Abstract
We have recently reported that mice born from dams stressed during pregnancy (PRS mice), in adulthood, have behavioral deficits reminiscent of behaviors observed in schizophrenia (SZ) and bipolar (BP) disorder patients. Furthermore, we have shown that the frontal cortex (FC) and hippocampus of adult PRS mice, like that of postmortem chronic SZ patients, are characterized by increases in DNA-methyltransferase 1 (DNMT1), ten-eleven methylcytosine dioxygenase 1 (TET1) and exhibit an enrichment of 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) at neocortical GABAergic and glutamatergic gene promoters. Here, we show that the behavioral deficits and the increased 5MC and 5HMC at glutamic acid decarboxylase 67 (Gad1), reelin (Reln) and brain-derived neurotrophic factor (Bdnf) promoters and the reduced expression of the messenger RNAs (mRNAs) and proteins corresponding to these genes in FC of adult PRS mice is reversed by treatment with clozapine (5 mg kg−1 twice a day for 5 days) but not by haloperidol (1 mg kg−1 twice a day for 5 days). Interestingly, clozapine had no effect on either the behavior, promoter methylation or the expression of these mRNAs and proteins when administered to offspring of nonstressed pregnant mice. Clozapine, but not haloperidol, reduced the elevated levels of DNMT1 and TET1, as well as the elevated levels of DNMT1 binding to Gad1, Reln and Bdnf promoters in PRS mice suggesting that clozapine, unlike haloperidol, may limit DNA methylation by interfering with DNA methylation dynamics. We conclude that the PRS mouse model may be useful preclinically in screening for the potential efficacy of antipsychotic drugs acting on altered epigenetic mechanisms. Furthermore, PRS mice may be invaluable for understanding the etiopathogenesis of SZ and BP disorder and for predicting treatment responses at early stages of the illness allowing for early detection and remedial intervention.
Introduction
Evidence from neuroimaging, neuropathology and epidemiological studies has led to the conclusion that schizophrenia (SZ) and bipolar (BP) disorder are neurodevelopmental disorders with an etiology that likely originates before birth.1, 2, 3, 4, 5 Related to this, it has been reported that prenatal or perinatal exposure to stress, inflammation or trauma disrupts brain development and increases the risk for developing SZ and related psychiatric disorders including BP disorder, anxiety and schizoaffective disorders.5, 6, 7, 8, 9, 10
Young adult (postnatal day (PND) 75) male mice, born from mothers stressed during pregnancy, here defined as PRS mice, exhibit a behavioral phenotype characterized by hyperactivity, stereotyped and compulsive behaviors, deficits in social interaction, pre-pulse inhibition, fear conditioning, object recognition and hypersensitivity to N-methyl d-aspartate receptor blockers.1 PRS mice also show molecular alterations in cortical GABAergic interneurons,1, 11, 12 and altered GABA/glutamate neuron interactions13 in a manner that is reminiscent of changes reported in the brains of SZ and BP patients.14, 15, 16, 17, 18 Evidence suggests19 that a cortical GABAergic dysfunction has a major role in the pathogenesis of altered synchronization of pyramidal neuronal firing rate that is crucial for inducing cognitive dysfunction in SZ and BP disorder patients20, 21 and in rodents studied in a neurodevelopmental animal model of SZ.22
Molecularly, PRS mice show altered DNA methylation profiles and a disrupted chromatin structure at genes typically expressed in GABAergic neurons, such as those encoding gamma aminobutyric acid 67 (Gad1) and reelin (Reln), and in glutamatergic neurons, such as the gene encoding brain-derived neurotrophic factor (Bdnf).1, 2, 23
The molecular epigenetic profile common to the frontal cortex (FC) and hippocampus of adult PRS mice and SZ and BP disorder patients1, 2, 19, 24, 25, 26, 27, 28, 29, 30 includes an increase in DNA methylating enzymes: DNA methyltransferase 1 (DNMT1) and ten-eleven methylcytosine dioxygenase-1 (TET1) messenger RNAs (mRNAs) and proteins and an enrichment of 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) at the GAD1, RELN and BDNF promoters.
