Abstract
The budding yeast E3 SUMO ligase Mms21, also known as Nse2, is a component of the Smc5/6 complex, which regulates sister chromatid cohesion, DNA replication, and repair. Our study shows that the mms21RINGΔ mutant exhibits (1) reduced ribosomal RNA production; (2) nuclear accumulation of ribosomal proteins; (3) elevated Gcn4 translation, indicating translational stress; and (4) upregulation of Gcn4 targets. Genes involved in ribosome biogenesis and translation are downregulated in the mms21RINGΔ mutant. We identified RPL19A as a novel genetic suppressor of the mms21RINGΔ mutant. Deletion of RPL19A partially suppresses growth defects in both smc5-6 and mms21RINGΔ mutants as well as nuclear accumulation of ribosome subunits in the mms21RINGΔ mutant. Deletion of a previously identified strong suppressor, MPH1, rescues both the accumulation of ribosome subunits and translational stress. This study suggests that the Smc5/6 complex supports nucleolar function.
Keywords: MMS21, NSE2, MPH1, nucleolus, RPL19A, Smc5/6, translational stress
MMS21 is a SUMO ligase that is a subunit of the Smc5/6 complex. The Smc5/6 complex has been shown to regulate sister chromatid segregation, DNA replication, and DNA damage repair (Lindroos et al. 2006; Rai et al. 2011; Almedawar et al. 2012; McAleenan et al. 2012; Wu et al. 2012; Bermudez-Lopez et al. 2015). The structural maintenance of chromosome (SMC) complexes are conserved from prokaryotes to humans and they promote chromosome integrity. Previous reports have shown that Mms21 SUMO ligase activity is required for the Smc5/6 complex to function in those cellular processes (Zhao and Blobel 2005; Murray and Carr 2008; Takahashi et al. 2008; Potts 2009; Stephan et al. 2011; Bermudez-Lopez et al. 2015).
One of the striking phenotypes of loss of Mms21 sumoylation activity is an aberrant nucleolus (Zhao and Blobel 2005). The nucleolus is the nuclear body where ribosomal DNA (rDNA) resides and ribosomes begin to be assembled from their RNA and protein components. Mms21 has been shown to target some nucleolar proteins for sumoylation (Albuquerque et al. 2013). The Smc5/6 complex binds to the rDNA repeats, suggesting it could play a role in the formation of the nucleolus (Torres-Rosell et al. 2005b). Furthermore, the Smc5/6 complex is required for rDNA integrity (Torres-Rosell et al. 2005a) and segregation (Torres-Rosell et al. 2005b). The aberrant nucleolar structure observed in cohesin loss-of-function mutants was a harbinger of defects in ribosome biogenesis (Bose et al. 2012). Given the defect in nucleolar structure in MMS21 mutants, we speculated that the sumoylation activity of Mms21 might be required for nucleolar function.
Faithful regulation of ribosome biogenesis is important for translation and cell proliferation. In proliferating cells, there is a high demand for protein production. To meet this demand, cells must produce ribosomes at a rapid rate (Montanaro et al. 2008; Lempiainen and Shore 2009). Defects in ribosome biogenesis have been shown to correlate with reduced or altered protein translation and growth defects (Zanchin et al. 1997; Yamada et al. 2007; Jack et al. 2011; Bose et al. 2012; Xu et al. 2013). More importantly, deregulation of ribosome biogenesis has been correlated with tumorigenesis and developmental syndromes, including cohesinopathies and ribosomopathies (Montanaro et al. 2008; Ruggero 2013; Zakari et al. 2015), demonstrating that proper ribosome biogenesis is critical for human health.
Ribosome biogenesis requires the assembly and transport of many different RNA and protein components. This process includes ribosomal RNA (rRNA) transcription, processing, ribosome assembly, and export. The budding yeast 35S rRNA precursor is transcribed by RNA polymerase I in the nucleolus and undergoes a series of cleavages and modifications to generate the 25S, 18S, and 5.8S rRNAs. The 25S, 5.8S, and 5S (transcribed by RNA polymerase III) rRNAs and the 18S rRNA assemble the 60S and the 40S preribosome particles, respectively, with both large and small ribosomal proteins and transacting factors (Strunk and Karbstein 2009; Kressler et al. 2010; Woolford and Baserga 2013). After assembly of the preribosome particles, the 60S and the 40S preribosome particles undergo maturation and are exported to the cytoplasm to form functional ribosomes.
We find that mutations in the Smc5/6 complex allow ribosomal proteins to accumulate in the nucleus. In addition, rRNA is produced at a reduced rate and translational stress is detected in the mms21RINGΔ mutant. The gene expression profile in the mms21RINGΔ mutant is consistent with the idea that translation could be negatively affected. Deletion of RPL19A or the previously identified suppressor MPH1 partially rescues growth and the accumulation of ribosomal proteins in the nucleus. Deletion of the gene encoding the Mph1 helicase also partially rescues rRNA production and translational stress in the mms21RINGΔ mutant. Our study suggests that the Smc5/6 complex supports nucleolar function.
Materials and Methods
Spot growth assay
Strains (all listed in Supplemental Material, Table S2) were grown to midlog phase at 30° and spotted in 10-fold serial dilutions onto YPD plates. The plates were incubated at 30° and 37° for 1–3 days.
Confocal microscopy
Images of live cells were taken with a 100× Plan Apochromat 1.46 N.A. oil objective using a confocal microscope (Perkin Elmer Ultraview Spinning Disk) and the Volocity 6.3 software program. Scale bars were created using ImageJ software.
RNA-sequencing analysis
Both WT and mms21RINGΔ cells were grown in triplicate to OD600 = 0.6 at 30°. Total RNA was isolated using the hot phenol method, subjected to ribodepletion, and libraries were made using the library construction kit Ribo-Zero (Epicentre) for sequencing. Reads were aligned to sacCer3 using Tophat v2.0.8 with default alignment parameters. The resulting binary alignment/map (BAM) files were sorted and indexed using SAMtools. Because there were >25 million reads per sample, the BAM files were downsampled to take 5 million reads at random for expression analysis.
Gcn4 site enrichment
Promoters consisting of 600 bases upstream of the transcription start site were extracted for all genes from sacCer3 in R. The consensus Gcn4 binding site TGASTCW (Natarajan et al. 2001) was used to query each promoter for the presence or absence of sites. The International Union of Pure and Applied Chemistry codes are S = G,C and W = A,T. The number of promoters in the differentially expressed gene sets scoring positive for a Gcn4 binding site was tabulated (Table S1). The degree of enrichment in the up- and downregulated genes compared to the background set (genes not differentially expressed) was compared and tested for statistical significance using a Fisher exact test.
Gene group enrichment
Various gene groups were examined for higher rankings in terms of differential expression relative to random groups of genes using a mean-rank gene set test (Wu and Smyth 2012).
3H-uridine incorporation assay
Strains were grown in SD −Ura medium supplemented with 6.7 ng/μl uracil in triplicate to OD600 = 0.3 at 30°. Five μCi of 3H-uridine was added to the cultures and incubated at 30° for 5 min. Samples were treated with 2.5 ml of 10% trichloroacetic acid (TCA) containing 2.5 mg/ml uridine. The samples were filtrated onto nitrocellulose membrane using a vacuum and washed with 5% TCA. The membranes were air dried and analyzed via scintillation counting (Beckman LS 6500).
Gcn4–lacZ reporter assay
Strains transformed with a p180–Gcn4–lacZ reporter plasmid and were grown in SD −Ura medium to midlog phase at 30°. Strains with the Gcn4–lacZ reporter integrated at TRP1 were grown in YPD. The cells were lysed in breaking buffer (100 mM Tris-Cl, Ph 8.0, 1 mM DTT, 20% glycerol, 1 mM PMSF) by vortexing with glass beads for 4 min at 4°. The supernatant was incubated with O-nitrophenyl-β-D-galactoside (ONPG) for 60 min. β-Galactosidase activity was measured at OD420.
