Abstract
Centromeres are responsible for the correct segregation of chromosomes during mitosis and meiosis. Holocentric chromosomes, characterized by multiple centromere units along each chromatid, have particular adaptations to ensure regular disjunction during meiosis. Here we show by detecting CENH3, CENP-C, tubulin, and centromeric repeats that holocentromeres may be organized differently in mitosis and meiosis of Rhynchospora pubera. Contrasting to the mitotic linear holocentromere organization, meiotic centromeres show several clusters of centromere units (cluster-holocentromeres) during meiosis I. They accumulate along the poleward surface of bivalents where spindle fibers perpendicularly attach. During meiosis II, the cluster-holocentromeres are mostly present in the midregion of each chromatid. A linear holocentromere organization is restored after meiosis during pollen mitosis. Thus, a not yet described case of a cluster-holocentromere organization, showing a clear centromere restructuration between mitosis and meiosis, was identified in a holocentric organism.
Keywords: holocentric chromosomes, CENH3, CENP-C, centromere structure/organization, inverted meiosis
THE centromere is the chromosome site responsible for spindle fiber attachment and faithful chromosome segregation during mitosis and meiosis. In general, every eukaryotic chromosome has a centromere on which the kinetochore complex assembles (Cleveland et al. 2003; Burrack and Berman 2012). In most eukaryotes, centromeric nucleosomes contain CENH3 (also known as CENP-A, a histone H3 variant that replaces canonical H3 at the centromere), and usually spans several hundred kilobase pairs often in association with centromere-specific repeats (Steiner and Henikoff 2015).
Centromere organization and dynamics vary between mitosis and meiosis (Duro and Marston 2015; Ohkura 2015). During mitosis, sister chromatids are held together by centromere cohesion until metaphase. Simultaneous with the disruption of cohesion, sister chromatids are pulled to opposite poles during anaphase. In contrast, during meiosis, sister centromere cohesion is ensured until metaphase II (Ishiguro and Watanabe 2007). The stepwise regulation of cohesion release during meiosis I (MI) and II (MII) is well studied in organisms with one primary constriction per chromosome (monocentric), ensuring the segregation of homologs at MI followed by the segregation of sister chromatids at MII (Duro and Marston 2015).
Contrary to monocentrics, the centromeres of holocentric chromosomes are distributed almost over the entire chromosome length and cohesion occurs along the entire associated sister chromatids (Maddox et al. 2004). Although this does not imply much difference during mitotic divisions, the presence of a holokinetic centromere (holocentromere) imposes obstacles to the dynamics of chromosome segregation in meiosis. Due to their alternative chromosome organization, species with holocentric chromosomes cannot perform the two-step cohesion loss during meiosis typical for monocentric species that requires the distinction between chromosome arms and sister centromeres (Haarhuis et al. 2014). In addition, the extended holocentric kinetochore increases the risk of a stable attachment to microtubules from both poles of the spindle (merotelic attachment), and hence an aberrant segregation of chromosomes may occur. As adaptation, species with holocentric chromosomes have evolved different solutions during meiosis, such as a restricted kinetochore activity, ensuring canonical meiosis order, and “inverted meiosis,” where a reverse order of sister chromatid and homolog separation occurs (see below) (reviewed in Viera et al. 2009 and Cuacos et al. 2015).
In the nematode Caenorhabditis elegans, the chromosomes form a single chiasma per bivalent at one of their termini that has the capacity to form crossovers (COs). The crossover location triggers the redistribution of proteins along the bivalent axis. Kinetochore components uniformly coat each half bivalent but are excluded from the so-called midbivalent region where COs occur (Albertson et al. 1997; Martinez-Perez et al. 2008). Although there are differences between male and female meiosis in regard to microtubule organization and attachment (Shakes et al. 2009; Wignall and Villeneuve 2009; Dumont et al. 2010), in both cases, one pair of sister chromatids faces one spindle pole and the other pair belonging to the second homolog faces the opposite pole. Finally, the sister chromatids remain attached via one chromosome end and become separated during the second meiotic division (Albertson and Thomson 1993; Martinez-Perez et al. 2008; Dumont et al. 2010).
Meiotic adaptations are also observed in other holocentric organisms such as in Heteroptera (Hughes-Schrader and Schrader 1961; Perez et al. 2000; Viera et al. 2009) and Parascaris species (Pimpinelli and Goday 1989), where spindle fibers attach to a restricted kinetochore region at a single chromosome end of each homolog during MI (telokinetic meiosis). Thus, this type of meiosis acts functionally as in monocentric species, since the homologs segregate to opposite poles already during MI. Remarkably, during MII the same telokinetic behavior is observed, although it seems to be random as to which one of the chromosomal termini acquires the kinetic activity in both divisions (Melters et al. 2012). These findings indicate a high plasticity for the centromere/kinetochore structures during meiotic divisions in holocentric organisms.
The holocentric plant species Rhynchospora pubera and Luzula elegans evolved an alternative strategy to deal with meiosis. They are characterized by showing the so-called inverted meiosis (Cabral et al. 2014; Heckmann et al. 2014), which means that sister chromatids segregate already at anaphase I, while the segregation of homologs is postponed to MII (also called postreductional meiosis). They also display individual chromatids at prophase II, indicating the complete loss of sister chromatid cohesion during MI. However, meiosis is not truly inverted in these species; instead, terminal chiasmata result in the exchange of some genetic material between homologous nonsister chromatids. Therefore, the segregating sister chromatids in MI still consist of a part of homologous nonsister chromatids. Furthermore, in contrast to the restriction of the kinetochore activity found in other holocentric species, L. elegans chromosomes show their holocentromere structure and activity also throughout meiosis. They interact individually and biorientate with the meiotic spindle. This results in the separation of partially recombined sister chromatids already during MI. To ensure a faithful haploidization, the homologous nonsister chromatids remain linked at their termini by chromatin threads after metaphase I until metaphase II, and separate at anaphase II. Thus, an inverted sequence of meiotic sister chromatid separation occurs (Heckmann et al. 2014).
Similarly, in the Cyperaceae species R. pubera, multiple spindle fibers amphitelically attach to the sister chromatids during MI (Guerra et al. 2010; Cabral et al. 2014). In mitosis, the chromosomes exhibit a linear holocentromere organization comprising CENH3-containing centromere units enriched in centromeric tandem repeats (named Tyba) and centromeric retroelements. In interphase, the holocentromeres dissociate and form multiple individual centromere units. During chromosome condensation toward mitotic metaphase, the centromeric units rejoin and form a linear distinct longitudinal centromere within a groove to ensure faithful chromosome segregation (Marques et al. 2015).
In contrast to mitotic chromosomes, where the (peri)centromeric histone marker H2AThr120ph is also organized in a linear manner, a dispersed distribution was found in meiotic chromosomes of R. pubera. In addition, multiple CENH3 patches enhanced at the poleward chromosome surface of highly condensed metaphase I bivalents were reported (Cabral et al. 2014). This suggests a deviating centromere organization during meiosis of R. pubera. However, the lack of simultaneous CENH3 and tubulin localization in other meiotic stages, and the limited microscopic resolution hampered a comprehensive characterization of the kinetic activity and centromere organization throughout the meiosis of this species.
In order to shed more light in the meiotic centromere organization of Rhynchospora, we labeled centromeric proteins (CENH3 and CENP-C), repeats (Tyba), and α-tubulin, and applied super-resolution microscopy to characterize the organization and dynamics of R. pubera holocentromeres throughout meiosis. We report that the holocentromere organization of R. pubera differs significantly between mitosis and meiosis, providing the identification of a not yet reported meiotic centromere organization among eukaryotes.
Materials and Methods
Plant material
R. pubera (Vahl) Boeckler plants were cultivated under humid conditions at the Experimental Garden of the Laboratory of Plant Cytogenetics and Evolution (Recife, Brazil) and in a greenhouse at the Leibniz Institute of Plant Genetics and Crop Plant Research (Gatersleben, Germany).