A reliable animal model of SZ should predict responsiveness to currently available antipsychotic drugs. Previous reports showed that the SZ-like behavioral alterations present in adult rodent offspring of prenatally stressed mothers, or mothers prenatally exposed to gestational immune activation, are corrected by the administration of clozapine1, 31, 32, 33 and valproate (VPA).1 Hence, we tested whether the epigenetic PRS mouse model possesses predictive validity by assessing the reversal of the behavioral and molecular epigenetic abnormalities by comparing the action of clozapine to that of haloperidol.
Clozapine, a high affinity monoaminergic receptor ligand preferring HTR1A and HTR2A receptors34, 35, 36, 37 and haloperidol, a monoaminergic receptor antagonist with selectivity for D2 receptors are both effective in treating some of the worst symptoms of psychosis, specifically the hallucinations, delusions and paranoia. However, only clozapine is effective in treating ‘positive symptoms' in antipsychotic ‘treatment-resistant SZ patients' and in attenuating ‘negative symptoms' and the ‘cognitive performance impairment' observed in SZ patients.38, 39
Clozapine is an antipsychotic with DNA demethylating properties,34, 35 whereas haloperidol is an antipsychotic that does not have the same chromatin remodeling properties as clozapine.34, 35, 36, 40 Here, we show that protracted (5 days+20 h washout) treatment with clozapine and VPA, but not haloperidol, normalize the DNA hypermethylation and behavioral alterations characteristic of PRS mice without inducing changes in nonstressed pregnant mice (NS mice). Mice born from non-stressed pregnant mothers, here defined as NS mice.
Materials and methods
Animals and PRS procedure
All procedures were performed according to NIH guidelines for animal research (Guide for the Care & Use of Laboratory Animals, NRC, 1996) and were approved by the Animal Care Committee of the University of Illinois at Chicago. Pregnant mice (Swiss albino ND4, Harlan, Indianapolis, IN, USA) were individually housed with a 12-h light–dark cycle, and food and water ad libitum. Control dams were left undisturbed throughout gestation, whereas stressed dams were subjected to repeated episodes of restraint stress, as described previously.1 The stress procedure consisted of restraining the pregnant dam in a transparent tube (12 × 3 cm) under a bright light for 45 min three times per day from the seventh day of pregnancy until delivery. After weaning (PND 21), male mice were selected for the study and housed four to five per cage separately by condition.
Drug treatment
Clozapine (Sandoz Pharmaceuticals, Princeton, NJ, USA) and haloperidol (Sigma, St Louis, MO, USA) were dissolved with a drop of glacial acetic acid brought to pH 6 with the addition of NaOH. The drugs (clozapine 5 mg kg−1 and haloperidol 1 mg kg−1) were administered to PRS and NS mice at PND 75 subcutaneously twice daily for five consecutive days. Vehicle was administered to corresponding control groups using a similar regimen. Behavioral tests were conducted on the day before starting treatment and were repeated 18 h after the last administration of antipsychotic or vehicle. Where indicated, animals were killed 2 h after the last behavioral test.
Behavioral tests
Locomotor activity and social interaction were examined in succession on the same day at PND 75 between 1000 and 1500 h. We selected PND 75 for behavioral testing because at this postnatal time, the performance of the offspring was more reproducible and stable than the performance measured at earlier developmental time points.
Locomotor activity
A computerized Animal Activity Monitoring System with VersaMax software (AccuScan Instruments, Columbus, OH, USA) was used for the quantification and tracking of locomotor activity in mice as described previously.1 Each activity cage consisted of a Perspex box (20 × 20 × 20 cm divided into quadrants) surrounded by horizontal and vertical infrared sensor beams. The total number of interruptions of the horizontal sensors (counts) was taken as a measure of horizontal activity, whereas that of vertical sensors was used as a measure of vertical activity. The activity was recorded for 15 min.