Rps2–GFP and Rpl25–GFP quantification
Strains were transformed with either pRS315–Rps2–GFP plasmid or pRS315–Rpl25–GFP plasmid. Cells were grown in SD −Leu or SD −Ura medium containing 0.02 mg/ml adenine to midlog phase at 30°. Mean peak GFP fluorescence intensity was measured using the MACSQuant analyzer (Miltenyi Biotec). The distances between biological replicates vs. the distances between samples with different genotypes was determined as previously described and the Kolmogorov–Smirnov (KS) statistic test was applied (Bose et al. 2012; Kim 2016).
Strain and data availability
Strains are available upon request. Table S2 contains a complete list of all strains used in this study. The Gene Expression Omnibus accession number for RNA-sequencing (RNA-seq) data is GSE69826. Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1027.
Results
RPL19A is a genetic suppressor of the mms21RINGΔ mutant
In order to gain insight into the function of Mms21 SUMO ligase activity, we performed a synthetic genetic array (SGA) screen for novel genetic suppressors of the mms21RINGΔ mutant in which its SUMO ligase activity is deficient. Since the mms21RINGΔ mutant exhibits a growth defect at 37°, we took advantage of this temperature sensitivity to identify potential genetic suppressors. From this SGA analysis, we found that deletion of RPL19A partially suppresses the temperature-sensitive phenotype of the mms21RINGΔ mutant.
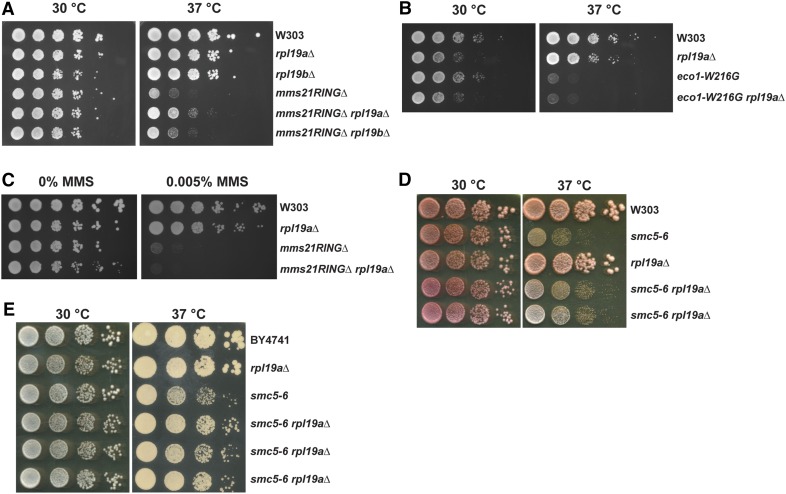
RPL19A is a ribosomal protein gene and has a paralog, RPL19B. In order to test the specificity of RPL19A as a genetic suppressor of the mms21RINGΔ mutant, we compared the growth rate of the mms21RINGΔ mutant to the mms21RINGΔ rpl19aΔ double mutant at both 30° and 37°. As shown in Figure 1A, the growth defect of the mms21RINGΔ mutant at 37° was partially rescued by rpl19aΔ but not by rpl19bΔ. In our previous report, we showed that a mutant in the cohesin acetyltransferase (eco1–W216G) exhibits a growth defect at the nonpermissive temperature of 37° (Lu et al. 2010). Therefore, we wanted to test whether rpl19aΔ can also suppress the temperature sensitivity of the eco1–W216G mutant via a spot growth assay (Figure 1B). This assay showed that the deletion of RPL19A does not suppress the growth phenotype of the eco1–W216G mutant at 37°, suggesting that genetic suppression by rpl19aΔ is specific to the mms21RINGΔ mutant. Then, we examined whether deletion of RPL19A could also suppress the MMS sensitivity of the mms21RINGΔ mutant (Figure 1C). Interestingly, the MMS sensitivity of the mutant was not rescued by deletion of RPL19A, indicating that this genetic suppression must act by a mechanism that does not rescue DNA damage. For growth curves, see Figure S1.
Figure 1.
Deletion of RPL19A partially suppresses the temperature sensitivity of the mms21RINGΔ mutant but not of the eco1–W216G mutant. (A) The growth defect of the mms21RINGΔ mutant at 37° is partially suppressed by deletion of RPL19A. The indicated strains were grown to midlog phase at 30° and were spotted in 10-fold serial dilutions onto YPD plates. (B) Deletion of RPL19A does not suppress the growth defect of the eco1–W216G mutant at 37°. The indicated strains were grown to midlog phase at 30° and were spotted in 10-fold serial dilutions onto YPD plates. (C) MMS sensitivity of the mms21RINGΔ mutant is not suppressed by deletion of RPL19A. The indicated strains were grown to midlog phase at 30° and were spotted in 10-fold serial dilutions onto YPD or YPD containing 0.005% MMS plates. (D and E) The growth defect of the smc5-6 mutant at 37° is partially suppressed by deletion of RPL19A. The indicated strains were grown as in A.
SMC5 encodes another subunit of the Smc5/6 complex. We next checked whether deletion of RPL19A would rescue the temperature-sensitive phenotype of the smc5-6 mutant. The growth of this mutant was also partially rescued by the deletion of RPL19A in both the W303 and BY4741 strain backgrounds (Figure 1, D and E), suggesting that rpl19aΔ may be a suppressor for Smc5/6 loss of function in general and is not specific to the mms21RINGΔ mutant.
Genes involved in ribosome biogenesis and protein translation are downregulated in the mms21RINGΔ mutant
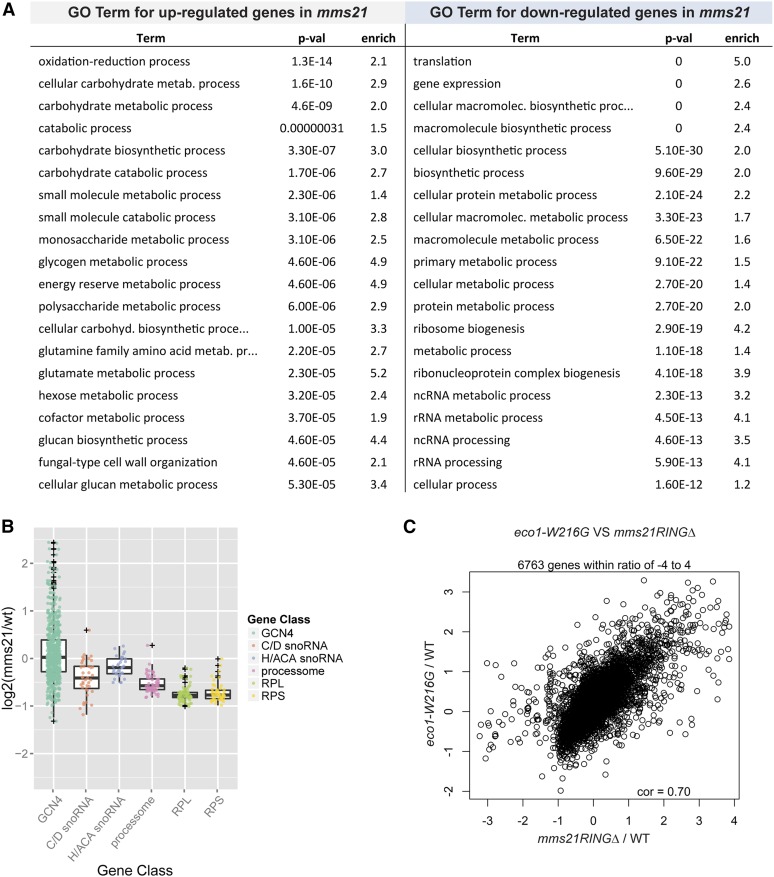
It has been previously shown that nucleolar morphology of the mms21RINGΔ mutant is aberrant (Zhao and Blobel 2005). Recent work from our lab has shown that the SMC complex known as cohesin is important for gathering the rDNA repeats into a functional nucleolar structure (Bose et al. 2012; Harris et al. 2014). Like cohesin, the Smc5/6 complex binds to rDNA repeats (Torres-Rosell et al. 2005b). Since the nucleolus serves as a ribosome factory, we hypothesized that the mms21RINGΔ mutant may have defects in ribosome biogenesis. In order to gain insight into the physiology of the mms21RINGΔ mutant, we performed RNA-seq on cells grown in YPD at 30°, the permissive temperature. The number of differentially expressed genes for the mms21RINGΔ mutant at an adjusted P-value of ≤0.05 was 1921. However, this set contains genes with ratios as low as 1.2. After imposing an additional criteria of fold change >1.5, the number of genes decreases to 1191. We carried out Gene Ontology term analysis on 657 upregulated genes and 337 downregulated genes (Figure 2A). The most significantly upregulated biological processes were carbohydrate metabolism and oxidation reduction. The most significantly downregulated biological processes in the mms21RINGΔ mutant were translation, ribosome biogenesis, and related processes.
