Abstract
Repair of DNA damage is essential to the preservation of genomic stability. During repair of double-strand breaks, several helicases function to promote accurate repair and prevent the formation of crossovers through homologous recombination. Among these helicases is the Fanconi anemia group M (FANCM) protein. FANCM is important in the response to various types of DNA damage and has been suggested to prevent mitotic crossovers during double-strand break repair. The helicase activity of FANCM is believed to be important in these functions, but no helicase activity has been detected in vitro. We report here a genetic and biochemical study of Drosophila melanogaster Fancm. We show that purified Fancm is a 3ʹ to 5ʹ ATP-dependent helicase that can disassemble recombination intermediates, but only through limited lengths of duplex DNA. Using transgenic flies expressing full-length or truncated Fancm, each with either a wild-type or mutated helicase domain, we found that there are helicase-independent and C-terminal-independent functions in responding to DNA damage and in preventing mitotic crossovers.
Keywords: biochemistry, genetics, DNA helicase, homologous recombination, synthesis-dependent strand annealing, ATP activity, crossing over
DNA helicases are a diverse group of enzymes that separate the two strands of duplex DNA. Using the free energy derived from the hydrolysis of a 5ʹ nucleoside triphosphate, generally ATP, the helicase catalyzes the unwinding of duplex DNA to yield single-stranded DNA (ssDNA), a process that is required in replication, transcription, recombination, and repair. Thus, helicases are involved in essentially all metabolic pathways that require the separation of duplex DNA (Brosh 2013; Khan et al. 2015).
Helicases exhibit a diversity of structure and mechanism that may be related to the often unique and specialized roles that these enzymes can play in the cell (Brosh 2013; Daley et al. 2013). Importantly, distinct helicases can interact with specific DNA substrates. For example, during repair of DNA damage, different helicases often act within particular pathways and on unique DNA intermediates that are generated as repair progresses, such as Holliday junctions (HJs) or displacement loops (D-loops). This can be observed in the requirement for helicases to recognize and act on specific DNA structures during the process of double-strand break (DSB) repair via homologous recombination (HR).
DSB repair by HR is a complex process with several key events: resection of the 5ʹ end at the strand break; invasion of the Rad51-coated 3ʹ ssDNA tail into a homologous duplex sequence, generating a D-loop; DNA synthesis primed from the invading 3ʹ end; and resolution into one of either two types of recombination product—crossovers (COs) or noncrossovers (NCOs). The formation of COs during DSB repair in mitotically dividing cells can be hazardous as they can result in loss of heterozygosity and gross chromosomal rearrangements (Lorenz and Whitby 2006; Andersen and Sekelsky 2010). Therefore, prevention of CO pathways through the activation and promotion of NCO pathways is favored in mitotic cells undergoing HR to ensure genomic stability.
To prevent CO generation, helicases can act on several DNA intermediates generated during DSB repair via HR. During HR, an invading DNA strand from the homologous chromosome forms a D-loop as described above. After synthesis, the invading strand can be unwound from the template and annealed to the other resected end, resulting in an NCO; a process known as synthesis-dependent strand annealing (SDSA) (Adams et al. 2003). Alternatively, the displaced strand can anneal to the other resected end, leading to the formation of an entwined structure referred to as a double-Holliday junction (dHJ). The dHJ can be processed by structure-specific endonucleases, possibly giving rise to a CO, or acted upon by a helicase/topoisomerase complex in a process known as dissolution, generating a NCO (Daley et al. 2013). Thus, helicases are essential in the promotion of NCO products either through promotion of D-loop disassembly through SDSA or the dissolution of the dHJ, thereby preventing the formation of potentially deleterious COs during repair (Andersen and Sekelsky 2010; Heyer et al. 2010; Daley et al. 2013).
One family of conserved DNA helicases/translocases whose members are involved in HR regulation is related to archaeal helicase-associated endonuclease for fork-structured DNA (Hef) (Komori et al. 2002; Prakash et al. 2009; Zheng et al. 2011; Lorenz et al. 2012). Pyrococcus furiosus Hef contains a conserved DEAD-box helicase domain and an ERCC4 C-terminal endonuclease domain. Hef functions as a homodimer in cleaving DNA forks and processing HJs into splayed arms, indicating roles for this protein during DNA replication and repair (Komori et al. 2004). The domain structure of Hef is similar to that of the eukaryotic structure-specific endonucleases MUS81 and XPF, but they have inactive helicase domains (Nishino et al. 2005). Conversely, in Fanconi anemia group M (FANCM), the nuclease domain is inactive (Meetei et al. 2005; Ciccia et al. 2007; Ciccia et al. 2008).
Mutations in FANCM cause Fanconi anemia (FA), a hereditary disorder characterized by an increased incidence of cancer, developmental abnormalities, and bone marrow failure (Meetei et al. 2005). A classic hallmark of cells from FA patients is a heightened sensitivity to DNA interstrand cross-linking (ICL) agents, including the chemotherapeutic agents cisplatin and mitomycin C. The primary role of FANCM appears to be to target disrupted replication forks and promote CO avoidance by processing DNA intermediates that occur during DSB repair via HR (Prakash et al. 2005; Prakash et al. 2009; Nandi and Whitby 2012).
The Saccharomyces cerevisiae FANCM ortholog, Mph1, has also been shown to be involved in preventing COs (Prakash et al. 2009), and mph1 mutants show sensitivity to DNA-damaging agents such as ionizing radiation (IR) and methyl methanesulfonate (MMS) (Scheller et al. 2000). Biochemical studies using purified Mph1 show that it is a 3ʹ to 5ʹ DNA helicase capable of unwinding Rad51-coated D-loops (Prakash et al. 2005; Prakash et al. 2009), and that it can process DNA intermediates that form later in repair, including HJs (Prakash et al. 2005; Prakash et al. 2009; Kang et al. 2012). Unwinding of HJs and D-loops has also been observed using the S. pombe ortholog Fml1 (Sun et al. 2008). In contrast, no helicase unwinding activity has been detected for human FANCM (Meetei et al. 2005; Gari et al. 2008). Together, genetic and biochemical studies suggest roles for FANCM and its orthologs in HR that are dependent upon their ability to use ATP hydrolysis to unwind or remodel DNA structures so as to prevent COs (Prakash et al. 2009; Lorenz et al. 2012; Mazón and Symington 2013; Mitchel et al. 2013; Kuo et al. 2014).
FANCM and orthologs may also have roles that are not dependent on catalytic activity. The C-terminal of human FANCM, like its Hef ancestor, has an ERCC4-like endonuclease domain. Although this domain is considered to be catalytically dead, it is involved in protein-protein interactions (Huang et al. 2010; Wang et al. 2013; Yang et al. 2013). Yeast and human FANCM have several motifs in the C-terminal that facilitates interaction with chromatin, additional FA proteins, and repair complexes (Deans and West 2009; Vinciguerra and D’andrea 2009). In human FANCM, two specific motifs (MM1 and MM2) have been shown to allow for interaction with the FA complex and the Bloom syndrome helicase (BLM) complex, which is involved in DSB repair via HR (Deans and West 2009). While these two motifs are not detected in yeast and Drosophila orthologs, there is still the potential for C-terminal interactions with other proteins involved in HR or DNA repair complexes.
