI/HindIII fragment was transferred from the Jc1-378-1 plasmid into phCMV-3-5B.
The lentivirus-based short hairpin RNA (shRNA) plasmids were generated with a pLKO.1 (Sigma) backbone. The oligonucleotides targeting the VCP coding region (GCA TTA CTG GTA ATC TCT TCG) and an irrelevant target (IRR) (TCT CGC TTG GGC GAG AGT AAG) were synthesized and ligated into pLKO.1.
For CRISPR-mediated knockdown (Kd), the oligonucleotides targeting VCP sg1 (GAT GAA TTG CAG TTG TTC CG), sg2 (GAC ACA GTG ATC CAC TGC GA), sg3 (AAA CTC ACC TCT CGT TTG AT), and sg4 (GCG TAT CGA CCC ATC CGG AA) and a scrambled oligonucleotide (IRR) (ATA GCG ACT AAA CAC ATC AA) were synthesized and ligated into LentiCRISPR-V2 (Addgene plasmid no. 52961).
To generate the retroviral vectors Lenti-RFP and Lenti-VCP, PCR amplicons were digested with XbaI/BamH1 and ligated into the Lenticas9-BLAST plasmid (Addgene plasmid no. 52962) to replace the Cas9 ORF. The VCP gene fragment was amplified from Huh7 cDNA, which was generated by reverse transcription by superscript III (Invitrogen). The red fluorescent protein (RFP) fragment was amplified from plasmid pTRIP-RFP (15). To generate Lenti-RFP-VCP, in which RFP is inserted in frame upstream of the VCP coding region, the amplified RFP-encoding gene was digested with BamHI and ligated into similarly digested Lenti-VCP. The retroviral vector pTRIP, which was reported previously (15), was modified to contain a blasticidin resistance gene (BSD) and a multicloning site and renamed pTRIP-BSD. PCR-amplified green fluorescent protein (GFP) and VCP-es, which contains shRNA-resistant mutations (GG ATA ACA GGA AAC CTG TTT G), were digested with XbaI and BamHI and ligated into similarly digested pTRIP-BSD to get plasmids pTRIP-BSD-GFP and pTRIP-BSD-VCP-es. The dominant negative mutant construct pTRIP-VCP-es-DN (E305Q/E578Q) was generated by site-directed mutagenesis. All of the constructs were proofed by DNA sequencing. Detailed information is available upon request.
Cell lines.
Human hepatoma cell lines Huh7 (Cell Bank of the Chinese Academy of Sciences, Shanghai, China, http://grbio.org/institution/cell-bank-type-culture-collection-chinese-academy-sciences) and Huh7.5 (kindly provided by Charles Rice) were routinely maintained in Dulbecco modified medium supplemented with 10% fetal bovine serum (Biological Industries catalog no. 04-001-1), 25 mM HEPES (Gibco), and nonessential amino acids (Gibco). Replicon cells were generated by electroporation of in vitro-transcribed RNAs into Huh7.5 cells and selected in conditioned medium supplemented with 5 μg/ml blasticidin (Invitrogen). The resulted replicon cells were pooled and maintained with the conditioned medium supplemented with 0.5 μg/ml blasticidin. Huh7.5 cells harboring pTRIP-BSD-GFP, pTRIP-BSD-VCP-es, and pTRIP-BSD-VCP-es-DN were generated by transducing Huh7.5 cells with vesicular stomatitis virus G protein (VSV-G)-pseudotyped lentiviral particles and growing cells in conditioned medium supplemented with 5 μg/ml blasticidin. The surviving cells were pooled and maintained in conditioned medium with 5 μg/ml blasticidin. For HCVcc infection, these three stable cell lines with exactly the same number of passages were used to avoid the effect of cell passage variation on HCV replication (16).
Antibodies and chemicals.
Anti-β-actin antibody from Sigma (A1978) was used in Western blotting analyses at a 1:5,000 dilution. Anti-c-Myc antibody from Santa Cruz Biotechnology (sc-40) was utilized at a 1:1,000 dilution for Western blotting. Anti-NS5A antibody (9E10) was used at a 1:2,000 dilution for Western blotting analyses. Anti-GFP antibody (sc-9996; Santa Cruz) was used for Western blotting at a 1:2,000 dilution. Rabbit anti-VCP antibody (ABclonal; A2795) was used at a 1:2,000 dilution for Western blotting. A mouse monoclonal anti-VCP antibody (Abcam; ab11433) was used at a 1:50 dilution for immunostaining. An anti-His antibody (Abcam; ab18184) was used at a 1:2,000 dilution for Western blotting. An anti-HA antibody (Abcam; ab130275) was used at a 1:1,000 dilution for Western blotting. An anti-HCV NS3 antibody (Virogen; 217-A) was used at a 1:1,000 dilution for Western blotting. Horseradish peroxidase (HRP)-conjugated anti-mouse (Santa Cruz; sc-2005) and anti-rabbit IgG secondary antibodies (Santa Cruz; sc-2004) were used at a 1:2,000 dilution for Western blotting. Cy3-conjugated anti-mouse IgG (Jackson, 115-165-062) was used at a 1:200 dilution for immunostaining. EerI was purchased from Tocris Bioscience (catalog no. 3922) and dissolved in dimethyl sulfoxide to make a 10 mM stock.
Virus.
HCVcc was generated as described previously, by electroporation of in vitro-transcribed Jc1G RNA into Huh7.5 cells, and the virus titer was determined in Huh7.5 cells by limiting dilution (17). VSV-G-pseudotyped lentiviral particles were generated by cotransfection of HEK293T cells with plasmids encoding VSV-G and HIV gag-pol and with the lentiviral provirus plasmids. The medium overlying the cells was harvested at 48 to 72 h after transfection, filtered through a 0.45-μm filter, and stored at −80°C. Cells were transduced with the pseudoparticles in the presence of 8 μg/ml Polybrene. The pseudoparticle dosage was determined by limiting dilution to produce about 60% transduced-cell survival in conditioned medium supplemented with 5 μg/ml puromycin (for pLKO.1 and LentiCRISPR_v2) or 5 μg/ml blasticidin (for pTRIP-BSD).
Affinity purification of protein complex.
For small-scale experiments, HCV replicon cells harboring sgJFH1 and sgJFH1-NS5A.HA.NS5B.His were seeded onto a 10-cm tissue culture dish. When subconfluent, the cells were scraped into Dulbecco's phosphate-buffered saline (PBS) (DPBS; Biological Industries, catalog no. 02-023-1) and centrifuged at 500 × g for 5 min. Cell pellets were resuspended in 500 μl of ice-cold lysis buffer (50 mM Tris Cl [pH 7.5], 1 mM EDTA, 15 mM MgCl2, 10 mM KCl, 1% Triton X-100, proteinase inhibitor cocktail [Roche]). After disruption by being passed through a 27-gauge needle 20 times and clarification by centrifugation at 15,000 × g for 10 min at 4°C, the soluble fraction was incubated with 30 μl of anti-HA antibody-coated beads (Sigma, A 2095) for 4 h at 4°C. The beads were collected by centrifugation and then washed four times with 1.2 ml of washing buffer (50 mM Tris Cl [pH 7.5], 1 mM EDTA, 15 mM MgCl2, 10 mM KCl, 1% Triton X-100, 20 mM imidazole). After being washed, the bound proteins were eluted with 50 μl of elution buffer (50 mM Tris Cl [pH 7.5], 1 mM EDTA, 15 mM MgCl2, 10 mM KCl, 1% Triton X-100, 250 μg/ml HA peptide [Sigma, I2149]) by incubation in a Thermomixer R (Eppendorf) at room temperature for 15 min (1,000 rpm). After centrifugation at 3,000 × g for 5 min, the supernatants were collected and diluted to 300 μl by lysis buffer supplemented with 20 mM imidazole. A 20-μl volume of Ni-Sepharose (GE Healthcare, 17-5268-01) was then added to the supernatants. After incubation at 4°C for 1 h and clarification, the Sepharose was washed four times with 600 μl of washing buffer containing 20 mM imidazole. The captured proteins were eluted in 30 μl of washing buffer containing 240 mM imidazole and mixed with 30 μl of 2×SDS loading buffer (100 mM Tris Cl [pH 6.8], 4% [wt/vol] SDS, 0.2% [wt/vol] bromophenol blue, 20% [vol/vol] glycerol, 10% 2-mercaptoethanol). After being boiled for 5 min, the proteins samples were separated by SDS-PAGE and transferred to a nitrocellulose membrane for immunoblotting.
For large-scale experiments, the cells from 10 150-mm tissue culture dishes were harvested and pooled. After centrifugation, the cells were resuspended in 3 ml of lysis buffer and broken by 20 strokes in a Dounce tissue grinder. After clarification, the soluble fraction was incubated by head-to-tail rotation with 200 μl of anti-HA antibody-coated beads (Sigma, A2095) for 4 h at 4°C. The beads were collected by centrifugation and then washed four times with 8 ml of washing buffer. The proteins were eluted in 300 μl of washing buffer supplemented with HA peptide (250 μg/ml), followed by a second elution with 100 μl of washing buffer supplemented with HA peptide (250 μg/ml). The eluted solutions were combined and diluted into 1.2 ml of lysis buffer containing 20 mM imidazole, and 40 μl of Ni Sepharose was added. After incubation at 4°C for 1 h, the beads were washed four times with 1.2 ml of washing buffer. The captured proteins were eluted in 40 μl of washing buffer containing 240 mM imidazole and mixed with 10 μl of 5× SDS loading buffer (250 mM Tris Cl [pH 6.8], 30% glycerol, 10% SDS, 0.02% bromophenol blue). After the addition of 2-mercaptoethanol to a final concentration of 5%, the eluted solution was boiled for 5 min. Proteins were resolved with NuPAGE Novex Bis-Tris Mini Gels (Invitrogen), and protein bands were visualized by silver staining.