Social interaction
Apparatus: Social approach of PRS and NS mice was measured using the ‘Three-Chambers Apparatus' which is a rectangular, transparent three-chambered box with each chamber measuring 20 cm long × 40.5 cm wide × 22 cm high. Small openings in the clear Plexiglas walls (10 cm wide × 5 cm high) divide the center compartment from the two side compartments. Two identical wire cups were placed in the left and right chambers, one for enclosing a stranger (novel) mouse and one for a control object. Between tests, the apparatus was thoroughly washed with 70% ethanol and then distilled water. Tests were performed under dim and even lighting, and sessions were recorded on video tape for later data analysis.
Test: The test mouse was first placed in the center chamber and habituated by allowing it to freely explore the entire apparatus for 5 min. Then the mouse was gently coerced into the center chamber and confined by closing the openings to the side chambers. The stranger mouse was then placed in the wire cup of one of the side chambers and the test was initiated by opening both doorways allowing the test mouse to again explore all the three chambers freely for 10 min. Social approach or interaction was defined as the ratio of the sniffing time for the wire cup enclosing the stranger mouse vs the empty cup. The reliability of the measurements was assessed by correlating the scores of two raters. For additional details, see Dong et al.2
Biochemical measurements
Methyl-DNA immunoprecipitation, chromatin immunoprecipitation (ChIP) and western blot assays each required around 10 mg of tissue and were carried out using two separate tissue punches from each FC hemisphere. FC coronal sections were dissected 2 mm anterior to the Bregma line and the sampling sublocation was consistent from animal to animal.
Quantitative real-time PCR
The quantitative PCR measurements were carried out using the Applied Biosystems Real-Time PCR System with a SYBR green master mix (Fermentas, Glen Burnie, MD, USA). After behavioral tests, total RNA from the frontal lobe (FC) of PRS and NS mice, was isolated using TRIZOL reagent (Life Technologies, Grand Island, NY, USA), and was further purified using the RNeasy kit (QIAGEN, Valencia, CA, USA). Reverse transcription was performed as previously described.2 The primer sequences used to amplify the genes analyzed are summarized in Supplementary Table 1. Each sample was run in duplicate and repeated twice. For normalizing mRNA expression, several housekeeping mRNAs (neuron-specific enolase (Nse), neuronal nuclear antigen (NeuN) and ActB) were chosen as internal controls. For each housekeeping gene, we measured the gene stability ranking using the NormFinder algorithm.41 This procedure allows for the identification of the housekeeping gene best suited for normalization. Because each of the genes studied yielded similar results when normalized to either Nse, NeuN or ActB, and because ActB had the highest housekeeping gene stability (NormFinder), we normalized our data ActB.
Western blot analysis
For protein quantification, we conducted measurements as described in detail elsewhere.2 Anti-DNMT1 monoclonal antibodies (Imagenex, San Diego, CA, USA), anti-TET1 polyclonal antibodies, (Millipore, Billerica, MA, USA), anti-GAD65/GAD67 polyclonal antibodies (Chemicon, Temecula, CA, USA), anti-RELN G-10 monoclonal antibodies (a generous gift of AM Goffinet, University of Namur, Brussels, Belgium) and anti-BDNF polyclonal antibodies (Santa Cruz, CA, USA) were used to detect DNMT1, TET1, GAD65, GAD67, RELN and BDNF proteins, respectively. The levels of these proteins in PRS vs NS were normalized by ActB protein levels using Act-B monoclonal antibodies (Sigma).
Methylated DNA immunoprecipitation
Methylated and hydroxymethylated DNA at the Gad1, Reln, and Bdnf-iv, -vi and -ix promoters were assessed using MeDIP and hMeDIP kits, respectively (Diagenode, Denville, NJ, USA) followed by quantitative PCR. Sample treatment and immunoprecipitation were performed as described by the manufacturer. We used selective immunoprecipitation to measure DNA methylation because bisulfite and many enzyme-dependent methods are incapable of distinguishing 5MC from the ~14% of methylcytosines in the brain that are 5HMC. Recent studies demonstrate that 5MC and 5HMC have very different functions and genomic locations.42, 43, 44 The percentage of methylated (or hydroxymethylated) vs unmodified promoter was calculated using the following equation: % [(me, hme DNA−IP/total input)]=2[(Ct(10% input)−3.32)−Ct(meDNA−IP)] × 100%.