A previous genetic study in our laboratory has shown that Drosophila Fancm, like its orthologs, is involved in the prevention of COs (Kuo et al. 2014). This study tested the response of Fancm in CO prevention and response to DNA-damaging agents. To better understand the role of the Fancm helicase activity in directing homologous recombination toward a NCO product, we tested the ability of the purified Fancm helicase to act on HR repair intermediates in vitro. We generated Fancm ATP hydrolysis mutants in vivo to examine the role of the helicase in responding to DNA damage and CO prevention. We also sought to understand the role, if any, of the C-terminal of Fancm in regulating repair events in Drosophila. We generated C-terminal truncations of Fancm in vivo and analyzed how these mutants respond to various DNA-damaging agents and their function in CO prevention.
Here we show that purified Fancm can unwind duplex DNA in a 3ʹ to 5ʹ direction in an ATP-dependent manner. Further, we provide evidence that Fancm can disassemble the HR D-loop intermediate. In vivo work used to study the role of the helicase activity and the C-terminal domain of Fancm reveals that Fancm lacking either helicase activity or the C-terminal is able to prevent some mitotic COs and respond to DNA damage.
Materials and Methods
Expression and purification of Drosophila FANCM
Truncated FANCM, lacking 840 C-terminal residues (FancmΔ), was cloned into pLIC-His- maltose binding protein (MBP) using InFusion cloning (Clontech), with primers FAM1 and FAM2 (Supplemental Material, Table S1) and complementary DNA (Drosophila Genomics Resource Center). The K84M (FancmΔKM) mutation was introduced into FancmΔ using the QuickChange Site-Directed Mutagenesis Kit (Agilent Technologies) with the pLIC-HisMBP-FancmΔ construct as the template and the KMQC primer (Table S1). The protein expression plasmid was maintained in Escherichia coli BL21DE3/pLysS and protein expression was induced by auto-induction (Studier 2005, 2014). Bacterial cultures were grown in 3 liters of ZYM5052 auto-induction medium (Studier 2005) at 25° for 24 hr. Cells were harvested by centrifugation, washed with 20 ml of sodium chloride-Tris-EDTA buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 100 mM NaCl), harvested again by centrifugation and stored as a cell pellet at −80° until use.
Drosophila FancmΔ and FancmΔKM were purified to near homogeneity (Figure S1) using Ni-NTA Resin (QIAGEN, Valencia, CA) and Amylose Resin (New England Biolabs, Beverly, MA) to take advantage of the two affinity tags present on the fusion protein. Cells were lysed in Buffer L (500 mM NaCl, 50 mM Tris-HCl, pH 7.0, 10% glycerol) with 100 mM PMSF, EDTA-free protease inhibitor cocktail, 0.1% triton X-100 and 1 mg/ml lysozyme by incubation at 4° for 45 min and then sonicated to reduce viscosity in 10-sec bursts. Cleared lysate, isolated by centrifugation, was incubated with 3-ml Ni-NTA resin, and 12-column volumes of Buffer L were flowed through the column. Protein was eluted using 300 mM imidazole in Buffer L and protein was detected using a Bradford assay (Bio-Rad, Hercules, CA). Peak fractions were concentrated and the buffer was exchanged with Buffer M (200 mM NaCl, 20 mM Tris-HCl, pH 7.4, 1 mM EDTA) using Amicon Ultra, Ultracel 50K Centrifugal Filters (Millipore, Bedford, MA). The protein was then bound to a 1.5-ml Amylose column, washed with 10-column volumes of Buffer M, and the protein was eluted in Buffer M with 50 mM maltose and 10 mM dextrose. Protein was detected by Bradford assay and dialyzed against storage buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.0, 10% glycerol, 0.1 mM EDTA) and stored at −20°. Protein purity was evaluated using SDS-PAGE.
DNA substrates
Synthetic oligonucleotides (Table S1) used for DNA substrate preparation were PAGE purified by the supplier (Integrated DNA Technologies). Radioactively labeled substrates were prepared by incubating 10 pmol oligonucleotide with 3 μM [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs) at 37° for 50 min, followed by a 20 min incubation at 70° to inactivate the enzyme. Labeled oligonucleotide was then annealed to its complement oligonucleotide in a ratio of 1:1.3 labeled:unlabeled oligonucleotide for fork substrates, or 1:1.3:1.3 labeled:unlabeled oligonucleotide for D-loop substrates. Annealing occurred in Buffer A (50 mM NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM MgCl2) by heating at 95° for 5 min and slowly cooling to room temperature. Hybridized DNA substrates were separated from unannealed oligonucleotide and free [γ32P]ATP using a Sephadex G-50 column (Pharmacia LKB, Piscataway, NJ).
ATPase assays
ATPase reactions were conducted using 212 nM of either FancmΔ or FancmΔKM. Reaction mixtures (20 µl) contained Buffer C (25 mM Tris-HCl, pH 7.5, 20 mM NaCl, 5 mM 2-mercaptoethanol, 10 μg/ml BSA), M13mp18 ssDNA titrated from 0 to 120 nM (nucleotide phosphate), and 3 mM MgCl2. For ATPase reactions that used dsDNA, pET15b plasmid that was cut with the restriction enzyme HpaI, extracted using phenol:chloroform, and precipitated with NaOAC, was used. All reagents except ATP were mixed and allowed to incubate on ice. ATP with a concentration of 3 mM and with trace amounts (∼60 nCi/μl) of [γ-32P]ATP was added to initiate the reaction, and incubation was at 37° for 5 min. Aliquots (5 μl) were removed, and stop solution (5 μl) was added to a final concentration of 17 mM EDTA, 3.4 mM ATP, and 3.4 mM ADP. Of this mixture, 2 μl was spotted onto a cellulose matrix TLC- polyethylene terephthalate plate (Sigma Chemical, St. Louis, MO) and developed in a 0.8 M LiCl/1 M formic acid solution. Plates were allowed to dry, exposed on a phosphor storage screen, and imaged using a Phosphorimager (Amersham, Piscataway, NJ). All images were quantified using ImageQuant software.
Helicase assays
Steady-state helicase unwinding reaction mixtures (20 μl) contained 0.1 nM radiolabeled DNA substrate (Table S1), 25 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 20 mM NaCl, 5 mM 2-mercaptoethanol (βME) and 10 μg/ml BSA. Protein was titrated from a concentration of 0.5 to 212 nM. Reactions were initiated by the addition of 3 mM ATP, incubated at 37° for 15 min and stopped with the addition of 10 μl of helicase stop solution (37.5% glycerol, 50 mM EDTA, 0.3% SDS, 0.5× TBE, 0.1% bromophenol blue). All reactions were resolved on 7.5% nondenaturing polyacrylamide gels containing 0.5× TBE and 0.1% SDS, at room temperature for 2 hr at 180 V. Gels were transferred to Whatman paper, allowed to soak for 30 min in drying buffer (40% methanol, 10% acetic acid, 3% glycerol), and dried for 6 hr using a gel dryer. Dried gels were exposed on a phosphor storage screen and imaged using a Phosphorimager (Amersham). All images were quantified using ImageQuant software.
