Abstract
Bipolar disorder (BD) is a severe and highly heritable neuropsychiatric disorder with a lifetime prevalence of 1%. Molecular genetic studies have identified the first BD susceptibility genes. However, the disease pathways remain largely unknown. Accumulating evidence suggests that microRNAs, a class of small noncoding RNAs, contribute to basic mechanisms underlying brain development and plasticity, suggesting their possible involvement in the pathogenesis of several psychiatric disorders, including BD. In the present study, gene-based analyses were performed for all known autosomal microRNAs using the largest genome-wide association data set of BD to date (9747 patients and 14 278 controls). Associated and brain-expressed microRNAs were then investigated in target gene and pathway analyses. Functional analyses of miR-499 and miR-708 were performed in rat hippocampal neurons. Ninety-eight of the six hundred nine investigated microRNAs showed nominally significant P-values, suggesting that BD-associated microRNAs might be enriched within known microRNA loci. After correction for multiple testing, nine microRNAs showed a significant association with BD. The most promising were miR-499, miR-708 and miR-1908. Target gene and pathway analyses revealed 18 significant canonical pathways, including brain development and neuron projection. For miR-499, four Bonferroni-corrected significant target genes were identified, including the genome-wide risk gene for psychiatric disorder CACNB2. First results of functional analyses in rat hippocampal neurons neither revealed nor excluded a major contribution of miR-499 or miR-708 to dendritic spine morphogenesis. The present results suggest that research is warranted to elucidate the precise involvement of microRNAs and their downstream pathways in BD.
Introduction
Bipolar disorder (BD) is a severe neuropsychiatric disorder with an estimated lifetime prevalence of 1%.1 BD is characterized by recurrent episodes of mania and depression, and shows a heritability of ~70%.2 Molecular genetic candidate studies and—more recently—genome-wide association studies (GWAS) have identified the first BD susceptibility genes.3, 4, 5, 6, 7 However, the disease pathways and underlying regulatory networks remain largely unknown.8
Accumulating evidence suggests that microRNAs (miRNAs) are implicated in the biological pathways that regulate brain development and synaptic plasticity.9, 10 This in turn suggests their possible involvement in the pathogenesis of several psychiatric disorders,11, 12 including BD.13, 14 Studies of the post-mortem brain tissue of BD patients have demonstrated altered miRNA expression profiles in the prefrontal cortex.13, 14
The miRNAs are a class of 21–25-nucleotide small noncoding RNAs. In the nucleus they are transcribed by RNA polymerase II to primary miRNA (pri-miRNA) transcripts, which are double-stranded stem loop structures comprising 100–1000 nucleotides.15, 16 Approximately 50% of all vertebrate miRNAs are processed from the introns of protein-coding genes or from genes encoding other noncoding RNA classes. However, miRNAs can also be encoded in intergenic regions.17
The pri-miRNAs are then processed by the Drosha-DGCR8 complex to precursor miRNAs.18, 19 These precursor miRNAs are 60–70 nucleotides in length. The precursor miRNAs are exported to the cytoplasm, where they are cleaved into ∼20-base pair (bp) mature miRNAs by the Dicer enzyme.16, 20 The mature miRNAs are incorporated into the RNA-induced silencing complex, which then targets distinct sets of messenger RNAs (mRNAs).21
The miRNAs control the expression of their target genes by binding to target sites within the mRNAs, typically in their 3′ untranslated regions.22, 23 A region of 2–7 or 2–8 consecutive nucleotides from the 5′ end of the mature miRNA forms the seed region, which is crucial for the recognition of the target genes.24 In general, each miRNA controls up to several hundred target mRNAs, whereas one mRNA target can be subjected to synergistic regulation by multiple miRNAs.25, 26 In consequence, miRNAs integrate different intracellular signals and regulate a number of signaling pathways.27, 28 Interestingly, the miRNA regulatory effect itself has been shown to be a heritable trait in humans.29
The hypothesis that miRNAs are implicated in BD is also supported by the results of the largest GWAS of BD to date.6 In this study, a single-nucleotide polymorphism (SNP) in an intergenic region flanking MIR2113 on chromosome 6q16.1 was the eighth strongest finding. However, no significant enrichment of BD-associated genes within the known or predicted targets of MIR2113 was observed.6
Several studies have investigated the role of single miRNAs in the development of psychiatric disorder,30, 31, 32 including BD.33 However, to our knowledge, no systematic, genome-wide analysis of miRNA-coding genes has yet been performed. The aim of the present study was, thus, to determine whether common variants at any of the known miRNA loci contribute to the development of BD.
Materials and methods
Sample description
The gene-based tests were performed using data from our previous GWAS of BD (9747 patients and 14 278 controls).6 This GWAS data set combined data from Canada, Australia and four European countries (MooDS) with the GWAS results of the multinational Psychiatric Genomics Consortium (PGC).3 The study was approved by the respective local Ethics Committees. Written informed consent was obtained from all participants.6
Genome-wide miRNA association analysis
For the gene-based analyses, a set-based testing approach adapted from the versatile gene-based test for GWAS34 was used. This algorithm is obtainable upon request. The chromosomal positions of all miRNAs (n=718) were obtained from miRBase release 13.0.35 This release contains a high confidence set of miRNAs for which detailed information about miRNA function and predicted target genes is available. Using the summary statistics, gene-wide P-values were calculated for all 636 autosomal miRNAs and their ±20 kilobase (kb) flanking sequences. Twenty-seven of these miRNA loci contained no common SNP. Therefore, gene-wide P-values were obtained for 609 miRNAs.