Quantitative reverse transcription (RT)-PCR.
RNAs were purified by TRIzol extraction and reverse transcribed with the PrimeScript RT reagent kit with gDNA Eraser (Perfect Real Time) from TaKaRa according to the manufacturer's instructions. The cDNA samples were subjected to real-time PCR (SYBR Premix Ex Taq Tli RNase H Plus) with the following primers for specific genes: GAPDH, (s [sense]) GGT ATC GTG GAA GGA CTC ATG A and (as [antisense]) ATG CCA GTG GCT TCC CGT TCA GC; HCV, (s) CCC TGT GAG GAA CTA CTG TCT TCA CGC and (as) GCT CAT GGT GCA CGG TCTA CGA GAC CT; ANKFY1, (s) CAG AGA AGC GCTG CGC TC and (as) CTC CAG CTG TCA CTG CGG; SHROOM3, (s) GGC GCA GTG AGC CTG C and (as) TGG TCA GGG CTC CGC C; SEC16A, (s) GAC ACA GCC CAG AGC ACA and (as) CTG CAG ACC AGG CTG AGG; VCP, (s) CCC AGC CCA AGA TGG ATG and (as) ATC CCC TAG GCG TAC ACG; SEC24C, (s) CCC CGC CAT TCC CTA TGG and (as) CTG AGA CCC AGG CAG CC; MOV10, (s) GAC AGA TCG CGA GCG GC and (as) CTC GACC GCT GGG CCG A; UPF1, (s) GCC AAG ACC AGC CAG TTG and (as) GCA GGG TCA CCT CTT TGC. The relative RNA levels were calculated by the 2−ΔΔCT method (18). The gene for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene for loading normalization.
HCV RNA transfection.
Before transfection, 1.25 × 105 Huh7.5 cells were seeded into a 24-well plate. When the cells had grown to nearly 80 to 90% confluence, 0.5 μg of RNA per well was transfected with the TransIT-mRNA Transfection kit (Mirus) at a ratio of 0.5:1:1 (RNA to boost to transfection reagent) according to the manufacturer's protocol.
Luciferase activity.
Supernatants were taken from each well and mixed with an equal volume of 2× passive lysis buffer (Promega). Luciferase activity was measured with Renilla luciferase substrate (Promega) according to the manufacturer's protocol, or cultured cells, after being washed with DPBS, were lysed with 1× passive lysis buffer and luciferase activity was measured with Renilla luciferase substrate.
Western blotting.
Cells were directly lysed with 2× SDS loading buffer (100 mM Tris-Cl [pH 6.8], 4% [wt/vol] SDS, 0.2% [wt/vol] bromophenol blue, 20% [vol/vol] glycerol, 10% 2-mercaptoethanol) and boiled for 5 min. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (GE Healthcare Life Sciences, catalog no.10401396). The membrane was incubated in blocking buffer (PBS, 0.05% Tween 20, 5% dried milk) and then incubated with primary antibody diluted in blocking buffer. The membrane was washed three times in PBS supplemented with 0.05% Tween 20 and incubated with an HRP-conjugated secondary antibody. After three washes, the membrane was visualized with Western Lightning Plus-ECL substrate (PerkinElmer, catalog no. NEL10500). Blots were reprobed with another antibody after being stripped with restore stripping buffer (Thermo, 21059), if necessary. Protein bands were quantified by densitometry with ImageJ, if necessary.
Gene Kd and HCV replication assay.
For small interfering RNA (siRNA) screening, Huh7.5 cells were seeded into 24-well plates at a density of 1 × 105 cells/ml. After overnight growth, the cells were transfected with Silencer siRNAs targeting ANKFY1 (Ambion, s28200), SHROOM3 (Ambion, s33455), SEC16A (Ambion, s19236), VCP (Ambion, s14765), SEC24C (Ambion, s18515), MOV10 (Ambion, s8922), and UPF1 (Ambion, s11927) at a final concentration of 30 nM by RNAiMAX (Invitrogen) according to the manufacturer's protocol. At 24 h posttransfection, the cells were retransfected essentially as described above. After another 24 h, cell viability was determined with Cell Counting Kit-8 (Dojinodo Laboratories, CK04) according to the manufacturer's protocol. The cells were then washed once with DPBS and infected with Jc1G at a multiplicity of infection (MOI) of 0.1. At 2 days postinfection, the supernatants were collected and mixed with an equal volume of 2× passive lysis buffer (Promega) and luciferase activity was measured with Renilla luciferase substrate (Promega, E2820) according to the manufacturer's protocol. Cells from triplicate wells were washed with DPBS and lysed in TRIzol (Ambion) for RNA analysis.
For shRNA-mediated gene Kd, Huh7.5 or Huh7.5 cells harboring pTRIP-BSD were seeded into six-well plates at a density of 1 × 105/ml. After overnight growth, the cells were transduced with pLKo.1 pseudoparticles. When near confluence, the cells were split 1:3 with conditioned medium supplemented with 5 μg/ml puromycin. Two days later, cells were infected with Jc1G or transfected with Jc1G RNA. The supernatants were collected, and the cells were harvested at the time points indicated.
For CRISPR single guide RNA (sgRNA)-mediated gene Kd, similarly, Huh7.5 cells were first seeded into six-well plates and transduced with LentiCRISPR_v2 pseudoparticles. When near confluence, the cells were split 1:3 with conditioned medium supplemented with 5 μg/ml puromycin. Two days later, the cells were infected with Jc1G, the supernatants were collected, and the cells were harvested at the time points indicated.
Immunofluorescence staining and confocal microscopy.
Cells were fixed with 4% formaldehyde in PBS at room temperature for 15 min. After being washed with DPBS, the coverslips were immunostained as described previously (15) or directly mounted with Mowiol mounting medium (0.1 M Tris HCl [pH 8.5], 25% glycerol, 10% Mowiol 4-88 [Calbiochem 475904]) and observed with a Leica TCS SP8 confocal laser microscope with a 63× numerical aperture 1.3 oil immersion objective. Images were captured with the LAS software and processed with Image J. For colocalization assessment, Pearson's correlation coefficients were quantified for the selected regions with the ImageJ plugin as previously described (19).
Statistical analysis.
Statistical analysis was performed with the GraphPad Prism 6 software. Specific tests are described in the figure legends.
Proteomic studies. (i) Reagents for proteomic studies.
Ammonium bicarbonate, dithiothreitol, iodoacetamide, and SDS were obtained from Sigma-Aldrich (St. Louis, MO). Acetonitrile (ACN, 99.9%), trifluoroacetic acid (TFA, 99.8%), and formic acid (FA) were purchased from Merck (Darmstadt, Germany). Sequencing grade modified trypsin was from Promega (Madison, WI, USA). The deionized water used for all experiments was obtained from a Milli-Q system (Millipore, Bedford, MA, USA). All other chemicals and reagents were of analytical grade and were obtained from the Shanghai Chemical Reagent Company.
(ii) Gel staining and protein in-gel digestion.
The gels were subjected to silver staining as described previously (20) and then scanned with a GS-800 calibrated densitometer (Bio-Rad). Overall, 10 bands were selectively sliced from the silver-stained gels as indicated in the figures. Each band was then cut into slices, and in-gel digestion was performed as previously described (21). Gel pieces were washed twice with a buffer including 15 mM ferricyanide potassium and 50 mM sodium hyposulfite and then washed with water until the gel pieces were destained completely. The gel pieces were reduced with dithiothreitol and alkylated with iodoacetamide. Afterwards, gel pieces were dehydrated with 100% ACN and dried at 37°C for 10 min. The dried gel pieces were subjected to trypsin digestion by the addition of a solution containing 25 mM ammonium bicarbonate and 12.5 ng/μl trypsin and then incubated at 37°C overnight for digestion. After digestion, 0.1% TFA in 50% ACN was added to extract peptides from gels. The extracted peptides were dried completely in a SpeedVac.
(iii) LC-MS analysis.
Samples were resuspended with buffer A (5% ACN containing 0.1% FA) and separated on a reverse-phase analytical column (Eksigent, C18, 3 μm, 150 mm by 75 μm) by nano-high-performance liquid chromatography (Eksigent Technologies). Peptides were subsequently eluted under the following gradient conditions with phase B (98% ACN with 0.1% FA) from 5 to 45% B (5 to 100 min), and the total flow rate was maintained at 300 nl/min. An electrospray voltage of 2.5 kV versus the inlet of the mass spectrometer was used. The TripleTOF 4600 mass spectrometer used was operated in information-dependent data acquisition mode to switch automatically between the mass spectrometry (MS) and tandem MS (MS/MS) acquisition modes. MS spectra were acquired across a mass range of 350 to 1,250 m/z with a 250-ms accumulation time per spectrum. MS/MS spectra were scanned from 100 to 1,250 m/z in high-sensitivity mode with rolling collision energy. The 25 most intense precursors were selected for fragmentation per cycle with a dynamic exclusion time of 25 s.