Fluorescence anisotropy
Reaction mixtures (50 μl) contained 10 nM 5′ fluorescent-labeled 6-carboxyfluorescein DNA substrate (Table S1), 25 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 20 mM NaCl, 5 mM βME, and 10 μg/ml BSA. Fluorescence anisotropy was measured as a function of Fancm concentration from 1 to 212 nM. Reactions were incubated at 25° for 5 min. Fluorescence anisotropy was measured using a Jobin Yvon Horiba Fluorolog-3 fluorometer with a Wavelength Electronics temperature-control box. Labeled dsDNA substrates were excited at 495 nm and emission was measured at 520 nm. Fluorescence anisotropy was calculated using the software provided by the instrument.
Drosophila stocks
Fly stocks were maintained at 25° with standard medium. Fancm0693 is a nonsense mutation previously described in Kuo et al. (2014). Deletion of endogenous Fancm (Fancmdel) was generated using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology (Bassett et al. 2013; Gratz et al. 2013). Oligonucleotides (Integrated DNA Technologies) used for guide RNA (Del1 and Del2; Table S1) were cloned into pU6 Bbs1 chimeric RNA vector, and this was injected into Cas9(X) (BestGene). Fancmdel deletes 3R: 21480913 to 3R: 21487017. In experiments reported here, Fancm mutants were st Fancm0693/Sb Fancmdel, or st Fancm0693/w+transgene Sb Fancmdel, expressed under the endogenous Fancm promoter. Plasmids used for injections of transformants were generated from a PCR-amplified genomic fragment (F1 and F2, Table S1). The K84M mutation was introduced using the QuickChange Site-Directed Mutagenesis Kit (Agilent Technologies) and Primer KMQC (Table S1). The truncated Fancm1-645 construct was generated from the full-length (FL) construct using endogenous MfeI sites. Infusion reaction was used to add the C-terminal and 3ʹ UTR with a PCR reaction and primer FA (Table S1) from the original construct. Fancm1-645K84M was generated in the same way, using the FL Walker A mutant (FLKM) construct. RT-PCR (QIAGEN) was used to determine expression (Figure S1) using primers RT (Table S1).
Mitotic CO assay
Mitotic COs were measured in the male germline as previously described (McVey et al. 2007), using the genetic markers st and Sb for each genotype indicated. At least 20 individual males were assayed for each genotype indicated. Statistical analyses and graphing were done in Prism 6 (GraphPad) using the Kruskal–Wallace test. P-values reported are corrected for multiple comparisons.
DNA damage sensitivity assays
Sensitivity to DNA-damaging agents was determined as previously described (Yıldız et al. 2004). Briefly, an aqueous solution of either MMS or mechlorethamine (HN2) at the indicated concentrations was added to the food during larval feeding. Adults in untreated vials were allowed to mate and lay eggs for 3 days before being transferred into fresh vials, allowed to lay eggs for 2 days, and treated with DNA-damaging agents. For IR, larvae were exposed to gamma rays in an irradiator at 1500 rad. At least 10 biological replications were performed for each genotype indicated. Relative survival was calculated for each vial as the ratio between mutant to control flies in treated vials and normalized to the ratio of mutant to control flies in the untreated vial. Vials with <20 progeny were discarded. Statistical analyses were performed as described above.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results and Discussion
Fancm is an ssDNA-dependent ATPase
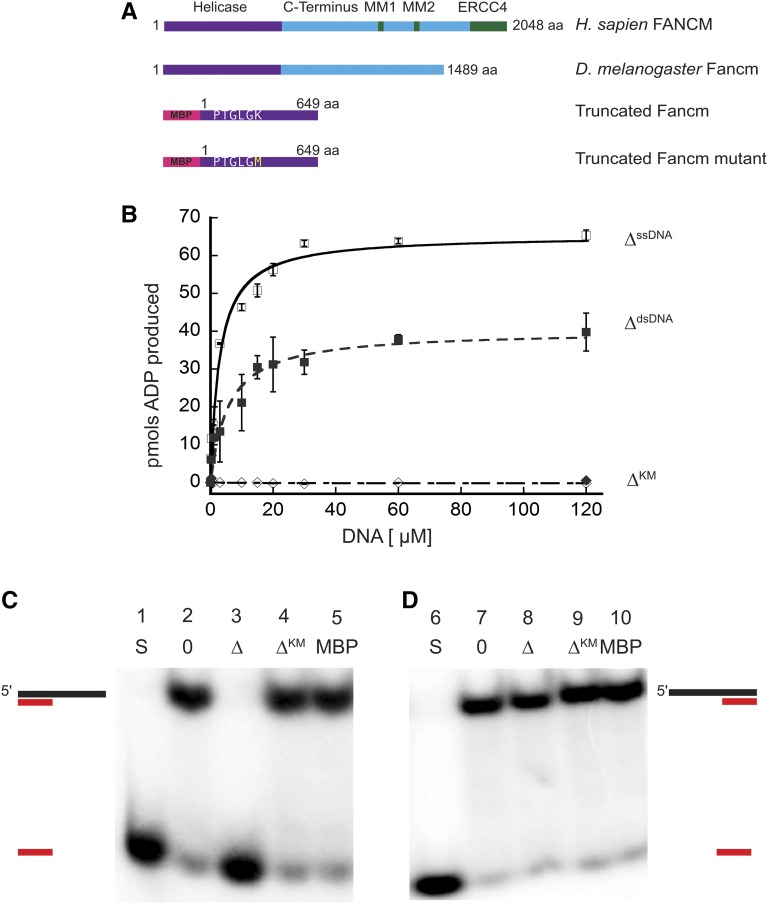
Previous genetic studies of Drosophila Fancm indicated a role for the protein in SDSA and in preventing mitotic COs (Kuo et al. 2014). To further understand roles of Fancm in DNA repair, we investigated the biochemical properties of purified Fancm. Superfamily 2 helicases, including Fancm, are characterized by several conserved motifs, including a Walker A motif that binds the triphosphate tail of ATP and consequently plays a role in ATP hydrolysis (Walker et al. 1982; Koonin 1993). We were unable to express and purify FL Fancm, so a truncated form of Fancm (FancmΔ) and a truncated form with a mutation in the Walker A motif (FancmΔKM) were overexpressed as His6x-MBP tagged proteins (Figure 1A) in E. coli, and each was purified to near homogeneity (Figure S2). This truncation was generated to encompass the entire helicase domain and is based off of purified truncations of the fission yeast ortholog, Fml1 (Sun et al. 2008).
Figure 1.