Identification and validation of the CENP-C gene and generation of CENP-C antibodies
The CENP-C gene was identified in silico by BLAST search from the transcriptome data of R. pubera (accession no. PRJEB9645, http://www.ebi.ac.uk/ena/). For the validation of expression, semiquantitative RT-PCR was performed with DNase-treated total RNA isolated from root, leaf, and anther tissue of R. pubera using the SpectrumTM Plant Total RNA kit (Sigma, St. Louis, MO). The complementary DNA (cDNA) was synthesized from 1 µg of total RNA using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, MA). PCR reactions were performed with the primer sequences: forward 5′-AATGACTTCACCCTCACCCG-3′ and reverse 5′-CCTTCTTGCAGGTCTAGTGC-3′. Primers for the constitutively expressed GAPDH gene (Banaei-Moghaddam et al. 2013), GAPDH-F CAATGATAGCTGCACCACCAACTG and GAPDH-R CTAGCTGCCCTTCCACCTCTCCA, were used as control for applying equal amounts of genomic DNA (gDNA) and cDNA. The amplified fragments were cloned into the StrataClone PCR Cloning Vector pSC-A-amp/kan (Agilent Technologies, Santa Clara, CA). Sequences of 10 randomly selected clones revealed only one CENP-C variant (GenBank, accession no. KU516997).
The peptide VRVKSFMSDEHADLIAKLAK was used to generate R. pubera CENP-C-specific (RpCENP-C) polyclonal antibodies. Peptide synthesis, immunization of rabbits, and peptide affinity purification of antisera were performed by LifeTein (http://www.lifetein.com).
Phylogenetic analysis of plant CENP-C sequences
Reference IDs for all CENP-C sequences used in this study are available in Supplemental Material, Table S2. A multiple alignment of protein sequences encoding the entire CENP-C sequences was generated using MAFFT (Katoh and Standley 2013) and refined manually. Evolutionary analyses were conducted with IQ-TREE (Nguyen et al. 2015) using ultrafast bootstrap (Minh et al. 2013). Phylogenetic history was inferred using the maximum likelihood method using the best-fit model: JTT + I + G4 acquired automatically with IQ-TREE. The analysis involved 30 protein sequences. The alignments and trees are stored in the CyVerse Data Store and can be downloaded at http://de.iplantcollaborative.org/dl/d/5EA7332F-1374-4BED-BD4C-BC69E41CA530/RpCENPC.rar.
Immunostaining of somatic and meiotic cells
Immunostaining for CENH3 and CENP-C was performed as described in Cabral et al. (2014) with some modifications. Anthers were fixed in ice-cold 4% paraformaldehyde in 1× PBS buffer pH 7.5 (1.3 M NaCl, 70 mM Na2HPO4, 30 mM NaH2PO4) for 1 hr and 30 min and squashed in a drop of the same buffer. Alternatively, anthers were treated with colchicine 0.05% for 24 hr at 10° and fixed as above. Tapetum cells of young anthers were used for the preparation of mitotic cells. Then, the slides were washed in 1× PBS and blocked with 3% BSA for 30 min at 37°. The antibodies used were rabbit anti-RpCENH3 (Marques et al. 2015) directly labeled with FITC and rabbit anti-RpCENP-C, both diluted 1:500 in 1% BSA in 1× PBS. The detection of anti-RpCENP-C was done with goat anti-rabbit-Cy3 (Sigma, no. F9887), diluted 1:200 in 1× PBS containing 1% BSA. The slides were counterstained with 2 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield H-1000.
For the simultaneous detection of CENH3 and tubulin, the anthers were fixed in methanol:acetic acid (3:1) for 2–24 hr. Then, the anthers were rinsed three times in 1× PBS for 5 min, and the pollen mother cells were squeezed out from the anthers and squashed in a drop of 1× PBS. The coverslips were removed after freezing in liquid nitrogen. Then, the material was washed in 1× PBS and immersed in 1× citric buffer for 1 min in a microwave at 800 W. Afterward, the slides were immediately washed in 1× PBS. The immunostaining procedure was conducted as described above. The CENH3 antibodies were detected by Cy3 or Alexa 488 goat anti-rabbit antibodies. Mouse anti-α-tubulin antibodies (Sigma, no. T5168) were diluted 1:50 in 1× PBS containing 1% BSA and detected with Alexa 488 or Cy5 goat anti-mouse antibodies (Thermo Fisher Scientific, no. A-11001) diluted 1:100 in the same buffer.
CENH3 fluorescence measurements
Comparative CENH3 fluorescence signal intensity measurements of degenerative and functional cells in pseudomonads were performed using ImageJ 1.48s (http//:imagej.nih.gov/ij). For measurements, we used the previously described formula (Gavet and Pines 2010) as follows: whole-cell signal = sum of the intensity of the pixels for one cell; background signal = average signal per pixel for a region selected just beside the cell; whole-cell signal corrected = whole-cell signal − (number of pixels for the selected cell = surface selected × background).
FISH
The centromere-specific repeat Tyba was detected with directly labeled 5′-Cy3 oligonucleotides (Tyba1: ATTGGATTATACATGGTAATTACGCATATAAAGTGCAAATAATGCAATTC; Tyba2: ACAGATTCTGAGTATATTTGAGCATTTCAAGCGATTTTGCATT) (Eurofins MWG Operon, http://www.eurofinsdna.com). FISH after immunostaining was performed as described by Ishii et al. (2015). 45S rDNA FISH was performed as described in Sousa et al. (2011).
Wide-field and super-resolution fluorescence microscopy
Wide-field fluorescence images were recorded using a Leica DM5500B microscope equipped with a Leica DFC FX camera and a deconvolution system. To analyze the substructures and spatial arrangement of immunosignals and chromatin beyond the classical Abbe/Raleigh limit (super-resolution), spatial structured illumination microscopy (3D-SIM) was applied using a Plan-Apochromat 63×/1.4 oil objective of an Elyra PS.1 microscope system and the software ZEN (Carl Zeiss, Thornwood, NY). The images were captured using 405-, 488-, 561-, and 642-nm laser lines for excitation and the appropriate emission filters and merged using the ZEN software (Weisshart et al. 2016). The Imaris 8.0 (Bitplane) software was used to measure the degree of colocalization between CENH3 and CENP-C. Briefly, after loading ZEN SIM image stacks, the Imaris colocalization tool was applied. An automatic threshold defined by the point spread function (PSF) was calculated and used to establish a new colocaliztion channel originating from the CENH3 and CENP-C channels. This resulting channel contains the channel statistics, including the degree of colocalization (in percentage) and the Pearson’s and Mander’s coefficients. Imaris 8.0 was also applied to produce 3D movies.
Data availability
Antibodies are available upon request. Sequence data are available at GenBank and the accession numbers are listed in the Materials and Methods section and in Table S2. The GenBank accession no. of R. pubera CENP-C is KU516997.
Results
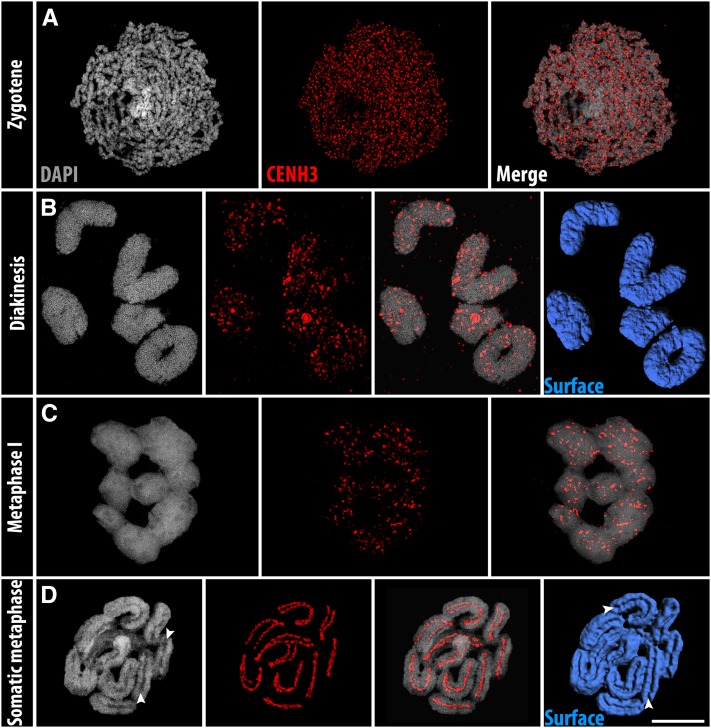
By applying a specific antibody against R. pubera CENH3, we detected a chromosome-wide random distribution of CENH3 from early prophase I until diakinesis in R. pubera (Figure 1, A and B). At metaphase I, multiple clustered CENH3 signals appeared (Figure 1C), and 3D surface rendering of the whole chromatin confirmed the absence of a centromere groove during meiosis (Figure 1B and File S1). These results strongly contrast to the linear holocentromere formation in mitosis, where the chromosomes exhibit a distinct longitudinal centromere groove (Marques et al. 2015) (Figure 1D and File S2).
