Abstract
Lateral gene transfer is an important mechanism for evolution among bacteria. Here, genome-wide gene insertion and deletion rates are modeled in a maximum-likelihood framework with the additional flexibility of modeling potential missing data. The performance of the models is illustrated using simulations and a data set on gene family phyletic patterns from Gardnerella vaginalis that includes an ancient taxon. A novel application involving pseudogenization/genome reduction magnitudes is also illustrated, using gene family data from Mycobacterium spp. Finally, an R package called indelmiss is available from the Comprehensive R Archive Network at https://cran.r-project.org/package=indelmiss, with support documentation and examples.
Keywords: gene insertion/deletion, indel rates, maximum likelihood, unobserved data
LATERAL gene transfer is an important, yet traditionally underestimated, mechanism for microbial evolution (McDaniel et al. 2010; Treangen and Rocha 2011). Whole gene insertions/deletions, referred to as indels here in the context of lateral gene transfer, can be deduced from examining gene presence/absence patterns on a phylogenetic tree of closely related taxa. Systematic investigation of the rates of such indels can be done via several methods. Parsimony methods can be used (Hao and Golding 2004); however, these are known to underestimate the number of events in phylogeny reconstruction (Felsenstein 2004). Sequence characteristics such as codon usage bias and G+C content have also been investigated in the past, but these are not always reliable (Koski and Golding 2001). Alternatively, phylogenies can also be constructed for individual genes, and a comparison of trees among individual genes can yield insights on the acquisition of foreign genes.
Maximum-likelihood techniques have previously been used to estimate gene indel rates (Hao and Golding 2006; Marri et al. 2006; Cohen and Pupko 2010). Traditionally, such likelihood-based analyses have required that the closely related sequences being investigated have complete genome sequences available. This ensures that no genome rearrangement masks a homolog (Hao and Golding 2006). Here, likelihood-based models are investigated that can also account for potentially unobserved or missing data.
The term “missing” here is used in a loose and informal sense. It is meant to measure the degree to which unobserved data may nevertheless contribute to a taxon’s data set, given the inferred rates from related taxa. Two different kinds of missing data are used here for illustration purposes.
In the first example, consider a taxon or a few taxa that have only subsets of their genome sampled. This could be due to genome degradation, errors in sequencing, errors in assembly, or incomplete next generation sequencing (NGS) studies. In bacterial evolution, genes are continually being inserted or deleted and the goal here is to estimate how much of the data has been missed within this background of continuous gene insertion/deletion.
As a second example, consider an intracellular pathogenic bacteria. It is well known that such species will adapt to their host by deleting unnecessary genes. Here, the missing data allude to the magnitude of genome reduction beyond the normal levels of genome flux. The goal, in this case, is to estimate this reduction while simultaneously estimating phylogenetic insertion/deletion rates. Not accounting for these missing data will bias estimates of indel rates.
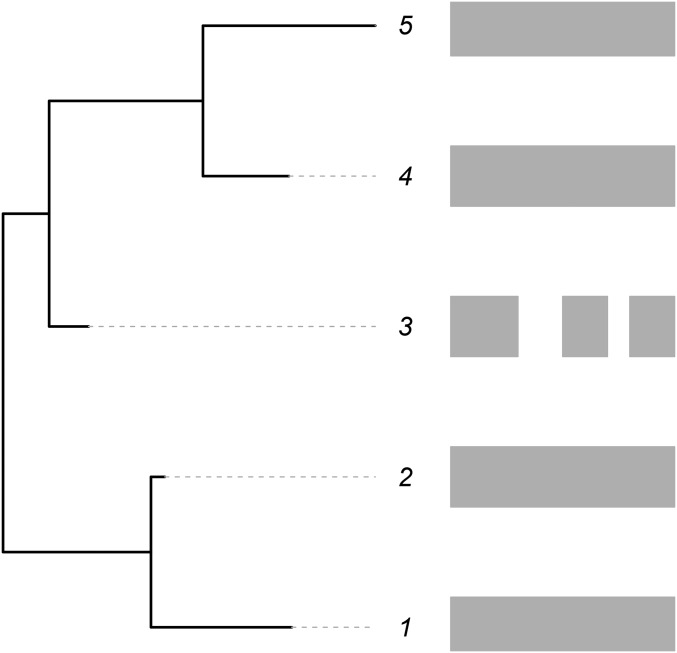
As an illustration, see Figure 1. Here, sequences for coding genes (shaded rectangles) are available for five closely related taxa. Unexpectedly, the data available for the third taxon seem to differ from those for the other taxa, and this taxon appears to be missing some genes that are present in the others. If these are closely related taxa, they should have approximately similar amounts of coding information. Hence, the data recorded for the third taxon seem to be unusual in comparison to related taxa. Indeed, it seems that more deletion has occurred in this taxon relative to the others. In this case, assuming that the third taxon has missing data as discussed above (via either of the two scenarios), modeling of the insertion and deletion rates for genes for all five of the taxa directly would lead to an overestimate of the deletion rate for the entire clade. However, accounting for this unexpected event would provide better estimates for the insertion/deletion rates and at the same time give an estimate of the proportion of missing data. Such a methodology would permit the separation of the confounded effects of missing data from normal gene gain and loss over time. The method cannot determine the reason that the data are missing but can estimate the magnitude. Note that while methods exist for handling missing data, ambiguous states, and sequencing error for nucleotides (Felsenstein 2004; Kuhner and McGill 2014; Yang 2014), this article is the first to propose dealing with missing data using such models on gene family membership data and to illustrate their performance.
Figure 1.
An illustration of the scenarios being modeled. The shaded bars on the right indicate gene content (presence/absence) within the genomes of five species related according to the phylogeny given on the left. The third species is missing some gene blocks that are present in the other species.
As evolutionary rates can vary among different clades or lineages on a tree, the analyses presented also include results from models that relax the assumption of homogeneity of gene insertion and deletion rates across all branches on the phylogeny. Such models can yield unique estimates of insertion and deletion rates for specific clades (or branch and node groupings) chosen based on evolutionary time or prior information. An R package called indelmiss (insertion deletion analysis while accounting for missing data) is provided that allows for efficient fitting of all models discussed (Appendix D). The rest of this article is structured as follows. Materials and Methods includes details on the likelihood calculations and the formulation of a model that incorporates missing data. Results illustrates model performance using simulations and data based on gene phyletic patterns from Gardnerella vaginalis and Mycobacterium spp. Finally, some conclusions and ideas for future work are discussed in the Discussion section.
Materials and Methods
To model gene evolution, a two-state (presence or absence) continuous-time Markov chain is used. Genes are assumed to be inserted or deleted independently of other genes and at constant rates. To eliminate the problem of paralogs, only the presence or absence of gene families is considered in the fashion of Hao and Golding (2004, 2006). Any paralogs are clustered as a single gene family and only one member of a family is retained. The criteria for being considered as belonging to a gene family are given in Results. For the Markov chain, an operational taxonomic unit (OTU) having a gene family present or absent is represented by a 1 or a 0.
Let the instantaneous rates of insertion and deletion be and respectively. Then, the rate matrix can be written as where the rows (and columns) represent presence and absence in the current (future) state, respectively. The transition rate matrix representing the probabilities of a transition from one state to the next can be easily derived (cf. Hao and Golding 2006),
where, as for the rows (and columns) represent presence and absence in the current (future) state, respectively. For example, the probability of gene presence in a descendant OTU given that it was also present in the ancestral OTU is given here by Hence, the values of and measure the rate of gene insertions (deletions) per gene family. This method requires a known phylogenetic tree to be provided as it is not designed to generate a tree. It is also possible to have different indel rates assigned to different parts of the phylogenetic tree if desired.
Evaluating the likelihood on a given phylogenetic tree is straightforward (Felsenstein 1973, 1981). As in Yang (2014), is defined as the conditional probability of observing data at the tips that are descendants of node i, given that the state at node i is Here, can be either gene presence (P) or absence (A). Traditionally, if is observed at node i and 0 otherwise. However, are not restricted to sum to one because they are not probabilities of different outcomes but rather probabilities of the same observation conditional on different events (Felsenstein 2004). Here, the “missingness” of the data is accommodated using this definition. If a gene is recorded as absent at tip i, the vector of conditional probabilities is i.e., the probability of observing gene absence given a gene is truly present (absent) is δ In effect, without such a correction, the conditional probability of observing gene presence given true gene presence is being underestimated. Similarly, if a gene is recorded as present at tip i, the vector of conditional probabilities is i.e., the probability of observing gene presence given a gene is truly present (absent) is Here, δ is the proportion of data that is unexpectedly missing compared to closely related taxa on the phylogenetic tree. Extending this formulation to multiple taxa, a vector of missing proportions can be constructed as for s number of taxa, where Table 1 summarizes the corrections necessary for the conditional probabilities.