Figure 2.
Genes involved in ribosome biogenesis and protein translation are downregulated in the mms21RINGΔ mutant. (A) The top 20 biological process terms for upregulated and downregulated genes in mms21RINGΔ mutant are shown. (B) Groups of genes involved in ribosome biogenesis and protein translation are downregulated in the mms21RINGΔ mutant. Various gene groups were examined for differential expression relative to random groups of genes using a mean-rank gene set test (Wu and Smyth 2012). The P-values for each gene group are shown as follows: Gcn4 P-value = 0.0049, size = 699; C/D snoRNAs P-value = 5.17E-09, size = 42; H/ACA snoRNAs P-value = 0.0038, size = 34; Processome P-value = 6.64E-21, size = 63; RPL P-value = 6.79E-36, size = 78; and RPS P-value = 4.65E-25, size = 56. (C) Gene expression profiles of the eco1–W216G and mms21RINGΔ mutants are significantly correlated. Reads per kilobase of transcript per million mapped reads (RPKM) values were calculated from the read counts for the eco1–W216G mutant and WT and converted to log2 ratios for comparison to the mms21RINGΔ mutant. Genes common to both experiments are compared. A total of 6763 genes exhibit a Pearson correlation of 0.70. The log2 ratios for mms21RINGΔ/WT and eco1–W216G/WT were compared to each other. Values with a z score of >5 in the data set were excluded.
In order to further analyze the up- and downregulated genes, we examined individual gene groups (Figure 2B). We found that transcripts for both the large and small ribosomal proteins were significantly downregulated in the mms21RINGΔ mutant (mean rank gene set test; large 6.79E-36, small 4.65E-25). We also observed a significant downregulation in small nucleolar RNAs (snoRNAs) (box CD 5.17E-09 and box H/ACA 0.0038). The box CD snoRNAs are hosted in introns of the ribosomal protein genes, so their downregulation correlates with the expression of these genes. Many genes involved in the processing steps of ribosome biogenesis and the post-transcriptional processing of noncoding RNAs (ncRNAs) were also downregulated (Processome 6.64E-21).
Gcn4 is a transcriptional activator that is translated when cells are under stress or nutritional starvation. Many targets of Gcn4 were upregulated in the mms21RINGΔ mutant (Figure 2B). Of the 699 genes with a Gcn4 binding site, 99 were present in the upregulated gene group, while only 40 were present in the downregulated gene group (Fisher exact test 0.0049, see Materials and Methods and Table S1). We previously characterized a mutant in the cohesin ECO1 acetyltransferase (eco1–W216G) that had similarities in its gene expression profile (Bose et al. 2012). For example, many targets of Gcn4 were upregulated (Bose et al. 2012). The Pearson correlation between eco1–W216G and mms21RINGΔ gene expression is 0.7 (Figure 2C). These results are consistent with the idea that the Smc5/6 complex is necessary for nucleolar function and suggest that Mms21 is important for a normal gene expression profile with respect to ribosome biogenesis and translation.
The mms21RINGΔ mutant exhibits defects in ribosome subunit export
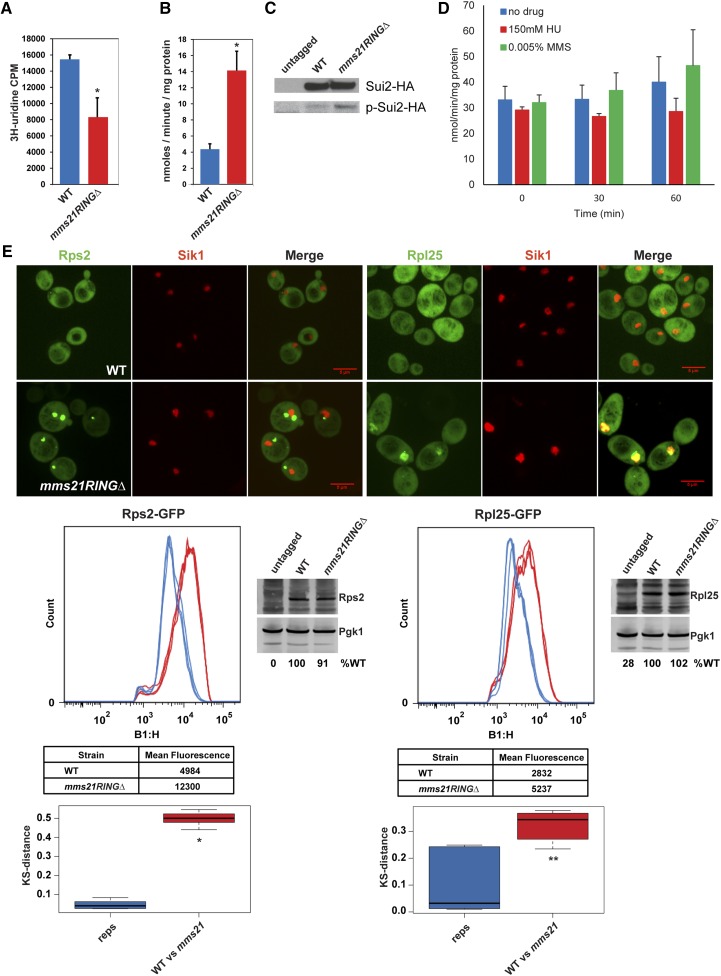
We next examined rRNA synthesis in the mms21RINGΔ mutant. We measured rRNA synthesis in the mms21RINGΔ mutant by performing a 3H-uridine incorporation assay. As shown in Figure 3A, total rRNA production was significantly reduced in the mutant. This result is consistent with defects in rRNA biogenesis in the mutant. Despite the reduced level of rRNA and messenger RNAs (mRNAs) encoding ribosomal proteins, we could not detect a reduction in global protein synthesis in the mutant (data not shown).
Figure 3.