Figure 1.
Contrasting holocentromere formation between meiosis and mitosis of R. pubera. CENH3 localization at the chromosomes during (A) zygotene, (B) diakinesis, (C) metaphase I, and (D) somatic metaphase. Arrowheads in D indicate mitosis-specific centromere grooves. Bar, 5 µm.
To confirm this contrasting centromere organization, we used the inner kinetochore protein CENP-C as an additional centromere marker. CENP-C is a key component of most eukaryotic centromeres and links the inner and outer (microtubule binding) components of the kinetochore (Earnshaw 2015). It has been shown that CENP-C colocalizes to CENH3, thus defining active centromere chromatin (Carroll et al. 2010; Kato et al. 2013; Falk et al. 2015). A single CENP-C candidate (RpCENP-C) was identified in an in silico analysis of the pollen mother cell transcriptome of R. pubera. The alignment of a RT-PCR-generated 713-bp partial transcript with the CENP-C sequences of other species supported the correct identification (Figure S1A). Phylogenetic analysis grouped RpCENP-C as a sister branch of Juncaceae and both as sister branches to the Poaceae clade (Figure S1B). Based on the identified sequence, RpCENP-C antibodies were generated.
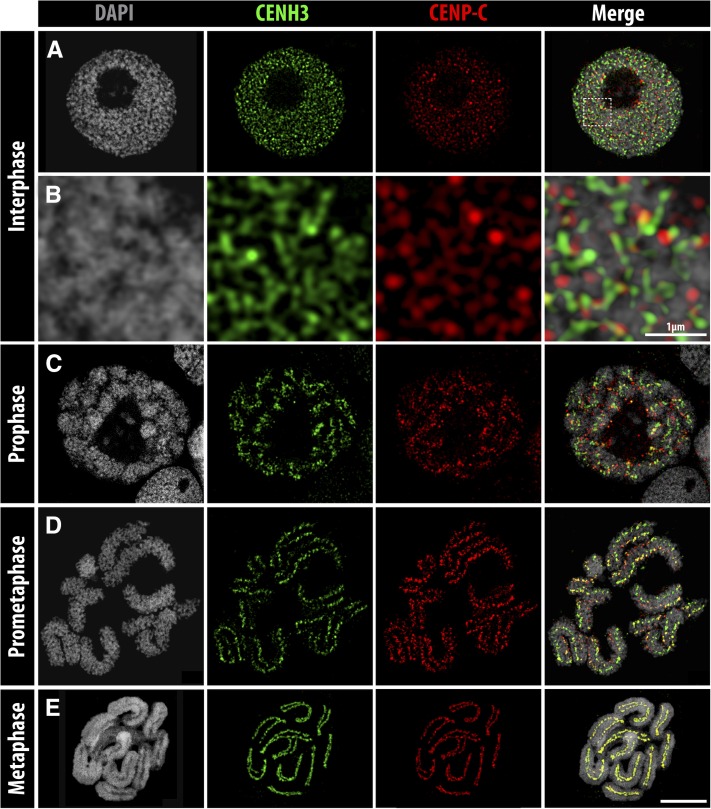
Rhynchospora pubera CENP-C- and CENH3-specific centromeric signals were observed in interphase nuclei as dispersed dot-like structures not so well colocalized (Figure 2, A and B). A progressive colocalization of both centromere marks was observed during mitotic prophase and prometaphase when chromosomes displayed interrupted linear CENH3/CENP-C signals (Figure 2, C and D). Finally, at metaphase onset, chromosomes showed both CENP-C and CENH3 signals colocalized along the mitotic groove of all chromosomes (Figure 2E and File S3). Based on ultrastructural analyses by super-resolution microscopy at a lateral resolution of ∼140 nm, the overlap between CENP-C and CENH3 signals was quantified. Compared to interphase, the degree of colocalization nearly doubled in prophase and further increased in metaphase (Table S1). This indicates the presence of CENP-C in addition to CENH3 at the centromeres of R. pubera at different mitotic stages and a progressive cell-cycle-dependent colocalization of both proteins.
Figure 2.
CENH3 and CENP-C distribution during the mitotic cell cycle of R. pubera, obtained from tapetum cells. (A and B) Interphase, (B) enlargement of A (squared), (C) prophase, (D) prometaphase, and (E) metaphase. Colocalized CENH3 and CENP-C signals are visible in yellow in the merge images. Bar, 5 µm, except when indicated.
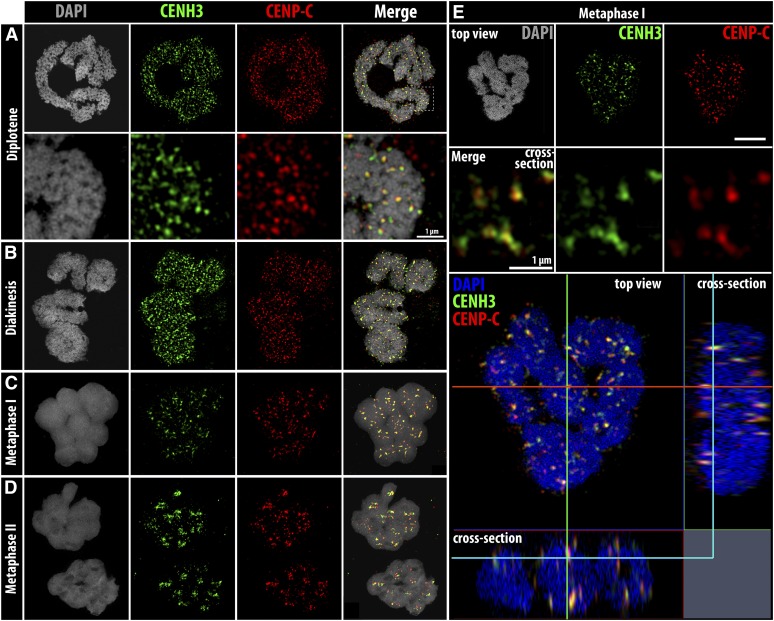
To validate the contrasting centromere organization observed on meiotic chromosomes, again we performed co-immunostaining with CENH3 and CENP-C antibodies. From early prophase I until diakinesis, CENH3 and CENP-C are evident as partially colocalized dispersed dot-like signals all over the chromosomes (Figure 3, A and B and File S4). At metaphase I, the bivalents are arranged at the equatorial plate and both CENH3 and CENP-C cluster along the poleward surface of the chromatids (Figure 3, C and E and File S5). Similar to somatic tissue, a clearly increased association between CENH3 and CENP-C was observed during meiosis compared to interphase (Table S1). At metaphase II, CENH3 and CENP-C are also highly clustered mostly occupying the midregion of each chromatid (Figure 3D). Hence, in contrast to the linear holocentromere organization observed during mitosis, a deviating assembly of centromere units occurs during meiosis, forming the so-called cluster-holocentromeres.
Figure 3.
Distribution of CENH3 and CENP-C during different meiotic stages. (A) Diplotene, (B) diakinesis, (C) metaphase I, and (D) metaphase II. (E) Metaphase I cell showing the colocalization of CENH3 and CENP-C in the cluster-holocentromeres. Overlapping signals are yellow in the merged images. Bar (in A), 5 µm for all images, except when indicated.
The mitotic holocentromeres of R. pubera are composed of centromeric tandem repeats called Tyba [centromeric DNA (cenDNA)] (Marques et al. 2015). The colocalization of CENH3 and cenDNA is also evident throughout MI and MII (Figure S2, A–C). Thus, despite a different centromere organization, the DNA composition of the centromere units does not differ between meiosis and mitosis, and Tyba repeats can be used as additional markers for tracking the centromere organization during meiosis.