The applied statistical algorithm is described in more detail in the article by Liu et al.34 Briefly, SNPs within these boundaries were grouped together, and a set-based test statistic was calculated as the sum of the χ2 one degree of freedom association P-values within the miRNA. The test statistic was compared with simulated test statistics from the multivariate normal distribution. An empirical miRNA-based P-value was calculated as the proportion of simulated test statistics above the observed test statistic. For the purposes of the present study, the 10% most significant SNPs for each miRNA were summarized. The calculated gene-based P-values were Bonferroni-corrected for multiple testing according to the number of investigated miRNAs (n=609).
As different reference panels were used for the imputation of the MooDS and PGC genotype data (1000 Genomes Project, February 2012 release, and HapMap phase 2 CEU, respectively), we used simulated test statistics on the basis of an intermarker linkage disequilibrium (LD) structure as derived from the HapMap phase 2 population genotypes. However, for miRNAs that showed a significant association with BD after Bonferroni correction, we also calculated gene-based tests based on 1000 Genomes Project phase 3 population genotypes.
Inflation of the observed and expected P-values for different SNP subcategories (SNPs in miRNA loci, SNPs in genes and intergenic SNPs) was defined as the degree of deviation from the expected uniform distribution in the quantile–quantile (Q–Q) plot and tested for significance using Fisher's exact test (one-sided) for different P-value thresholds. Only LD-pruned SNPs (r2<0.8) were used for the enrichment analysis.
Follow-up of miRNA association results—regional association plots
A window-based approach that included common variants in miRNAs and flanking sequences was applied. To determine whether the signal was associated with any of the miRNAs of interest, visual inspection of the regional association plots was performed.
Regional association results from our BD GWAS6 were plotted for all associated miRNAs and their ±500-kb flanking regions using LocusZoom.36 A signal was considered miRNA-associated if the top SNP of the region was located at, or was in high or moderate LD (r2>0.6) with, the miRNA locus.
Follow-up of miRNA association results—miRNA brain expression
To investigate expression of the associated miRNAs in the human brain, data from a recent study of miRNA expression patterns in the developing human brain were re-analyzed.37 A miRNA was defined as showing brain expression if it had a total read count of >120 across all investigated samples.37
In addition, miRNA expression was measured in rat cortical neurons and forebrain. All procedures involving animals followed the guidelines of the German Animal Protection Legislation and the experiments were approved by the Local Committee for Animal Health (RP Gießen). Total RNA was isolated from the postnatal day 15 rat forebrain or synaptosomes, as described elsewhere.38 Briefly, the total RNA from the forebrain of postnatal day-15 Sprague–Dawley rat pups was extracted using peqGOLD TriFast reagent (Peqlab, Erlangen, Germany) in accordance with the manufacturer's instructions. Small RNA libraries were constructed and sequenced at the EMBL genomic core facility (Heidelberg, Germany) using the HiSeq platform (Illumina, San Diego, CA, USA). The web-based software MiRanalyzer was used to determine miRNA expression levels (http://bioinfo2.ugr.es/miRanalyzer/miRanalyzer.php.).39
miRNA target gene analysis
Targets of the associated miRNAs 499, 708 and 1908 were obtained from TargetScan (Release 6.2).40 The Allen human brain atlas (http://www.brain-map.org/)41 was consulted to determine whether predicted target genes are expressed in the human brain. Target genes were considered brain-expressed if they had shown expression in the hippocampal formation in at least four of the six donor brains. Gene-based P-values for all brain-expressed miRNA targets were calculated using versatile gene-based test for GWAS,34 and our BD GWAS data set.6 To capture regulatory regions, the default settings in versatile gene-based test for GWAS were used. Enrichment of associated targets was calculated as follows: the number of associated target genes for each miRNA was compared with the number of associated genes from 100 000 random target sets of brain-expressed genes. Each target gene set comprised the same number of genes as the miRNA target genes itself.
Pathway analysis of target genes
The subsequent analyses were restricted to brain-expressed target genes of miR-499, miR-708 and miR-1908, with a gene-based association P-value of <0.05. If the chromosomal distance between two target genes was below 100 kb or if the top SNPs of two target genes were in strong or moderate LD (D'>0.4), only the target gene with the lowest gene-based P-value was retained in the pathway analysis to ensure the independency of association signals. In total, 107 target genes were included in the pathway analyses (Supplementary Box 1). Gene ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes pathway testing was performed using the WebGestalt (Web-based Gene Set Analysis Toolkit) for the brain-expressed, BD-associated target genes of the three associated miRNAs. Bonferroni correction was used to adjust for multiple testings. Significant pathways were filtered to achieve a minimum of three genes per set.