Drosophila Fancm is a 3′ to 5′ DNA helicase dependent on ATP hydrolysis. (A) Schematic of Fancm. Domains and motifs present in human FANCM are marked. Conserved domains or motifs in D. melanogaster are noted. Truncated forms depicted are with an N-terminal MBP tag. (B) ATP hydrolysis by Fancm. Fancm ATPase activity was examined as a function of DNA concentration using either M13mp18 ssDNA (□⋄) or dsDNA (▪♦) as the DNA cofactor. All reactions were incubated at 37° for 5 min. □ ▪ 212 nM Fancm∆ on ssDNA (∆ssDNA); ⋄♦ 212 nM Fancm∆KM (∆KM). The average values from at least three independent experiments were plotted. Error bars represent SEM (ssDNA) or SD of the mean (dsDNA). (C) Fancm unwinds duplex DNA. Protein (212 nM) was incubated with a 5′ radiolabeled 15-bp partial duplex with a 25-nt 3′ overhang (15/40). (D) Fancm is a 3′ to 5′ DNA helicase. Protein (212 nM) was incubated with a 5′ radiolabeled 15-bp partial duplex with a 25-nt 5′ overhang (−15/40). Lane 1 and 6 (S) are boiled loading controls indicating ssDNA. Lanes 2 and 7 (0) are no-protein controls. Fancm∆ in lane 3 and 8 (∆), Fancm∆KM in lane 4 and 9 (∆KM), and MBP in lane 5 and 10 (MBP). Colored strand represents radiolabeled strand. Substrate oligonucleotides are in Table S1.
We confirmed the ATPase activity of purified FancmΔ and measured several biochemical parameters to characterize this activity (Figure S3). There was no detectable ATP hydrolysis in the absence of DNA, whereas the ATPase activity of the purified protein was higher in the presence of circular M13 ssDNA compared to that of dsDNA, confirming that the protein is a DNA-dependent ATPase (Figure 1B). In addition, we measured the effective rate constant (Keff) and the Vmax for ssDNA (Keff, 2.8 µM; Vmax, 65.3 pmol) and dsDNA (Keff, 5.7 µM; Vmax, 40.1 pmol), under these conditions, further confirming that ssDNA stimulates ATPase activity more strongly than dsDNA. As expected, the FancmΔKM mutant lacked ATPase activity (Figure 1B). Taken together, these results indicate that Fancm is a DNA-dependent ATPase and this activity is dependent on the lysine residue found in the canonical helicase motif I (Figure 1A). ATPase activity stimulated by ssDNA as well as dsDNA has been reported for human FANCM and yeast Fml1, while Mph1 only exhibits ssDNA-dependent activity (Meetei et al. 2005; Prakash et al. 2005; Nandi and Whitby 2012). Fluorescence anisotropy was used to determine if differences in ATPase stimulation were a result of DNA binding (Figure S4). No significant differences in binding to ssDNA vs. dsDNA were detected for the truncated protein.
Fancm is a 3ʹ to 5ʹ DNA helicase
To determine if Drosophila Fancm is active as a helicase, unwinding assays were performed using partial duplex substrates under steady-state conditions. Purified protein was incubated with DNA substrate and the reaction was initiated by the addition of ATP. The wild-type (FancmΔ) helicase completely unwound a 15-bp partial duplex substrate with a 25-bp 3ʹ ssDNA tail (15/40) (Figure 1C, third lane). There was no detectable unwinding of the substrate at an equal concentration of mutant protein FancmΔKM (Figure 1C, fourth lane). When the same reaction was conducted with a 15-bp partial duplex with 25-bp 5ʹ ssDNA tail (−15/40), the wild-type helicase failed to unwind the substrate (Figure 1D). Fluorescence anisotropy was used to determine if there was a difference in binding of FancmΔ to these structures. No significant difference in binding affinity was detected when Fancm was incubated with partial duplex structures with either a 3ʹ or 5ʹ ssDNA tail (Figure S4); indicating that unwinding of the protein is a result of a directional bias and classifies Fancm as a 3ʹ to 5ʹ helicase, consistent with previous work on orthologs (Prakash et al. 2005). These data also support the conclusion that Fancm cannot unwind blunt-ended duplex DNA, as no unwinding of the −15/40 substrate was detected even at longer incubation times.
As shown in Figure S5, no unwinding of the 15/40 substrate was detected when either ATP or MgCl2 were omitted from the reaction. Moreover, unwinding was undetectable when the nonhydrolyzable ATP analog AMP-PNP was substituted for ATP. Taken together, these data indicate that unwinding by the Fancm helicase is dependent upon the ability of the protein to hydrolyze ATP and the FancmΔKM mutant is a “helicase-dead” protein.
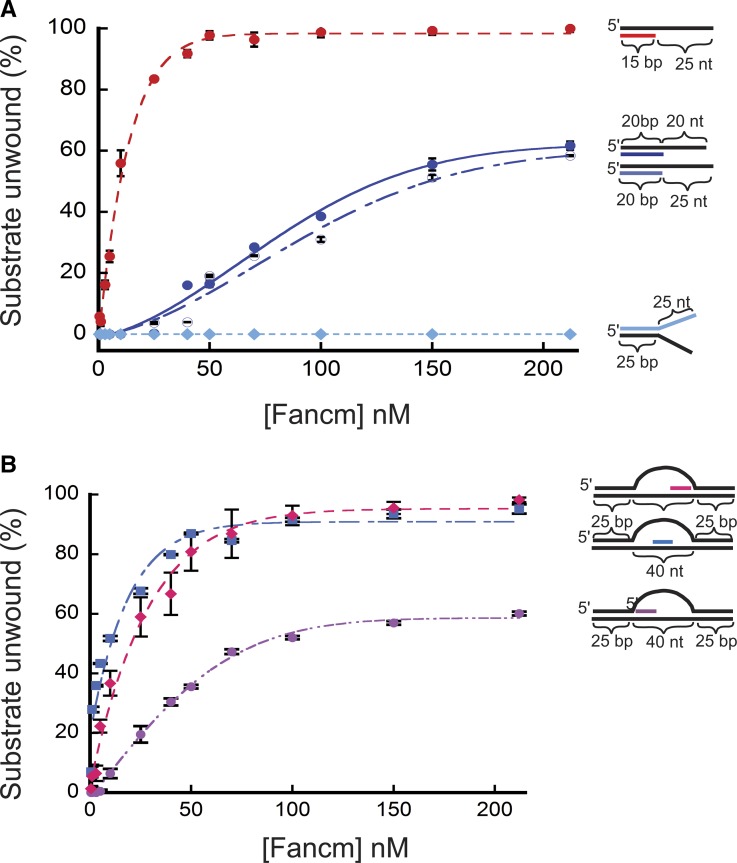
Fancm has limited unwinding capability
Further testing of the helicase activity of Fancm revealed a limit in unwinding longer regions of duplex DNA. A substantial decrease in unwinding activity was observed using a 20-bp partial-duplex substrate with a 20-bp 3ʹ ssDNA tail (20/40). Only 60% of the DNA substrate was unwound by the wild-type helicase at a concentration of protein that unwound all of the 15/40 partial duplex substrate (Figure 2A). To exclude the possibility that the reduced length of the free 3ʹ tail was responsible for this result, we generated a 20-bp partial-duplex substrate with a 25-bp 3ʹ ssDNA tail (20/45). As seen with the 20/40 substrate, Fancm was only able to unwind 60% of the 20/45 substrate (Figure 2A). We also measured unwinding activity using two splayed-arm substrates, one with a 3ʹ single-stranded region of 25 bp, and one with a 3ʹ single-stranded region of 20 bp; both substrates had a 15-bp duplex region. Both substrates were completely unwound, indicating that neither the length of the 3ʹ tail nor the complexity of the substrate affects unwinding (Figure S6A). An additional splayed-arm substrate with a 25-bp duplex region and 25-nt 5ʹ and 3ʹ ssDNA arms was also tested (Figure 2A), with no detectable unwinding.