(iv) Database search and data exploration.
MS database searching was performed as previously described, with some modifications (22). Peak lists were first generated from raw files with ProteoWizard v3.0 at default settings. Searching of a custom database that contains 20,264 human protein entries and 130,224 HCV protein entries was performed with Mascot v2.3.2 (Matrix Science, United Kingdom). These protein sequences were extracted from the UniProtKB/Swiss-Prot database (release 2014_03) by using the species names as keyword constraints with an in-house perl script. The search parameters were set as follows. The digestion enzyme selected was trypsin (fully enzymatic). A maximum of two missed cleavages was allowed. The mass value was set as monoisotopic, and peptide charge states of 2+ and 3+ were taken into account. Carbamidomethyl (C) was configured as a fixed modification, and oxidation (M) was configured as a variable modification. Mass tolerances for precursor and product ions were 25 ppm and 0.2 Da, respectively. The searching results from Mascot were parsed into Scaffold QS (Proteome Software, v4.3.4). All of the search results were combined automatically by Scaffold, and data evaluation was performed. The Scaffold local false-discovery rate algorithm assigned probabilities to Mascot identifications. Proteins were grouped by the legacy-independent sample protein grouping method. Protein identifications with a threshold of >99.0% probability assigned by the ProteinProphet algorithm (23), at least one unique peptide, and a peptide threshold of >95% probability assigned by the PeptideProphet algorithm (24, 25) were accepted.
RESULTS
Tagging of HCV nonstructural proteins in the context of viral replication.
To uncover the host factors that are intimately associated with HCV replication, we attempted to purify the viral replicase formed during authentic viral replication and identify the associated host factors. We aimed to affinity tag two replicase components with different tags to facilitate tandem affinity purification. Two viral replicase components, NS5A and NS4B, were successfully tagged. Tagging of NS5A does not affect viral replication, whereas tagging of NS4B attenuates viral replication (10, 14). In addition to NS5A, we aimed to tag a replicase component other than NS4B without obviously attenuating viral replication. We selected NS5B because of its constitutive role in replicase assembly rather than for its enzymatic activity (26, 27). It is tolerant to replacement with exogenous sequences and to insertion mutations (28, 29). We utilized the HCV replicon system where the self-replicating bicistronic viral RNA contains an antibiotic resistance gene and encodes the viral polypeptide encompassing NS3 to NS5B (30). In the first attempt with the BB7 subgenomic replicon (genotype con1b) (13), we selected a tolerant site for 1D4 peptide insertion with attenuated viral replication (data not shown).
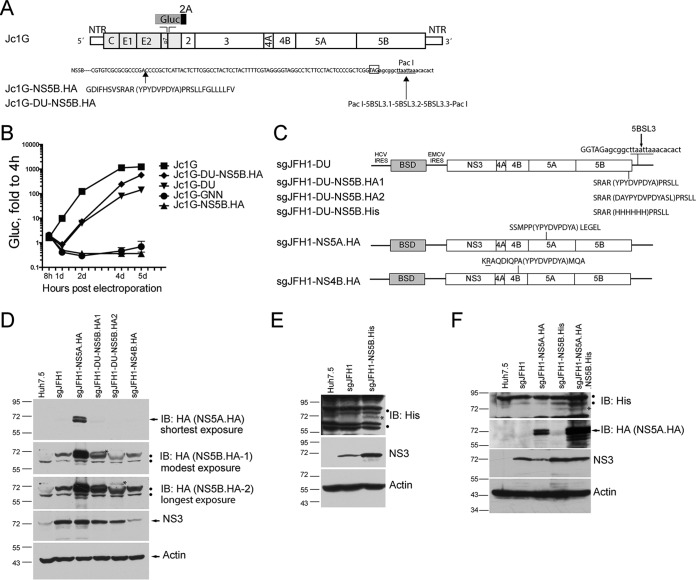
We next sought to modify this tolerant site to increase viral replication. First, the HA-encoding sequence was inserted at the equivalent site in the backbone of the Jc1G genotype 2a HCV infectious clone containing a secreted Gaussia luciferase reporter (12) (Fig. 1A). The recombinant infectious clone, named Jc1G-NS5B.HA, could not replicate (Fig. 1B), possibly because the insertion may disrupt the RNA kissing loop between 5BSL3 and 3′SL2, which is essential for RNA replication (31, 32). Introduction of a duplicated NS5B stem-loop into an artificially introduced PacI site downstream of the stop codon of NS5B to restore the kissing loop (Fig. 1A) resulted in efficient replication of Jc1G-DU-NS5B.HA (Fig. 1B). It was noted that insertion of the duplicated region (Jc1G-DU) attenuated viral replication compared with that of the wild-type virus (Fig. 1B). Further insertion of HA into the NS5B region in Jc1G-DU enhanced viral replication (Fig. 1B). Although attenuated, Jc1G-DU-NS5B.HA replicated to a level similar to that of wild-type Jc1G at late time points (5 days posttransfection) (Fig. 1B). We then generated a subgenomic replicon construct of Jc1G-DU-NS5B.HA and subgenomic replicon constructs with HA insertion in NS5A (14) and NS4B (10) as reported, respectively. The resulting replicon, sgJFH1-DU-NS5B.HA1 (Fig. 1C), along with sgJFH1-NS5A.HA, replicated similarly to the nontagged sgJFH1 replicon, whereas sgJFH1-NS4B.HA (Fig. 1C), as reported, replicated to a lesser extent, as judged by the NS3 protein levels (Fig. 1D). The addition of amino acids to the ends of the HA peptide for epitope accessibility optimization (sgJFH1-DU-NS5B.HA2) also attenuated viral replication, as evidenced by the lower NS3 protein levels (Fig. 1D). There was a lower level of NS5B-HA than NS5A-HA, as judged by anti-HA antibody blotting (Fig. 1D), which is most likely due to the lower intrinsic stability of NS5B protein (33). We could not detect NS4B-HA in sgJFH1-NS4B.HA replicon cells (data not shown), most likely because of attenuated viral replication (10). Thus, we used sgJFH1-DU-NS5B-HA1 in the subsequent experiments. We then replaced the HA peptide in NS5B in this construct with a His6 peptide (Fig. 1C), and the resulting sgJFH1-NS5B.His replicon also replicated similarly to sgJFH1 (Fig. 1E). Insertion of the His peptide enhanced viral replication, as higher NS3 levels were detected (Fig. 1E), although this enhancement disappeared after passaging (data not shown). Finally, we introduced HA and His peptides into NS5A and NS5B simultaneously to obtain double-affinity-tagged replicon sgJFH1-NS5A.HA.NS5B.His. The expression of NS5A-HA and NS5B-His was confirmed via Western blotting with the anti-HA and anti-His antibodies, respectively (Fig. 1F).
FIG 1.
Tagging of HCV nonstructural proteins in the context of viral replication. (A) Schematic of HCV Jc1G constructs. Duplicated 5BSL3 is inserted into the artificially introduced PacI site in Jc1G-DU. The stop codon of NS5B (boxed) and the artificially introduced PacI site (underlined) are shown. The HA peptide sequence (bracketed) is inserted into the NS5B protein sequence. Gluc, Gaussia luciferase; NTR, UTR; 2A (black bar), the foot-and-mouth disease virus 2A autoproteolytic peptide. (B) Huh7.5 cells were transfected with in vitro-transcribed RNAs. Luciferase activity in supernatants was measured at various times and normalized to the value obtained at 4 h. Mean values ± standard deviations are shown (n = 3). Gluc, relative light units. Similar results were obtained in multiple independent experiments. (C) Schematic of HCV sgJFH1 constructs. Peptide sequences inserted into viral nonstructural proteins are shown in parentheses. (D) Western blotting (immunoblotting [IB])of pooled replicon cells with the antibodies indicated. Arrows indicate the specific protein bands. Dots indicate nonspecific bands. The asterisks indicate specific NS5B-HA bands. (E) Western blotting of sgJFH1-NS5B.His replicon cells. Dots indicate nonspecific bands. The asterisk indicates a specific NS5B-His band. (F) Western blotting of sgJFH1-NS5A.HA.NS5B.His replicon cells. The arrow indicates specific NS5A-HA bands. Dots indicate nonspecific bands. The asterisk indicates a specific NS5B-His band. The migration of size standards (in kilodaltons) in SDS-PAGE is indicated to the left of the gels in panels D to F.
Affinity purification of the HCV replicase.