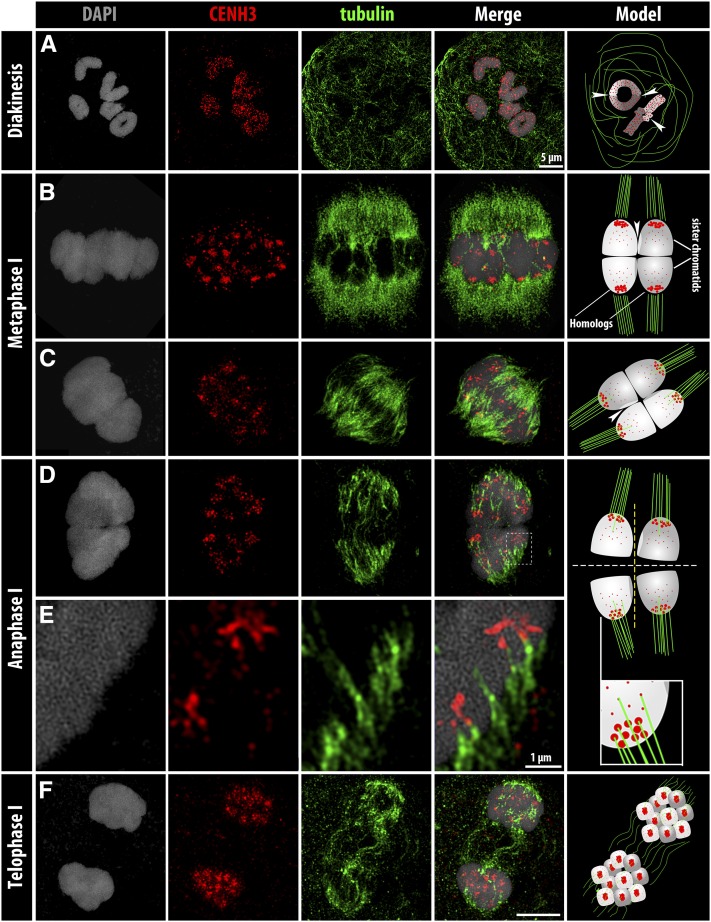
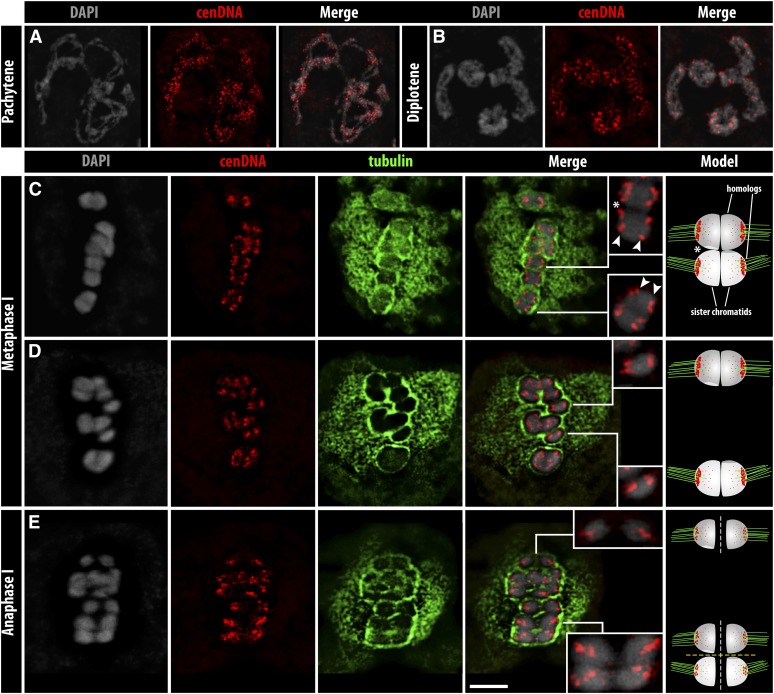
To check how and when the spindle fibers attach to the centromere units, the distribution of α-tubulin and CENH3/cenDNA were analyzed throughout meiosis. From early prophase I until diakinesis, no colocalization was found between spindle fibers and centromeres (Figure 4A), which were scattered all over the chromosomes (Figure 5, A and B, and File S6). At diakinesis, the bivalents are visible as typical rod and ring bivalents, corresponding to one and two chiasmata, respectively (Figure 4A). At early metaphase I, the bivalents are equatorially oriented and clustered CENH3/cenDNA signals are mostly enriched along the poleward surface of the bivalents, showing a bipolar orientation of the sister chromatids (Figure 4B, Figure 5C, and Figure S2D). At late metaphase I, the centromere units become less clustered and the sister cluster-holocentromeres colocalize with the spindle fibers from opposite poles (amphitelic attachment) (Figure 4C, Figure S2D, and File S7). Univalents are often (3.5%) found in R. pubera (Cabral et al. 2014) and they always show the same amphitelic attachment (Figure 5D). At anaphase I, the sister cluster-holocentromeres are pulled by spindle fibers from opposite poles, resulting in the separation of sister chromatids (Figure 4D and Figure 5E). At this stage, the spindle fibers are clearly colocalized with fewer clustered centromere units (Figure 4E and File S8), likely a result of centromere tension. Chromatids migrate as single chromatids in both univalents and bivalents (Figure 5E), supporting the early loss of sister chromatid cohesion and chiasmata resolution. At telophase I, the cluster-holocentromeres are mainly accumulated in the midregion of each chromatid and show less colocalization with the spindle fibers (Figure 4F). Thus, despite of the different centromere organization during MI, the centromere units colocalize with the spindle fibers during meiosis.
Figure 4.
CENH3 and α-tubulin arrangement during meiosis. (A) Diakinesis, (B) early and (C) late metaphase I, (D) anaphase I, (E) enlargement of D (squared), and (F) telophase I. Interpretation models are illustrated at the last right column; sister chromatids are indicated by equal greyscales; dark and light gray indicate homologs. Putative crossovers are indicated by exchanged light and dark gray chromatin (arrowheads). While in A, rod bivalents have one chiasma, ring bivalents have two of them. The dashed white and yellow lines indicate early sister chromatid cohesion loss and chiasmata resolution, respectively. Bar (in F), 5 µm for all images, except when indicated.
Figure 5.
cenDNA (Tyba) and α-tubulin arrangement during MI. (A and B) Detection of cenDNA during prophase I. (C–E) cenDNA and α-tubulin distribution in (C and D) metaphase I and (E) anaphase I. Insets in C show the biorientation of the sister centromeres (arrowheads) at metaphase I. Insets in D show the biorientation of the sister centromeres from univalents. The upper and lower insets in E show the sister chromatids separating from each other from a univalent and a bivalent, respectively. Interpretation models are illustrated in the last right column. The sister chromatids are indicated by identical gray scales, while dark and light gray indicate homologs. Putative crossovers are indicated by exchanged light and dark gray chromatin (asterisk). Dashed white and yellow lines indicate early sister chromatid cohesion loss and chiasmata resolution, respectively. Low quality of tubulin staining is due to the immuno-FISH method. Bar (in E), 5 µm.