Functional analyses of miR-499 and miR-708 in rat hippocampal neurons
To test the possible involvement of miR-499 or miR-708 in the regulation of synaptic function, experiments were performed to investigate the effect of miR-499 and miR-708 overexpression on dendritic spine morphogenesis in primary rat hippocampal neurons. We initially focused on overexpression, as this can be easily achieved by the transfection of expression plasmids containing pri-miRNA cassettes. miRNA overexpression constructs were generated by inserting the respective pri-miRNA sequences into the 3'-untranslated repeat of the luciferase reporter gene within pmiRGLO (Promega, Madison, WI, USA). Thereby, luciferase reporter assays could be used to monitor the efficiency of pri-miRNA processing. To investigate the potential involvement of miR-499-5p and miR-708-5p in dendritic spine morphogenesis, hippocampal neurons of embryonic day-18 Sprague–Dawley rats (Charles River Laboratories, Sulzfeld, Germany) were transfected with miRNA-overexpressing constructs for 6 days before fixation. Images with a resolution of 1024 × 1024 pixels were obtained using a LSM5 Zeiss Pascal confocal microscope (Jena, Germany) and in a magnification of × 63 /1.4. A maximum projection was reconstructed with the Zeiss LSM 510 Meta software from a z-stack consisting of seven optical slices at 0.45-μm interval. The average intensity of an area of 2180 nm2 containing 250–300 spines per cell was measured using the ImageJ 1.48v software (National Institutes of Health, Bethesda, MD, USA), as described elsewhere.38 During imaging and analysis, the investigator was blind to the transfection condition.
Results
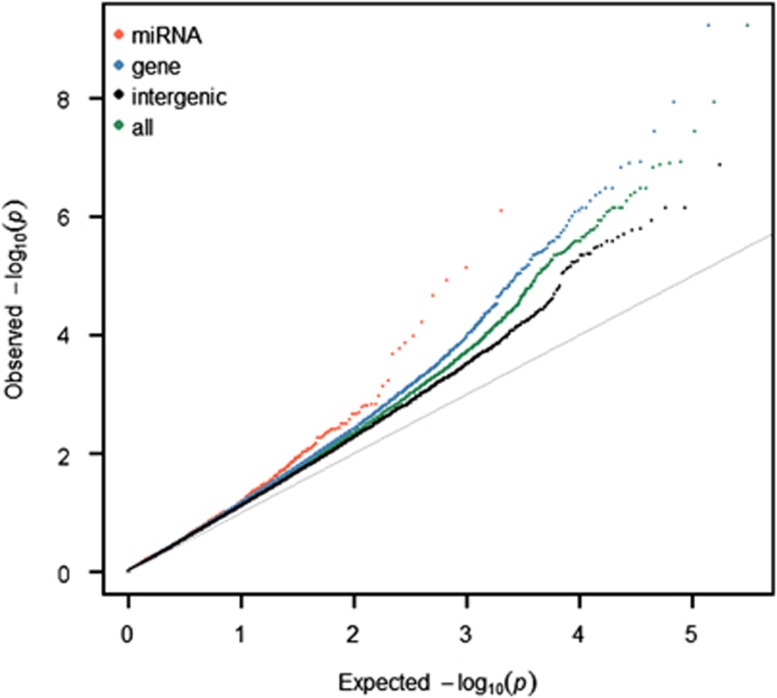
Overall, the nominal P-values of SNPs at miRNA loci were enriched with lower values than would be expected with a uniform P-value distribution (Figure 1). This deviation from the expected normal Q–Q plot distribution indicates a general enrichment for miRNAs among BD-associated SNPs. Category testing for different P-value thresholds revealed a significant enrichment for BD-associated SNPs in miRNA loci for P-values<1 × 10−4 (Supplementary Table 1). This deviation was also observed among SNPs in genes but not for intergenic SNPs.
Figure 1.
Quantile–quantile (Q–Q) plot of single-nucleotide polymorphism (SNP) P-values. The −log10 of the observed genome-wide association studies (GWAS) P-values for linkage disequilibrium (LD)-pruned SNPs (on the y axis) are plotted versus the −log 10 of the expected P-values (under null, on the x axis). The solid line represents expected uniform distribution. Red dots represent the data distribution of P-values of SNPs at microRNA loci; blue dots represent SNPs in genes; black dots represent P-values of intergenic SNPs; and green dots represent the data distribution of all SNPs.
Gene-based analysis in our BD GWAS data6 generated nominally significant P-values for 98 of the 609 miRNAs. These included miR-2113, which was located at the genome-wide significant locus on chromosome 6q16.1 in the original BD GWAS analyses.6 After correction for multiple testing, nine miRNAs showed a significant association with BD (Table 1). The additional calculation of gene-based tests for these nine miRNAs on the basis of 1000 Genomes LD structure generated nominal P-values of ⩽7.20 × 10−5 (Supplementary Table 2).
Table 1. Results of the gene-based tests for the nine microRNAs that withstood Bonferroni correction.
| miRNA | Chr | nSNPs | Top SNP | p Top SNP | p Corr Gene | miRNA-assoc. signal | Expr. hum. brain |
|---|---|---|---|---|---|---|---|
| miR-499 | 20 | 27 | rs3818253 | 6.58 × 10−7 | 0.0012 | Yes | Yes |
| miR-640 | 19 | 21 | rs2965184 | 7.23 × 10−7 | 0.0012 | Yes | No |
| miR-708 | 11 | 72 | rs7108878 | 3.45 × 10−7 | 0.0012 | Yes | Yes |
| miR-581 | 5 | 36 | rs697112 | 3.61 × 10−6 | 0.0073 | Yes | No |
| miR-644 | 20 | 12 | rs7269526 | 1.22 × 10−5 | 0.0104 | No | No |
| miR-135a-1 | 3 | 20 | rs9311474 | 2.16 × 10−5 | 0.0122 | No | Yes |
| let-7 g | 3 | 9 | rs6445358 | 2.23 × 10−5 | 0.0305 | No | Yes |
| miR-1908 | 11 | 16 | rs174575 | 2.85 × 10−5 | 0.0353 | Yes | Yes |
| miR-611 | 11 | 23 | rs174535 | 5.03 × 10−5 | 0.0457 | No | No |
Abbreviations: Chr, chromosome; expr. hum. brain, expression in the human brain according to Ziats and Rennert;37 miRNA, microRNA; miRNA-assoc. signal, specificity of the associated finding in the regional association plot; p Corr Gene, Bonferroni-corrected gene-based P-value; p Top SNP, P-value of the Top SNP within gene; nSNPs, number of investigated SNPs; SNP, single-nucleotide polymorphism.