Figure 2.
Unwinding of partial duplex DNA substrates by Fancm. Helicase reactions were performed as described in Materials and Methods. The indicated concentrations of Fancm were incubated with 0.1 nM of the indicated substrate for 15 min. Colored strand on each substrate represents radiolabeled 5′ strand. Quantitative data from at least three experiments were plotted as the average for each protein concentration. Error bars represent the SEM. Oligonucleotides used to make these substrates can be found in Table S1. (A) Comparison of the fraction of substrate unwound with partial duplex substrates of different duplex lengths. Pink ●, 15-bp duplex region with a 25-nt overhang. Blue ●, 20-bp duplex region with a 20-nt overhang. Blue ○, 20-bp duplex region with a 25-nt overhang; Blue ♦, 25-bp duplex region with 25-nt single-stranded arms. (B) Unwinding of D-loop intermediate substrates by Fancm. Pink ●, front; blue ▪, middle; pink ♦, end. Bubble structures were made using a two 90-nt oligonucleotides with 25 bp of complementary ends with a 40-nt noncomplementary middle (A1/A2). Substrate oligonucleotides are in Table S1.
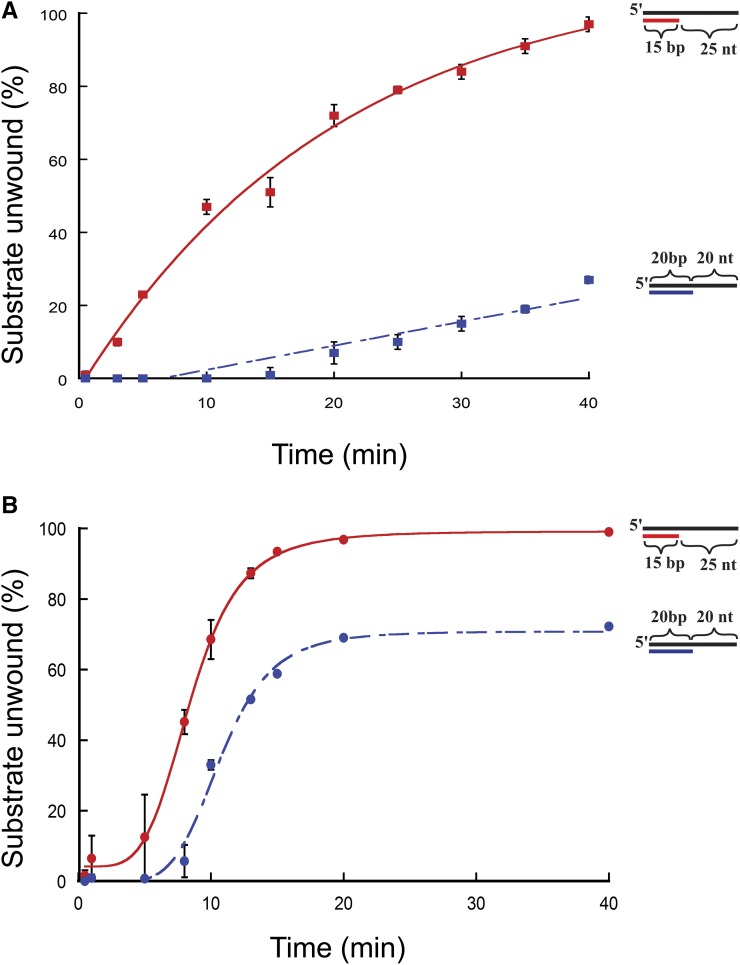
To test if the initial rate of the reaction or the duration of the reaction affected unwinding, unwinding for each substrate was determined using 10 and 150 nM protein at various time points for the 15/40 and 20/40 substrates used (Figure 3, A and B). At 10 nm protein concentration (Figure 3A), FancmΔ was able to fully unwind the 15/40 substrate over the course of the experiment, but could unwind only 37% of the 20/40 substrate. The same is true for reactions using 150 nm protein (Figure 3B). These data indicate that the inability to unwind the 20/40 substrate, even after extended incubation, is not due to the presence of inactive protein in the preparation or to loss of activity during the course of the reaction.
Figure 3.
Time-course unwinding of partial-duplex DNA substrates by Fancm. Helicase reactions were performed as described in Materials and Methods. The indicated concentrations of Fancm were incubated with 0.1 nM of the indicated substrate for the indicated time. Colored strand on each substrate represents radiolabeled 5′ strand. Quantitative data from at least three experiments were plotted as the average for each protein concentration. Error bars represent the SEM. Oligonucleotides used to make these substrates can be found in Table S1. (A and B) Comparison of the fraction of substrate unwound with 10-nm FancmΔ on partial-duplex substrates at the indicated time points of different duplex lengths. (A) Red ▪, 15-bp duplex region with a 25-nt overhang (15/40); blue ▪, 20-bp duplex region with a 20-nt overhang (20/40). (B) Red ●, 15-bp duplex region with a 25-nt overhang (15/40); blue ●, 20-bp duplex region with a 20-nt overhang (20/40). Substrate oligonucleotides are in Table S1.
The inability of Fancm to catalyze unwinding of greater lengths of duplex DNA may result from the use of a truncated protein or missing factors such as post-translational modifications or interacting proteins. Alternatively, the limited helicase may be an intrinsic property of this protein. Similar truncation of Fml1 did not appear to limit the length of duplex unwinding (Nandi and Whitby 2012). Conversely, there are cases where inclusion of sequences predicted to be unstructured, like the C-terminal of Fancm, impedes activity in vitro. An example is Blm helicase, where the FL protein cannot unwind nucleosomal DNA but a protein that has only the conserved helicase domain can (Fujimoto et al. 2009). Thus, while we cannot exclude the possibility that Drosophila Fancm unwinds longer duplex regions in vivo, we hypothesize that the limited unwinding ability we observed is reflective of the protein’s functions in DNA repair.
Based on in vivo data (Kuo et al. 2014), we hypothesized that Fancm may be involved in SDSA by displacing D-loops. In previous yeast studies it was shown that Mph1 can unwind the D-loop structures generated during recombination (Prakash et al. 2009). To test the ability of Fancm to unwind complex structures, we constructed substrates resembling a recombination D-loop intermediate. We incubated Fancm with a 40-nt bubble-like structure containing an “invading” homologous strand in which the duplex region was limited to 15 bp. To determine whether position of the invading strand had an effect on unwinding, the invading strand was positioned at the “front”, “middle”, and “end” of the homologous template strand within the bubble (Figure 2B). Fancm catalyzed robust unwinding of substrates with the invading strand positioned in the middle and at the end of the bubble. However, Fancm unwound the substrate with the invading strand positioned at the front with much lower efficiency. The decrease in substrate unwound as the position of the duplex region is moved is most likely not a result of the length of the duplex region, but rather of the ability of Fancm to access the duplex region. This is possibly a result of a lack of an ssDNA region to which the helicase can bind to initiate unwinding. The middle and end both have regions that mimic the partial duplex with an ssDNA 3ʹ tail. However, the front position substrate does not have a partial duplex with an ssDNA 3ʹ tail, but instead has a 5ʹ ssDNA tail. As shown above, Fancm does not catalyze unwinding of a substrate with a 5ʹ ssDNA tail (see Figure 1D). However, in this more complex substrate, there is an open ssDNA region on the opposite strand of the bubble. Fancm most likely unwinds enough of the duplex arm, generating a 3ʹ tail, and thereby catalyzing unwinding of the invading strand. When a 5ʹ ssDNA tail was added to more closely mimic an invading strand, no difference in unwinding was detected (Figure S6B).