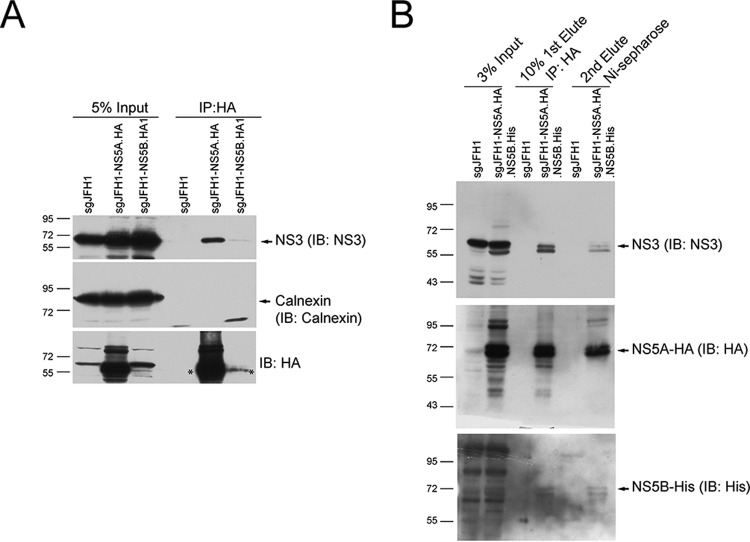
We optimized the conditions for solubilizing the replicase from cells and meanwhile maintaining the interactions between the replicase components. After optimization, we tried to affinity capture NS5A-HA and NS5B-HA in sgJFH1-NS5A.HA and sgJFH1-NS5B.HA1 replicon cells and examined if another replicase component, NS3, could be captured. In both cell types, we detected NS3 but not the ER membrane protein calnexin in the captured protein complexes (Fig. 2A), indicating that the interactions between the viral replicase components were maintained and not due to a membrane-dependent nonspecific association. Detection of only a weak NS3 protein band in anti-NS5B-HA captured protein complexes was probably due to the low level of NS5B-HA protein (Fig. 2A and 1D). We then used sgJFH1-NS5A.HA.NS5B.His replicon cells and performed two-step affinity purification. Solubilized cell lysates were sequentially purified by using the HA and His tags (Fig. 3A). At each step, NS3 could be detected in the captured complexes (Fig. 2B). In addition, after two-step affinity purification, NS3, NS5A, and NS5B were detected in the captured complexes (Fig. 2B). Thus, by affinity tagging HCV NS5A and NS5B in a replicon system, we could purify the solubilized viral replicase that contains at least NS3, NS5A, and NS5B.
FIG 2.
Affinity purification of the solubilized HCV nonstructural protein complex. (A) Solubilized sgJFH1-NS5A.HA and sgJFH1-NS5B.HA1 cell lysates were incubated with anti-HA antibody-coated beads. The captured proteins were analyzed by Western blotting (immunoblotting [IB]) with the antibodies indicated. The cell lysate from sgJFH1 was used as a negative control. The arrows indicate specific protein bands. The asterisks indicate HA-fused NS5A and NS5B proteins. The NS5B-HA bands are likely not visible in the Input lanes because of low protein levels. Representative data from multiple experiments with similar results are shown. IP, immunoprecipitation. (B) Two-step affinity purification of the HCV nonstructural protein complex. Lysates from cells expressing the replicons indicated were subjected to two-step affinity purification. The proteins eluted at each step were collected and analyzed by Western blotting with the antibodies indicated. The arrows indicate specific protein bands. Migration of size standards (in kilodaltons) is indicated to the left of the gels in both panels. Representative data from multiple experiments with similar results are shown.
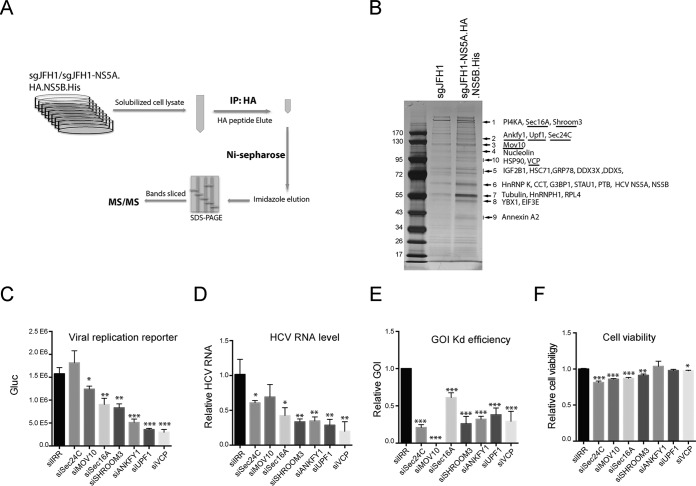
FIG 3.
Identification of host factors associated with the HCV protein complex. (A) Schematic of two-step affinity purification of the HCV protein complex and identification of associated host proteins. IP, immunoprecipitation. (B) After two-step affinity purification, the eluted proteins were resolved by SDS-PAGE. Specifically, enriched protein bands (arrows) in the sgJFH1-NS5A.HA.NS5B.His sample were identified by MS. Previously reported HCV-interacting host proteins and identified viral proteins in the bands indicated are shown. Host factors selected for further study are underlined. (C to F) siRNA screening of selected host factors. Huh7.5 cells were transfected twice with siRNAs against host factors at a final concentration of 30 nM and then infected with Jc1G at an MOI of 0.1 for 2 days. The values to the left are molecular sizes in kilodaltons. (C) Gaussia luciferase (Gluc) activity in the supernatants from infected cells are shown. Mean values ± standard deviations are shown (n = 3). (D) HCV RNA levels in the infected cells were quantified by quantitative RT-PCR and normalized against GAPDH RNA levels. HCV RNA levels relative to siIRR (irrelevant target) are shown. Mean values ± standard deviations are shown (n = 3). (E) RNA levels of specific host factors were quantified and normalized against GAPDH RNA levels. RNA levels relative to siIRR are shown. Mean values ± standard deviations are shown (n = 3). (F) Before HCV infection, cell viability was measured and normalized against siIRR. Mean values ± standard deviations are shown (n = 3). Statistical analysis was performed by comparing the siGOI and siIRR groups (*, P < 0.05; **, P < 0.01; ***, P < 0.001, two-tailed, unpaired t test).
Identification of host factors associated with the HCV replicase.
We then scaled up the affinity purification experiment by using untagged sgJFH1 as a negative control. After two-step affinity purification, the eluted protein complexes were resolved by SDS-PAGE and the protein bands were visualized by silver staining (Fig. 3A). Overall, 10 sgJFH1-NS5A.HA.NS5B.His-specific or -enriched bands were sliced from the sgJFH1-NS5A.HA.NS5B.His lane and the proteins they contained were identified by MS (Table 1). The identified host proteins included proteins that were associated with intracellular transport, cytoskeleton components, RNP complex components, and chaperones. Among these proteins, numerous known HCV-interacting proteins like PI4KA, HnRNPK, PTB, HSP90, and nucleolin were unambiguously identified (Fig. 3B and Table 1).
TABLE 1.
Categories of host factors found to be associated with the HCV replicase
| Category and band no. | prot_score | prot_mass | Gene name | Protein description | Total no. of nonredundant peptides identified | No. of distinct peptides identified | Reference(s) |
|---|---|---|---|---|---|---|---|
| HCV protein | |||||||
| 6 | 110 | 49,784 | NS5A | HCV NS5A | 3 | 3 | |
| 6 | 90 | 65,668 | NS5B | HCV NS5B | 3 | 3 | |
| Transport | |||||||
| 1 | 2,246 | 233,622 | PI4KA | Phosphatidylinositol 4-kinase alpha | 17 | 14 | 54 |
| 1 | 153 | 234,855 | SEC16A | Protein transport protein Sec16A | 3 | 3 | |
| 2 | 562 | 117,330 | SEC16B | Protein transport protein Sec16B | 5 | 5 | |
| 2 | 397 | 119,789 | SEC24C | Protein transport protein Sec24C | 4 | 4 | |
| 2 | 245 | 120,472 | SEC24A | Protein transport protein Sec24A | 3 | 3 | |
| 10 | 43 | 89,950 | VCP | Transitional endoplasmic reticulum ATPase | 1 | 1 | |
| 9 | 89 | 36,031 | SEC13 | Protein SEC13 homolog | 1 | 1 | |
| 9 | 150 | 38,808 | ANXA2 | Annexin A2 | 3 | 1 | 55 |
| 10 | 25 | 87,393 | SEC23B | Protein transport protein Sec23B | 1 | 1 | |
| 10 | 49 | 87,018 | SEC23A | Protein transport protein Sec23A | 2 | 2 | |
| 2 | 649 | 129,915 | ANKFY1 | Ankyrin repeat and FYVE domain-containing protein 1 | 4 | 4 | |
| 4 | 24 | 102,704 | PRRT3 | Proline-rich transmembrane protein 3 | 1 | 1 | |
| Cytoskeleton | |||||||
| 1 | 38 | 218,321 | SHROOM3 | Shroom3 | 1 | 1 | |
| 4 | 17 | 108,561 | AP2A1 | AP-2 complex subunit alpha-1 | 3 | 2 | |
| 7 | 1,500 | 50,255 | TUBB4B | Tubulin beta-4B chain | 8 | 1 | 56 |
| 7 | 468 | 50,788 | TUBA1A/B/C | Tubulin alpha-1A chain | 4 | 0 | |
| 8 | 153 | 50,865 | TUBB1 | Tubulin beta-1 chain | 3 | 1 | |
| 10 | 53 | 103,563 | ACTN1 | Alpha-actinin-1 | 1 | 1 | 57 |
| RNP complex | |||||||