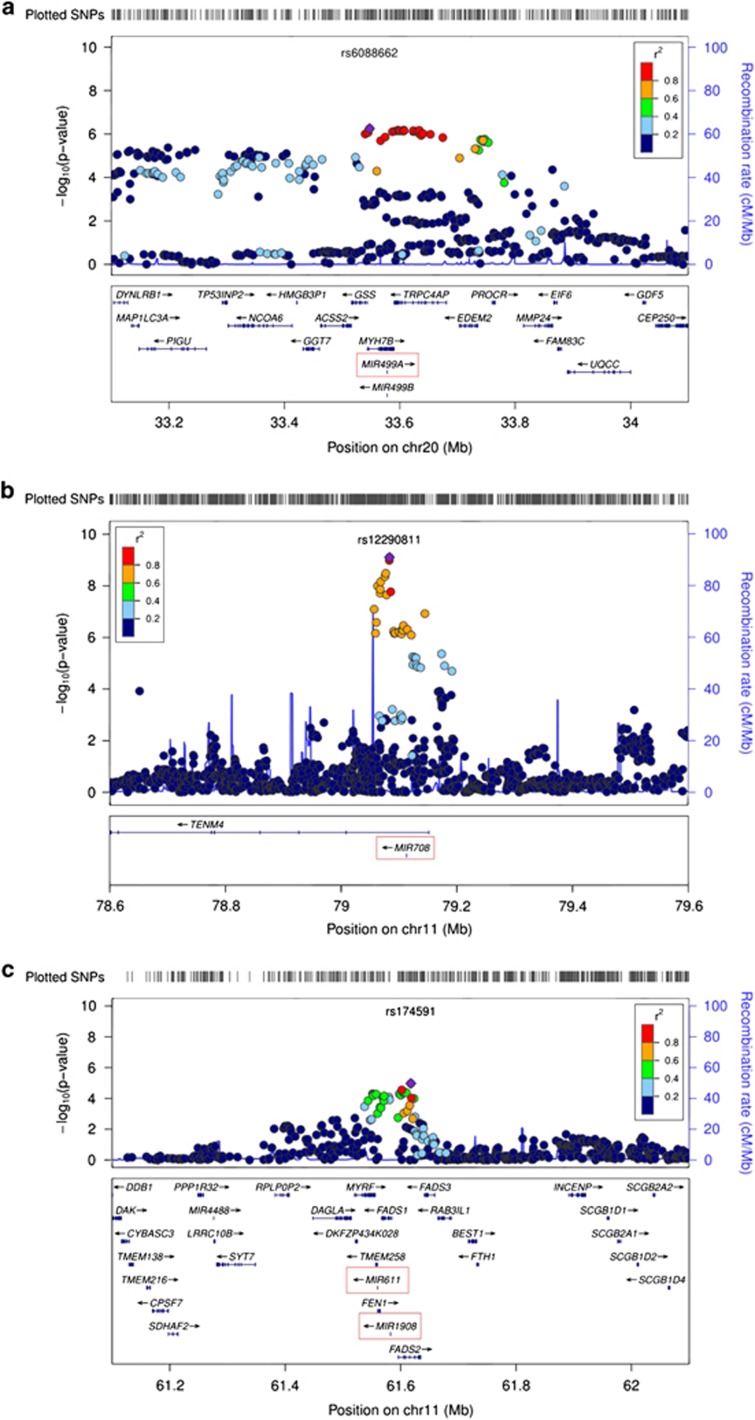
Visual inspection of the regional association plots revealed a miRNA-associated signal for five of the nine miRNAs (Figure 2, Supplementary Figures 1–4).
Figure 2.
Regional association plots of miR-499, miR-708 and miR-1908. Regional association results for the three most promising associated microRNAs miR-499 (a), miR-708 (b) and miR-1908 (c), and their ±500-kb flanking regions were plotted using LocusZoom (Pruim et al.36). The plot of miR-1908 (c) includes miR-611, which is also localized at the depicted chromosomal locus.
The re-analysis of the expression data from Ziats and Rennert37 revealed that five of the nine miRNAs were expressed in the human brain (Table 1).
Three of these (miR-499, miR-708 and miR-135a-1) were also found to be expressed in the rat forebrain. This method could not be used to investigate the expression of the other miRNAs, as they are not expressed in rats.35
The regional association plots and the miRNA expression data in human brain tissue suggest that the three brain-expressed miRNAs, that is, miR-499, miR-708 and miR-1908, are the most promising candidates for further analyses. The three miRNAs had 296, 181 and 67 target genes, respectively. Of these 286, 174 and 56 showed brain expression (Table 2).