ChIP assays
We performed ChIP assays based on protocols previously described.2 The percentages of immunoprecipitated DNA were calculated as described for MeDIP/hMeDIP. ChIP grade anti-DNMT1 (Imagenex) and anti-MeCP2 polyclonal antibodies (Millipore) were used to precipitate cross-linked chromatin.
Statistical analysis
Results are expressed as mean±s.e.m. Experimental differences were assessed by Student's t-test or by one-way analysis of variance followed by Student–Newman–Keuls post hoc comparisons, or two-way analysis of variance for repeated measurements using Predictive Analytics Software v.18 (SPSS, Chicago, IL, USA). The criterion for significance was P<0.05, two tailed.
Results
Clozapine and VPA, but not haloperidol, corrects the behavioral alterations in PRS mice
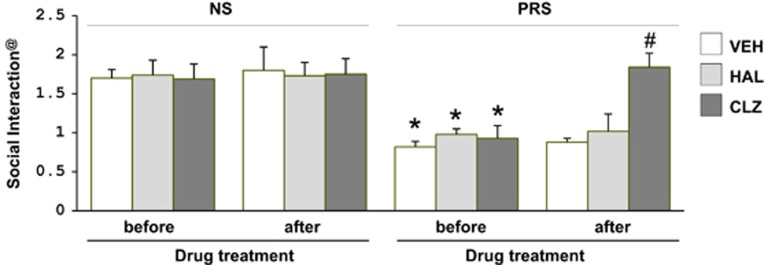
Consistent with our previous reports,1, 2, 23 we show that young adult (PND 75) PRS mice exhibit locomotor hyperactivity in an open-field arena (Table 1) and demonstrate a marked social interaction deficit (Figure 1). These behavioral alterations were corrected by protracted treatment with clozapine (5 mg kg−1 subcutaneously, twice a day for 5 days, followed by 18 h of washout) but not by haloperidol (1 mg kg−1 subcutaneously twice a day for 5 days, followed by 18 h washout; Table 1 and Figure 1). It should be noted that the doses of clozapine and haloperidol selected for this study elicited marked sedation (clozapine) and catalepsy (haloperidol) 1 h after injection in both PRS and NS mice. However, there were no signs of sedation or catalepsy after 18 h washout. These drug doses were chosen on the basis of preliminary studies showing that 5 mg kg−1 of clozapine elicited demethylation of hypermethylated Reln promoters in mice treated protractedly with large doses of methionine.38, 39
Table 1. Increased locomotor activity in PRS mice is reduced by clozapine but not haloperidol.
| Treatment |
Horizontal activity (Counts per 15 min) |
|
|---|---|---|
| NS | PRS | |
| Vehicle | 3656±182 | 4229±132a |
| Haloperidol | 3323±226 | 4040±223 |
| Clozapine | 3538±147 | 3866±217b |
P<0.05 when vehicle-treated PRS mice are compared with vehicle-treated NS mice.
P<0.05 when clozapine-treated PRS mice are compared with vehicle-treated PRS mice. One-way analysis of variance (F5,30=4.0, P=0.002) followed by Student–Newman–Keuls multiple-comparison procedures.
Seventy-five-day-old PRS (prenatally stressed) or NS (nonstressed) male mice were subjected to locomotor activity test and divided in groups of six mice with comparable locomotor activity values. Mice were then treated subcutaneously, twice a day for 5 days, with vehicle, 5mg kg−1 clozapine or 1 mg kg−1 haloperidol, respectively. The locomotor activity was measured again on the sixth day, 18 h after the last treatment. The data are expressed as mean±s.e.m. of six mice per group.
Figure 1.
Social interaction deficit in PRS mice is reversed by treatment with clozapine but not haloperidol. Seventy-five-day-old PRS (prenatally stressed) or NS (nonstressed) male mice were treated subcutaneously, twice a day for 5 days, with vehicle (VEH), 5 mg kg−1 clozapine (CLZ) or 1 mg kg−1 haloperidol (HAL). @The social interaction activity (ratio of sniffing time to the wire cup with vs without the stranger mouse) was measured the day before initiating the antipsychotic treatment and 18 h after the last antipsychotic treatment. The data are expressed as mean±s.e.m. of six mice per group. #P<0.001 when clozapine treated PRS mice are compared with PRS mice receiving vehicle or to the social interaction values of the same mice before initiating treatment. *P<0.05 when PRS mice are compared with NS mice. Repeated measures followed by Bonferroni post hoc multiple comparisons.