Table 1. The proportion of data that is unexpectedly missing in the data for species i compared to closely related taxa on the phylogenetic tree.
| Observed | ||
|---|---|---|
| True | “0” | “1” |
| “0” | 1 | 0 |
| “1” | ||
Then, for a node i with two daughter nodes j and k times and apart, respectively,
This conditional probability vector can be calculated for each node in the tree in a postorder tree traversal fashion. Finally, the probability vector is weighted by the root probability of the states at the root of the tree. This yields the probability of the observed gene family presence/absence data given the tree as
The log-likelihood for the n observed gene family patterns can then be calculated as Typically, when the same rates are being fitted to the entire tree (homogeneous rates), stationarity is assumed. However, the probability of the observed gene family presence can also be estimated at the root within the maximum-likelihood framework. This improves the fit if the process of gain and loss has not reached stationarity (Spencer and Sangaralingam 2009).
However, genes that are not observed as present in any of the taxa, e.g., ancient genes that have been lost, are obviously omitted in the data. This reflects the sampling bias. A correction for the sampling bias (Felsenstein 1992; Lewis 2001; Hao and Golding 2006; Cohen and Pupko 2010) can be imposed such that the probability is conditional on observing the gene in at least one species,
where is the probability of gene h being absent in all taxa. Here, computed by calculating likelihood on the tree of interest using a vector of zeros as observed data, is the same for all genes.
Certain assumptions are made here that can be relaxed in future work. First, genes can be regained after having been deleted. Note that removing this assumption did not improve the results in Hao and Golding (2006). Next, paralogues are excluded in the construction of gene families. Finally, it is assumed that each gene has an equal probability of not being recorded as present; i.e., the missingness is equally prevalent among different sites in the genome.
Four models are used for the analyses here. These four models estimate indel rates (where the deletion rate is the same as the insertion rate, i.e., ), indel rates with proportions of missing data for taxa of interest unique insertion and deletion rates, and unique insertion and deletion rates with proportions of missing data for taxa of interest, respectively. These models are referred to as models 1–4 hereon. This is the first article to investigate fitting and estimating proportions of missing data (models 2 and 4) on gene family membership data. The models were implemented in R (R Core Team 2014) and are available as a package (Appendix D). Parameter estimates and standard errors are obtained from numerical optimization, using PORT routines (Gay 1990) as implemented in the nlminb function in R and the hessian function in package numDeriv (Gilbert and Varadhan 2012), respectively.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and the {\sf R} package available online.
Results
The likelihood-ratio test is known to favor parameter-rich models in large molecular data sets (Yang 2014, p. 146). However, choosing a best-fitted model from a set of models can be done conveniently with penalized likelihood-based model selection criteria. Here, we make use of both the Akaike information criterion (AIC) (Akaike 1973) and the Bayesian information criterion (BIC) (Schwarz 1978) to pick a model with superior fit to the data,
where is the log-likelihood at the maximum-likelihood estimates, N is the number of gene phyletic patterns, and m is the number of parameters estimated for the model.
Simulations
Multiple simulations were conducted to evaluate parameter recovery and judge the efficacy of the model selection criteria.
Simulation set 1:
One hundred random samples of 5000 gene presence/absence phyletic patterns were simulated for six taxa with using the phangorn package (Schliep 2011) in R. For simulating each sample, a tree was randomly generated using the APE package (Paradis et al. 2004) in R with the branch lengths sampled from a beta distribution with shape parameters 1 and 4 (to simulate closely related taxa). Here, missingness was not simulated. Models 1–4 were run on these. All models yielded insertion and deletion rate estimates close to generating values. A Kruskal–Wallis rank sum test for the μ estimates and ν estimates from the four models yielded P-values of 0.645 and 0.781, respectively (note that ANOVA with a Welch correction for homogeneity also does not yield enough evidence to reject the null). This implies that the models fitting a missing data proportion yielded rate estimates that were similar to the estimates from models 1 and 3. The BIC and AIC picked the generating model (model 1) 100 and 76 times, respectively. For models 2 and 4, a missing data parameter was fitted for the OTU at tip 1 in each of the 100 runs. Model 4, which fitted insertion and deletion rates along with a missing data proportion, yielded on average and (median ) across the 100 runs (Table 2).
Table 2. Mean estimates for indel rates and proportion of missing data along with the ranges across 100 runs for simulation set 1.
| Recovered | ||||
|---|---|---|---|---|
| Expected | Model 1 | Model 2 | Model 3 | Model 4 |
| 1.00 (0.94, 1.08) | 1.00 (0.93, 1.06) | 1.00 (0.93, 1.08) | 1.00 (0.93, 1.06) | |
| 1.00 (0.94, 1.08) | 1.00 (0.93, 1.06) | 1.01 (0.90, 1.39) | 1.00 (0.90, 1.39) | |
| — | 0.00 (0.00, 0.02) | — | 0.01 (0.00, 0.03) | |
| Best (AIC) | 76 | 8 | 14 | 2 |
| Best (BIC) | 100 | 0 | 0 | 0 |
No missing data were simulated but possible missing data were estimated for tip 1 (of six tips). The last two rows give the number of times the AIC or the BIC selected the model in the column.
This simulation was repeated with unequal deletion and insertion rates: and respectively. Models 3 and 4 were run on these (Table 3). The BIC and AIC picked the generating model (model 3) 99 and 92 of 100 times, respectively. Both model 3 and model 4 yielded average estimates of μ and ν that were very close to the values used in the simulation. Model 4 estimated an average (for the OTU at tip 1) with a median Welch two-sample two-sided t-tests (not assuming equal variance) comparing μ and ν estimates (from models 3 and 4) yielded P-values of 0.476 and 0.824, respectively.
Table 3. Mean estimates for indel rates and proportion of missing data along with the ranges across 100 runs for simulation set 1.
| Recovered | ||
|---|---|---|
| Expected | Model 3 | Model 4 |
| 0.66 (0.62, 0.71) | 0.66 (0.62, 0.70) | |
| 1.98 (1.80, 2.09) | 1.98 (1.80, 2.09) | |
| — | 0.00 (0.00, 0.02) | |
| Best (AIC) | 92 | 8 |
| Best (BIC) | 99 | 1 |
No missing data were simulated but possible missing data were estimated for tip 1 (of six). Unequal values of μ and ν are used. The last two rows give the number of times the AIC or the BIC selected the model in the column.
Again, this last simulation was repeated with unequal deletion and insertion rates: and respectively. But, after the phyletic patterns were generated, a proportion of genes that were originally present for a single taxon (tip 1; same across runs) were recorded as absent in each run. This proportion was sampled in each simulation from a uniform distribution between 0 and 0.6. Note that the upper limit of 0.6 is arbitrary and used for convenience; a higher upper limit could easily have been used. The BIC and AIC picked the generating model (model 4) 95 and 97 times, respectively (Table 4). As expected, the model selection criteria picked model 3 on those occasions where the estimated missing data proportion was very small and so model 4 did not result in a substantially better fit to the data. Model 3, which cannot account for a missing data proportion, yielded an average and across the 100 runs. On the other hand, model 4 averaged and for tip 1 (median ). Clearly, model 4 yields insertion and deletion rate estimates close to the generating values while providing a reasonable estimate of the proportion of missing data. Table 4 also shows tighter ranges for the estimates for model 4 across the 100 runs. Welch two-sample two-sided t-tests comparing μ and ν estimates (from models 3 and 4) yielded p-values equal to and respectively. A Welch two-sample one-sided t-test comparing deletion rate estimates from models 3 and 4 suggests that model 3 yields, on average, a higher estimate for deletion rate than model 4 (p-value ). This supports our thesis that artificially inflated deletion rates are inferred if missing data are not explicitly accounted for in these indel rate models.
Table 4. Mean estimates for indel rates and proportion of missing data along with the ranges across 100 runs for simulation set 1.
| Recovered | ||
|---|---|---|
| Expected | Model 3 | Model 4 |
| 1.13 (0.63, 5.09) | 0.67 (0.62, 0.70) | |
| 2.77 (1.74, 14.72) | 1.98 (1.80, 2.12) | |
| — | 0.00 ( 0.01) | |
| Best (AIC) | 97 | 3 |
| Best (BIC) | 95 | 5 |
For models 3 and 4, missing data were simulated for tip 1 (of six) with a random proportion sampled from a uniform distribution between 0 and 0.6. The last two rows give the number of times the AIC or the BIC selected the model in the column.