Table 2. Target gene and pathway analysis for miR-499, miR-708 and miR-1908.
| MicroRNA | No. of brain-expressed target genes | No. of brain-expressed target genes, P<0.05 | P enrichment | No. of significant targets (corr) | No. of significant pathways |
|---|---|---|---|---|---|
| miR-499-5p | 286 | 59 | 0.7172 | 4 | 12 |
| miR-708-5p | 174 | 37 | 0.9265 | 2 | 1 |
| miR-1908-5p | 56 | 17 | 0.1422 | 1 | 5 |
Abbreviations: No. of significant pathways, number of significant pathways at P≤0.05; No. of significant targets (corr), number of significant target genes after Bonferroni correction for multiple testing; P enrichment, P-value of the enrichment analysis (X2-test).
Results of the target gene analysis for the three brain-expressed microRNAs miR-499, miR-708 and miR-1908 that were associated with bipolar disorder after correction for multiple testing.
The target gene enrichment analysis showed no significant enrichment of BD-associated genes within the targets of miR-499, miR-708 or miR-1908 (Table 2). After Bonferroni correction, miR-1908 had one (KLC2) and miR-708 had two significant target genes (NRAS and CREB1), whereas miR-499 had four significant target genes (GPC6, C16orf72, WDR82 and CACNB2).
Pathway testing revealed 18 significant canonical pathways that are driven by brain-expressed target genes of the three miRNAs (Table 2). For each miRNA, the results of the GO analysis are presented as directed acyclic graphs (Supplementary Figure 5). The target genes that drive a particular pathway are listed in Supplementary Table 3.
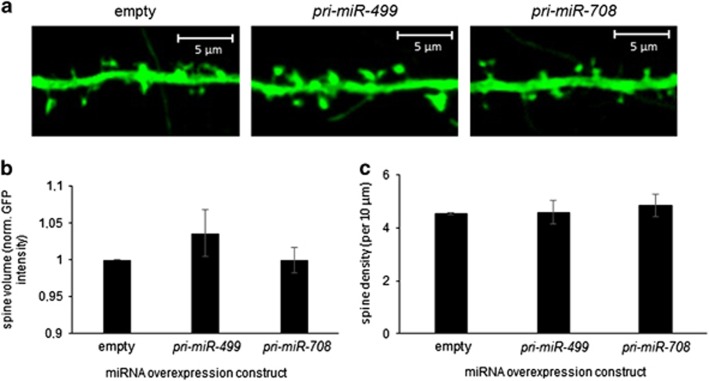
Luciferase assays revealed efficient processing of pri-miR-499, but not pri-miR-708, upon transfection of the respective constructs in neurons (Supplementary Figure 6). Overexpression of miR-499 led to a small and statistically nonsignificant increase in spine volume (Figure 3), but no effect on spine density was observed. As expected, transfection of the non-effective miR-708 expression construct had no significant effect on spine morphological parameters. Taken together, these results suggest that increasing levels of the BD-associated miR-499 have no—or only minimal—modulatory function during dendritic spine morphogenesis.
Figure 3.
Effect of the overexpression of miR-499 and miR-708 on dendritic spine size and density in primary rat hippocampal neurons. DIV14 primary hippocampal neurons were transfected with: (i) empty pmirGLO (250 ng) or (ii) pmirGLO (250 ng) containing pri-miR-499 or pri-miR-708 in the 3'-untranslated repeat of the Firefly luciferase gene and green fluorescent protein (GFP). The transfected neurons were then cultured until DIV19 and fixed for fluorescence microscopy. (a) Representative images for cells transfected with the indicated pmirGLO constructs or GFP only. A three-dimensional reconstruction was made from seven 45-μm stacks; scale bars, 5 μm. (b) Spine volume quantification of hippocampal neurons transfected with the indicated pmirGLO constructs. Values are represented as means±s.d. (n=3; 24 neurons per condition with a 200–250 spine count per cell). (c) Spine density of hippocampal neurons transfected with the indicated pmirGLO constructs. Values are represented as means±s.d. per 10 μm dendritic length (n=3; 24 neurons per condition). Data are presented as the mean of three independent transfections normalized to the empty pmirGLO condition±s.d.
Discussion
The present genetic association results for miRNA-coding genes suggest that miRNAs and their target genes may be implicated in the development of BD. The nominal P-values of SNPs at miRNA loci showed early deviation from the expected null line in the Q–Q plot, and this leftward shift reflects an enrichment of BD-associated SNPs at miRNA loci.
For the nine miRNAs that withstood Bonferroni correction, we additionally calculated the gene-based tests on the basis of the 1000 Genomes LD structure. This analysis revealed nominally gene-based P-values ⩽7.20 × 10−5 for all nine miRNAs, indicating that the results of gene-based tests on the basis of either HapMap phase 2 or 1000 Genomes Project data are highly comparable using our BD GWAS data.
Eight of the nine associated miRNAs were located in a host gene, including the three brain-expressed miRNAs miR-499, miR-708 and miR-1908. Recent studies have reported a high correlation between the expression of a host gene and the resident miRNA.15, 42 Previous authors have hypothesized that this finding may be because of the fact that miRNAs residing in introns are likely to share their regulatory elements and primary transcript with their host gene.24 Some authors point out that host genes and their resident miRNAs may even have synergistic effects, which would have important implications for the fine-tuning of gene expression patterns in the genome.43, 44 On the basis of the present genetic association results, it is impossible to determine whether the association was attributable to the host gene, the miRNA or both. Further analyses are therefore warranted to clarify this, which was beyond the scope of the present analysis. However, the general enrichment of BD-associated SNPs at miRNA loci (Figure 1) and the results of our target gene analyses support the hypothesis that the majority of the associated miRNAs are implicated in BD etiology.