The mms21RINGΔ mutant exhibits ribosome biogenesis defects. (A) rRNA production is reduced in mms21RINGΔ mutant. Both WT and mms21RINGΔ mutant cells were grown to midlog phase in SD −Ura medium containing 6.7 ng/μl uracil. The cultures were labeled with 3H-uridine for 5 min at 30°. Total rRNA level was measured by scintillation counting. Error bars indicate the standard deviation from three independent experiments. P = 0.0034. (B) Gcn4 translation is increased in the mms21RINGΔ mutant. Both WT and mms21RINGΔ cells were transformed with the p180–Gcn4–LacZ reporter plasmid and were grown to midlog phase at 30°. β-Galactosidase activity was measured using ONPG as a substrate. Error bars indicate the standard deviations from three independent experiments, P = 0.0012. (C) Sui2 is more phosphorylated in the mms21RINGΔ mutant compared to WT. The indicated strains were grown to midlog phase at 30°. Phosphorylation level of Sui2HA, in both WT and mms21RINGΔ strains was analyzed using anti-phospho eIF2α (Serine 51, Cell Signaling) and anti-HA antibodies via Western blotting. (D) Gcn4 translation is not elevated by treatment of WT cells with either 150 mM HU or 0.005% MMS for 30 or 60 min. WT cells were grown to midlog phase and treated with either 150 mM HU or 0.005% MMS for 30 or 60 min at 30°. Gcn4 translation level was measured as shown in B. (E) Rps2 and Rpl25 ribosomal proteins accumulate in mms21RINGΔ mutant. Both WT and mms21RINGΔ mutants were transformed with either the Rps2–GFP or Rpl25–GFP reporter plasmid. Live cell images were taken using confocal microscopy with a ×100 objective (Perkin Elmer Ultraview Spinning Disk) and the Volocity 6.3 software program. Bars, 5 μm. For Rps2–GFP or Rpl25–GFP quantification, the strains carrying Rps2–GFP or Rpl25–GFP were grown to midlog phase at 30° in SD −Leu medium supplemented with 0.02 mg/ml adenine. Mean peak GFP fluorescence intensity was measured by performing cytometry analysis. The number of cells or “count” is shown on the y-axis. GFP intensity histograms were plotted using the Macsquant B1-H::FITC-H detector (B1-H, x-axis). The x-axis represents the log scaled pulse height parameter from detector B1, which uses 488-nm excitation and collects fluorescence emission using a 525/50 bandpass filter. The KS test was used to measure the distances between three biological replicates (reps) and the distances between WT and mutants (WT vs. mms21). P-values were measured using a t-test. * P = 1.8953e-13, ** P = 0.00322.
Gcn4 translation is upregulated to promote the transcription of its target genes such as genes involved in amino acid biosynthetic pathways. Gcn4 translation has been extensively used as an indicator of translational stress (Hinnebusch 1997). Thus, we decided to analyze whether Gcn4 translation was affected in the mms21RINGΔ mutant at permissive temperature. The translation of Gcn4 was assessed from both a WT strain and the mms21RINGΔ mutant harboring the p180–Gcn4–lacZ plasmid by measuring β-galactosidase activity. This assay demonstrated that the β-galactosidase activity in the mms21RINGΔ mutant was elevated by threefold, compared to WT (Figure 3B), similar to previous observations in the eco1–W216G mutant (Bose et al. 2012). Consistent with translational stress, we found that Sui2 (eif2α) was more phosphorylated in the mms21RINGΔ mutant as compared to WT (Figure 3C).
We were concerned that the elevation in Gcn4 might be due to the constitutive activation of the S-phase checkpoint in the mms21RINGΔ mutant, so we examined β-galactosidase activity in a WT strain treated with either 150 mM HU or 0.005% MMS. In neither case did the treatment elevate the levels of β-galactosidase activity (Figure 3D), suggesting that DNA damage or replication slow down alone is not sufficient to promote Gcn4 translation. A previous study with a higher concentration of MMS found that MMS could induce Gcn4, but independently of the DNA damage checkpoint, suggesting Gcn4 translational activation can be separated from checkpoint activation (Natarajan et al. 2001). Furthermore, γ-irradiation, which causes DNA damage, does not affect Gcn4 levels, further suggesting that DNA damage and Gcn4 activation are not necessarily linked (Jelinsky et al. 2000). Therefore, the translational stress in the mms21RINGΔ mutant is not necessarily due to the activation of the DNA damage checkpoint.
Next, we monitored the localization of both the large and small subunits of ribosomal proteins in both WT and the mms21RINGΔ mutant at permissive temperature. The Rps2– and Rpl25–GFP reporter plasmids have been previously used to visualize the localization of the 40S and 60S subunit ribosomal proteins, respectively (Bose et al. 2012). Both Rps2– and Rpl25–GFP proteins are evenly distributed throughout the cytoplasm in WT cells. Accumulation of these proteins into foci is often indicative of a defect in ribosome biogenesis. Although endogenous ribosomal protein transcripts are downregulated overall, Rps2–GFP and Rpl25–GFP expressed from the plasmids are present at normal levels in whole cell extracts. These proteins accumulate in the nucleus in the mms21RINGΔ mutant (Figure 3E), suggesting that ribosome assembly or export might be impaired in this mutant. The Rpl25–GFP protein colocalized with Sik1–RFP in this mutant, indicating that Rpl25, a 60S ribosomal protein, accumulates in the nucleolus. Interestingly, the Rps2–GFP protein did not colocalize with Sik1–RFP, a nucleolar marker, in the mutant; but instead seemed to be predominantly nuclear. In order to quantify the ribosomal protein accumulations in the mms21RINGΔ mutant, we carried out cytometry analysis for peak fluorescence for the reporters (Kim 2016). These results showed that the accumulation of both the 40S and the 60S ribosomal proteins in the nucleus and nucleolus was significantly increased in the mms21RINGΔ mutant (Figure 3E), suggesting a defect in ribosome biogenesis. These results suggest that Mms21 supports ribosome biogenesis and prevents translational stress.