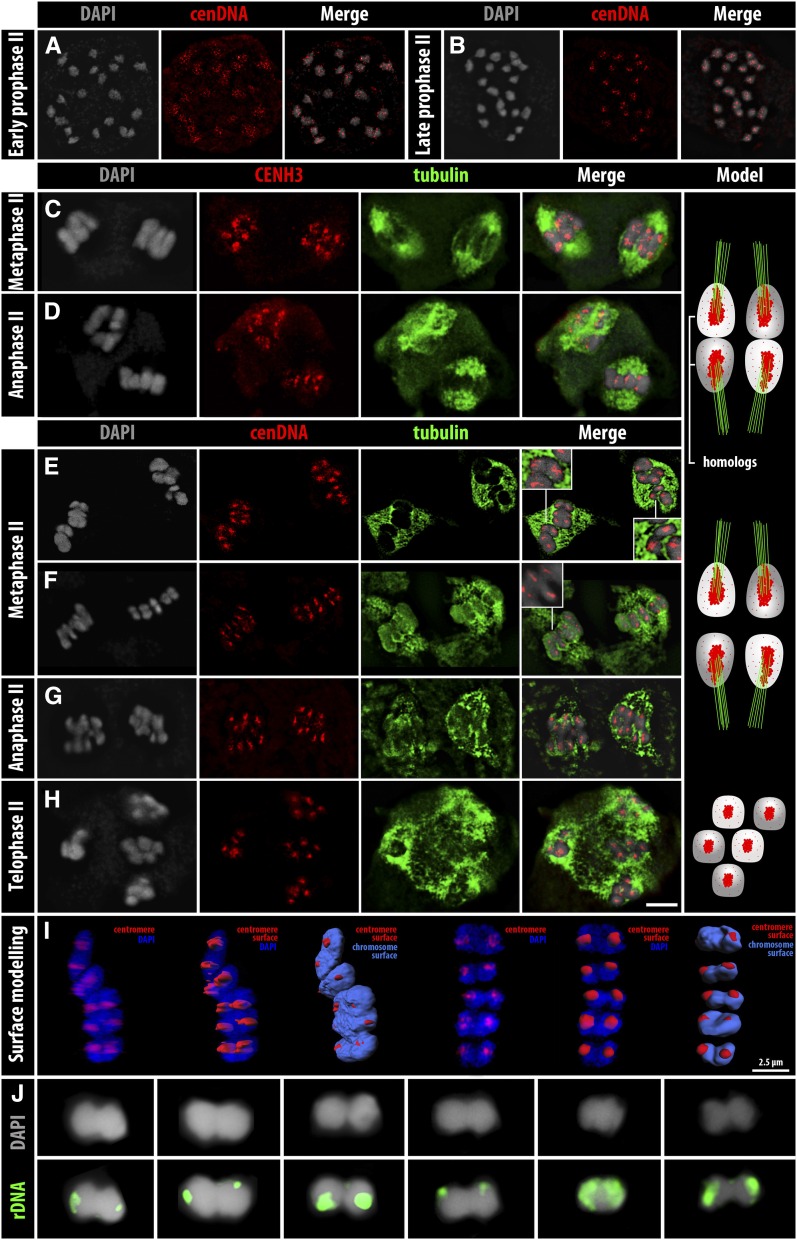
During early MII and at prophase II, in each cell a diploid number (2n = 10) of individualized round-shaped chromatids is present. They display dispersed centromere signals (Figure 6A). Then, when homologous nonsister chromatids associate in pairs toward metaphase II, the centromeric signals become visible as few cluster signals in the midregion of each chromatid (Figure 6B). Tubulin staining, especially during MII, is challenging in Rhynchospora; thus, the distribution of spindle fibers is difficult to visualize. At metaphase II onset, the pairs of homologous nonsister chromatids show mostly a single cluster-holocentromere in the midregion of each chromatid, which is stretched by spindle fibers from opposite poles (Figure 6, C, E, and F insets). The chromatids are of drop-like shape due to the tension caused by the spindle fibers (Figure 6F). Surface rendering of metaphase II cells confirmed that the cluster-holocentromeres are mostly organized as a single cluster in the midregion in each chromatid, occupying external and internal domains (Figure 6I, File S9, and File S10). During anaphase II the stretched homologous nonsister chromatids are then pulled to opposite poles (Figure 6, D and G). Finally, at telophase II, the tetrads contain four haploid nuclei with five chromatids each, showing five clustered centromeric signals (Figure 6H). Thus, in contrast to the numerous cluster-holocentromeres observed in metaphase I, at metaphase II mostly a single cluster-holocentromere is present, occupying a specific domain extending from the internal to external midregion of each chromatid. Colchicine treatment did not disturb the patterns of cluster-holocentromere formation during MI and MII (Figure S2, E and F).
Figure 6.
Cluster-holocentromere arrangement and homologous nonsister chromatid orientation during MII. (A and B) Localization of the cenDNA (Tyba) during prophase II. (C and D) CENH3 and tubulin arrangement in (C) metaphase II and (D) anaphase II cells. (E–H) α-Tubulin and cenDNA arrangement during (E and F) metaphase II, (G) anaphase II, and (H) telophase II. (I) Surface rendering of metaphase II chromosomes showing the centromeres. (J) The 45S rDNA localization on pairs of homologous nonsister chromatids. Low quality of tubulin staining is due to the immuno-FISH method. Bar (in H), 5 µm for A–H images.
Due to the unusual arrangement of homologous nonsister chromatids at metaphase II, we asked whether the chromatid orientation is influenced by the telomeres. Since the 45S ribosomal DNA (rDNA) clusters are located terminally on three chromosome pairs of R. pubera (Sousa et al. 2011), we performed FISH with a 45S rDNA probe. The presence of the FISH signals always at the pole sides (n = 27) (Figure 6J) supports the finding of Cabral et al. (2014), that preferentially the non-rDNA telomeres of the homologous nonsister chromatids associate. This indicates that the homologous nonsister chromatids are axially oriented during metaphase II, contrasting with the equatorial orientation of the bivalents at metaphase I.
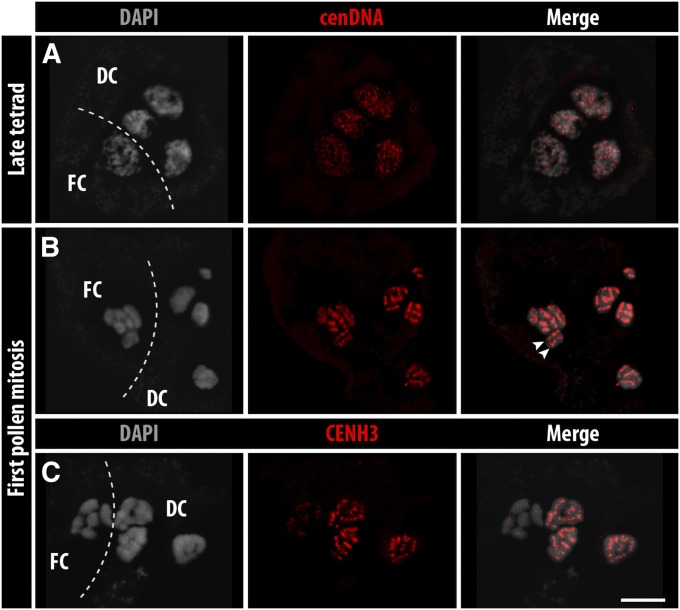
To test whether a linear centromere structure becomes reestablished after meiosis, the subsequent pollen mitosis was analyzed. In most plants, all four male haploid products produce pollen. In contrast, in R. pubera a selective microspore abortion occurs, leading to pollen dispersal as pseudomonads (San Martin et al. 2013; Rocha et al. 2016). Thus, at the end of meiosis, three of four haploid spores degenerate and a single one remains functional to develop the mature pollen. At late tetrad stage, the four haploid nuclei decondense and the cluster-holocentromeres dissociate into smaller centromere units (Figure 7A). Finally, a linear holocentromere organization appears at first pollen mitosis in all four cells of the pseudomonad, as identified after FISH with cenDNA (Figure 7B). However, no groove-like structure is evident at this stage (File S11), perhaps due to differences in cell-type-specific chromosome condensation. Remarkably, only the functional cell replicates, as indicated by double linear cenDNA signals. Instead, the degenerative cells possess only single chromatids (Figure 7B). CENH3 linear signals were clearly present in the three degenerative nuclei, while the functional cells showed only weak, indistinct CENH3 signals (Figure 7C). Whole-cell CENH3 fluorescence signal intensity measurements revealed that functional cells have approximately half of the CENH3 content compared to degenerative cells (Table S3).
Figure 7.
Reestablishment of a linear holocentromere structure in the R. pubera chromosomes during pseudomonad development. Centromeres labeled by (A and B) cenDNA (Tyba) and (C) CENH3. FC, functional cell; DC, degenerative cells. The arrowheads in B indicate both holocentromeres of a single replicated chromosome.