Regional association plots and expression data suggest that the miRNAs miR-499, miR-708 and miR-1908 are the most promising candidates in terms of the development of BD.
The miRNA miR-499 is located in a region on chromosome 20q11 that showed genome-wide significant association in a previous GWAS of BD.45 As miR-499 is located in a region of high LD, which includes the genes GSS, MYH7B and TRPC4AP (Figure 2), further analyses of this chromosomal region are required to refine the association signal.45 However, miR-499 represents a very promising candidate in this region.
MiR-499 regulates apoptotic pathways involving the calcium-dependent protein phosphatase calcineurin.46 A recent study demonstrated an upregulation of miR-499 in the prefrontal cortex of patients with depression.47 In a study of exosomal miRNA expression, miR-499 showed differential expression in the post-mortem brains of BD patients compared with controls.48 When considering a possible pathomechanism, it is important to note that a common SNP (rs3746444) is located in the seed region of the mature miR-499-3p.49 This seed region is crucial for both the recognition of the target sites and the binding of the target genes. The SNP rs3746444 was not among the 2 267 487 SNPs analyzed in our large BD meta-analysis.6 However, rs3746444 achieved a nominally significant P-value of 0.0023 (risk allele: rs3746444-G) in a combined analysis of the seven MooDS samples (2266 patients and 5028 controls),6 which excluded the PGC data set.3 Furthermore, the allele rs3746444-G has been associated with hallucinations and lack of motivation in schizophrenia patients.50 This suggests that this SNP may confer susceptibility to BD by influencing depressive and psychotic endophenotypes. However, it may only partly explain the association signal at this locus.
Our target gene analysis revealed that miR-499 had four significant target genes, including the previously reported genome-wide significant risk gene for psychiatric disorders CACNB2.51
Brain-expressed target genes of miR-499-5p exhibited an enrichment in biological processes related to cerebral development, which might however, at least partly, reflect the fact that our pathway analysis was restricted to brain-expressed genes. In addition, our pathway analysis indicates a potential role of miR-499 in the regulation of the actin cytoskeleton. Interestingly, this pathway has been identified in a previous investigation of differentially and concordantly expressed genes enriched in association signals for schizophrenia and BD.52 Substantial research evidence suggests that the rearrangement of the cytoskeleton is crucial for neuronal cell migration and maturation, neurite outgrowth and maintenance of synaptic density and plasticity.53, 54, 55, 56 These combined data suggest that miR-499 is an interesting candidate for BD pathogenesis.
The miRNA miR-708 is located in the first intron of ODZ4 (odd Oz/ten-m homolog 4, TENM4), which has been reported as a genome-wide significant susceptibility gene for BD.3
A recent study of postpartum psychosis—a disorder that often heralds the incipient onset of BD57—suggested differential expression of miR-708 in the monocytes of affected patients compared with controls.58 In another study, Xu et al.59 demonstrated an altered expression profile for miR-708 in mouse hippocampal neurons and showed that this was mediated by oxidative stress. Another recent study found that miR-708 regulated the expression of neuronatin, which is a membrane protein in the endoplasmic reticulum. Interestingly, the neuronatin-mediated regulation of intracellular Ca2+ levels has been implicated in cell migration and neural induction within embryonic stem cells.60
Our target gene analysis revealed that miR-708 had two significant target genes. These include CREB1 that has previously been identified as a susceptibility gene for major depressive disorder.61, 62, 63 In addition, CREB1 was found to be associated with BD in a recent study of large-scale BD samples64 that included 8403 patients and 11 588 controls of our BD GWAS.6 However, the present pathway analysis provided no strong evidence for an enrichment of biological processes of relevance to psychiatric disorder.
MiR-1908 is located in the first intron of the fatty acid desaturase 1 (FADS1) gene on chromosome 11. To date, few published studies have investigated the function of miR-1908. One recent study implicated miR-1908 as a cancer biomarker.65 A further study found that miR-1908 belonged to a miRNA cluster that downregulates the MARK1 signaling pathway, thus altering cell proliferation and differentiation.66
Pathway analysis results for miR-1908 indicate a potential role of the miRNA-regulated target gene network in key neuronal processes (GO subcategories: neuron projection and nervous system development). As these pathways showed the strongest enrichment, further research into miR-1908 and its regulated network appears to be warranted.
Although initial efforts have been made to elucidate the regulation of miRNA expression,67 the manner in which miRNA expression and processing are regulated remains largely unknown. Given that pri-miRNAs have a length of 100–1000 bp,16 the present study investigated common variants at the miRNA loci and ±20 kb flanking sequences in order to capture possible regulatory regions. However, further analyses of the regulation of miRNA expression by common variants are required to determine whether, and how, the presently described association signals influence the expression levels and function of the implicated miRNAs. The present approach did not allow investigation of SNPs with trans-expression quantitative trait loci (eQTL) effects on miRNAs. As recent studies suggest that ~50% of the identified miRNA eQTLs are trans-eQTLs,68 investigations into the association between miRNA trans-eQTLs and BD are indicated.