| 2 | 143 | 125,578 | UPF1 | Regulator of nonsense transcripts 1 | 3 | 3 | |
| 3 | 402 | 114,512 | MOV10 | Putative helicase MOV-10 | 6 | 6 | |
| 3 | 82 | 91,269 | HNRNPU | Heterogeneous nuclear ribonucleoprotein U | 2 | 2 | |
| 5 | 1,586 | 63,783 | IGF2BP1 | Insulin-like growth factor 2 mRNA-binding protein 1 | 9 | 7 | 58 |
| 5 | 509 | 64,008 | IGF2BP3 | Insulin-like growth factor 2 mRNA-binding protein 3 | 5 | 3 | |
| 5 | 435 | 66,195 | IGF2BP2 | Insulin-like growth factor 2 mRNA-binding protein 2 | 2 | 1 | 59 |
| 5 | 316 | 73,597 | DDX3X | ATP-dependent RNA helicase DDX3X | 4 | 1 | 59–61 |
| 5 | 49 | 69,618 | DDX5 | Probable ATP-dependent RNA helicase DDX5 | 2 | 1 | 59, 62 |
| 5 | 31 | 70,854 | PABPC1 | Polyadenylate-binding protein 1 | 1 | 1 | |
| 6 | 1,500 | 51,230 | HNRNPK | Heterogeneous nuclear ribonucleoprotein K | 3 | 3 | 59, 63 |
| 6 | 104 | 63,428 | STAU1 | Double-stranded RNA-binding protein Staufen homolog 1 | 2 | 1 | 59, 64 |
| 6 | 47 | 52,189 | G3BP1 | Ras GTPase-activating protein-binding protein 1 | 1 | 1 | 13, 59 |
| 6 | 38 | 57,357 | PTBP1 | Polypyrimidine tract-binding protein 1 | 1 | 1 | 65 |
| 7 | 364 | 49,484 | HNRNPH1 | Heterogeneous nuclear ribonucleoprotein H | 5 | 1 | 66 |
| 8 | 153 | 35,903 | YBX1 | Nuclease-sensitive element-binding protein 1 | 1 | 1 | 67 |
| 4 | 429 | 76,625 | NCL | Nucleolin | 8 | 8 | 68 |
| Chaperon | |||||||
| 5 | 1,386 | 71,082 | HSPA8 | Heat shock cognate 71 kDa protein | 10 | 2 | 59 |
| 5 | 742 | 72,402 | HSPA5 | 78 kDa glucose-regulated protein | 6 | 4 | 59 |
| 6 | 294 | 60,153 | CCT8 | T-complex protein 1 subunit theta | 1 | 1 | |
| 6 | 215 | 61,066 | CCT3 | T-complex protein 1 subunit gamma | 1 | 1 | |
| 6 | 117 | 60,819 | TCP1 | T-complex protein 1 subunit alpha | 1 | 1 | 59 |
| 6 | 114 | 60,089 | CCT5 | T-complex protein 1 subunit epsilon | 1 | 1 | 69 |
| 6 | 79 | 59,842 | CCT7 | T-complex protein 1 subunit eta | 2 | 2 | |
| 10 | 83 | 85,006 | HSP90AA1 | Heat shock protein HSP 90-alpha | 2 | 1 | 70 |
| Translation machines | |||||||
| 4 | 171 | 96,246 | EEF2 | Elongation factor 2 | 2 | 2 | |
| 7 | 19 | 47,953 | RPL4 | 60S ribosomal protein L4 | 1 | 1 | 59 |
| 8 | 294 | 52,587 | EIF3E | Eukaryotic translation initiation factor 3 subunit E | 4 | 4 | 59 |
| 8 | 294 | 37,654 | EIF3F | Eukaryotic translation initiation factor 3 subunit F | 3 | 3 | |
| 8 | 196 | 46,353 | EIF4A1 | Eukaryotic initiation factor 4A-I/II | 3 | 0 | |
| 8 | 153 | 46,609 | RPL3L | 60S ribosomal protein L3-like | 3 | 1 | |
| 9 | 410 | 34,423 | RPLP0 | 60S acidic ribosomal protein P0 | 5 | 1 | |
| 9 | 323 | 32,947 | RPSA | 40S ribosomal protein SA | 4 | 4 | |
| 9 | 90 | 34,569 | RPL5 | 60S ribosomal protein L5 | 1 | 1 | |
| Nuclear pore complex | |||||||
| 6 | 20 | 58,168 | KPNA2 | Importin subunit alpha-1 | 1 | 1 | |
| 10 | 25 | 74,380 | LMNA | Prelamin-A/C | 1 | 1 |
Identification of VCP as an active factor required for viral replication.
We then selected host factors that have not been reported, on the basis of a PubMed search, to be involved in HCV replication and performed siRNA screening to determine the effect of each factor on viral replication. We selected COPII vesicle coat subunit Sec24C (34) and regulating protein Sec16A (35, 36); Shroom3, which is required for apical constriction and epithelial invagination (37) and is probably involved in cytoskeleton organization (38); ANKFY1 (also known as rabankyrin-5), which is a Rab5 effector (39); AAA+ATPase VCP, which is well known for its role in ER-associated degradation (ERAD) (40); RNA helicase MOV10, which is a putative RNA helicase and a component of the RNA-induced silencing complex (41); and UPF1, which is involved in the nonsense-mediated decay (NMD) pathway (42). Huh7.5 cells were transfected with siRNAs targeting the genes of interest (GOIs) and then infected with HCV Jc1G. The supernatants were collected to measure reporter luciferase activity (Fig. 3C). Intracellular viral RNA levels were examined by quantitative RT-PCR (Fig. 3D). The Kd efficiency of selected genes was confirmed by quantitative RT-PCR with gene-specific primers (Fig. 3E), and cell viability upon gene Kd was measured (Fig. 3F). Kd of Sec16A, Shroom3, ANKFY1, UPF1, and VCP significantly reduced viral replication, as determined by viral replication reporter and viral RNA quantification (Fig. 3C and D). Among the seven selected genes, that for VCP was found among the top hits affecting viral replication (Fig. 3C and D).
VCP is involved in HCV genome replication.
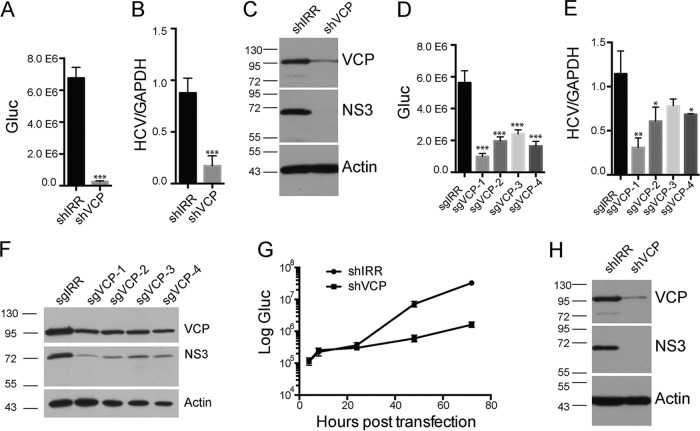
We further investigated the role of VCP in HCV replication. To complement siRNA-mediated silencing, we performed shRNA-mediated Kd in Huh7.5 cells and infected the shRNA-transduced cells with Jc1G. Kd of VCP dramatically reduced HCV replication, as assessed via luciferase reporter activity, viral RNA quantification, and viral protein expression (Fig. 4A to C). We also performed CRISPR (clustered regularly interspaced short palindromic repeats)-mediated gene Kd. All sgRNAs targeting VCP in Huh7.5 cells reduced VCP protein expression (Fig. 4F) and significantly reduced Jc1G viral replication, as assessed via the luciferase reporter activity (Fig. 4D), viral RNA level (Fig. 4E), and viral protein level (Fig. 4F).
FIG 4.
VCP is involved in HCV replication. (A to C) HCV infection of VCP-Kd cells. Huh7.5 cells were transduced with lentiviruses expressing shRNA against an irrelevant target (IRR) or VCP and then infected with Jc1G at an MOI of 0.01. (A) Gaussia luciferase (Gluc) activity in the supernatants at 3 days postinfection is shown. Mean values ± standard deviations are shown (n = 4). (B) HCV RNA levels quantified by quantitative RT-PCR are shown. Mean values ± standard deviations are shown (n = 3). (C) Western blot analysis of infected cells with antibodies against the proteins indicated. (D to F) Huh7.5 cells were transduced with lentiviruses expressing the sgRNAs indicated for CRISPR-mediated targeting and then infected with Jc1G at an MOI of 0.01 for 2 days. (D) The Gluc activity in supernatants was measured. Mean values ± standard deviations are shown (n = 4). (E) The HCV RNAs from infected cells were quantified by RT-PCR and normalized to the level of GAPDH RNA. Mean values ± standard deviations are shown (n = 3). (F) Western blotting of the infected cell lysates indicated. Statistical analysis comparing the sgVCP and sgIRR groups was performed. (G, H) HCV RNAs were transfected into VCP-Kd cells. (G) Gluc activity in the supernatants at various time points posttransfection is shown. Mean values ± standard deviations are shown (n = 3). (H) Western blotting of transfected cells at 72 h posttransfection. *, P < 0.05; **, P < 0.01; ***, P < 0.001, two-tailed, unpaired t test. Representative data from multiple experiments with similar results are shown. The values to the left of the blots in panels C, F, and H are molecular sizes in kilodaltons.