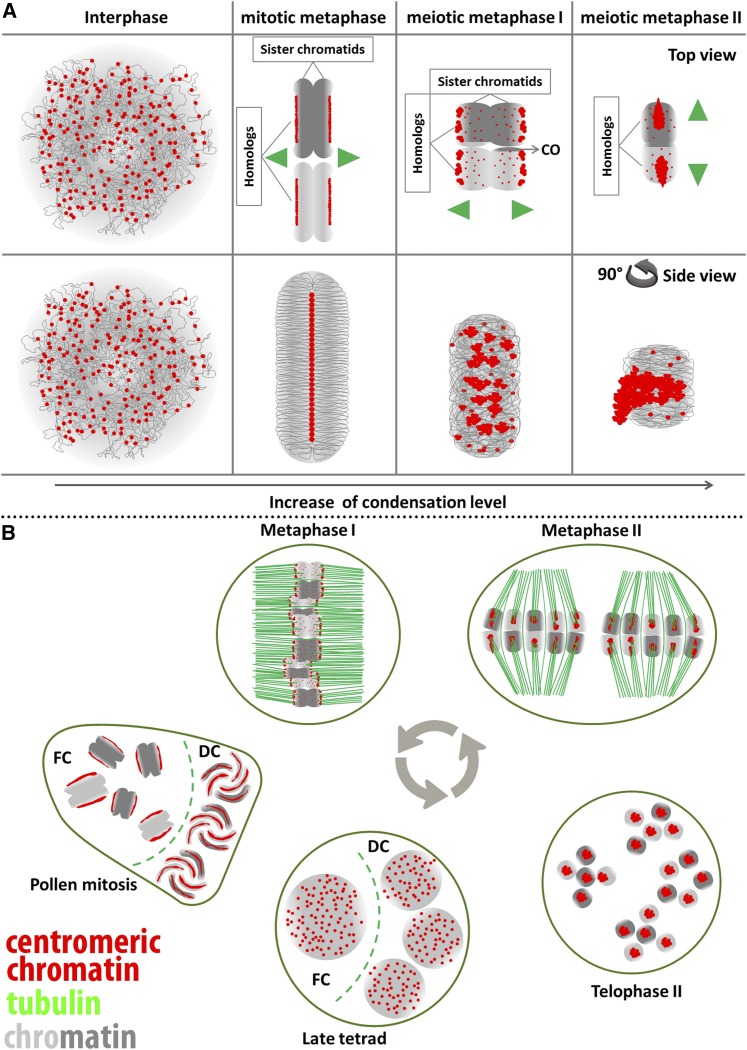
In summary, we conclude that the centromere unit arrangement differs between mitosis and meiosis in R. pubera. There is a transition from the mitotic linear organization within a groove to the cluster-holocentromere arrangement at meiosis as summarized in Figure 8. Finally, a linear holocentromere organization is reestablished at first pollen mitosis, but without groove formation (Figure 8B).
Figure 8.
Model of differential holocentromere organization in the holocentric plant R. pubera. (A) Top and side (90° left turn) views of the centromere organization during mitosis and meiosis. During interphase, the centromere units are genome-wide dispersed in both somatic and meiotic cells. While the process of chromosome condensation occurs, striking differences exist between mitotic and meiotic chromosomes. In mitotic chromosomes, linear holocentromeres are formed within a groove, whereas both MI and MII chromosomes show a cluster-holocentromere organization and no grooves are visible. (B) Cell cycle dynamics of cluster-holocentromere organization and spindle fiber arrangement. During MI, cluster-holocentromeres are oriented along the poleward surface of equatorially oriented bivalents, and the sister chromatids colocalize with spindle fibers from opposite poles (amphitelic attachment) causing their separation in anaphase I. During MII, the cluster-holocentromeres are localized in the midregion of each chromatid. At this stage, pairs of homologous nonsister chromatids are axially orientated and adopt a drop-like shape most likely due to the tension caused by the spindle forces at anaphase II. This causes the segregation of homologous chromatids. At telophase II, each chromatid adopts a spheric shape with a strongly condensed cluster holocentromere in the midregion. During decondensation at late tetrads, the centromere units dissociate. Then, during first pollen mitosis they reassociate in such a way that a linear holocentromere is reestablished. At this stage only the functional cell shows double centromere DNA signals caused by replication, whereas the CENH3 amount is clearly reduced compared to the degenerative cells.
Discussion
The mitotic holocentromere structures of R. pubera are not present during meiosis
Although R. pubera and L. elegans belong to sister families in the same order Poales, these holokinetic species show strikingly different meiotic centromere structures. While both species possess a linear holocentromere organization during mitotic metaphase, only L. elegans chromosomes exhibit the same structure also during meiosis (Heckmann et al. 2014). In contrast, R. pubera centromere units cluster during meiosis, but no distinct linear holocentromere within a groove is formed. A restoration of the linear holocentromere organization occurs after meiosis, during first pollen mitosis, although no groove is formed, in agreement with recent observations during pseudomonad development (Rocha et al. 2016).
Why does the centromere organization differ between mitotic and meiotic chromosomes in R. pubera? The alternative association of centromeric units during meiosis may be due to a stronger degree of chromosome condensation and/or the absence of factors required for the linear arrangement of the holocentromeres. A deviating composition and dynamics of SMC proteins, such as cohesins and condensins, could explain the striking divergences between mitosis and meiosis (Zamariola et al. 2014). Indeed, during R. pubera meiosis, the chromatids lose their elongated shape, become round-shaped, and do not form a groove. In contrast, similar chromatid and groove structures were found during mitosis and meiosis of L. elegans (Heckmann et al. 2014). Poleward clustering of centromeres in R. pubera might help avoid merotelic attachments to spindle microtubules. Clustering, however, is not likely a consequence of attached spindle microtubules pulling toward opposite poles, since a colchicine treatment of meiotic cells did not seem to disturb the formation of cluster-holocentromeres. In addition, a differential CENH3 loading dynamic during meiosis may act as adaptation to deal with holocentricity during meiosis. Indeed, the meiotic CENH3 loading may differ from mitosis in plants (Ravi et al. 2011; Schubert et al. 2014). In contrast to mitosis, CENH3 deposition is biphasic during meiosis in rye and apparently linked with a quality check of CENH3 (Schubert et al. 2014).
A different centromere structure during meiosis has been reported for a number of holocentric species. In C. elegans, the kinetochore activity involves a mechanism independent of CENH3 and CENP-C during MI and MII (Monen et al. 2005), and the chromosomes are ensheathed by microtubule bundles running laterally along their sides during female meiosis (Wignall and Villeneuve 2009; Schvarzstein et al. 2010). However, in male meiosis, the microtubule bundles are enriched at the bivalent ends facing polewards, indicating a telokinetic-like activity (Wignall and Villeneuve 2009). The holocentric worm Parascaris univalens restricts the kinetic activity of the microtubules to the heterochromatic terminal regions during male meiosis. These regions lack kinetochore structures and interact directly with the spindle fibers (Goday and Pimpinelli 1989; Pimpinelli and Goday 1989). Also in holocentric Heteroptera species, a restricted localized kinetic activity during MI and MII was reported (Perez et al. 2000; Papeschi et al. 2003). In most cases of telokinetic meiosis, a mechanism seems to be involved where both chromatid termini can acquire kinetic activity. This demonstrates a special case of kinetochore plasticity.
In the hemipteran genus Oncopeltus, the presence of a holokinetic kinetochore plate during mitosis, but its absence during meiosis, was identified by electron microscopy. Additionally, multiple microtubule attachment sites were found at the meiotic chromosomes (Comings and Okada 1972). Similar findings were reported for other holocentric organisms, i.e., the nematode Ascaris lumbricoides (Goldstein 1977), the hemiptera Rhodnius prolixus (Buck 1967) and Graphosoma italicum (Rufas and Gimenez-Martin 1986), and the Lepdoptera Bombyx mori (Friedlander and Wahrman 1970). In contrast, in the holocentric scorpion Tityus bahiensis, a kinetochore plate throughout meiosis was found, while in the spiders Dysdera crocata and Segestria florentina, kinetochore plates were evident only during MII (Benavente 1982). Thus, the absence of a kinetochore plate during meiosis seems to occur rather frequently among holocentric organisms and was postulated to be related to the restriction of kinetic activity and terminalization of chiasmata necessary for a normal progression of meiosis in those organisms (Comings and Okada 1972; Pimpinelli and Goday 1989). In addition, it is interesting to notice that all holocentric insects lacking kinetochore plates during meiosis also lack CENH3 and CENP-C genes, and occasionally some other inner kinetochore proteins, whereas most of the outer kinetochore genes were still present (Drinnenberg et al. 2014). Whether the lack of CENH3 and CENP-C causes a misassembly of kinetochore plates during meiosis in these organisms is still unknown.
Thus, the meiotic holocentromeres in R. pubera are unique as it is the only holocentric species so far showing a differential centromere organization in mitosis and meiosis, while spindle fibers attach to its centromere units composed of CENH3 and CENP-C. As discussed above, most organisms showing differential centromere organization either lack CENH3 and CENP-C (Drinnenberg et al. 2014) or these proteins do not play a role in chromosome segregation during meiosis (i.e., C. elegans). In contrast, a similar organization of mitotic and meiotic holocentromeres was found in L. elegans, although no CENP-C antibody has been generated and tested for this species (Heckmann et al. 2014).