Dosage of RPL19A regulates ribosomal protein accumulation in the mms21RINGΔ mutant
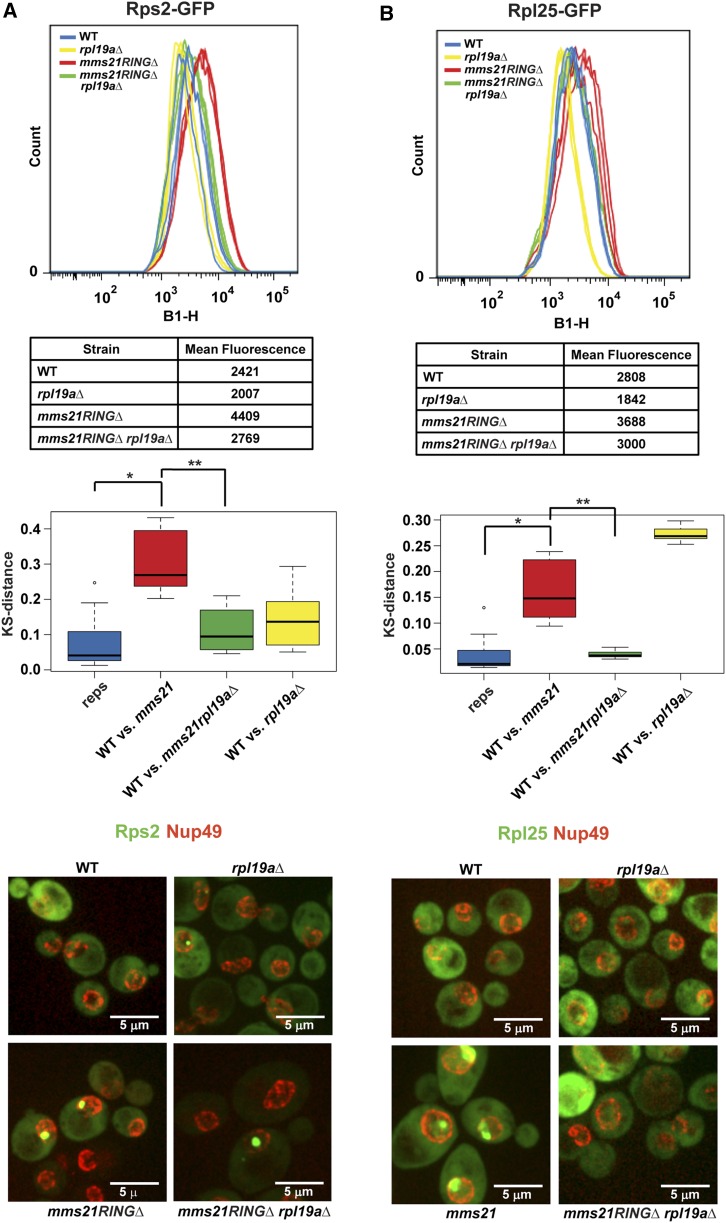
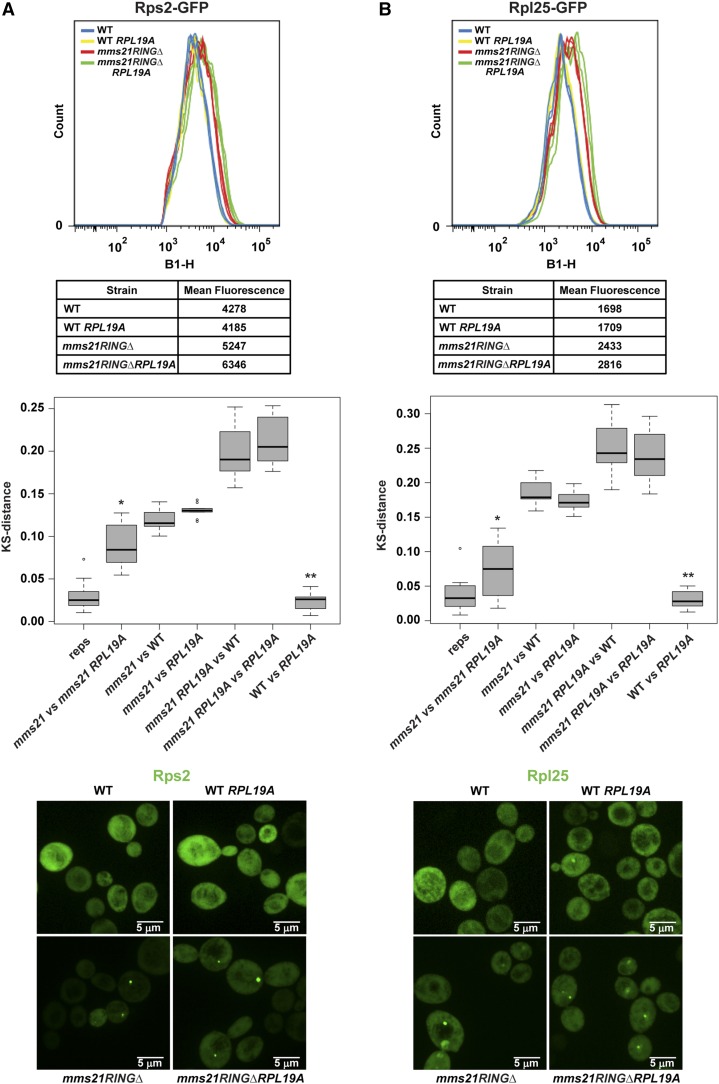
Since the mms21RINGΔ mutant accumulates both the 40S and the 60S ribosomal subunits in the nucleus and the nucleolus (Figure 3E), we examined whether this phenotype was rescued by deletion of RPL19A. The cytometry analysis, which was also verified by microscopy, indicated that nuclear/nucleolar accumulation of the 40S and the 60S ribosomal proteins in the mms21RINGΔ mutant were significantly reduced by the deletion of RPL19A (Figure 4). Taken together, these data suggest that the deletion of RPL19A suppresses defective localization of the two ribosomal proteins examined in the mms21RINGΔ mutant.
Figure 4.
Deletion of RPL19A partially suppresses ribosomal protein accumulation in the mms21RINGΔ mutant. (A) Deletion of RPL19A partially suppresses Rps2–GFP accumulation in the mms21RINGΔ mutant. Measurement of mean peak GFP fluorescence intensity was performed as described in Figure 3E using the indicated strains. * P = 1.9141E-05, ** P = 6.258E-06. For microscopy images in both A and B, the nuclear periphery is marked with Nup49–mCherry. (B) Deletion of RPL19A partially suppresses Rpl25–GFP accumulation in the mms21RINGΔ mutant. Mean peak GFP fluorescence intensity was measured as shown in Figure 3E. * P = 7.5033E-05, ** P = 0.0006041.
Given that the deletion of RPL19A suppresses the accumulation of both the 60S and the 40S subunits in the mms21RINGΔ mutant, we wondered whether overexpression of RPL19A would exacerbate the accumulation of ribosomal proteins in the mutant. To test this idea, RPL19A was overexpressed under the GAL1–10 promoter at permissive temperature. Under these conditions, there is no additional growth defect in the mutant and levels of β-galactosidase from the Gcn4–lacZ reporter are not further elevated (not shown). Rps2– and Rpl25–GFP mean peak fluorescence intensity was measured in WT and the mms21RINGΔ strains by conducting a cytometry assay. Overexpression of RPL19A did not increase the peak fluorescence intensity of Rps2–GFP in WT cells (Figure 5A). However, RPL19A overexpression further increased the mean fluorescence intensity of Rps2–GFP in the mms21RINGΔ mutant, suggesting that the 40S ribosomal subunit accumulation is exacerbated. We also examined the effect of the RPL19A overexpression on the Rpl25–GFP protein in both WT and the mms21RINGΔ mutant (Figure 5B). Unlike the strong effect on Rps2 accumulation, the overexpression of RPL19A only modestly increased Rpl25–GFP accumulation in the mms21RINGΔ mutant. These experiments indicate that overproduction of a ribosomal subunit can exacerbate the nuclear accumulation of ribosome components in the mms21RINGΔ mutant, suggesting that export of some ribosomal proteins may become more compromised.
Figure 5.
Overexpression of RPL19A exacerbates Rps2–GFP accumulation in the mms21RINGΔ mutant. (A) Accumulation of Rps2–GFP in the mms21RINGΔ mutant is enhanced by overexpression of RPL19A. Both WT and mms21RINGΔ cells were transformed with either empty plasmid or pBY011–RPL19A plasmid containing the GAL1,10 promoter. The indicated strains were used to measure mean peak GFP fluorescence intensity as shown in Figure 3E. * P = 6.10E-05, ** P = 0.3972. (B) Overexpression of RPL19A does not significantly increase accumulation of Rpl25–GFP in the mms21RINGΔ mutant. The indicated strains were transformed with either empty plasmid or pBY011–RPL19A plasmid carrying the GAL1,10 promoter. Rpl25–GFP mean peak fluorescence intensity was measured as shown in Figure 3E. * P = 0.0437, ** P = 0.3751.