To determine the viral life cycle in which VCP was involved, we overcame viral entry by transfecting viral RNAs into VCP-Kd cells. Incoming viral RNA translation was comparable in shIRR control cells and shVCP cells at 4 and 8 h posttransfection, suggesting that VCP was not involved in viral translation (Fig. 4G). In contrast, luciferase activity and viral protein abundance were dramatically reduced in shVCP cells at later time points (Fig. 4G and H), suggesting that VCP was mainly involved in viral genome amplification.
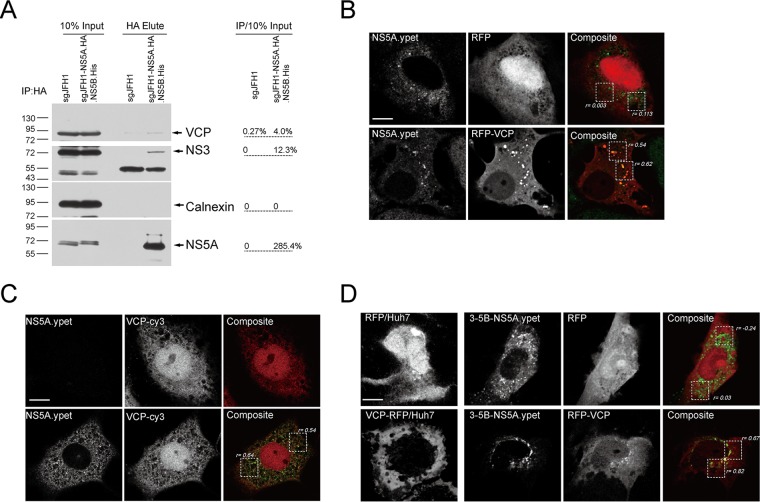
VCP associates with the HCV replicase.
We then examined the association between VCP and viral replicase. First, HA affinity purification of NS5A in sgJFH1-NS5A.HA.NS5B.His replicon cells showed that VCP was enriched in the pulled replicase complexes, similarly to the replicase component NS3, whereas the ER marker calnexin was not (Fig. 5A). We then examined the colocalization of RFP-VCP with the replicase marker NS5A in sgJFH1-NS5A.ypet replicon cells in which ypet is fused in frame to NS5A. We observed RFP-VCP (Pearson's colocalization coefficient [rp] = 0.54 and 0.62) but not RFP (rp = 0.003 and 0.113) colocalized with NS5A.yet clustering sites in these cells (Fig. 5B). We also examined the colocalization of endogenous VCP with NS5A in sgJFH1-NS5A.ypet replicon cells. Endogenous VCP exhibited cytoplasmic and nuclear distribution and colocalized with NS5A.yet in the cytoplasm (rp = 0.54 and 0.64) (Fig. 5C). Further, we used the HCV RC surrogate system in which the HCV polypeptide NS3-5B is expressed and RC assembly occurs independently of viral replication (5) to examine the association of VCP with the viral replicase marker. As shown in Fig. 5D, RFP-VCP changed its localization and relocalized to the NS5A.ypet clustering sites in the NS3-5B.ypet-expressing cells in which ypet is fused in frame to NS5A (rp = 0.67 and 0.82). Thus, VCP associates with the HCV RC and is required for viral replication.
FIG 5.
VCP associates with HCV RC. (A) Anti-HA antibody affinity purification with lysates from the replicon cells indicated. The bands were quantified and normalized to the input. Similar results were obtained in another independent experiment. IP, immunoprecipitation. The values to the left are molecular sizes in kilodaltons. (B to D) Colocalization of VCP with HCV NS5A. (B) RFP- or RFP-VCP-expressing plasmids were transfected into sgJFH1-NS5A.ypet subgenomic replicon cells. (C) Endogenous VCPs in sgJFH1-NS5A.ypet subgenomic replicon cells were immunostained. (D) Huh7 cells were transfected with constructs expressing RFP or RFP-VCP alone (left panels) or cotransfected with a 3-5B-NS5A.ypet-expressing construct (remaining panels). Cells were fixed and visualized by confocal microscopy. The Pearson correlation coefficients for the boxed regions were quantified. Representative images of multiple independent experiments are shown. Scale bars, 10 μm.
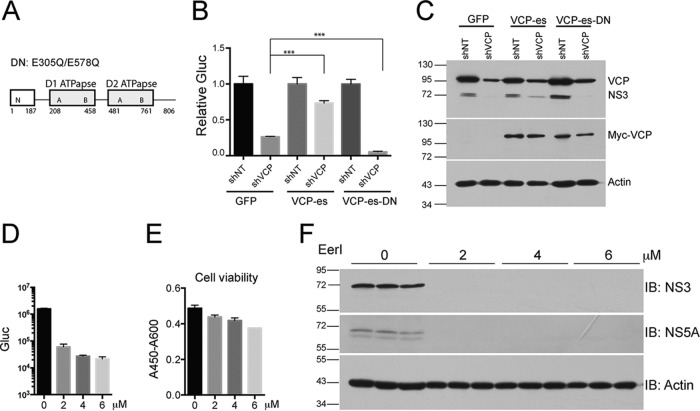
ATPase activity is required for HCV replication.
VCP contains two ATP-binding (AAA) domains (Fig. 6A). Double mutation (E305Q/E578Q) of these two domains completely abolishes ATPase activity (43). To determine whether the ATPase activity of VCP is required for HCV replication, we performed a rescue experiment by expressing the shRNA-resistant version of VCP and dominant negative mutant VCP (E305Q/E578Q) in VCP-Kd cells. We first generated cell lines stably expressing shRNA-resistant, myc-tagged VCP and VCP-DN and then knocked down the endogenous VCP in these cells by using shRNA. Expression of exogenous myc-tagged VCP was confirmed by Western blotting with anti-myc antibody (Fig. 6C). Although exogenously expressed VCP did not completely restore VCP expression in VCP-Kd cells, most likely because of the high abundance of endogenous VCP, it partially restored HCV replication (Fig. 6B and C). In contrast, VCP-DN did not rescue HCV replication but synergistically reduced viral replication in VCP-Kd cells (Fig. 6B), demonstrating that it acted as a dominant negative mutant protein. We then used a VCP inhibitor to block its ATPase activity and examined the effect on HCV replication. EerI inhibits VCP-dependent ERAD, most likely by irreversibly binding to VCP (44). EerI treatment at 2 μM dramatically reduced HCV replication and viral protein levels (Fig. 6D and F) with only about 10% cell viability reduction (Fig. 6E). EerI treatment at 4 μM completely blocked viral replication (Fig. 6D and F) with only about 15% cell viability reduction (Fig. 6E). Taken together, these data indicate that the ATPase activity of VCP is required for HCV replication.
FIG 6.
ATPase activity of VCP is required for HCV replication. (A) Schematic of VCP domains. Mutated amino acids in the dominant negative (DN) form of the protein are indicated. (B, C) Stable cell lines expressing GFP, VCP-es (shRNA escape), or VCP-es-DN were transduced with lentiviruses expressing shRNAs against irrelevant target (IRR) or VCP and then infected with Jc1G at an MOI of 0.01. (B) Gaussia luciferase (Gluc) activity in supernatants at 3 days postinfection. Gluc activity was normalized against each shIRR group. Mean values ± standard deviations are shown (n = 3). (C) Western blotting of the cell lysates indicated; proteins and blotting antibodies are indicated to the right. Representative data from multiple experiments with similar results are shown. (D to F) VCP inhibitor EerI inhibited HCV replication. (D) Huh7.5 cells were infected with Jc1G at an MOI of 0.5 for 8 h. After washing, medium with various concentration of EerI was added. One day later, the Gluc activity in the supernatants was measured. Mean values ± standard deviations are shown (n = 3). (E) Cell viability was measured. Mean values ± standard deviations are shown (n = 3). (F) Western blot (immunoblot [IB]analysis of cell lysates with the antibodies indicated. Representative data from multiple experiments with similar results are shown. The values to the left of the blots in panels C and F are molecular sizes in kilodaltons.
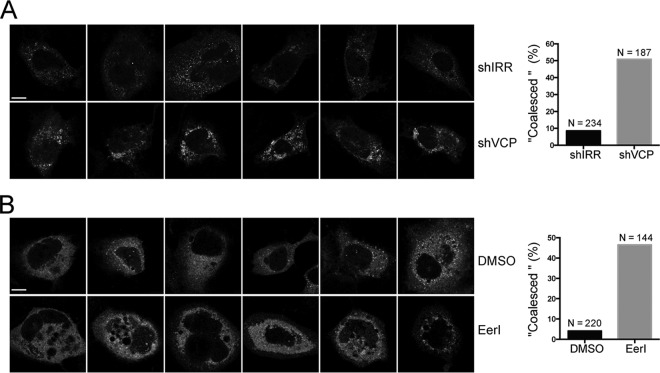
Abolishing VCP function resulted in aberrant HCV NS5A distribution.