Simulation set 2:
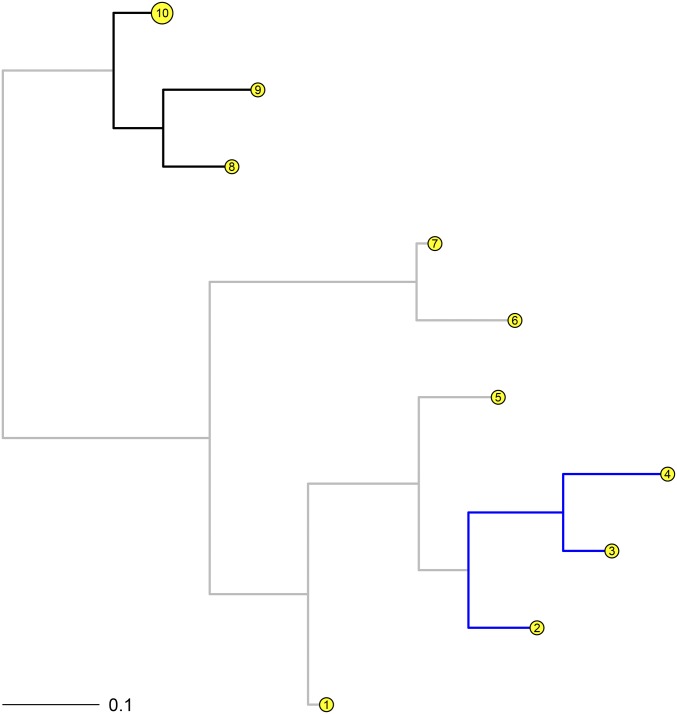
As noted in the Introduction, evolutionary rates can vary among different clades or lineages on a tree and so here, heterogeneous gene insertion and deletion rates among different lineages are simulated and analyzed in the presence of missing data. Five hundred random samples of 5000 gene presence/absence phyletic patterns were simulated for 10 taxa (Figure 2). First, a tree with 10 taxa was generated with branch lengths sampled from a beta distribution with shape parameters 1 and 8 (to simulate closely related taxa). The patterns simulated using the phangorn package were based on base deletion rates sampled from between 0.625 and 1.167 with the insertion rates exploring the interval between 0.875 and 2.500. The branch lengths for the clades with the branches in blue and black in Figure 2 were multiplied by scaling factors sampled independently from the interval respectively. As a result, the deletion (insertion) rates for the blue clade over the 500 samples were generated from Similarly, the deletion (insertion) rates for the black clade over the 500 samples were generated from Missing data were simulated at tips by randomly and independently sampling from a uniform distribution between 0 and 0.6. For the analyses, tips were allowed a missing data proportion.
Figure 2.
Phylogram for the simulated data set 2. Here, colored clades were simulated with different indel rates.
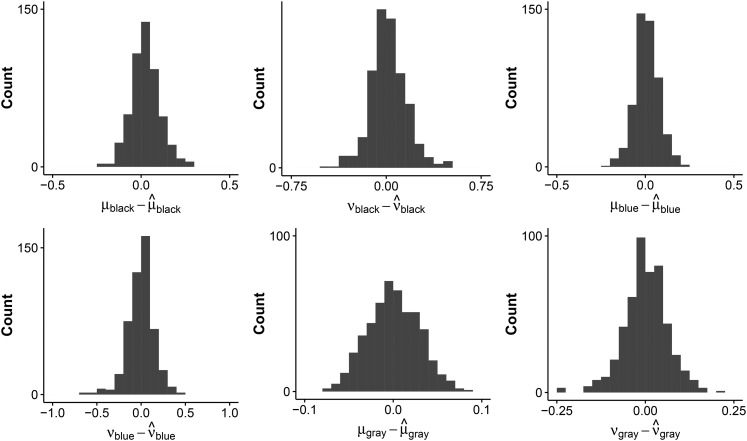
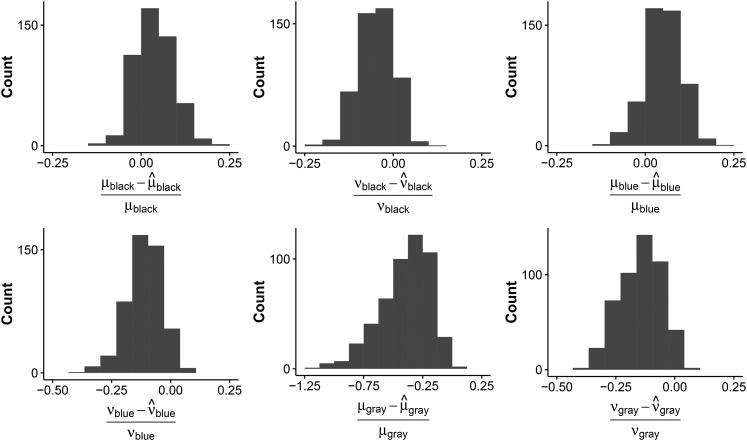
Model 4 was run on all 500 samples. Moreover, the probability of gene family presence was also estimated at the root. Both models, with and without estimating the probability of gene family presence at the root, perform well on average. The parameter estimates are close to the sampled parameters. For example, for the model that did not estimate the probability of gene family presence at the root, the difference in given and estimated parameters for the unique insertion and deletion rates for the three colored clades centered on 0.009 (median 0.006) for the 500 samples (see Figure 3).
Figure 3.
Histograms of the difference between the given and the estimated insertion (ν) and deletion (μ) rates for each clade for simulation set 2. The three color annotations correspond to the colors in Figure 2.
Moreover, the estimated proportions of missing data are also close to the given parameters (Table 5). A note of caution for the reader: If the same 500 random samples are fitted with a missing data proportion for the 10th tip (where 1000 genes are then recorded as absent during data simulation) instead of the 9th tip, the overall estimates do not vary by much. However, if the same samples are fitted with missing data proportions for all tips (1–10), the models become overparameterized and while the recovered parameter estimates are reasonable on average, parameter estimates can deviate wildly, especially for longer trees (see Discussion). In the context of accounting for sequence errors in nucleotide models, Yang (2014) recommends that at least one genome be free of sequence errors. Here, we err on the side of caution, and for the real data analyses, a minimum of three taxa are assumed to not possess any missing data and are modeled without missing data proportions.
Table 5. Ranges for differences between simulated and estimated proportion of missing data for the corresponding taxa over 500 samples from simulation data set 2.
| Tip labels | |||||||
|---|---|---|---|---|---|---|---|
| Difference | 1 | 2 | 3 | 5 | 6 | 8 | 9 |
| ( 0.02) | ( 0.00) | ( 0.02) | ( 0.02) | ( 0.02) | ( 0.00) | ( 0.03) | |
The average difference for each taxon was 0.00. The tree in Figure 2 is used with μ, ν, and sampled from a range of values (see text).
In Appendix E, we test cases where only the lineages with the highest apparent gene data loss are modeled with a proportion of missing data. This is a type of model misspecification. Nevertheless, the results are reasonably good despite the inappropriateness of the test.
Two examples
G. vaginalis data:
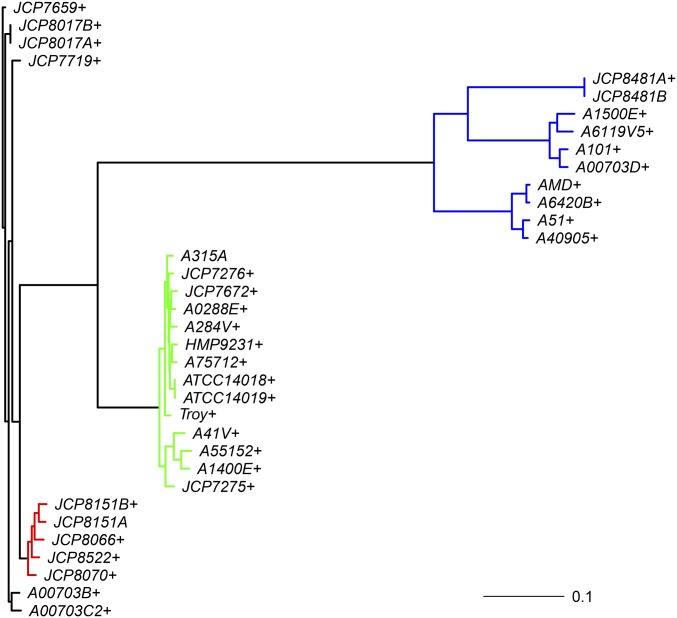
Data containing gene presence/absence measurements on 2036 genes for 35 OTUs of G. vaginalis (Figure 4) are analyzed. G. vaginalis is known to be associated with bacterial vaginosis (Verhelst et al. 2004; Menard et al. 2008). One of these OTUs is a draft genome (labeled Troy), generated from the remains of a fossilized concretion from Troy, Western Anatolia (present-day Turkey). The tree was rooted on the branch leading to JCP7659, using Figtree (Rambaut 2014). These data yielded 746 distinct phyletic patterns of gene presence and absence. Of the total 2036 genes, 558 genes were present in all OTUs. But another 151 genes were present in all OTUs except the ancient genome Troy (see Appendix A).
Figure 4.
Phylogram for the G. vaginalis data. The coloring of the branches corresponds to the grouping for model 4 from method C. The + signs indicate that a missing data proportion was fitted for the associated taxa. Appendix A gives references and strain information for these taxa.