The smc5-6 mutant accumulates both the small and large subunits of ribosomal proteins
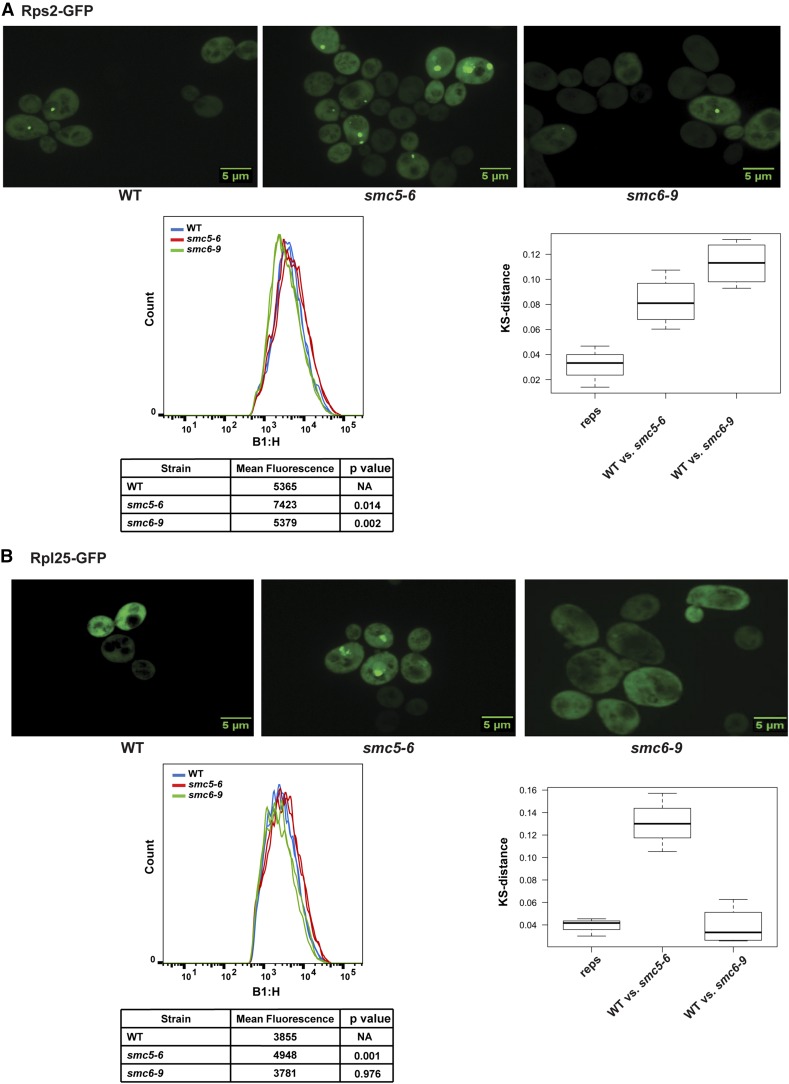
Our data show that the mms21RINGΔ mutant, in which SUMO ligase activity is deficient, exhibits defects in the export of ribosomal subunits. A recent report suggests that the SUMO ligase activity of Mms21 is required for Smc5/6 complex-mediated DNA repair (Bermudez-Lopez et al. 2015). Thus, we wanted to test whether other subunits of the Smc5/6 complex also contribute to regulation of ribosome biogenesis. We examined the localization of both small and large subunits of ribosomal proteins in the smc5-6 mutant at permissive temperature. Both Rps2– and Rpl25–GFP proteins accumulated in the smc5-6 mutant compared to WT, but not in the smc6-9 mutant (Figure 6). The phenotype of the smc5-6 mutant was milder than in the mms21RINGΔ mutant and elevated β-galactoside levels were not detected using the Gcn4–lacZ reporter (not shown). Unlike the mms21RINGΔ mutant, the smc5-6 mutant has no detectable growth defect at 30°, which may partially explain the mild phenotype of this mutant at 30°. However, the accumulation of ribosomal proteins in the smc5-6 mutant is consistent with the idea that Smc5/6 complex function may be required for normal ribosome biogenesis. While it is possible that the Mms21 SUMO ligase acts independently from the Smc5/6 complex for nucleolar function, it seems more likely that deletion of the RING domain of MMS21 may provide a surgical removal of the SUMO ligase activity compared to temperature-sensitive mutations in SMC5 or SMC6, accounting for why the RING deletion provides the strongest phenotype.
Figure 6.
Both Rps2–GFP and Rpl25–GFP accumulate in the smc5-6 mutant. (A and B) Both the 40S and 60S subunits of ribosomal protein reporters accumulate in the smc5-6 mutant but not the smc6-9 mutant. Strains were transformed with either the Rps2–GFP or Rpl25–GFP reporter plasmid. Images were taken as described in Figure 3E. For either Rps2–GFP (A) or Rpl25–GFP (B) quantification, the strains carrying either Rps2–GFP or Rpl25–GFP were grown to midlog phase at 28° in dropout medium supplemented with 0.02 mg/ml adenine. Mean peak Rps2–GFP (A) or Rpl25–GFP (B) fluorescence intensity was measured by performing cytometry analysis as described in Figure 3E. P-values were measured using a t-test.
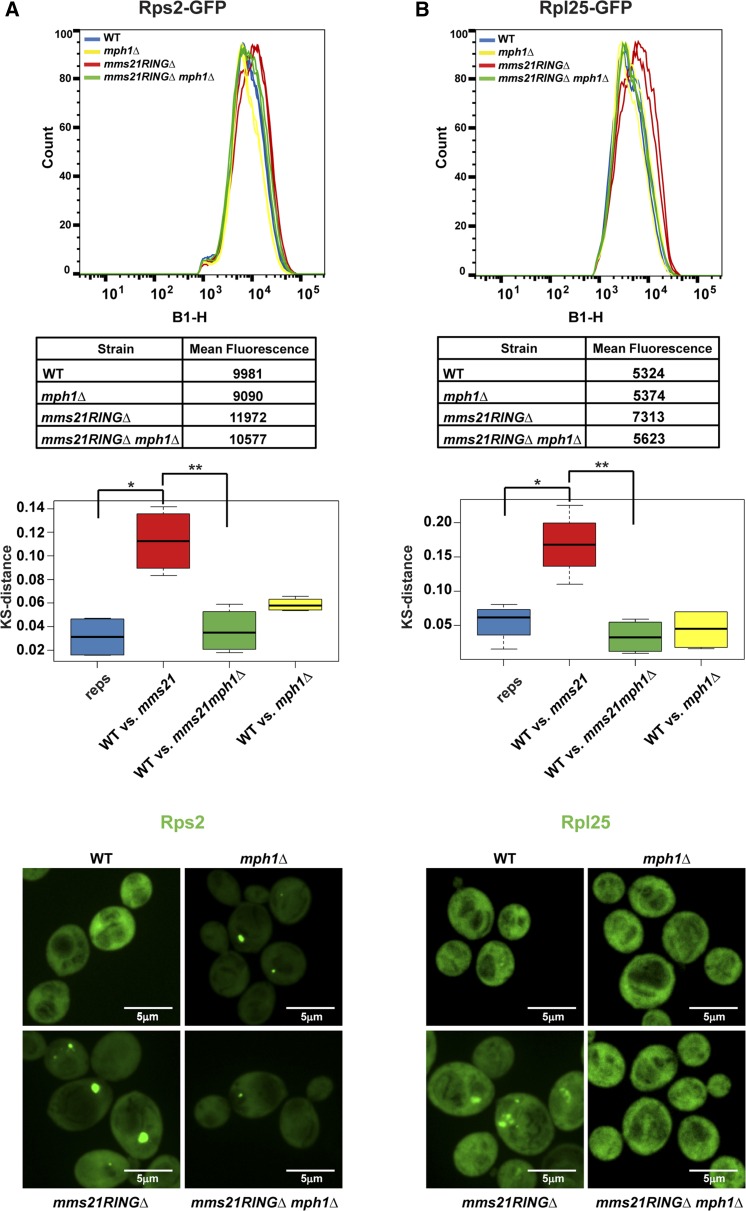
Deletion of MPH1 partially rescues ribosomal protein accumulation, rRNA production, and translational stress in the mms21RINGΔ mutant
Deletion of MPH1 has previously been shown to rescue the MMS and temperature sensitivity of the mms21RINGΔ mutant (Chen et al. 2009). MPH1 encodes a helicase that operates during replication-associated recombination. The deletion of MPH1 in the context of the mms21RINGΔ mutation has been shown to reduce the accumulation of unresolved toxic recombination intermediates. We verified that deletion of MPH1 or RAD54, a DNA-dependent ATPase, suppresses the mms21RINGΔ mutant growth phenotypes, with MPH1 having a stronger effect (Figure S2). In addition, we found that deletion of MPH1 could rescue the nuclear accumulation of Rps2–GFP and Rpl25–GFP (Figure 7).