We took advantage of the fact that HCV replication was dramatically reduced in VCP-DN-expressing cells in which endogenous VCP was further knocked down by shRNA. We attempted to examine if there was a change in NS5A distribution in the context of NS3-5B in these cells, which may indicate alteration of viral replicase assembly. We cotransfected NS3-5B.ypet with VCP-DN into the shRNA-transduced cells and monitored NS5A-ypet distribution by confocal microscopy. Compared with shIRR cells with NS5A-ypet, shVCP cells with NS5A-ypet exhibited a “coalesced” phenotype in which NS5A proteins were clustered and condensed (Fig. 7A). We also examined the NS5A-ypet distribution when cells were treated with EerI. We treated NS3-5B.ypet-expressing cells with 2 μM EerI for 24 h. At this dosage, EerI dramatically reduced viral replication (Fig. 6D). As in shVCP cells, EerI treatment altered the distribution of NS5A-ypet and resulted in a “coalesced” phenotype (Fig. 7B). Thus, abolishing VCP function resulted in aberrant HCV NS5A distribution in the replicase assembly surrogate system.
FIG 7.
Abolishing VCP function resulted in aberrant HCV NS5A distribution. (A) Huh7 cells were transduced with shIRR and shVCP lentiviruses. Two day later, the cells were cotransfected with phCMV-3-5B.ypet and VCP-es-DN at a ratio of 1:1 (microgram per microgram). At 1 day posttransfection, the cells were fixed and observed by confocal microscopy. The cells with a “coalesced” phenotype were enumerated, and the numbers were plotted (N is the total number of cells enumerated). Representative images are shown. (B) Huh7 cells were transfected with phCMV-3-5B.ypet. At 1 day posttransfection, the cells were treated with 2 μM EerI. One day later, the cells were fixed and observed by confocal microscopy. The cells with a “coalesced” phenotype were enumerated, and the numbers were plotted (N is the total number of cells enumerated). Representative images are shown. Scale bars, 10 μm. DMSO, dimethyl sulfoxide.
DISCUSSION
Almost all positive-strand RNA viruses use a replication strategy by which viral proteins modify the host membrane to assemble the membrane-associated viral replicase. Viruses hijack host factors to facilitate this energy-unfavorable process. Identification of the replicase-associated host factors and dissection of their roles in replicase assembly will help us understand the molecular mechanisms of RC biogenesis.
Purification of membrane-associated viral replicase remains technically challenging. By dual-affinity tagging two HCV replicase components, we tried to purify the solubilized HCV replicase. We detected the replicase components NS5A, NS5B, and NS3 in the purified complexes after sequential affinity purification (Fig. 2B and 3B). We found specifically enriched host proteins associated with viral replicase, including numerous known HCV-interacting proteins (Fig. 3B and Table 1).
Of the genes selected for further study, Kd of those for Shroom3, ANKFY1, UPF1, and VCP significantly reduced viral replication (Fig. 3C and D), suggesting potential roles for these proteins in viral replication. Shroom3, a cytoskeleton organization regulator (38), is required for apical constriction and epithelial invagination (37). HCV replication factories are protrusions from the ER membranes toward the cytosol (5) that may employ cytoskeleton reorganization via Shroom3. ANKFY1 (also known as rabankyrin-5), a Rab5 effector, is required for macropinocytosis (39). Rab proteins are required for HCV RC formation (9). Rab5 associated with HCV RC, and Kd of Rab5 reduced HCV replication (45, 46). As ANKFY1 was to be copurified with HCV replicase and Kd of ANFY1 significantly reduced viral replication (Fig. 3B to D), Rab5-ANKFY1 pairs may play a role in RC formation. UPF1 is well known for its involvement in the NMD pathway (42) and was recently reported to restrict Semliki Forest virus infection. In contrast to restriction of viral replication, UPF1 is required for HIV1 virion production (47). In this study, we found that UPF1 is required for HCV replication (Fig. 3C and D).
Among the seven genes selected, that for VCP was the top hit affecting viral replication (Fig. 3C and D) and is involved in HCV genome amplification (Fig. 4G). The ATPase activity of VCP was required for HCV replication (Fig. 6). VCP associated with viral replicase (Fig. 5A) and colocalized with viral RC components (Fig. 5B and C). In addition, in an RC surrogate system, VCP relocalized to the RC factories (Fig. 5D). Further, when VCP function was abolished by overexpression of VCP-DN and knocking down by shRNA or EerI, HCV NS5A in the RC surrogate system exhibited a “coalesced” phenotype (Fig. 7), indicating aberrant replicase assembly. These data indicate that VCP is hijacked to the RC and associates with the viral replicase. It may participate in replicase assembly via its AAA+ATPase activity.
The AAA+ proteins play an important role in diverse cellular functions that are dependent on their ATPase activity to induce conformational changes in diverse substrate proteins (48). AAA+ATPase proteins have been reported in the positive-strand RNA viral life cycle. VPS4 is involved in brome mosaic virus and tomato bushy stunt virus RC formation by cooperating with ESCRT complexes (49, 50) and in HCV virion production (51). VCP was identified in two RNA interference screening experiments to be involved in poliovirus replication (52) and in the regulation of Sindbis virus entry (53). VCP colocalizes with poliovirus nonstructural proteins in virus-infected cells (52). It is likely that hijacking host AAA+ATPases to modulate RC assembly or the viral life cycle is conserved among positive-strand RNA viruses.
In conclusion, we developed an HCV subgenomic replicon system by dual-affinity tagging two replicase components. Using this dual-affinity-tagged replicon system, we purified the viral replicase and identified VCP as a pivotal replicase-associated host factor that is required for viral genome replication. We propose that HCV hijacks host AAA+ATPase for its replicase assembly. Dissection of the molecular mechanism by which VCP regulates viral replicase assembly may lead to new insights into viral RC biogenesis and novel antiviral strategies.
ACKNOWLEDGMENTS
We are grateful to Yumei Wen for her warm encouragement, to Charles Rice for enlightening discussion and kindly providing the HCV reagents, and to Margaret R. MacDonald for her critical reading of the manuscript.
This work was supported in part by the National Basic Research Program of China (2015CB554301 to Z. Yi), Fudan University intramural funds (Zhuoxue fellowship to Z. Yi), the National Key Basic Research Program of China (2012CB519005 to Z. Yuan), and the Sino-German Transregional Collaborative Research and National Natural Science Foundation of China for International Collaboration (81461130019 to Z. Yuan).
REFERENCES
- 1.den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 2.Nagy PD, Pogany J. 2011. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 4.Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat Rev Microbiol 5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 5.Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu M, Rice CM. 2013. Structures of hepatitis C virus nonstructural proteins required for replicase assembly and function. Curr Opin Virol 3:129–136. doi: 10.1016/j.coviro.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul D, Madan V, Bartenschlager R. 2014. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe 16:569–579. doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yi ZG, Fang CY, Pan TT, Wang JD, Yang PY, Yuan ZH. 2006. Subproteomic study of hepatitis C virus replicon reveals Ras-GTPase-activating protein binding protein 1 as potential HCV RC component. Biochem Biophys Res Commun 350:174–178. doi: 10.1016/j.bbrc.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 9.Manna D, Aligo J, Xu C, Park WS, Koc H, Heo WD, Konan KV. 2010. Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology 398:21–37. doi: 10.1016/j.virol.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. 2013. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol 87:10612–10627. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung HY, Gu M, Buehler E, MacDonald MR, Rice CM. 2014. Seed sequence-matched controls reveal limitations of small interfering RNA knockdown in functional and structural studies of hepatitis C virus NS5A-MOBKL1B interaction. J Virol 88:11022–11033. doi: 10.1128/JVI.01582-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, Dustin LB. 2008. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology 48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi ZG, Pan TT, Wu XF, Song WH, Wang SS, Xu Y, Rice CM, MacDonald MR, Yuan ZH. 2011. Hepatitis C virus co-opts Ras-GTPase-activating protein-binding protein 1 for its genome replication. J Virol 85:6996–7004. doi: 10.1128/JVI.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J Virol 78:7400–7409. doi: 10.1128/JVI.78.14.7400-7409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi ZG, Yuan ZH, Rice CM, MacDonald MR. 2012. Flavivirus replication complex assembly revealed by DNAJC14 functional mapping. J Virol 86:11815–11832. doi: 10.1128/JVI.01022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol 77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. 2008. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3:619–628. doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- 20.Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem 68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 21.Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. 1999. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20:601–605. [DOI] [PubMed] [Google Scholar]