We have reported previously that adult PRS mice exhibit other SZ-related behavioral alterations that are normalized by clozapine or VPA administration.1 Here, we limited the behavioral studies to include locomotor activity and social interaction because the changes in these two tests are reproducible in PRS mice (Table 1 and Figure 1) and these tests are considered less stressful than fear conditioning, pre-pulse inhibition or dizocilpine treatment. As stressful tests could possibly alter biochemical parameters per se, we avoided these latter behavioral assays in the present study.
Clozapine and VPA, but not haloperidol, corrects promoter hypermethylation of psychiatric disorder-related genes in PRS mice
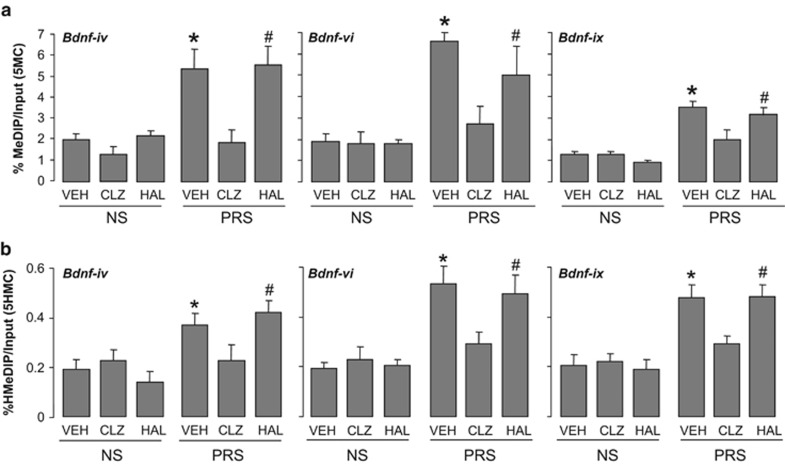
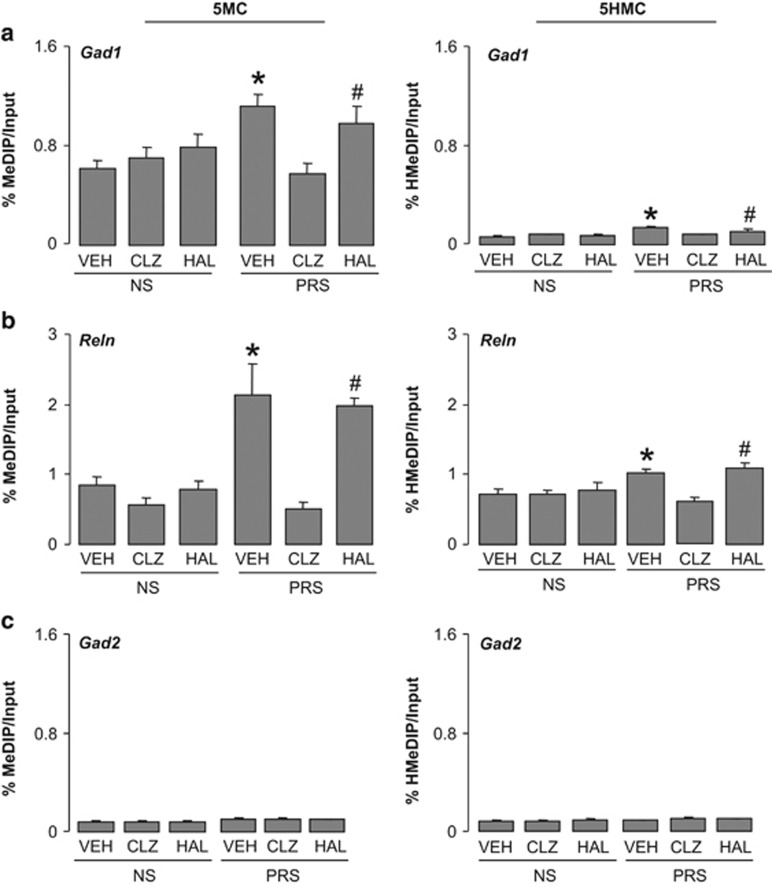
Next we tested whether cytosines proximal to the Gad1, Reln, Bdnf promoters are hypermethylated (5-methylation or 5-hydroxymethylation) in PRS mice. To distinguish between 5MC and 5HMC, we used MeDIP and hMeDIP,28 which are immunoprecipitation-based assays that use highly specific anti-5MC and 5HMC antibodies. We show that the Bdnf-iv, Bdnf-vi and Bdnf-ix (Figure 2), Gad1 and Reln promoters (Figure 3) are enriched in 5MC and/or 5HMC in FC of PRS mice. Importantly, the methylation and hydroxymethylation enrichment of Bdnf-iv, Bdnf-vi and Bdnf-ix, (Figure 2), Reln and Gad1 (Figure 3) promoters present in the cortex of PRS mice were consistently and significantly reduced in clozapine-treated mice but not in haloperidol-treated mice (Figures 2a and b; Figure 3). In NS mice, clozapine treatment failed to alter the level of promoter methylation or hydroxymethylation.
Figure 2.