Method A:
Models 1–4 were fitted to these gene phyletic patterns, assuming homogeneity of gene insertion and deletion rates across all branches on the phylogeny. In addition, for models 2 and 4, a missing data parameter was fitted for all taxa except three (cf. Figure 4). The estimate for is This implies that during the evolutionary time period required for one substitution per nucleotide site (on average), an entire gene could possibly have been inserted/deleted three times. Model 2, which fits a proportion of missing data as well, yielded Clearly, estimating the proportion of missing data leads to a lower indel estimate. As in the simulations, this implies that not fitting a proportion of missing data explicitly can lead to artificially inflated indel estimates. Twenty-seven of the missing data proportions were estimated to be between 0 and 0.05, with four taxa between 0.05 and 0.1 and the Troy strain yielding an estimate of Note that this model yielded superior AIC and BIC values to those of model 1. Moreover, the high missing data proportion for the Troy strain cannot be explained away by estimating unique insertion and deletion rates as in model 3. This latter model yielded and suggesting high rates of gene insertion and deletion; however, this model yielded inferior AIC and BIC values to those of model 2. This implies that accounting for possible missing data is more important to the model fit on these data than fitting deletion and insertion rates separately. Of all the models, model 4 results in the best fit to the data in terms of AIC and BIC values. After accounting for the missing data proportions, and Furthermore, note that i.e., the missing data proportion for the Troy strain remained much higher than expected (median estimated missing data proportion is 0.015). The insertion and deletion rates suggest that insertion is occurring at almost twice the rate of deletion after accounting for possible missing data. Accounting for the missing data proportions dramatically reduces the estimate (close to half) for deletion rate between models 3 and 4.
Method B:
Models 2 and 4 were also run assuming that all branches follow the same insertion and deletion rates but that the only taxon of interest with a missing data proportion is the Troy strain. This was done to check whether models 2 and 4 in method A were overparameterized and a simpler model could yield an equivalent fit. Similar results to those observed for method A were obtained. AIC and BIC values indicate that model 2 yields a superior fit to models 1 and 3 from method A. Estimates for from models 2 and 4 were and respectively. However, note that AIC and BIC values for method B models were consistently lower than for equivalent method A models.
Method C:
Models were fitted with different insertion and deletion rates for specific clades (cf. colored branches in Figure 4). Here, the root probability of gene family presence was also estimated. Not doing so, when fitting clade-specific insertion and deletion rates, resulted in inferior AIC and BIC values. The clades with different insertion and deletion rates were chosen based on the major distinct phylogenetic groupings (cf. colored branches in Figure 4). Ideally, this grouping should be informed by biological intuition. In this case, we have used clade membership. In other cases, branch lengths may also be used as a proxy to cluster branches. Preferably, a priori information should be used. As in method A, missing data proportions were also fitted. Model 4 yielded the best fit (Table 6). The probability of gene family presence at the root was estimated to be From this model, 28 of the missing data proportions were estimated to be between 0 and 0.05, with three taxa between 0.05 and 0.1 (ATCC14018, JCP7672, and A6420B all ∼0.063). The estimated missing data proportion for Troy was again much higher than the median missing data proportion (0.015). The estimate of 0.228 corresponds to ∼273 genes. This supports Devault (2014) who noted, based on genome size comparisons, that the true gene content of the G. vaginalis Troy strain may be underrepresented by between 200 and 300 genes. The clade with the Troy strain (green clade) has high estimated rates of insertion with an estimated rate of deletion after accounting for missing data proportions. Estimated rates of deletion and insertion for the red clade are and respectively. The blue clade is found to have a low deletion rate of with insertion rate estimated to be The branches and the taxa connected by black colored branches, on the other hand, have a low estimated rate of insertion with an estimated rate of deletion after accounting for missing data proportions. Moreover, the model selection criteria values suggest that the observed gene presence/absence data for Troy are not explained as well by a gene insertion/deletion model without accounting for potential missing data. Other variations on the model variables in terms of clades or groups of branches with unique rates were also run (results not shown). However, model 4 from method C discussed above yielded the best fit in terms of AIC and BIC values among all models and methods.
Table 6. Clade-specific rate estimates (and standard errors) from model 4.
| Branch grouping | ||||
|---|---|---|---|---|
| Rates | Black | Red | Green | Blue |
| 0.79 (0.05) | 1.89 (0.31) | 0.93 (0.16) | 0.18 (0.03) | |
| 0.91 (0.05) | 7.00 (0.39) | 10.54 (0.34) | 1.78 (0.07) | |
| 0.23 (0.01) | ||||
| 0.59 (0.01) | ||||
The colors of the clades correspond to Figure 4. All but three taxa have the proportion of missing data estimated. Only the estimates for Troy are shown here; the remainder are listed in Appendix A.
Pathogenic bacteria data:
The genus Mycobacterium includes several causative agents of important diseases in humans and animals. For example, Mycobacterium tuberculosis and M. leprae are known to be the causative agents of tuberculosis (Cole et al. 1998) and leprosy (Cole et al. 2001), respectively. M. ulcerans is known to cause Buruli ulcers in humans (Stinear et al. 2007). M. bovis causes tuberculosis in cattle and M. avium causes disease in immunocompromised individuals (Senaratne and Dunphy 2009).
The Mycobacterium genus contains many intracellular bacteria. Among these, M. leprae is a known obligate intracellular parasite. Most other species are facultative with the exception of the recently discovered M. lepromatosis that also causes leprosy (Han et al. 2009; Han and Silva 2014). Obligate intracellular bacteria are typically characterized by smaller genome sizes compared to facultative intracellular or free-living bacteria (Bordenstein and Reznikoff 2005). Furthermore, close to half of the genome of M. leprae is known to be pseudogenes and noncoding regions (Cole et al. 2001). An analysis of gene insertion/deletion rates can shed light on rates of lateral gene transfer while providing a maximum-likelihood estimate of how much coding genetic material has been discarded (or become nonfunctional) given the passage of evolutionary time and the evolutionary relationships between congeneric Mycobacterium species.
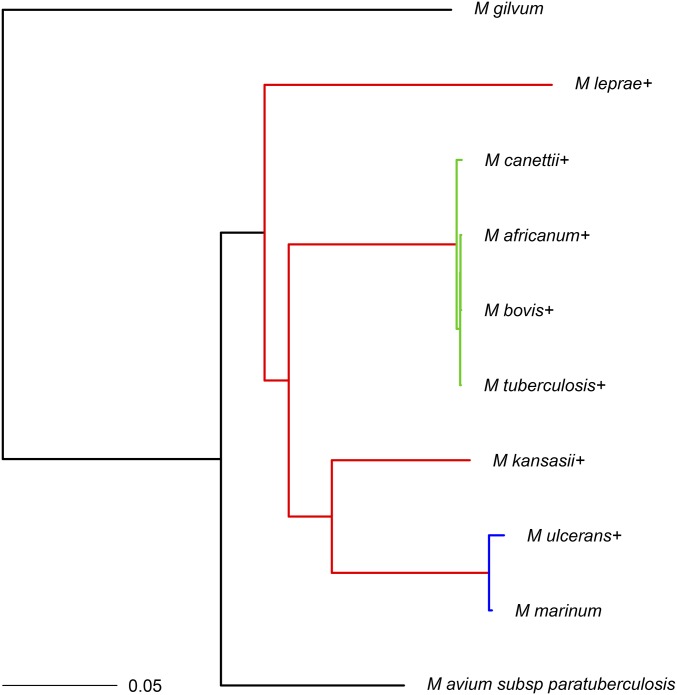
To identify gene families, a procedure similar to that of Hao and Golding (2004, 2006) was followed. This procedure was applied to 10 congeneric Mycobacterium genomes downloaded from the NCBI as outlined in Appendix B. A phylogeny for Mycobacterium species has been proposed in the literature (O’Neill et al. 2015). Details on the construction of the phylogenetic tree are provided in the above-mentioned article. Here, we use a pruned version of the tree (Figure 5) from O’Neill et al. (2015) and analyze the gene family presence/absence data as outlined in Appendix B. Note that the M. leprae genome used in the construction of their tree was different: O’Neill et al. (2015) used M. leprae Br4923 with accession no. NC_011896.1 while we used M. leprae TN (NC_002677.1) (Appendix C).
Figure 5.
Phylogram for the Mycobacterium spp. data from O’Neill et al. (2015). The coloring of the branches corresponds to the grouping for the model that best fitted the data. The + signs indicate that a missing data proportion was fitted for the associated taxa.