A linear holocentromere organization is not required for the reversion of the chromatid segregation events during meiosis in holokinetic species
We confirmed the previously reported unusual process of meiosis in R. pubera (Cabral et al. 2014) by showing a bipolar sister centromere orientation and their attachment to microtubules from opposite spindle poles in MI (amphitelic attachment), the segregation of the sister chromatids to opposite poles already during anaphase I, and the alignment and segregation of homologous nonsister chromatids only during the second meiotic division. Remarkably, a differential orientation of cluster-holocentromeres was observed from MI and MII. While during MI the cluster-holocentromeres were observed mostly accumulated along the poleward surface of the bivalents, in MII the cluster-holocentromeres were mostly visible as a single cluster in the midregion of each chromatid. Notably, the homologous nonsister chromatids are preferentially associated by their non-rDNA termini at metaphase II as already described by Cabral et al. (2014). The results indicate a distinct orientation and interaction of spindle fibers with the cluster-holocentromeres between MI and MII. While during metaphase I, the bivalents orient perpendicular to the spindle poles, during metaphase II the pairs of homologous nonsister chromatids orient with their longer axis in parallel to the spindle poles.
Moreover, our results show that a linear holocentromere organization as found in L. elegans is not required for the reversion of the segregation events of the sister/homologous chromatids during meiosis. Actually, considering an end-to-end interaction of the homologous nonsister chromatids in metaphase II, the linear structure is compatible with proper chromatid segregation toward opposite poles because Luzula chromosomes maintain a U-shape conformation in MII. In fact, the highly clustered holocentromere found at metaphase II and anaphase II in R. pubera seems to present an alternative solution to reduce the risk of merotelic attachment of microtubules. However, while no missegregation was found during MI in R. pubera, it was reported that 19.5% of all MII products had incorrect chromosome numbers (Cabral et al. 2014). In the nematode C. elegans, the chromokinesin KLP-19 counteracts persistent merotelic attachments (Powers et al. 2004). Whether in R. pubera a similar correction mechanism exists is unknown. Although merotelic attachments might cause missegregation during MII of R. pubera, Cabral et al. (2014) suggested that pairs of homologous nonsister chromatids may have failed to connect to each other, thus leading to missegregation in MII.
During first pollen mitosis, CENH3 signals were much stronger in the degenerative cells, while the functional cell showed a weak and indistinct labeling. These differences might be explained by the absence of de novo incorporation of CENH3 molecules after the exit from meiosis. Thus, preexisting CENH3 could be partitioned equally between duplicated sister centromeres as a result of cell replication, which occurs only in the functional cell (evidenced by double lines of cenDNA signals). Thereby, a fixed number of CENH3 molecules split between the generative and vegetative nucleus, which explains the 50% of CENH3 signal intensity found in functional cells compared to degenerative cells. Alternatively, active CENH3 removal in the functional haploid cell after meiosis exit could cause the reduction of CENH3 molecules as found in rye (Schubert et al. 2014). The latter is possible, since the removal of CENH3 has been observed in vegetative pollen cells of Arabidopsis thaliana (Schoft et al. 2009; Merai et al. 2014). Furthermore, the weak CENH3 signals observed in the functional cell suggests that a reduced amount of CENH3 is still sufficient for proper chromosome segregation (Liu et al. 2006; Lermontova et al. 2011; Karimi-Ashtiyani et al. 2015).
What does the unusual meiotic centromere arrangement of R. pubera imply?
The inappropriate occurrence of crossovers in the proximity of the primary constriction of monocentric chromosomes affects negatively the meiotic chromosome segregation by influencing the centromeric cohesion (Talbert and Henikoff 2010; Vincenten et al. 2015). Accordingly, the occurrence of very few crossovers is reported for holocentric organisms, generally one or two per rod and ring bivalent, respectively, mostly located at the noncentromeric terminal regions (Cuacos et al. 2015). This is also true for R. pubera, in which chiasmata occur terminally. In this case, recombined bivalents are resolved because of the loss of sister chromatid cohesion in anaphase I. Furthermore, R. pubera faces another challenge during meiosis, since its unusual centromere arrangement of meiotic chromosomes could cause a high risk of misorientation during MI. However, no chromosome fragmentation or anaphase bridges were observed during the meiosis of R. pubera. But, the unusual centromere organization might be associated with decrease in recombination, which could be the cause of the frequent (3.5%) occurrence of univalents in R. pubera. In fact, this could also explain the occurrence of univalents in the achiasmatic meiosis of R. tenuis (Cabral et al. 2014). Thus, it seems that the meiosis of Rhynchospora is adapted to solve potential meiotic errors due to the unusual centromere arrangement.
The Cf19 complex of yeast [also known as the constitutive centromere-associated network (CCAN) in other organisms] prevents meiotic double-strand breaks (DSBs) proximal to the centromeres, which are essential to initiate recombination (Vincenten et al. 2015). Nevertheless, although meiotic DSBs are suppressed at core centromeric regions in yeast, they frequently occur only a few kilobases away from the centromeres (Buhler et al. 2007; Pan et al. 2011). In Rhynchospora, meiotic DSBs are normally formed and processed in early prophase I, as evidenced by the presence of multiple RAD51 foci (Cabral et al. 2014). Other meiotic events typical of the first meiotic prophase, such as the meiotic axis formation, appear normal in R. pubera, since the axial element protein ASY1 showed the typical pattern known from monocentric species (Cabral et al. 2014). Therefore, it is interesting that the meiotic cluster-holocentromere arrangement of R. pubera does not disturb DSB formation, axis architecture, or synaptonemal complex formation. Thus, to deal with its centromere architecture during meiosis, a very accurate regulation of meiotic recombination is likely to exist in R. pubera.
In conclusion, the holocentromeres of R. pubera are unique with respect to their differential organization during mitosis and meiosis. Our results reinforce the idea of high centromere plasticity among holocentric organisms and offer a novel model for understanding centromere evolution and function among eukaryotes.
Acknowledgments
We thank Stefan Heckmann for helpful comments and discussion. We thank the Brazilian Agency Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior (CAPES) for the special visiting researcher grant and project funding for A.H. and a fellowship for A.M., The Brazilian National Council for Scientific and Technological Development (CNPq) for financial support for A.P.-H., and also the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) for support. The authors declare no conflict of interests.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.191213/-/DC1.