The data presented here indicate that Fancm as a 3ʹ to 5ʹ DNA helicase that is able to unwind up to 20 bp of partial-duplex DNA substrates in an ATP-dependent manner. In addition, the enzyme is able to dissociate short duplex regions in more complex D-loop-like structures. The failure of the protein to unwind longer duplex regions may be the result of in vitro conditions or lack of an important accessory protein or modification. Efforts to detect unwinding of longer duplex regions in the presence of an ssDNA binding protein (E. coli SSB) or under other conditions (e.g., different salt concentration) were unsuccessful.
Mph1 and Fml1 have both been shown to be active helicases unwinding up to 100 bp of duplex DNA (Prakash et al. 2005). On the other hand, human FANCM has been shown to migrate D-loops and HJs, but no unwinding activity has been reported (Meetei et al. 2005; Prakash et al. 2005; Gari et al. 2008; Sun et al. 2008; Zheng et al. 2011). The data presented here suggest that Drosophila Fancm, while similar to both the yeast and human orthologs, is unique. Unlike human FANCM, it is an active helicase, yet we could not detect unwinding of longer duplex regions like Mph1 and Fml1. Although we were not able to detect DNA unwinding by the protein of duplex regions of >20 bp, there may be other factors that can contribute to an increase in helicase activity. The unwinding activity of Mph1 is stimulated by the addition of replication protein A (RPA) (Prakash et al. 2005); although SSB did not stimulate unwinding activity of Fancm, it is possible that Drosophila RPA or other proteins would do so.
Helicase-dead and truncated Fancm are each able to prevent a subset of mitotic COs
The C-terminal region of Fancm in yeast and human orthologs contains motifs that facilitate protein-protein interactions. Human FANCM has a helix-hairpin-helix region in its ERCC4-like domain that allows for association with FAAP24, an interaction that helps stabilize the protein on chromatin (Huang et al. 2010; Wang et al. 2013). The presence of human FANCM of motifs 1 and 2 (MM1 and MM2) allow the interaction of FANCM with the FA core complex and the BLM complex, respectively (Deans and West 2009; Hoadley et al. 2012). It should be noted that Drosophila Fancm has neither the ERCC4 domain nor recognizable MM1 or MM2 motifs. The lack of these sequences is consistent with the fact that the interacting partners associated with these domains, FAAP24, FANCA, and RMI1, are not present in Drosophila (FAAP24 appears to be missing from all insects, FANCA from holometabolous insects, and RMI1 from Schizophoran flies; unpublished observations). Nonetheless, it is likely that the C-terminal region of Fancm, although lacking any recognizable motifs found in orthologs, may contribute to the regulation and function of the protein.
To identify the role the C-terminal has in regards to function of the protein in regulation of HR, we generated transgenic recombinant flies expressing either FL or truncated Fancm. The truncated transgenic recombinant flies are identical to the FancmΔ protein characterized in vitro, except that it lacks the His and MBP tags. We refer to the transgenic truncated Fancm as tr to distinguish it from FancmΔ that has these tags. To investigate the role of the helicase activity in CO prevention and DNA-damage response, we generated transgenic recombinant flies that express either FL or truncated Fancm with either a wild-type helicase domain or the helicase-dead mutation used in vitro (Figure 1A and Figure 3A).
Previous reports on functions of Drosophila Fancm used the nonsense mutation Fancm0693 (L78ter) in trans to Df(3R)ED6058, a 423.1-kb genomic deletion that removes >50 genes (Kuo et al. 2014). To ensure that any mutant phenotype described was an effect of loss of Fancm and not the heterozygous deletion of surrounding genes, we used CRISPR technology to generate a partial deletion of Fancm (Fancmdel) that should result in no protein being produced. The mutants used here were heteroallelic for Fancmdel and Fancm0693. Fancm transcript is undetectable in the null background, indicating that no endogenous Fancm is being produced in flies of this genotype (Figure S1). In all assays performed, no significant difference was observed between the previous results using Df(3R)ED6058 and our experiments using Fancmdel, allowing us to conclude that Fancmdel is a null allele and the previous experiments with the deficiency can be attributed to loss of Fancm. All subsequent experiments reported here used one copy of the transgene in the Fancmdel/Fancm0693 null background, and comparisons are made in reference to the null genotype.
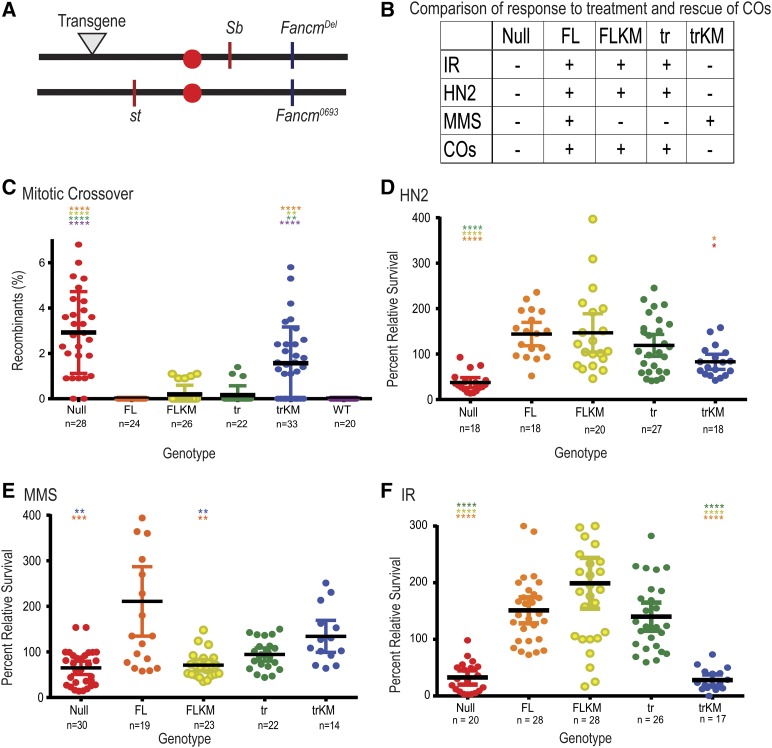
As previously reported, Fancm mutants exhibit a significant increase in the number of spontaneous mitotic COs (Kuo et al. 2014). We assayed spontaneous mitotic COs in the male germline since meiotic COs do not occur in males. COs were scored between the visible markers st and Sb (∼20% of the genome) (Figure 4A). No COs were detected in wild-type males or in Fancm null mutants with the FL transgene (Figure 4C), indicating that the transgenes used are fully functional and that Fancm is indeed involved in preventing mitotic COs. Flies with the truncated Walker A mutant (trKM) transgene showed an increase in COs similar to that of the null mutants. The presence of an active Fancm helicase domain without the C-terminal (tr) reduced the rate of spontaneous mitotic COs to near wild-type levels. Interestingly, the FLKM also reduced CO levels (Figure 4C). The fact that tr and FLKM reduced CO levels to near wild type yet trKM did not, indicates that there are at least two partially independent functions of Fancm in preventing COs: one that requires the helicase activity but not the C-terminal, and another that is dependent on the C-terminal but does not require helicase activity.