Models 1–4 were fitted to the gene phyletic patterns given the tree constructed above, assuming homogeneity of gene insertion and deletion rates across all branches on the phylogeny (method A). Missing data proportions were fitted for all taxa except three (cf. Figure 5). Note that as seen for the G. vaginalis analysis, models 2 and 4 fitted the data better in terms of AIC and BIC values compared to models 1 and 3, respectively. High missing data proportions were estimated for M. leprae and M. ulcerans compared to the median missing data proportions of 0.034 and 0.036 from models 2 and 4, respectively. Model 4, which fits both insertion and deletion rates, yielded estimates of and The high missing data proportion estimate for M. leprae is in line with findings of an extreme case of reductive evolution (Cole et al. 2001; Gómez-Valero et al. 2007).
The assumption of homogeneous indel rates across all branches was relaxed and different models were fitted according to different combinations of clades or branch groupings with unique indel rates. In terms of AIC and BIC values, model 4 with unique rates for each of the colored branches in Figure 5 fitted the best (method B; Table 7). For the best-fitting model, the probability of gene family presence at the root was also estimated (i.e., we do not assume that stationarity has been achieved), which improved the fit of the model. The probability of gene family presence at the root was estimated to be 0.066 (SE = 0.004). Using this model, missing data proportions estimated for M. leprae and M. ulcerans are and (median estimated missing data proportion is 0.024), corresponding to ∼1660 and 869 genes, respectively.
Table 7. Clade-specific rate estimates (and standard errors) from model 4.
| Branch grouping | ||||
|---|---|---|---|---|
| Rates | Black | Red | Green | Blue |
| 0.39 (0.04) | 1.10 (0.06) | 1.67 (0.91) | 0.89 (0.80) | |
| 0.22 (0.02) | 0.38 (0.02) | 3.92 (0.29) | 2.85 (0.22) | |
| 0.54 (0.01) | ||||
| 0.24 (0.01) | ||||
| 0.07 (0.00) | ||||
The colors of the clades correspond to Figure 5. All but three taxa have the proportion of missing data estimated. Only the estimates for M. leprae and M. ulcerans are shown here, the remainder are listed in Appendix B.
The branches in red in Figure 5 had an estimated rate of deletion and estimated rate of insertion The low rate of insertion and relatively low rate of deletion (high compared to the rate of insertion) corresponds well with M. leprae being a niche specialist. Indeed, a recent study found remarkable genomic conservation and that very few large insertions or deletions have taken place in a comparison of ancient and modern M. leprae strains (Schuenemann et al. 2013). Moreover, note also that M. kansasii had the largest number of genes (cf. Table B1) among the Mycobacterium spp. of interest here. The estimated indel rates along with the large genome size of M. kansasii also suggest a slow rate of evolution.
The blue group in Figure 5, on the other hand, yielded and The high missing data proportion estimated for M. ulcerans reconciles well with it evolving from M. marinum and undergoing reductive evolution for niche adaptation (Rondini et al. 2007; Stinear et al. 2007; Demangel et al. 2009). The surprisingly higher estimated insertion rate in the clade with M. ulcerans might be attributed to the relatively higher number of genes that are unique to M. ulcerans (cf. Table B1) and the short branch length leading to M. ulcerans compared to M. leprae, the other species that underwent rapid gene loss. This reflects the relatively older gene inactivation event of M. leprae (Gómez-Valero et al. 2007) vs. the relatively recent divergence of M. ulcerans (and reductive evolution) from M. marinum (Stinear et al. 2000). The branches in green in Figure 5 (M. tuberculosis complex) had an estimated rate of deletion and an estimated rate of insertion Finally, the black group in Figure 5 yielded and Model 3, which does not give an estimate of missing data proportion (and fitted the data poorly in terms of AIC and BIC values), yielded higher deletion rates for the red, blue, and green groups.
Finally, models were also fitted with the same branch grouping topology as above except that the branches leading to M. leprae, and M. ulcerans and M. marinum, were constrained to have the same insertion and deletion rates. This model also yielded similarly high values of estimated missing data. However, this model also gave an inferior fit in terms of AIC and BIC values, suggesting that even though M. leprae and M. ulcerans have both undergone genome reduction, these should not be grouped together given the relative times of emergence of M. leprae and M. ulcerans.
Often, when working with Mycobacterium data, the PE/PPE genes are filtered out. Such a subset of the gene family data set is analyzed in Appendix F. An alternate topology for Mycobacterium spp. is also used for the data set constructed in this section to show sensitivity of the models to the given phylogenetic trees. Results based on this are discussed in Appendix G. The results do not differ qualitatively in either analysis.
Discussion
Here, a maximum-likelihood method to estimate gene insertion/deletion rates with missing data is investigated. This variant allows for much better fitting of gene phyletic patterns when the data observed have unexpected patterns. This can be manifested in two different ways: first, where only a subset of the data was actually detected and second, when the data correctly show much fewer coding genes than closely related taxa. In these cases, the interpretation of the estimated missing data proportion differs. In the former case, the missing data could allude to genome degradation, or errors in sequencing, or issues in gene family creation, etc., while in the latter case, they would allude to the proportion of discarded or nonfunctional genes compared to closely related taxa. Accordingly, two examples based on G. vaginalis and Mycobacterium spp. gene families were analyzed. Simulations were conducted that illustrated good parameter recovery and showed that model selection criteria like AIC and BIC perform well.
An R package that implements all models discussed in this article is also announced (Appendix D). The phylogenetic comparative methods investigated are numerically stable and perform well on average for closely related taxa. Good parameter recovery is shown via simulations. Note that parameter estimates tend to be better and with tighter confidence intervals for shorter trees.
The user is also cautioned to be wary of overparameterization. Even though assigning missing data proportions to all tips on a tree can sometimes result in reasonable answers (especially on short trees, i.e., for very closely related OTUs), our simulations show that parameter estimates can also deviate wildly (especially for longer trees). This occurs because the model uses information from other taxa to determine the highest-likelihood parameters. If all taxa are potentially missing data, then it is difficult to determine the true underlying rates of indels relative to missing data. In our experience, this overparameterization situation is easily observable due to the resulting higher variances in parameter estimates.
To choose between competing models, model selection criteria like the AIC and BIC are utilized here. Overall, the BIC is found to exhibit superior performance to the AIC in picking the generating model in our simulations. On the rare occasions this is not true, it is because the estimated missing data proportion was very small and so the BIC value for the generating model (model 4 with a very small proportion of missing data simulated) did not result in a substantially better fit to the data. Here, the AIC can (rarely) perform better due to the smaller penalty in its functional form.
Future extensions include mixture model generalizations (Spencer and Sangaralingam 2009; Cohen and Pupko 2010) that can allow for heterogeneous rates among gene families. Gamma rate variation can also be incorporated for fitting variable rates among different gene families. Combining the models herein with partition models could also shed light on gene family-category specific pseudogenization, e.g., the proportion of nuclear genes vs. proportion of mitochondrial genes pseudogenized.
Acknowledgments
The authors thank the reviewers and an associate editor for their helpful comments. This material is based on work supported by the National Science Foundation Graduate Research Fellowship Program (DGE-1256259) and the National Institutes of Health National Research Service Award (T32 GM07215) (to T.D.M.). C.S.P. is supported by National Institutes of Health R01AI113287. U.J.D. and G.B.G. were supported by National Sciences and Engineering Research Council grant 140221-10 (to G.B.G.). The authors declare no conflict of interest.
Appendices
Appendix A: G. vaginalis Data
These data are presented in Devault (2014) and in A. M. Devault, T. D. Mortimer, A. Kitchen, H. Kiesewetter, J. M. Enk, G. B. Golding, J. Southon, M. Kuch, A. T. Duggan, W. Aylward, S. N. Gardner, J. E. Allen, A. M. King, G. D. Wright, M. Kuroda, K. Kato, D. E. G. Briggs, G. Fornaciari, E. C. Holmes, H. N. Poinar, and C. S. Pepperell (unpublished results), where the pipeline for creating the gene family and phylogeny is discussed at length. A brief summary of the pipeline follows. It was not feasible to construct a contiguous G. vaginalis genome due to lack of synteny in the ancient reads and high coverage variability. Hence, a de novo approach was used to construct the Troy gene content using reads assembled to the annotated coding regions of extant G. vaginalis strains (Table A1). Coding sequence (CDS) annotations extracted from 34 modern G. vaginalis full and scaffold annotated genome sequences were concatenated into a reference genome. Trimmed paired-end Nod1_1h-UDG reads were then assembled to the set of 34 CDS concatenated references. All paired and unpaired reads that mapped were extracted and assembled using Velvet (Zerbino and Birney 2008). The 1207 contigs generated were analyzed using the nucleotide–nucleotide basic local alignment search tool (BLASTN) (Altschul et al. 1997) against the nonredundant (NR) database to detect any non-G. vaginalis sequences. This step resulted in 20 contigs being excluded because the top hit in BLASTN was Staphylococcus saprophyticus (previously determined to be a primary constituent of the sample), leaving a final set of 1187 G. vaginalis contigs. This set of genomic contigs resulted in 972 genes based on annotation using Prokka (Seemann 2014). Paired-end assembly of Nod1_1h-UDG reads to the final set of contigs was done using Bowtie2 (Langmead and Salzberg 2012) followed by removal of duplicates using SAMtools (Li et al. 2009). OrthoMCL (Li et al. 2003) was used to group protein sequences from the annotations by Prokka, using an all vs. all BLAST. These groups were subsequently filtered for those containing all genomes of interest and such that no proteins were present in more than one copy. The nucleotide sequences for the corresponding genes were individually aligned with MAFFT (Katoh et al. 2002) and trimmed with trimAl (Capella-Gutiérrez et al. 2009). The alignments were concatenated and core SNPs were obtained, excluding regions corresponding to a gap in any strain. Finally, RAxML (Stamatakis 2014) was run on this core SNP alignment to create a phylogenetic tree. The support for the tree was calculated using 100 bootstrap replicates.