- 22.Cui SJ, Xu LL, Zhang T, Xu M, Yao J, Fang CY, Feng Z, Yang PY, Hu W, Liu F. 2013. Proteomic characterization of larval and adult developmental stages in Echinococcus granulosus reveals novel insight into host-parasite interactions. J Proteomics 84:158–175. doi: 10.1016/j.jprot.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 24.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Liu XH, Zheng WM, Zhang L, Yao J, Yang PY. 2014. Screening of missing proteins in the human liver proteome by improved MRM-approach-based targeted proteomics. J Proteome Res 13:1969–1978. doi: 10.1021/pr4010986. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz M, Chen J, Lee WM, Janda M, Ahlquist P. 2004. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc Natl Acad Sci U S A 101:11263–11268. doi: 10.1073/pnas.0404157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazakov T, Yang F, Ramanathan HN, Kohlway A, Diamond MS, Lindenbach BD. 2015. Hepatitis C virus RNA replication depends on specific cis- and trans-acting activities of viral nonstructural proteins. PLoS Pathog 11:e1004817. doi: 10.1371/journal.ppat.1004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arumugaswami V, Remenyi R, Kanagavel V, Sue EY, Ngoc Ho T, Liu C, Fontanes V, Dasgupta A, Sun R. 2008. High-resolution functional profiling of hepatitis C virus genome. PLoS Pathog 4:e1000182. doi: 10.1371/journal.ppat.1000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brass V, Gouttenoire J, Wahl A, Pal Z, Blum HE, Penin F, Moradpour D. 2010. Hepatitis C virus RNA replication requires a conserved structural motif within the transmembrane domain of the NS5B RNA-dependent RNA polymerase. J Virol 84:11580–11584. doi: 10.1128/JVI.01519-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray CL, Rice CM. 2011. Turning hepatitis C into a real virus. Annu Rev Microbiol 65:307–327. doi: 10.1146/annurev-micro-090110-102954. [DOI] [PubMed] [Google Scholar]
- 31.You SY, Stump DD, Branch AD, Rice CM. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol 78:1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friebe P, Boudet J, Simorre JP, Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J Virol 79:380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin C, Prágai BM, Grakoui A, Xu J, Rice CM. 1994. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol 68:8147–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stagg SM, LaPointe P, Razvi A, Gurkan C, Potter CS, Carragher B, Balch WE. 2008. Structural basis for cargo regulation of COPII coat assembly. Cell 134:474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharyya D, Glick BS. 2007. Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol Biol Cell 18:839–849. doi: 10.1091/mbc.E06-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whittle JR, Schwartz TU. 2010. Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat. J Cell Biol 190:347–361. doi: 10.1083/jcb.201003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plageman TF, Chauhan BK, Yang C, Jaudon F, Shang X, Zheng Y, Lou M, Debant A, Hildebrand JD, Lang RA. 2011. A Trio-RhoA-Shroom3 pathway is required for apical constriction and epithelial invagination. Development 138:5177–5188. doi: 10.1242/dev.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoder M, Hildebrand JD. 2007. Shroom4 (Kiaa1202) is an actin-associated protein implicated in cytoskeletal organization. Cell Motil Cytoskeleton 64:49–63. doi: 10.1002/cm.20167. [DOI] [PubMed] [Google Scholar]
- 39.Schnatwinkel C, Christoforidis S, Lindsay MR, Uttenweiler-Joseph S, Wilm M, Parton RG, Zerial M. 2004. The Rab5 effector rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol 2:E261. doi: 10.1371/journal.pbio.0020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer H, Bug M, Bremer S. 2012. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 41.Höck J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. 2007. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang YF, Imam JS, Wilkinson MF. 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 43.Dalal S, Rosser MF, Cyr DM, Hanson PI. 2004. Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol Biol Cell 15:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chapman E, Maksim N, de la Cruz F, La Clair JJ. 2015. Inhibitors of the AAA+ chaperone p97. Molecules 20:3027–3049. doi: 10.3390/molecules20023027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone M, Jia S, Heo WD, Meyer T, Konan KV. 2007. Participation of Rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J Virol 81:4551–4563. doi: 10.1128/JVI.01366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eyre NS, Fiches GN, Aloia AL, Helbig KJ, McCartney EM, McErlean CS, Li K, Aggarwal A, Turville SG, Beard MR. 2014. Dynamic imaging of the hepatitis C virus NS5A protein during a productive infection. J Virol 88:3636–3652. doi: 10.1128/JVI.02490-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serquiña AKP, Das SR, Popova E, Ojelabi OA, Roy CK, Gottlinger HG. 2013. UPF1 is crucial for the infectivity of human immunodeficiency virus type 1 progeny virions. J Virol 87:8853–8861. doi: 10.1128/JVI.00925-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanson PI, Whiteheart SW. 2005. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 49.Diaz A, Zhang JT, Ollwerther A, Wang XF, Ahlquist P. 2015. Host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog 11:e1004742. doi: 10.1371/journal.ppat.1004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barajas D, Martin IFD, Pogany J, Risco C, Nagy PD. 2014. Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of tomato bushy stunt virus replicase. PLoS Pathog 10:e1004087. doi: 10.1371/journal.ppat.1004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corless L, Crump CM, Griffin SDC, Harris M. 2010. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol 91:362–372. doi: 10.1099/vir.0.017285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arita M, Wakita T, Shimizu H. 2012. Valosin-containing protein (VCP/p97) is required for poliovirus replication and is involved in cellular protein secretion pathway in poliovirus infection. J Virol 86:5541–5553. doi: 10.1128/JVI.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panda D, Rose PP, Hanna SL, Gold B, Hopkins KC, Lyde RB, Marks MS, Cherry S. 2013. Genome-wide RNAi screen identifies SEC61A and VCP as conserved regulators of Sindbis virus entry. Cell Rep 5:1737–1748. doi: 10.1016/j.celrep.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Buhler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Backes P, Quinkert D, Reiss S, Binder M, Zayas M, Rescher U, Gerke V, Bartenschlager R, Lohmann V. 2010. Role of annexin A2 in the production of infectious hepatitis C virus particles. J Virol 84:5775–5789. doi: 10.1128/JVI.02343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lai CK, Jeng KS, Machida K, Lai MMC. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J Virol 82:8838–8848. doi: 10.1128/JVI.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lan SY, Wang H, Jiang H, Mao HX, Liu XY, Zhang XN, Hu YW, Xiang L, Yuan ZH. 2003. Direct interaction between alpha-actinin and hepatitis C virus NS5B. FEBS Lett 554:289–294. doi: 10.1016/S0014-5793(03)01163-3. [DOI] [PubMed] [Google Scholar]
- 58.Li Y, Masaki T, Shimakami T, Lemon SM. 2014. hnRNP L and NF90 interact with hepatitis C virus 5′-terminal untranslated RNA and promote efficient replication. J Virol 88:7199–7209. doi: 10.1128/JVI.00225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Upadhyay A, Dixit U, Manvar D, Chaturvedi N, Pandey VN. 2013. Affinity capture and identification of host cell factors associated with hepatitis C virus (+) strand subgenomic RNA. Mol Cell Proteomics 12:1539–1552. doi: 10.1074/mcp.M112.017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ariumi Y, Kuroki M, Abe KI, Dansako H, Ikeda M, Wakita T, Kato N. 2007. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol 81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Q, Pene V, Krishnamurthy S, Cha H, Liang TJ. 2013. Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly. Nat Med 19:722–729. doi: 10.1038/nm.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goh PY, Tan YJ, Lim SP, Tan YH, Lim SG, Fuller-Pace F, Hong WJ. 2004. Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J Virol 78:5288–5298. doi: 10.1128/JVI.78.10.5288-5298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsieh TY, Matsumoto M, Chou HC, Schneider R, Hwang SB, Lee AS, Lai MMC. 1998. Hepatitis C virus core protein interacts with heterogeneous nuclear ribonucleoprotein K. J Biol Chem 273:17651–17659. [DOI] [PubMed] [Google Scholar]
- 64.Blackham SL, McGarvey MJ. 2013. A host cell RNA-binding protein, Staufen1, has a role in hepatitis C virus replication before virus assembly. J Gen Virol 94:2429–2436. doi: 10.1099/vir.0.051383-0. [DOI] [PubMed] [Google Scholar]
- 65.Aizaki H, Choi KS, Liu MY, Li YJ, Lai MMC. 2006. Polypyrimidine-tract-binding protein is a component of the HCV RNA replication complex and necessary for RNA synthesis. J Biomed Sci 13:469–480. doi: 10.1007/s11373-006-9088-4. [DOI] [PubMed] [Google Scholar]
- 66.Lee JW, Liao PC, Young KC, Chang CL, Chen SSL, Chang TT, Lai MD, Wang SW. 2011. Identification of hnRNPH1, NF45, and C14orf166 as novel host interacting partners of the mature hepatitis C virus core protein. J Proteome Res 10:4522–4534. doi: 10.1021/pr200338d. [DOI] [PubMed] [Google Scholar]
- 67.Chatel-Chaix L, Germain MA, Motorina A, Bonneil E, Thibault P, Baril M, Lamarre D. 2013. A host YB-1 ribonucleoprotein complex is hijacked by hepatitis C virus for the control of NS3-dependent particle production. J Virol 87:11704–11720. doi: 10.1128/JVI.01474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shimakami T, Honda M, Kusakawa T, Murata T, Shimotohno K, Kaneko S, Murakami S. 2006. Effect of hepatitis C virus (HCV) NS5B-nucleolin interaction on HCV replication with HCV subgenomic replicon. J Virol 80:3332–3340. doi: 10.1128/JVI.80.7.3332-3340.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Inoue Y, Aizaki H, Hara H, Matsuda M, Ando T, Shimoji T, Murakami K, Masaki T, Shoji I, Homma S, Matsuura Y, Miyamura T, Wakita T, Suzuki T. 2011. Chaperonin TRiC/CCT participates in replication of hepatitis C virus genome via interaction with the viral NS5B protein. Virology 410:38–47. doi: 10.1016/j.virol.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 70.Ujino S, Yamaguchi S, Shimotohno K, Takaku H. 2009. Heat-shock protein 90 is essential for stabilization of the hepatitis C virus nonstructural protein NS3. J Biol Chem 284:6841–6846. doi: 10.1074/jbc.M806452200. [DOI] [PMC free article] [PubMed] [Google Scholar]