Figure 4.
Fancm has genetically separable functions. (A) Map of Fancm null allele (Fancm0693), CRISPR deletion (Fancmdel), transgene landing site (▾), and st and Sb genes. Schematic of transgenes generated are as seen in Figure 1A, without tags. (B) Table comparison of all transgenic Fancm genotypes and null genotype. − indicates no rescue of the null phenotype, + indicates rescue. (C) Spontaneous mitotic CO rates were measured between st and Sb. (D–F) Comparison of sensitivities of Fancm. Plots show the survival of the indicated phenotype relative to wild-type control flies in the same vial after exposure to (D) 0.002% HN2 (0.1 M), (E) 0.05% MMS (3.23 mM), or (F) IR (1500 rad). Red ●, null; Orange ●, FL; yellow ●, FLKM; green ●, tr, light blue ●, trKM; dark blue ●, wild type (WT). Each dot represents one vial, n measures number of vials. Mean percentage of progeny is represented by black horizontal bar. 95% C.I.s are represented by colored error bars. Statistical comparisons were done for Fancm compared to each other genotype. Statistically significant comparisons are indicated above error bars; **** P < 0.0001 by Kruskal–Wallace test, corrected for multiple comparisons.
Although both FLKM and the tr transgenes reduced the levels of spontaneous mitotic COs seen in the null mutant, COs were still detected above wild-type levels. The difference between these genotypes and wild type does not cross the threshold typically used to be considered statistically significant, but we have never detected a CO in any wild-type male (McVey et al. 2007; Kuo et al. 2014; Lafave et al. 2014); hence, we believe the elevation is biologically significant. These data indicate that Fancm must be both FL and catalytically active to prevent all mitotic COs. However, the presence of either the FL helicase-dead protein, or the absence of the C-terminal but with retention of ATPase activity, is sufficient to prevent most mitotic COs.
Separation of function in Fancm’s roles in the response to DNA damage
Since the mitotic CO assay measures spontaneous germline COs detected in progeny, we cannot determine how or when COs are occurring. We therefore cannot provide a mechanistic explanation for the difference between the COs seen in FLKM and the COs in tr. The difference in CO phenotype found among the transgenes of Fancm led us to investigate whether there was a difference in DNA damage response using the transgenes described above.
Previous sensitivity studies using Drosophila Fancm showed that Fancm mutants were sensitive to the cross-linking agent HN2, the alkylating agent MMS, and strand breakage induced by IR (Kuo et al. 2014). These types of damage engender a variety of DNA-repair mechanisms. Since HN2 can induce mono-adducts, intrastrand cross-links, and ICLs, the alkylating agent MMS was tested to distinguish between the role of Fancm in repair of ICLs vs. a broader role in damage repair. While both MMS and HN2 damage can involve replication fork impairment, the cross-links induced by HN2 could lead to DSBs (Muniandy et al. 2010; Clauson et al. 2013). IR was therefore used to determine if Fancm is involved in repair of DSBs.
As previously reported, Fancm null mutants were sensitive to all damaging agents tested. The sensitivities seen in the null mutants are rescued when the FL transgene is present (Figure 4, B and D–F). The FL mutant (FLKM) and tr transgene both rescued sensitivity to HN2 and IR, but not MMS (Figure 4, B and D–F). The trKM transgene failed to rescue sensitivity to HN2 and IR, but did appear to rescue MMS sensitivity (Figure 4, B and D–F). However, progeny with the trKM transgene have delayed developmental timing. If MMS in unstable after addition to the culture medium, it is possible that control larvae ingested food immediately after addition of MMS, whereas trKM larvae ingested food at a later time, after a substantial fraction of MMS was already degraded. Because of this complication, we cannot be confident that the higher relative survival of trKM flies reflects functional capacity of the truncated, helicase-dead Fancm protein.
The difference in rescue among the transgenes in response to damage by HN2 and IR compared to MMS may represent different functional roles of Fancm in various DNA-damage response pathways. The ability of both the FLKM and tr transgene to rescue the sensitivity to HN2 and IR (Figure 4, D and F) is reminiscent of the role of these transgenes in CO prevention (Figure 4C), and again hints at separable functions of Fancm: one that is dependent on the helicase and one that is dependent on the C-terminal. Taken together, we propose that Fancm regulates or participates in multiple DNA-damage responses.
The ability to rescue sensitivity to MMS (and HN2), is representative of Fancm having more than one role in repair. The difference in response to HN2 and MMS in the FLKM and tr may be a result of whether Fancm is functioning with other proteins or independently. Kuo et al. (2014) investigated functions of Fancm that are independent of the FA pathway by comparing phenotypes of Fancm mutants to those of FancI mutants. Differences in sensitivity suggested a role for Fancm in DSB repair that is independent of the FA response.
We hypothesize that Fancm not only acts separately from the FA repair response, but can act both catalytically and noncatalytically in repair of DSBs. A catalytic role in the formation of NCO products might be to unwind short D-loops or to initiate D-loop unwinding. A noncatalytic function might be to recruit HR repair proteins that direct repair toward NCO products, perhaps by extending unwinding of longer D-loops. These dual roles are seen in the FLKM and the tr genotypes. The lack of helicase activity in FLKM prevents it from unwinding D-loops, resulting in some COs being made after these progress to dHJ intermediates. The COs we see in the tr genotype could result from the lack of Fancm recruiting HR repair proteins, such as Blm. Fancm’s proposed interaction with Blm and involvement with HR and D-loop displacement is supported by studies in humans and yeast (Prakash et al. 2005; Nandi and Whitby 2012; Mitchel et al. 2013; Kuo et al. 2014). Blm mutants have more spontaneous mitotic COs than Fancm mutants (McVey et al. 2007; Kuo et al. 2014). Interestingly, Blm Fancm double mutants have the same number of mitotic COs as Fancm single mutants, consistent with the hypothesis that Fancm recruits Blm to prevent COs.
FANCM and its orthologs have been shown to dissociate D-loops, leading to the suggestion that they promote SDSA (Gari et al. 2008; Sun et al. 2008; Prakash et al. 2009). As shown above, Drosophila Fancm is also capable of unwinding D-loop-like structures. Use of a gap-repair assay directly demonstrated roles for both Fancm and Blm in SDSA in Drosophila (Adams et al. 2003; Kuo et al. 2014). Based on the data presented above, we propose that one role of Fancm might be to unwind short D-loops, leaving Blm to unwind D-loops that have been extended by additional synthesis or to continue unwinding those initiated by Fancm. In either case, it is possible that Fancm recruits Blm to D-loops. Unfortunately, Kuo et al. (2014) were unable to conduct this assay in Blm Fancm double mutants and genetic complications prevented us from using our Fancm transgenes in the SDSA assay, so these hypotheses cannot be tested with available reagents.