The number of genes and the number of unique genes present for each OTU in the gene database constructed with 2036 total number of genes are provided. A total of 558 genes were present in all OTUs. Note that the number of genes here refers to membership in gene families, not the total number of genes for each OTU. The missing data proportions are from method C, model 4 for the G. vaginalis analysis.
Appendix B: Mycobacterium Data
Even though the best BLAST hit is known to not always be the best indicator for the nearest phylogenetic neighbor or even an orthologue (Koski and Golding 2001), building gene families typically relies on using sequence similarity. Briefly, coding sequences were obtained for 10 congeneric Mycobacterium species from NCBI. These are M. leprae, M. ulcerans, M. africanum, M. kansasii, M. tuberculosis, M. marinum, M. canettii, M. avium, M. bovis, and M. gilvum (Table B1). In the literature, M. canettii, M. tuberculosis, M. bovis, and M. africanum are often grouped together as a M. tuberculosis complex. While most genomic sequences were available in the RefSeq database, M. gilvum and M. marinum were not (at the time of data collection: September 2014) and were obtained as GenBank files from NCBI. Insertion sequences, prophages, and transposases were filtered out before the creation of a gene family database. To identify gene families, potential homologues were measured according to sequence similarity, using BLASTP (Altschul et al. 1997). Here, the final alignment was built using the Smith–Waterman algorithm (Smith and Waterman 1981) and soft masking was also employed (cf. Moreno-Hagelsieb and Latimer 2008). Reciprocal hits with expect values <0.05 and match lengths (no gaps) that cover >85% of the query protein length were retained. Protein sequences satisfying these criteria were allocated to the same gene family. Furthermore, all potential paralogues were clustered in the same family under the conservative assumption that these were results of gene duplication and not an insertion of a similar gene. Genes without any identified homologues (according to the above criteria) were searched against the NR database. As above, genes that had hits with expect values <0.05 and match lengths that cover >85% of the query protein length were retained. As in Hao and Golding (2004, 2006), the “single link” method of Friedman and Hughes (2003) was used to group genes into gene families. This means that the allocation of genes to a family is associative in that if a query gene is similar to any of the genes in a gene family, the query gene belongs to the same family. Genes retained from both all-vs.-all BLASTP steps between the Mycobacterium spp. under consideration and from the search against the NR database were used to create a gene family database. Note that this database has been constructed based on sequence similarity and hence should be treated as conservative. For example, a laterally transferred gene that shares similarity to an existing gene might be treated as a paralogue when building the database. Our database construction method would also falter if a single gene in a specific taxon is split into two reading frames by a frameshift or premature stop codon. These genes may not be allocated to the same gene family as they might no longer cover 85% of the query. This is another mechanism of apparent gene gain, distinct from lateral gene transfer.
Appendix C: Gene Presence/Absence Patterns
The gene presence/absence patterns for the G. vaginalis and Mycobacterium spp. along with the respective trees are provided in a supplementary R package.
Appendix D: R Package
An R package named indelmiss is available from the Comprehensive R Archive Network (CRAN). The package can fit four models for estimating gene insertion/deletion rates. These four models estimate indel rates (where the insertion and deletion rates are forced to be equal), indel rates and proportions of missing data for taxa of interest, unique insertion and deletion rates, and unique insertion and deletion rates and proportions of missing data for taxa of interest, respectively (Table D1).
The package can also implement a correction for sampling bias, i.e, genes that were not observed for any of the OTUs. It is possible to simulate gene phyletic patterns for indel analysis as well as to provide custom trees and data. Furthermore, in lieu of weighting presence/absence contributions at the root of the tree by equilibrium frequencies (default behaviour), it is also possible to use equal weighting and user-specified probabilities and to estimate the probabilities at the root, using maximum likelihood. Moreover, clade or evolutionary-grade specific insertion and deletion rates can be estimated as well. The models have been optimized for speed with the recursive likelihood calculation written using the Rcpp package (Eddelbuettel et al. 2011). Note that packages markophylo (Dang and Golding 2016) and DiscML (Kim and Hao 2014) can fit models 1 and 3 (that do not account for missing data) as well.
Installation instructions
The following commands install indelmiss binaries from CRAN:
install.packages(“indelmiss”, dependencies = TRUE, repos = “http://cran.r-project.org”)
Source package from CRAN
install.packages(“indelmiss”, dependencies = TRUE, repos = “http://cran.r-project.org”, type = “source”)
If “-lgfortran” and/or “-lquadmath” errors are encountered on an OS X system, unpack fortran-4.8.2-darwin13.tar.bz2 from http://r.research.att.com/libs/into/usr/local. This issue has cropped up in the past with Rcpp/RcppArmadillo. Also, see the Rcpp FAQs vignette on https://cran.r-project.org/package=Rcpp/index.html.
Prior to installing an R package from source (download from CRAN) that requires compilation on Windows, Rtools needs to be installed from http://cran.r-project.org/bin/windows/Rtools/. Rtools contains MinGW compilers needed for building packages requiring compilation of Fortran, C, or C++ code. The PATH variable should be allowed to be modified during installation of Rtools. If this is not permitted, the PATH variable must be set to include “RTools/bin” and “Rtools/gcc-x.y.z/bin,” where “x.y.z” refers to the version number of gcc, following the installation of Rtools. Then,
install.packages(c(“Rcpp”,”ape”,”phangorn”, “numDeriv”, “testthat”), repos = “http://cran.r-project.org”)
install.packages(“indelmiss_1.0.7.tar.gz”, repos = NULL, type = “source”)
Make sure the version number of indelmiss reflects the latest version on CRAN.
Appendix E: Simulation Set 3
We investigated cases where only the lineages with the highest apparent gene data loss on a given phylogeny are modeled with a proportion of missing data. Similar to simulation set 2, heterogeneous gene insertion and deletion rates among different lineages are simulated and analyzed in the presence of missing data. Five hundred random samples of 5000 gene presence/absence phyletic patterns were simulated for 10 taxa on the same tree (with the different-colored clades following different rates) as used in simulation set 2 (Figure 2). The patterns simulated using the phangorn package followed the same parameters as those followed in simulation set 2. However, here, missing data were simulated at tips by randomly and independently sampling from a uniform distribution between 0.2 (0) and 0.6 (0.15). Hence, for tips 1 and 6, a smaller proportion of missing data were randomly simulated than that for tips 3, 5, and 9. As in simulation set 2, no missing data were simulated at tips 2 and 8. Only tips were allowed a missing data proportion. In effect, only those taxa are modeled with missing data proportions that have the highest apparent gene loss.
Model 4 was run on all 500 samples. The parameter estimates are close to the sampled parameters. We find that the results differ cladewise. For example, tips 1 and 6 in Figure 2 are in the gray clade, in which we saw the highest amount of bias for missing data proportion for tip 5 (Table E1; also note the ranges of the differences for the three tips). Missing data proportions for tips 3 and 9 are estimated very accurately but the missing data proportion for tip 5, although estimated with a reasonable magnitude, is somewhat biased. Figure E1 shows the differences between true and estimated rates, standardized by the true rates. Clearly, results based on the deletion rate for the gray clade (second row, middle column in Figure E1) are worse compared to the others. Deletion rates tend to be overestimated for the gray clade. While these results are intuitive, we urge caution in interpreting these results. Note that this is but one investigation of model misspecification. More work needs to be done to investigate this phenomenon in these phylogenetic comparative models.
Appendix F: PE/PPE Genes
When working with Mycobacterium spp. data, the PE/PPE genes are often left out of the analysis. Here, the gene family data set constructed in Two examples in the main text is filtered to remove such gene families. This is done by removing any gene families with constituent genes that were associated with PE or PPE gene families as evidenced by their annotation in the downloaded genomes. A total of 7683 gene families were left in the database. The best-fitting model from Two examples was rerun here. The results are summarized in Table F1. Clearly, the parameter estimates here are close to the estimates in Two examples.