The results of the functional analyses of miR-499 and miR-708 in rat hippocampal neurons revealed no major contribution of these miRNAs to the morphogenesis of dendritic spines, which represent the major sites of synaptic contact. However, only the results for miR-499 can be considered robust, as the miR-708 expression construct did not increase miR-708 in primary neurons effectively. Alternative strategies for miR-708 expression, together with miR-499/708 loss-of-function approaches, must be tested before definite conclusions regarding the role of these miRNAs in dendritic spine morphogenesis can be drawn. Moreover, to obtain more comprehensive insights into the potential effects of these miRNAs on synaptic function, future experiments should be complemented by immunocytochemistry analyses of synaptic marker proteins and electrophysiological recordings. Beyond a potential involvement in dendritic spine morphogenesis, these miRNAs could also regulate other aspects of neuronal morphology, such as dendrite arborization or axon growth, which could be tested in future studies.
Conclusion
The results of the present miRNA and target gene analyses suggest that the brain-expressed miRNAs miR-499, miR-708 and miR-1908 may contribute to the development of BD. Further research is warranted to elucidate the involvement of these miRNAs and their downstream pathways in BD.
Acknowledgments
We are grateful to all of the patients and control subjects who contributed to this study. The study was supported by the German Federal Ministry of Education and Research (BMBF) through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under the auspices of the e:Med Programme (grant 01ZX1314A to MMN and SC, grant 01ZX1314G to MR, grant 01ZX1314J to BMM), and through e:AtheroSysMed (Systems medicine of myocardial infarction and stroke, grant 01ZX1313B to BMM). MMN is a member of the DFG-funded Excellence-Cluster ImmunoSensation. MMN also received support from the Alfried Krupp von Bohlen und Halbach-Stiftung. The study was supported by the German Research Foundation (DFG; grant FOR2107; RI908/11-1 to MR; SCHR1136/3-1 to GS; NO246/10-1 to MMN). MG-S received the grant no. 89/2012 from UEFISCDI, Romania. Canadian patients were genotyped within the ConLiGen project (www.ConLiGen.org), with the support of a grant from the Deutsche Forschungsgemeinschaft to MR, MB and TGS (RI 908/7-1). Controls for Germany II were drawn from the Heinz Nixdorf Recall Study (HNR) cohort, which was established with the support of the Heinz Nixdorf Foundation. Recruitment of the Australian sample was supported by an Australian NHMRC program grant (number 1037196). The recruitment of the Canadian patients was supported by a grant from the Canadian Institutes of Health Research #64410 to MA. The study also used data generated by the GABRIEL consortium (controls for the sample Russia). Funding for the generation of these data was provided by the European Commission as part of GABRIEL contract number 018996 under the Integrated Program LSH-2004-1.2.5-1. Post-genomic approaches to understand the molecular basis of asthma aiming at a preventive or therapeutic control and the Wellcome Trust under award 084703. Canadian controls were drawn from the French Canadian study (SLSJ), which was supported in part by the Canada Research Chair Environment and genetics of respiratory diseases and allergy, the Canadian Institutes of Health Research (Operating grant No. MOP-13506) and the Quebec Respiratory Network of the Fonds de recherche en Santé du Québec (FRQS). Polish controls were recruited by the International Agency for Research on Cancer (IARC)/Centre National de Genotypage (CNG) GWAS Initiative. We thank the Bipolar Disorder Working Group of the PGC (PGC-BD) for providing access to the relevant data.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Craddock N, Sklar P. Genetics of bipolar disorder. Lancet 2013; 381: 1654–1662. [DOI] [PubMed] [Google Scholar]
- Nothen MM, Nieratschker V, Cichon S, Rietschel M. New findings in the genetics of major psychoses. Dialogues Clin Neurosci 2010; 12: 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry 2008; 13: 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet 2008; 40: 1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun 2014; 5: 3339. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012; 13: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI Jr., Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I et al. Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry 2014; 71: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron 2009; 64: 303–309. [DOI] [PubMed] [Google Scholar]
- Schratt G. MicroRNAs at the synapse. Nat Rev Neurosci 2009; 10: 842–849. [DOI] [PubMed] [Google Scholar]
- Forstner AJ, Degenhardt F, Schratt G, Nöthen MM. MicroRNAs as the cause of schizophrenia in 22q11.2 deletion carriers, and possible implications for idiopathic disease: a mini-review. Front Mol Neurosci 2013; 6: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Karayiorgou M, Gogos JA. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res 2010; 1338: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res 2010; 124: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry 2011; 69: 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godnic I, Zorc M, Jevsinek Skok D, Calin GA, Horvat S, Dovc P et al. Genome-wide and species-wide in silico screening for intragenic MicroRNAs in human, mouse and chicken. PLoS One 2013; 8: e65165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffioletti E, Tardito D, Gennarelli M, Bocchio-Chiavetto L. Micro spies from the brain to the periphery: new clues from studies on microRNAs in neuropsychiatric disorders. Front Cell Neurosci 2014; 8: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res 2004; 14: 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 2004; 18: 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003; 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11: 597–610. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005; 123: 631–640. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5: 522–531. [DOI] [PubMed] [Google Scholar]
- Meola N, Gennarino VA, Banfi S. microRNAs and genetic diseases. Pathogenetics 2009; 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3' UTR. RNA 2008; 14: 1297–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci 2007; 27: 8546–8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 2007; 318: 271–274. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 2003; 426: 845–849. [DOI] [PubMed] [Google Scholar]