While there are many similarities found between the orthologs of FANCM, there are also many differences that are informative. Some of these can be explained by the assay conditions, but they may also reveal functional divergence. The inability to detect conserved binding motifs is likely a consequence of coevolution between FANCM and other proteins. Regardless, it is clear through this study, as well as work done in other organisms, that FANCM has a broad and diverse role in DNA maintenance and repair.
Acknowledgments
We thank Danielle Rognstad for assistance with the fluorescence anisotropy and Dorothy Erie and members of the Sekelsky laboratory for helpful discussions. This work was supported by grants from the National Institutes of General Medical Sciences to J.S. under award numbers 5RO1 GM-099890 and 1R35 GM-118127. N.-E.R. was supported by grants from the National Institutes of Health Initiative for Maximizing Student Diversity (5R25 GM-055336), a National Science Foundation Graduate Research Fellowship (DGE-1144081), and The Royster Society of Fellows of The Graduate School at the University of North Carolina at Chapel Hill.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.192534/-/DC1.
Communicating editor: S. K. Sharan
Literature Cited
- Adams M. D., McVey M., Sekelsky J., 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. [DOI] [PubMed] [Google Scholar]
- Andersen S. L., Sekelsky J., 2010. Meiotic vs. mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic vs. mitotic DSB repair are reflected in different pathway usage and different outcomes. BioEssays 32: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P., Liu J. L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R. M., Jr, 2013. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 13: 542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Ling C., Coulthard R., Yan Z., Xue Y., et al. , 2007. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell 25: 331–343. [DOI] [PubMed] [Google Scholar]
- Ciccia A., Mcdonald N., West S. C., 2008. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 77: 259–287. [DOI] [PubMed] [Google Scholar]
- Clauson C., Scharer O. D., Niedernhofer L., 2013. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb. Perspect. Biol. 5: a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley J. M., Niu H., Sung P., 2013. Roles of DNA helicases in the mediation and regulation of homologous recombination. Adv. Exp. Med. Biol. 767: 185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans A. J., West S. C., 2009. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol. Cell 36: 943–953. [DOI] [PubMed] [Google Scholar]
- Fujimoto S., Tomschik M., Zlatanova J., 2009. Does BLM helicase unwind nucleosomal DNA? Biochem. Cell Biol. 87: 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K., Décaillet C., Stasiak A. Z., Stasiak A., Constantinou A., 2008. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell 29: 141–148. [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W. D., Ehmsen K. T., Liu J., 2010. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44: 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley K. A., Xue Y., Ling C., Takata M., Wang W., et al. , 2012. Defining the molecular interface that connects the Fanconi anemia protein FANCM to the Bloom syndrome dissolvasome. Proc. Natl. Acad. Sci. USA 109: 4437–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Kim J. M., Shiotani B., Yang K., Zou L., et al. , 2010. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol. Cell 39: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. H., Munashingha P. R., Lee C. H., Nguyen T. A., Seo Y. S., 2012. Biochemical studies of the Saccharomyces cerevisiae Mph1 helicase on junction-containing DNA structures. Nucleic Acids Res. 40: 2089–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I., Sommers J. A., Brosh R. M., Jr, 2015. Close encounters for the first time: Helicase interactions with DNA damage. DNA Repair (Amst.) 33: 43–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori K., Fujikane R., Shinagawa H., Ishino Y., 2002. Novel endonuclease in Archaea cleaving DNA with various branched structure. Genes Genet. Syst. 77: 227–241. [DOI] [PubMed] [Google Scholar]
- Komori K., Hidaka M., Horiuchi T., Fujikane R., Shinagawa H., et al. , 2004. Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of Archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 279: 53175–53185. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., 1993. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 21: 2541–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H. K., McMahan S., Rota C. M., Kohl K. P., Sekelsky J., 2014. Drosophila FANCM helicase prevents spontaneous mitotic crossovers generated by the MUS81 and SLX1 nucleases. Genetics 198: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafave M. C., Andersen S. L., Stoffregen E. P., Holsclaw J. K., Kohl K. P., et al. , 2014. Sources and structures of mitotic crossovers that arise when BLM helicase is absent in Drosophila. Genetics 196: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A., Whitby M. C., 2006. Crossover promotion and prevention. Biochem. Soc. Trans. 34: 537–541. [DOI] [PubMed] [Google Scholar]
- Lorenz A., Osman F., Sun W., Nandi S., Steinacher R., et al. , 2012. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 336: 1585–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazón G., Symington L. S., 2013. Mph1 and Mus81-Mms4 prevent aberrant processing of mitotic recombination intermediates. Mol. Cell 52: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., Sekelsky J., 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei A. R., Medhurst A. L., Ling C., Xue Y., Singh T. R., et al. , 2005. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37: 958–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K., Lehner K., Jinks-Robertson S., 2013. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet. 9: e1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniandy P. A., Liu J., Majumdar A., Liu S. T., Seidman M. M., 2010. DNA interstrand crosslink repair in mammalian cells: step by step. Crit. Rev. Biochem. Mol. Biol. 45: 23–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S., Whitby M. C., 2012. The ATPase activity of Fml1 is essential for its roles in homologous recombination and DNA repair. Nucleic Acids Res. 40: 9584–9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Komori K., Tsuchiya D., Ishino Y., Morikawa K., 2005. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure 13: 143–153. [DOI] [PubMed] [Google Scholar]
- Prakash R., Krejci L., Van Komen S., Anke Schurer K., Kramer W., et al. , 2005. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 280: 7854–7860. [DOI] [PubMed] [Google Scholar]
- Prakash R., Satory D., Dray E., Papusha A., Scheller J., et al. , 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 23: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller J., Schurer A., Rudolph C., Hettwer S., Kramer W., 2000. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155: 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41: 207–234. [DOI] [PubMed] [Google Scholar]
- Studier F. W., 2014. Stable expression clones and auto-induction for protein production in E. coli. Methods Mol. Biol. 1091: 17–32. [DOI] [PubMed] [Google Scholar]
- Sun W., Nandi S., Osman F., Ahn J. S., Jakovleska J., et al. , 2008. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell 32: 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P., D’andrea A. D., 2009. FANCM: A landing pad for the Fanconi Anemia and Bloom’s Syndrome complexes. Mol. Cell 36: 916–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J., 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1: 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Leung J. W., Jiang Y., Lowery M. G., Do H., et al. , 2013. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol. Cell 49: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Zhang T., Tao Y., Wang F., Tong L., et al. , 2013. Structural insights into the functions of the FANCM-FAAP24 complex in DNA repair. Nucleic Acids Res. 41: 10573–10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yıldız Ö., Kearney H., Kramer B. C., Sekelsky J. J., 2004. Mutational analysis of the Drosophila DNA repair and recombination gene mei-9. Genetics 167: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. F., Prakash R., Saro D., Longerich S., Niu H., et al. , 2011. Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein. DNA Repair (Amst.) 10: 1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.