Appendix G: Alternate Mycobacterium spp. tree
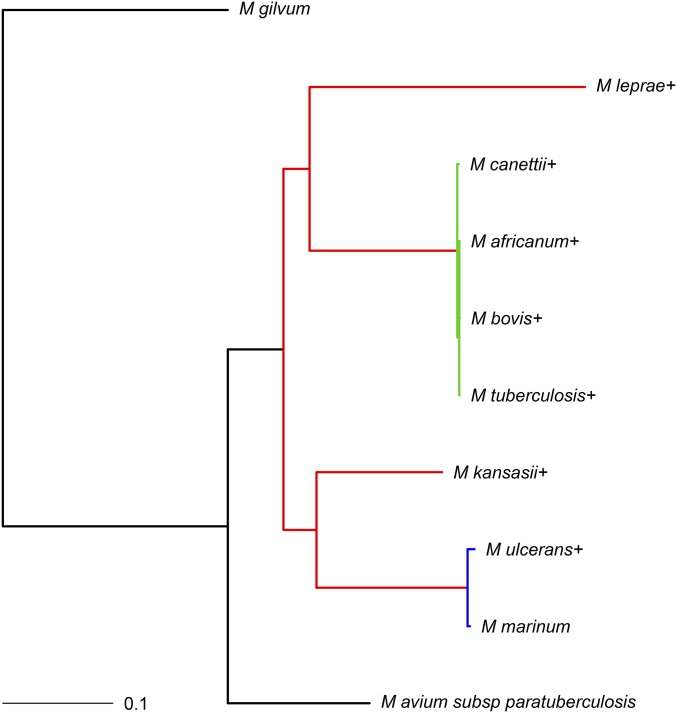
An alternate phylogenetic tree for Mycobacterium spp. was also constructed using MrBayes (Ronquist and Huelsenbeck 2003). This section illustrates the sensitivity of the studied models to the given phylogenetic trees. Multiple sequence alignments of nucleotide sequences of 50 genes found in each taxon were constructed using MAFFT (Katoh et al. 2002). These alignments were then concatenated and provided to MrBayes. A general time-reversible substitution model with gamma-distributed rate variation was run ( generations with 25% burn-in) with M. gilvum specified as the outgroup. A tree (without branch lengths) constructed using the PATRIC bacterial bioinformatics resource center (Wattam et al. 2014) was provided to MrBayes as a starting tree for the algorithm. Ten random perturbations of the start tree were specified in MrBayes. The unrooted result from MrBayes had 100% support for every branch. Using Figtree (Rambaut 2014), this tree was rooted on the branch leading to M. gilvum. The pruned tree used in Two examples differs in the placement of M. leprae (and branch lengths) from the tree constructed using MrBayes. Here, we analyze the gene family data set constructed in Two examples, using the tree constructed using MrBayes in detail. Overall, the relative rates for the different-colored branches (see colored branches in Figure G1) in the best-fitting model are in agreement with the analysis in Two examples.
Again, the best-fitting model from Two examples was rerun here. From this model, M. leprae and M. ulcerans had estimated missing data proportions of and (median missing data proportion is 0.026), respectively. The estimates of 0.547 and 0.215 correspond to ∼1687 and 758 genes, respectively. The branches in red in Figure G1 had an estimated rate of deletion and estimated rate of insertion
The blue group in Figure G1, on the other hand, yielded and The branches in green (M. tuberculosis complex) had an estimated rate of deletion and an estimated rate of insertion Finally, the black group yielded and Here, the probability of gene family presence at the root was estimated to be Overall, the parameter estimates follow mostly the same qualitative trend as in Two examples.
Figure E1.
Histograms of the difference between the given and the estimated insertion (ν) and deletion (μ) rates for each clade for simulation set 2. The three color annotations correspond to the colors in Figure 2.
Figure G1.
Phylogram for the Mycobacterium spp. data constructed using MrBayes with the branch lengths measured in expected substitutions per site. The coloring of the branches corresponds to the grouping for model 4 from method B. The + signs indicate that a missing data proportion was fitted for the associated taxa. Appendix B gives references and strain information for these taxa.
Table A1. NCBI accession numbers with genome sizes for 35 G. vaginalis strains.
| Name | Accession | Size (bp) | Genes | Unique | Missing |
|---|---|---|---|---|---|
| A00703C2 | ADEU00000000 | 1,546,682 | 1165 | 0 | 0.023 |
| A00703B | ADET00000000 | 1,566,055 | 1190 | 1 | 0.018 |
| JCP8070 | ATJK00000000 | 1,475,754 | 1125 | 0 | 0.002 |
| JCP8522 | ATJE00000000 | 1,470,487 | 1093 | 0 | 0.010 |
| JCP8066 | ATJL00000000 | 1,515,433 | 1130 | 0 | 0.004 |
| JCP8151A | ATJI00000000 | 1,556,353 | 1187 | 0 | — |
| JCP8151B | ATJH00000000 | 1,551,237 | 1186 | 0 | 0.000 |
| JCP7275 | ATJS00000000 | 1,560,434 | 1175 | 0 | 0.045 |
| A1400E | ADER00000000 | 1,716,325 | 1295 | 0 | 0.002 |
| A55152 | ADEQ00000000 | 1,643,189 | 1231 | 0 | 0.014 |
| A41V | AEJE00000000 | 1,659,370 | 1223 | 0 | 0.000 |
| Troy | NA | 1,435,761 | 924 | 0 | 0.228 |
| ATCC14019 | NC_014644 | 1,667,350 | 1251 | 0 | 0.006 |
| ATCC14018 | ADNB00000000 | 1,604,161 | 1189 | 0 | 0.062 |
| A75712 | ADEM00000000 | 1,672,968 | 1257 | 0 | 0.016 |
| HMP9231 | NC_017456 | 1,726,519 | 1293 | 0 | 0.010 |
| A284V | ADEL00000000 | 1,650,838 | 1235 | 0 | 0.012 |
| A0288E | ADEN00000000 | 1,708,773 | 1291 | 0 | 0.002 |
| JCP7672 | ATJP00000000 | 1,600,533 | 1194 | 0 | 0.064 |
| JCP7276 | ATJR01000000 | 1,659,589 | 1253 | 0 | 0.035 |
| A315A | AFDI00000000 | 1,653,275 | 1250 | 0 | — |
| A40905 | NC_013721 | 1,617,545 | 1186 | 0 | 0.038 |
| A51 | ADAN00000000 | 1,672,842 | 1208 | 0 | 0.018 |
| A6420B | ADEP00000000 | 1,493,594 | 1092 | 2 | 0.064 |
| AMD | ADAM00000000 | 1,606,758 | 1166 | 1 | 0.007 |
| A00703D | ADEV00000000 | 1,490,797 | 1103 | 0 | 0.002 |
| A101 | AEJD00000000 | 1,527,495 | 1141 | 0 | 0.010 |
| A6119V5 | ADEW00000000 | 1,499,602 | 1114 | 0 | 0.005 |
| A1500E | ADES01000000 | 1,548,244 | 1118 | 2 | 0.023 |
| JCP8481B | ATJF00000000 | 1,569,779 | 1170 | 0 | — |
| JCP8481A | ATJG00000000 | 1,567,375 | 1165 | 0 | 0.012 |
| JCP7719 | ATJO00000000 | 1,559,149 | 1213 | 0 | 0.026 |
| JCP8017A | ATJN00000000 | 1,605,521 | 1283 | 0 | 0.019 |
| JCP8017B | ATJM00000000 | 1,599,351 | 1273 | 0 | 0.027 |
| JCP7659 | ATJQ00000000 | 1,532,641 | 1186 | 0 | 0.037 |
Table B1. NCBI accession numbers with genome sizes for 10 Mycobacterium sequences.
| Name | Accession | Size (bp) | Genes | Unique | Missing |
|---|---|---|---|---|---|
| M. gilvumPYR-GCK | CP000656.1 | 5,547,747 | 5241 | 1239 | — |
| M. lepraeTN | NC_002677.1 | 3,268,203 | 1605 | 204 | 0.543 |
| M. canettiiCIPT 140010059 | NC_015848.1 | 4,482,059 | 3861 | 61 | 0.004 |
| M. africanumGM041182 | NC_015758.1 | 4,389,314 | 3830 | 28 | 0.023 |
| M. bovisAF2122/97 | NC_002945.3 | 4,345,492 | 3918 | 144 | 0.029 |
| M. tuberculosisH37Rv | NC_000962.3 | 4,411,532 | 3906 | 80 | 0.024 |
| M. kansasiiATCC 12478 | NC_022663.1 | 6,432,277 | 5712 | 1112 | 0.000 |
| M. ulceransAgy99 | NC_008611.1 | 5,631,606 | 4160 | 284 | 0.239 |
| M. marinumM | CP000854.1 | 6,636,827 | 5423 | 460 | — |
| M. aviumMAP4 | NC_021200.1 | 4,829,424 | 4326 | 504 | — |
The number of genes represents the number of coding sequences downloaded from NCBI. The number of genes unique to each OTU in the gene database is provided. There are 8034 total number of gene families in the database with a total of 959 genes present in all OTUs. The missing data proportions are from the best-fitting model 4 for the Mycobacterium spp. analysis.