Figure 7.
Deletion of MPH1 partially suppresses ribosomal protein accumulation in the mms21RINGΔ mutant. Measurement of Rps2–GFP (A) and Rpl25–GFP (B) fluorescence intensity was performed as described in Figure 3E using the indicated strains. For A, * P = 0.004005, ** P = 0.6924. For B, * P = 0.009664, ** P = 0.2997.
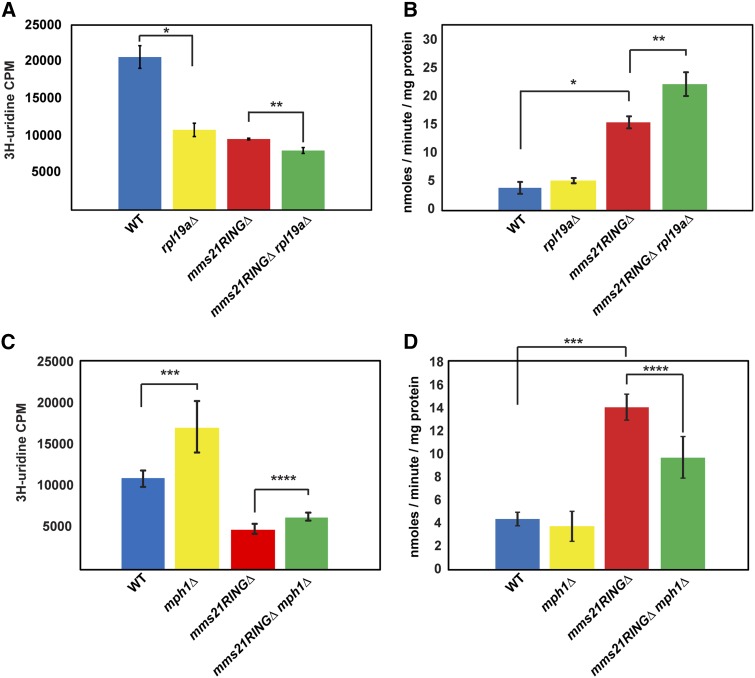
We further examined whether the deletion of RPL19A or MPH1 could rescue rRNA production in the mms21RINGΔ mutant at permissive temperature. Intriguingly, rRNA synthesis in the rpl19aΔ mutant was also reduced, compared to WT. rRNA production in the mms21RINGΔ rpl19aΔ double mutant was slightly but significantly decreased relative to the rpl19aΔ and the mms21RINGΔ single mutants, suggesting that the deletion of RPL19A does not rescue the rRNA production defect observed in the mms21RINGΔ mutant (Figure 8A). Deletion of MPH1 alone appears to increase rRNA synthesis and provides a slight but significant rescue of rRNA production in the context of the mms21RINGΔ mutant (Figure 8C).
Figure 8.
Deletion of MPH1 but not RPL19A partially rescues rRNA production and Gcn4 translation in the mms21RINGΔ mutant. (A) The rRNA production defect in the mms21RINGΔ mutant is not rescued by deletion of RPL19A. The indicated strains were used for a 3H-uridine incorporation assay as shown in Figure 3A. Error bars represent the standard deviations, * P = 0.0007, ** P = 0.0028 (t-test). (B) Elevated Gcn4 translation in the mms21RINGΔ mutant is not suppressed by deletion of RPL19A. β-Galactosidase assays were performed with the indicated strains as described for Figure 3B. Error bars indicate the standard deviations, * P = 4.43E-25, ** P = 1.58E-12 (t-test). (C) The rRNA production defect in the mms21RINGΔ mutant is slightly improved by deletion of MPH1. The indicated strains were used for a 3H-uridine incorporation assay as described in Figure 3A. Error bars represent the standard deviations, *** P = 0.0322, **** P = 0.0294 (t-test). (D) Elevated Gcn4 translation in the mms21RINGΔ mutant is partially reduced by deletion of MPH1. β-Galactosidase assays were performed with the indicated strains as described for Figure 3B. Error bars indicate the standard deviations, *** P = 4.02E-15, **** P = 8.61E-07 (t-test).
We next focused on translational stress. We examined Gcn4 translation in the mms21RINGΔ rpl19aΔ and mms21RINGΔ mph1Δ double mutants to determine whether either deletion could suppress the elevated β-galactosidase activity in the mms21RINGΔ mutant. The mms21RINGΔ rpl19aΔ double mutant had even higher levels of β-galactosidase activity than observed in the mms21RINGΔ mutant (Figure 8B), suggesting that translational stress was not relieved, even though the ribosomal proteins accumulate less. In contrast, we found that the mms21RINGΔ mph1Δ double mutant had significantly lower levels of β-galactosidase activity compared to the mms21RINGΔ mutant strain (Figure 8D), suggesting that in this case, nuclear accumulation of ribosomal proteins, rRNA production, and translational stress are partially relieved by MPH1 deletion. This suggests that the elimination of toxic recombination intermediates helps to partially restore normal translation in the mms21RINGΔ mutant.
Discussion
Previous reports have shown that Smc5/6 and its SUMO ligase activity provided by Mms21 are crucial for the roles of the complex in sister chromatid cohesion, restart of stalled DNA replication forks, and DNA damage repair (Rai et al. 2011; Almedawar et al. 2012; McAleenan et al. 2012; Wu et al. 2012; Bermudez-Lopez et al. 2015). While the mms21RINGΔ mutant had been previously reported to have aberrant nucleolar morphology (Zhao and Blobel 2005), and Mms21 had been reported to target some nucleolar proteins for sumoylation (Albuquerque et al. 2013), we now provide evidence that the Smc5/6 complex significantly contributes to nucleolar function. In this study, we have shown that Smc5/6, and in particular the SUMO ligase activity of the Mms21 subunit, are important to promote rRNA production, assembly and/or export of ribosomal proteins, and normal translation.
Our study identified RPL19A as a novel genetic suppressor of the mms21RINGΔ mutant. We observed reduced rRNA production, nuclear accumulation of ribosomal proteins, and elevated Gcn4 translation in the mms21RINGΔ mutant. We speculate that deletion of RPL19A may partially relieve an imbalance between ribosomal RNA and ribosomal protein that causes the accumulation of ribosomal protein–rRNA complexes in the nucleus in the mms21RINGΔ mutant. We speculate that overexpression of RPL19A increases this imbalance. However, it appears that the rescue provided by deletion of RPL19A is not sufficient to restore the production of rRNA, or to rescue translational stress.
We have previously shown that the related SMC complex cohesin is important for ribosome biogenesis. Mms21-dependent sumoylation of cohesin has been shown to be important for cohesion (Takahashi et al. 2008; McAleenan et al. 2012; Wu et al. 2012). Thus, one possible explanation for our findings is that mutations affecting Smc5/6 function are disrupting cohesion, which has already been shown to be important for nucleolar function. Other mechanisms are also possible, including that (1) Mms21 sumoylation of nucleolar proteins is critical for nucleolar function; (2) the role of Smc5/6 in DNA replication and DSB repair, in particular at the rDNA, is important for nucleolar function; or (3) the role of Mms21 in nuclear–cytoplasmic trafficking affects nucleolar function (Rothenbusch et al. 2012). Lending support to the second mechanism, deletion of MPH1, which blocks the formation of toxic recombination intermediates (Chen et al. 2009), partially suppressed translational stress in the mms21RINGΔ mutant. This result is reminiscent of the rescue provided by deletion of FOB1 in the eco1–W216G strain (Lu et al. 2014). Deletion of FOB1 allows bidirectional replication of the ribosomal DNA, which partially rescued low rRNA production and translational stress associated with the cohesin acetyltransferase mutant. In the future, it will be important to further determine the molecular mechanisms by which Smc5/6 contributes to nucleolar function.