- Geeleher P, Huang SR, Gamazon ER, Golden A, Seoighe C. The regulatory effect of miRNAs is a heritable genetic trait in humans. BMC Genomics 2012; 13: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstner AJ, Basmanav FB, Mattheisen M, Bohmer AC, Hollegaard MV, Janson E et al. Investigation of the involvement of MIR185 and its target genes in the development of schizophrenia. J Psychiatry Neurosci 2014; 39: 386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonte B et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med 2014; 20: 764–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazisar M, Cammaerts S, van der Ven K, Forero DA, Lenaerts AS, Nordin A et al. MIR137 variants identified in psychiatric patients affect synaptogenesis and neuronal transmission gene sets. Mol Psychiatry 2015; 20: 472–481. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang C, Huang J, Yuan C, Hong W, Chen J et al. MiRNA-206 and BDNF genes interacted in bipolar I disorder. J Affect Disord 2014; 162: 116–119. [DOI] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet 2010; 87: 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014; 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziats MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry 2014; 19: 848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol 2009; 11: 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg M, Rodriguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res 2011; 39: W132–W138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005; 11: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter D, Marr C, Krumsiek J, Lang EW, Theis FJ. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genomics 2010; 11: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A. Critical association of ncRNA with introns. Nucleic Acids Res 2011; 39: 2357–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EK, Hamshere M, Forty L, Gordon-Smith K, Fraser C, Russell E et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry 2013; 18: 1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP et al. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med 2011; 17: 71–78. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Zhang H, Rizavi H, Cook EH, Dwivedi Y. Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PLoS One 2014; 9: e86469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banigan MG, Kao PF, Kozubek JA, Winslow AR, Medina J, Costa J et al. Differential expression of exosomal microRNAs in prefrontal cortices of schizophrenia and bipolar disorder patients. PLoS One 2013; 8: e48814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Tong Y, Zhang HM, Wang K, Hu T, Shan G et al. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum Mutat 2012; 33: 254–263. [DOI] [PubMed] [Google Scholar]
- Zou M, Li D, Lv R, Zhou Y, Wang T, Liu J et al. Association between two single nucleotide polymorphisms at corresponding microRNA and schizophrenia in a Chinese population. Mol Biol Rep 2012; 39: 3385–3391. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Xu J, Chen J, Kim S, Reimers M, Bacanu SA et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry 2015; 20: 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer M, Hausott B, Klimaschewski L. Rho GTPases as regulators of morphological neuroplasticity. Ann Anat 2011; 193: 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellenchi GC, Gurniak CB, Perlas E, Middei S, Ammassari-Teule M, Witke W. N-cofilin is associated with neuronal migration disorders and cell cycle control in the cerebral cortex. Genes Dev 2007; 21: 2347–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Curtis I. Functions of Rac GTPases during neuronal development. Dev Neurosci 2008; 30: 47–58. [DOI] [PubMed] [Google Scholar]
- Kreis P, Barnier JV. PAK signalling in neuronal physiology. Cell Signal 2009; 21: 384–393. [DOI] [PubMed] [Google Scholar]
- Jones I, Craddock N. Familiality of the puerperal trigger in bipolar disorder: results of a family study. Am J Psychiatry 2001; 158: 913–917. [DOI] [PubMed] [Google Scholar]
- Weigelt K, Bergink V, Burgerhout KM, Pescatori M, Wijkhuijs A, Drexhage HA. Down-regulation of inflammation-protective microRNAs 146a and 212 in monocytes of patients with postpartum psychosis. Brain Behav Immun 2013; 29: 147–155. [DOI] [PubMed] [Google Scholar]
- Xu S, Zhang R, Niu J, Cui D, Xie B, Zhang B et al. Oxidative stress mediated-alterations of the microRNA expression profile in mouse hippocampal neurons. Int J Mol Sci 2012; 13: 16945–16960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S, McDonnell K, Choi H, Gao D, Hahn M, Joshi N et al. Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration. Cancer Cell 2013; 23: 63–76. [DOI] [PubMed] [Google Scholar]
- Carlezon WA Jr., Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci 2005; 28: 436–445. [DOI] [PubMed] [Google Scholar]
- Maher BS, Hughes HB 3rd, Zubenko WN, Zubenko GS. Genetic linkage of region containing the CREB1 gene to depressive disorders in families with recurrent, early-onset, major depression: a re-analysis and confirmation of sex-specific effect. Am J Med Genet B Neuropsychiatr Genet 2010; 153B: 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace TL, Stellitano KE, Neve RL, Duman RS. Effects of cyclic adenosine monophosphate response element binding protein overexpression in the basolateral amygdala on behavioral models of depression and anxiety. Biol Psychiatry 2004; 56: 151–160. [DOI] [PubMed] [Google Scholar]
- Li M, Luo XJ, Rietschel M, Lewis CM, Mattheisen M, Muller-Myhsok B et al. Allelic differences between Europeans and Chinese for CREB1 SNPs and their implications in gene expression regulation, hippocampal structure and function, and bipolar disorder susceptibility. Mol Psychiatry 2014; 19: 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings-Goss RA, Campbell MC, Tishkoff SA. Global population-specific variation in miRNA associated with cancer risk and clinical biomarkers. BMC Med Genomics 2014; 7: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS et al. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget 2014; 5: 5439–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Ziliak D, Im HK, LaCroix B, Park DS, Cox NJ et al. Genetic architecture of microRNA expression: implications for the transcriptome and complex traits. Am J Hum Genet 2012; 90: 1046–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel C, Deutsch S, Letourneau A, Migliavacca E, Montgomery SB, Dimas AS et al. Identification of cis- and trans-regulatory variation modulating microRNA expression levels in human fibroblasts. Genome Res 2011; 21: 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.