Communicating editor: J. A. Birchler
Literature Cited
- Albertson D. G., Thomson J. N., 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1: 15–26. [DOI] [PubMed] [Google Scholar]
- Albertson D. G., Rose A. M., Villeneuve A. M., 1997. Chromosome organization, mitosis, and meiosis, pp. 47–48 in C. elegans II, edited by Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Banaei-Moghaddam A. M., Meier K., Karimi-Ashtiyani R., Houben A., 2013. Formation and expression of pseudogenes on the B chromosome of rye. Plant Cell 25: 2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavente R., 1982. Holocentric chromosomes of arachnids: presence of kinetochore plates during meiotic divisions. Genetica 59: 23–27. [Google Scholar]
- Buck R. C., 1967. Mitosis and meiosis in Rhodnius prolixus: the fine structure of the spindle and diffuse kinetochore. J. Ultrastruct. Res. 18: 489–501. [DOI] [PubMed] [Google Scholar]
- Buhler C., Borde V., Lichten M., 2007. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 5: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack L. S., Berman J., 2012. Flexibility of centromere and kinetochore structures. Trends Genet. 28: 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral G., Marques A., Schubert V., Pedrosa-Harand A., Schlogelhofer P., 2014. Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat. Commun. 5: 5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C. W., Milks K. J., Straight A. F., 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189: 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Mao Y., Sullivan K. F., 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421. [DOI] [PubMed] [Google Scholar]
- Comings D. E., Okada T. A., 1972. Holocentric chromosomes in Oncopeltus: kinetochore plates are present in mitosis but absent in meiosis. Chromosoma 37: 177–192. [DOI] [PubMed] [Google Scholar]
- Cuacos M., Franklin F. C. H., Heckmann S., 2015. Atypical centromeres in plants: what they can tell us. Front. Plant Sci. 6: 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I. A., deYoung D., Henikoff S., Malik H. S., 2014. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife 3: e03676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., Oegema K., Desai A., 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12: 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duro E., Marston A. L., 2015. From equator to pole: splitting chromosomes in mitosis and meiosis. Genes Dev. 29: 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., 2015. Discovering centromere proteins: from cold white hands to the A, B, C of CENPs. Nat. Rev. Mol. Cell Biol. 16: 443–449. [DOI] [PubMed] [Google Scholar]
- Falk S. J., Guo L. Y., Sekulic N., Smoak E. M., Mani T., et al. , 2015. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348: 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander M., Wahrman J., 1970. The spindle as a basal body distributor. A study in the meiosis of the male silkworm moth, Bombyx mori. J. Cell Sci. 7: 65–89. [DOI] [PubMed] [Google Scholar]
- Gavet O., Pines J., 2010. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell 18: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goday C., Pimpinelli S., 1989. Centromere organization in meiotic chromosomes of Parascaris univalens. Chromosoma 98: 160–166. [DOI] [PubMed] [Google Scholar]
- Goldstein P., 1977. Spermatogenesis and spermiogenesis in Ascaris lumbricoides Var. suum. J. Morphol. 154: 317–337. [DOI] [PubMed] [Google Scholar]
- Guerra M., Cabral G., Cuacos M., Gonzalez-Garcia M., Gonzalez-Sanchez M., et al. , 2010. Neocentrics and holokinetics (holocentrics): chromosomes out of the centromeric rules. Cytogenet. Genome Res. 129: 82–96. [DOI] [PubMed] [Google Scholar]
- Haarhuis J. H., Elbatsh A. M., Rowland B. D., 2014. Cohesin and its regulation: on the logic of X-shaped chromosomes. Dev. Cell 31: 7–18. [DOI] [PubMed] [Google Scholar]
- Heckmann S., Jankowska M., Schubert V., Kumke K., Ma W., et al. , 2014. Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat. Commun. 5: 4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes-Schrader S., Schrader F., 1961. The kinetochore of the Hemiptera. Chromosoma 12: 327–350. [DOI] [PubMed] [Google Scholar]
- Ishiguro K., Watanabe Y., 2007. Chromosome cohesion in mitosis and meiosis. J. Cell Sci. 120: 367–369. [DOI] [PubMed] [Google Scholar]
- Ishii, T., N. Sunamura, A. Matsumoto, A. E. Eltayeb, and H. Tsujimoto, 2015 Preferential recruitment of the maternal centromere-specific histone H3 (CENH3) in oat (Avena sativa L.) × pearl millet (Pennisetum glaucum L.) hybrid embryos. Chromosome Res. 23: 709–718. [DOI] [PubMed] [Google Scholar]
- Karimi-Ashtiyani R., Ishii T., Niessen M., Stein N., Heckmann S., et al. , 2015. Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Natl. Acad. Sci. USA 112: 11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Jiang J., Zhou B. R., Rozendaal M., Feng H., et al. , 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340: 1110–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I., Koroleva O., Rutten T., Fuchs J., Schubert V., et al. , 2011. Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J. 68: 40–50. [DOI] [PubMed] [Google Scholar]
- Liu S. T., Rattner J. B., Jablonski S. A., Yen T. J., 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175: 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P. S., Oegema K., Desai A., Cheeseman I. M., 2004. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 12: 641–653. [DOI] [PubMed] [Google Scholar]
- Marques A., Ribeiro T., Neumann P., Macas J., Novak P., et al. , 2015. Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proc. Natl. Acad. Sci. USA 112: 13633–13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Schvarzstein M., Barroso C., Lightfoot J., Dernburg A. F., et al. , 2008. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22: 2886–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters D. P., Paliulis L. V., Korf I. F., Chan S. W., 2012. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 20: 579–593. [DOI] [PubMed] [Google Scholar]
- Merai Z., Chumak N., Garcia-Aguilar M., Hsieh T. F., Nishimura T., et al. , 2014. The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes. Proc. Natl. Acad. Sci. USA 111: 16166–16171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B. Q., Nguyen M. A., von Haeseler A., 2013. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30: 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J., Maddox P. S., Hyndman F., Oegema K., Desai A., 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 7: 1248–1255. [DOI] [PubMed] [Google Scholar]
- Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., 2015. Meiosis: an overview of key differences from mitosis. Cold Spring Harb. Perspect. Biol. 7: a015859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H. G., et al. , 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papeschi A. G., Mola L. M., Bressa M. J., Greizerstein E. J., Lia V., et al. , 2003. Behaviour of ring bivalents in holokinetic systems: alternative sites of spindle attachment in Pachylis argentinus and Nezara viridula (Heteroptera). Chromosome Res. 11: 725–733. [DOI] [PubMed] [Google Scholar]
- Perez R., Rufas J. S., Suja J. A., Page J., Panzera F., 2000. Meiosis in holocentric chromosomes: orientation and segregation of an autosome and sex chromosomes in Triatoma infestans (Heteroptera). Chromosome Res. 8: 17–25. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S., Goday C., 1989. Unusual kinetochores and chromatin diminution in Parascaris. Trends Genet. 5: 310–315. [DOI] [PubMed] [Google Scholar]
- Powers J., Rose D. J., Saunders A., Dunkelbarger S., Strome S., et al. , 2004. Loss of KLP-19 polar ejection force causes misorientation and missegregation of holocentric chromosomes. J. Cell Biol. 166: 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M., Shibata F., Ramahi J. S., Nagaki K., Chen C., et al. , 2011. Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet. 7: e1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha D. M., Marques A., Andrade C. G. T. J., Guyot R., Chaluvadi S. R., et al. , 2016. Developmental programmed cell death during asymmetric microsporogenesis in holocentric species of Rhynchospora (Cyperaceae). J. Exp. Bot. Epub ahead of print. doi: 10.1093/jxb/erw300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufas J. S., Gimenez-Martin G., 1986. Ultrastructure of the kinetochore in Graphosoma italicum (Hemiptera, Heteroptera). Protoplasma 132: 142–148. [Google Scholar]
- San Martin J. A. B., Andrade C. G. T. D., Mastroberti A. A., Mariath J. E. D., Vanzela A. L. L., 2013. Asymmetric cytokinesis guide the development of pseudomonads in Rhynchospora pubera (Cyperaceae). Cell Biol. Int. 37: 203–212. [DOI] [PubMed] [Google Scholar]
- Schoft V. K., Chumak N., Mosiolek M., Slusarz L., Komnenovic V., et al. , 2009. Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep. 10: 1015–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V., Lermontova I., Schubert I., 2014. Loading of the centromeric histone H3 variant during meiosis-how does it differ from mitosis? Chromosoma 123: 491–497. [DOI] [PubMed] [Google Scholar]
- Schvarzstein M., Wignall S. M., Villeneuve A. M., 2010. Coordinating cohesion, co-orientation, and congression during meiosis: lessons from holocentric chromosomes. Genes Dev. 24: 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes D. C., Wu J. C., Sadler P. L., Laprade K., Moore L. L., et al. , 2009. Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 5: e1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A., Barros e Silva A. E., Cuadrado A., Loarce Y., Alves M. V., et al. , 2011. Distribution of 5S and 45S rDNA sites in plants with holokinetic chromosomes and the “chromosome field” hypothesis. Micron 42: 625–631. [DOI] [PubMed] [Google Scholar]
- Steiner F. A., Henikoff S., 2015. Diversity in the organization of centromeric chromatin. Curr. Opin. Genet. Dev. 31: 28–35. [DOI] [PubMed] [Google Scholar]
- Talbert P. B., Henikoff S., 2010. Centromeres convert but don’t cross. PLoS Biol. 8: e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viera A., Page J., Rufas J. S., 2009. Inverted meiosis: the true bugs as a model to study. Genome Dyn. 5: 137–156. [DOI] [PubMed] [Google Scholar]
- Vincenten N., Kuhl L. M., Lam I., Oke A., Kerr A. R., et al. , 2015. The kinetochore prevents centromere-proximal crossover recombination during meiosis. eLife 4: e10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshart, K., J. Fuchs, and V. Schubert, 2016 Structured Illumination Microscopy (SIM) and Photoactivated Localization Microscopy (PALM) to analyze the abundance and distribution of RNA Polymerase II molecules on flow-sorted Arabidopsis nuclei. Bio-protocol 6: e1725. [Google Scholar]
- Wignall S. M., Villeneuve A. M., 2009. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat. Cell Biol. 11: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamariola L., Tiang C. L., De Storme N., Pawlowski W., Geelen D., 2014. Chromosome segregation in plant meiosis. Front. Plant Sci. 5: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Antibodies are available upon request. Sequence data are available at GenBank and the accession numbers are listed in the Materials and Methods section and in Table S2. The GenBank accession no. of R. pubera CENP-C is KU516997.