Given that both cohesin and Smc5/6 promote nucleolar function, a picture starts to emerge in which stress at the rDNA may trigger a translational stress response regulated by this ancient group of SMC complexes. Interestingly, the role of cohesion in nucleolar function and translation appears to be evolutionarily conserved and an important part of the etiology of a group of developmental syndromes known as the cohesinopathies (Xu et al. 2013; Xu et al. 2015). Increasing translation in zebrafish models for the cohesinopathies had dramatic rescue effects on developmental phenotypes. A recent report demonstrated that mutations in human NSMCE2/MMS21 lead to primordial dwarfism and insulin resistance (Payne et al. 2014). We speculate that nucleolar dysfunction caused by mutations in MMS21 might contribute to this human syndrome.
Acknowledgments
We thank Malcolm Moses for his technical contributions to the experiments and Richard Shrock for assistance with manuscript preparation and submission. We also thank the Molecular Biology Core at the Stowers Institute for Medical Research for RNA-sequencing libraries and for sequencing and Hua Li for the Kolmogorov–Smirnov statistic test.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.181750/-/DC1.
Communicating editor: O. Cohen-Fix
Literature Cited
- Albuquerque C. P., Wang G., Lee N. S., Kolodner R. D., Putnam C. D., et al. , 2013. Distinct SUMO ligases cooperate with Esc2 and Slx5 to suppress duplication-mediated genome rearrangements. PLoS Genet. 9: e1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almedawar S., Colomina N., Bermudez-Lopez M., Pocino-Merino I., Torres-Rosell J., 2012. A SUMO-dependent step during establishment of sister chromatid cohesion. Curr. Biol. 22: 1576–1581. [DOI] [PubMed] [Google Scholar]
- Bermudez-Lopez M., Pocino-Merino I., Sanchez H., Bueno A., Guasch C., et al. , 2015. ATPase-dependent control of the Mms21 SUMO ligase during DNA repair. PLoS Biol. 13: e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose T., Lee K. K., Lu S., Xu B., Harris B., et al. , 2012. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet. 8: e1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. H., Choi K., Szakal B., Arenz J., Duan X., et al. , 2009. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc. Natl. Acad. Sci. USA 106: 21252–21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris B., Bose T., Lee K. K., Wang F., Lu S., et al. , 2014. Cohesion promotes nucleolar structure and function. Mol. Biol. Cell 25: 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., 1997. Translational regulation of yeast GCN4: a window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 272: 21661–21664. [DOI] [PubMed] [Google Scholar]
- Jack K., Bellodi C., Landry D. M., Niederer R. O., Meskauskas A., et al. , 2011. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 44: 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky S. A., Estep P., Church G. M., Samson L. D., 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 20: 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H, A. C. Box, H. Li, and J. L. Gerton, 2016 Using fluorescent reporters in conjunction with cytometry and statistics to assess nuclear accumulation of ribosomal proteins in Methods in Molecular Biology, Vol. 1515, edited by K. Yokomori and K. Shirahige. Humana Press (in press). [DOI] [PubMed]
- Kressler D., Hurt E., Bassler J., 2010. Driving ribosome assembly. Biochim. Biophys. Acta 1803: 673–683. [DOI] [PubMed] [Google Scholar]
- Lempiainen H., Shore D., 2009. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 21: 855–863. [DOI] [PubMed] [Google Scholar]
- Lindroos H. B., Strom L., Itoh T., Katou Y., Shirahige K., et al. , 2006. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 22: 755–767. [DOI] [PubMed] [Google Scholar]
- Lu S., Goering M., Gard S., Xiong B., McNairn A. J., et al. , 2010. Eco1 is important for DNA damage repair in S. cerevisiae. Cell Cycle 9: 3315–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Lee K. K., Harris B., Xiong B., Bose T., et al. , 2014. The cohesin acetyltransferase Eco1 coordinates rDNA replication and transcription. EMBO Rep. 15: 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleenan A., Cordon-Preciado V., Clemente-Blanco A., Liu I. C., Sen N., et al. , 2012. SUMOylation of the alpha-kleisin subunit of cohesin is required for DNA damage-induced cohesion. Curr. Biol. 22: 1564–1575. [DOI] [PubMed] [Google Scholar]
- Montanaro L., Trere D., Derenzini M., 2008. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 173: 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. M., Carr A. M., 2008. Smc5/6: A link between DNA repair and unidirectional replication? Nat. Rev. Mol. Cell Biol. 9: 177–182. [DOI] [PubMed] [Google Scholar]
- Natarajan K., Meyer M. R., Jackson B. M., Slade D., Roberts C., et al. , 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21: 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne F., Colnaghi R., Rocha N., Seth A., Harris J., et al. , 2014. Hypomorphism in human NSMCE2 linked to primordial dwarfism and insulin resistance. J. Clin. Invest. 124: 4028–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P. R., 2009. The Yin and Yang of the MMS21–SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair (Amst.) 8: 499–506. [DOI] [PubMed] [Google Scholar]
- Rai R., Varma S. P., Shinde N., Ghosh S., Kumaran S. P., et al. , 2011. Small ubiquitin-related modifier ligase activity of Mms21 is required for maintenance of chromosome integrity during the unperturbed mitotic cell division cycle in Saccharomyces cerevisiae. J. Biol. Chem. 286: 14516–14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenbusch U., Sawatzki M., Chang Y., Caesar S., Schlenstedt G., 2012. Sumoylation regulates Kap114-mediated nuclear transport. EMBO J. 31: 2461–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D., 2013. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 5: pii: a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A. K., Kliszczak M., Morrison C. G., 2011. The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett. 585: 2907–2913. [DOI] [PubMed] [Google Scholar]
- Strunk B. S., Karbstein K., 2009. Powering through ribosome assembly. RNA 15: 2083–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Dulev S., Liu X., Hiller N. J., Zhao X., et al. , 2008. Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet. 4: e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J., Machin F., Aragon L., 2005a Smc5-Smc6 complex preserves nucleolar integrity in S. cerevisiae. Cell Cycle 4: 868–872. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J., Machin F., Farmer S., Jarmuz A., Eydmann T., et al. , 2005b SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 7: 412–419. [DOI] [PubMed] [Google Scholar]
- Woolford J. L., Jr, Baserga S. J., 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Smyth G. K., 2012. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40: e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Kong X., Ji Z., Zeng W., Potts P. R., et al. , 2012. Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl. Genes Dev. 26: 1473–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Lee K. K., Zhang L., Gerton J. L., 2013. Stimulation of mTORC1 with L-leucine rescues defects associated with Roberts syndrome. PLoS Genet. 9: e1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Sowa N., Cardenas M. E., Gerton J. L., 2015. L-leucine partially rescues translational and developmental defects associated with zebrafish models of Cornelia de Lange syndrome. Hum. Mol. Genet. 24: 1540–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Horigome C., Okada T., Shirai C., Mizuta K., 2007. Yeast Rrp14p is a nucleolar protein involved in both ribosome biogenesis and cell polarity. RNA 13: 1977–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakari M., Yuen K., Gerton J. L., 2015. Etiology and pathogenesis of the cohesinopathies. Wiley Interdiscip. Rev. Dev. Biol. 4: 489–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanchin N. I., Roberts P., DeSilva A., Sherman F., Goldfarb D. S., 1997. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol. Cell. Biol. 17: 5001–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Blobel G., 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]