Table D1. Gene insertion/deletion models available in indelmiss.
| Model | δ | |
|---|---|---|
| M1 | E | |
| M2 | E | ✓ |
| M3 | V | |
| M4 | V | ✓ |
Here, and are deletion rate(s), insertion rate(s), and proportion(s) of missing data for taxa of interest, respectively. Here, E implies that parameters and are equal, while V implies that they are free to vary.
Table E1. Averages and ranges for differences between simulated and estimated proportions of missing data for the corresponding taxa over 500 samples from simulation data set 3.
| Tip labels | |||
|---|---|---|---|
| Difference | 3 | 5 | 9 |
| 0.00 ( 0.03) | 0.03 ( 0.08) | 0.00 ( 0.02) | |
The tree in Figure 2 is used with μ, ν, and sampled from a range of values (see text). While missing data are simulated on five tips, only three of those are modeled with a proportion of missing data.
Table F1. Inferred insertion and deletion rates on a subset of the gene family data (without PE/PPE genes).
| Group | ||
|---|---|---|
| Red | ||
| Green | ||
| Blue | ||
| Black |
The branch group colors refer to the sets of branches fitted with unique insertion and deletion rates in Figure G1. This subset had a total of 7683 gene families. Missing data proportions estimated for M. leprae and M. ulcerans are and corresponding to ∼1610 and 765 genes, respectively. The median estimated missing data proportion was 0.023. The probability of gene family presence at the root was estimated to be 0.073 (SE = 0.004).
Footnotes
Communicating editor: J. Lawrence
Literature Cited
- Akaike H., 1973. Information theory and an extension of the maximum likelihood principle, pp. 267–281 in Proceeding of the Second InternationalSymposium on Information Theory, edited by Petrov B. N., Caski F. Springer, Budapest. [Google Scholar]
- Altschul S. F., Madden T. L., Schffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein S. R., Reznikoff W. S., 2005. Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 3: 688–699. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O., Pupko T., 2010. Inference and characterization of horizontally transferred gene families using stochastic mapping. Mol. Biol. Evol. 27: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., et al. , 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Eiglmeier K., Parkhill J., James K. D., Thomson N. R., et al. , 2001. Massive gene decay in the leprosy bacillus. Nature 409: 1007–1011. [DOI] [PubMed] [Google Scholar]
- Dang U. J., Golding G. B., 2016. markophylo: Markov chain analysis on phylogenetic trees. Bioinformatics 32: 130–132. [DOI] [PubMed] [Google Scholar]
- Demangel C., Stinear T. P., Cole S. T., 2009. Buruli ulcer: reductive evolution enhances pathogenicity of Mycobacterium ulcerans. Nat. Rev. Microbiol. 7: 50–60. [DOI] [PubMed] [Google Scholar]
- Devault, A., 2014 Genomics of ancient pathogenic bacteria: novel techniques & extraordinary substrates. Ph.D. Thesis, McMaster University, Hamilton, Ontario, Canada. [Google Scholar]
- Eddelbuettel D., François R., Allaire J., Chambers J., Bates D., et al. , 2011. Rcpp: seamless R and C++ integration. J. Stat. Softw. 40: 1–18. [Google Scholar]
- Felsenstein J., 1973. Maximum likelihood and minimum-steps methods for estimating evolutionary trees from data on discrete characters. Syst. Zool. 22: 240–249. [Google Scholar]
- Felsenstein J., 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17: 368–376. [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 1992. Phylogenies from restriction sites: a maximum-likelihood approach. Evolution 46: 159–173. [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 2004. Inferring Phylogenies. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Friedman R., Hughes A. L., 2003. The temporal distribution of gene duplication events in a set of highly conserved human gene families. Mol. Biol. Evol. 20: 154–161. [DOI] [PubMed] [Google Scholar]
- Gay D. M., 1990. Usage Summary for Selected Optimization Routines. AT&T Bell Laboratories, Murray Hill, NJ. [Google Scholar]
- Gilbert, P., and R. Varadhan, 2012 numDeriv: Accurate Numerical Derivatives (R package version 2012.9-1). Available at: https://cran.r-project.org/web/packages/numDeriv/index.html.
- Gómez-Valero L., Rocha E. P., Latorre A., Silva F. J., 2007. Reconstructing the ancestor of Mycobacterium leprae: the dynamics of gene loss and genome reduction. Genome Res. 17: 1178–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. Y., Silva F. J., 2014. On the age of leprosy. PLoS Negl. Trop. Dis. 8: e2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. Y., Sizer K. C., Thompson E. J., Kabanja J., Li J., et al. , 2009. Comparative sequence analysis of Mycobacterium leprae and the new leprosy-causing Mycobacterium lepromatosis. J. Bacteriol. 191: 6067–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W., Golding G. B., 2004. Patterns of bacterial gene movement. Mol. Biol. Evol. 21: 1294–1307. [DOI] [PubMed] [Google Scholar]
- Hao W., Golding G. B., 2006. The fate of laterally transferred genes: life in the fast lane to adaptation or death. Genome Res. 16: 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T., 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Hao W., 2014. Discml: an R package for estimating evolutionary rates of discrete characters using maximum likelihood. BMC Bioinformatics 15: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski L. B., Golding G. B., 2001. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 52: 540–542. [DOI] [PubMed] [Google Scholar]
- Kuhner M. K., McGill J., 2014. Correcting for sequencing error in maximum likelihood phylogeny inference. G3 4: 2545–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. O., 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50: 913–925. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J., Roos D. S., 2003. Orthomcl: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri P. R., Hao W., Golding G. B., 2006. Gene gain and gene loss in Streptococcus: Is it driven by habitat? Mol. Biol. Evol. 23: 2379–2391. [DOI] [PubMed] [Google Scholar]
- McDaniel L. D., Young E., Delaney J., Ruhnau F., Ritchie K. B., et al. , 2010. High frequency of horizontal gene transfer in the oceans. Science 330: 50. [DOI] [PubMed] [Google Scholar]
- Menard J.-P., Fenollar F., Henry M., Bretelle F., Raoult D., 2008. Molecular quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clin. Infect. Dis. 47: 33–43. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G., Latimer K., 2008. Choosing blast options for better detection of orthologs as reciprocal best hits. Bioinformatics 24: 319–324. [DOI] [PubMed] [Google Scholar]
- O’Neill M. B., Mortimer T. D., Pepperell C. S., 2015. Diversity of Mycobacterium tuberculosis across evolutionary scales. PLoS Pathog. 11(11): e1005257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K., 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- R Core Team , 2014. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Rambaut, A., 2014 FigTree, version 1.4.1 Available at: http://tree.bio.ed.ac.uk/software/figtree/.
- Rondini S., Käser M., Stinear T., Tessier M., Mangold C., et al. , 2007. Ongoing genome reduction in Mycobacterium ulcerans. Emerg. Infect. Dis. 13: 1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P., 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Schliep K. 2011. Phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuenemann V. J., Singh P., Mendum T. A., Krause-Kyora B., Jäger G., et al. , 2013. Genome-wide comparison of medieval and modern Mycobacterium leprae. Science 341: 179–183. [DOI] [PubMed] [Google Scholar]
- Schwarz G., 1978. Estimating the dimension of a model. Ann. Stat. 6: 461–464. [Google Scholar]
- Seemann T., 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- Senaratne R. H., Dunphy K. Y., 2009. Sulphur metabolism in Mycobacteria, in Mycobacterium: Genomics and Molecular Biology, pp. 149–170. edited by Parish T., Brown A. Caister Academic Press, Norfolk, UK. [Google Scholar]
- Smith T. F., Waterman M. S., 1981. Identification of common molecular subsequences. J. Mol. Biol. 147: 195–197. [DOI] [PubMed] [Google Scholar]
- Spencer M., Sangaralingam A., 2009. A phylogenetic mixture model for gene family loss in parasitic bacteria. Mol. Biol. Evol. 26: 1901–1908. [DOI] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T. P., Jenkin G. A., Johnson P. D. R., Davies J. K., 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182: 6322–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T. P., Seemann T., Pidot S., Frigui W., Reysset G., et al. , 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17: 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T. J., Rocha E. P. C., 2011. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7: e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhelst R., Verstraelen H., Claeys G., Verschraegen G., Delanghe J., et al. , 2004. Cloning of 16s rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A. R., Abraham D., Dalay O., Disz T. L., Driscoll T., et al. , 2014. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42: D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2014. Molecular Evolution: A Statistical Approach. Oxford University Press, Oxford. [Google Scholar]
- Zerbino D. R., Birney E., 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and the {\sf R} package available online.