Increased levels of 5MC (a) and 5HMC (b) on Bdnf-iv, -vi, -ix promoter regions in the frontal cortex of PRS mice are reduced by treatment with clozapine (CLZ) but not haloperidol (HAL). Seventy-five-day-old PRS (prenatally stressed) or NS (nonstressed) male mice were treated subcutaneously twice a day for 5 days with vehicle (VEH), 5 mg kg−1 clozapine or 1 mg kg−1 haloperidol. The enrichment of 5MC and 5HMC was measured on the sixth day, 20 h after the last treatment and 2 h after the last behavioral test. The data are expressed as mean±s.e.m. and analyzed statistically with one-way ANOVA followed by Student–Newman–Keuls multiple comparisons. ANOVA for 5MC: Bdnf-iv (F5,30=8.9, P<0.001), Bdnf-vi (F5,30=9.51, P<0.001), Bdnf-ix (F5,30=7.13, P<0.001). ANOVA for 5HMC: Bdnf-iv (F5,30=2.8, P<0.02), Bdnf-vi (F5,30=13.9, P<0.001), Bdnf-ix (F5,30=8.7, P<0.001). *P<0.05 when vehicle-treated PRS samples are compared with PRS clozapine-treated samples or with treated or untreated NS samples. Student–Newman–Keuls multiple comparisons. #P<0.05 when haloperidol-treated PRS samples are compared with PRS clozapine-treated samples or with treated and untreated NS samples. Student–Newman–Keuls multiple comparisons. ANOVA, analysis of variance; BDNF, brain-derived neurotrophic factor; 5HMC, 5-hydroxymethylcytosine; 5MC, 5-methylcytosine.
Figure 3.
Increased levels of 5MC and 5HMC on Gad1 (a) and Reln (b) promoter regions in the frontal cortex of PRS mice are reduced by treatment with clozapine (CLZ) but not haloperidol (HAL). Note there is no enrichment of 5MC or 5HMC on Gad2 (c) promoter region in the PRS and NS mice. Dose and schedule of drug treatment are as described in Figure 1. Data are expressed as mean±s.e.m. and analyzed with one-way ANOVA followed by Student–Newman–Keuls multiple comparisons. ANOVA for Gad1 5MC (F5,41=5.8, P<0.001), Gad1 5HMC (F5,42=5.8, P<0.001); Reln 5MC (F5,37=5.4, P<0.001), Reln 5HMC (F5,42=5.8, P<0.001); Gad2 5MC (F5,42=0.64, P=0.2), Gad2 5HMC (F5,42=1,49, P=0.24). *P<0.05 when vehicle (VEH)-treated PRS samples are compared with PRS clozapine-treated samples or with treated or untreated NS samples. #P<0.05 when haloperidol-treated PRS samples are compared with PRS clozapine-treated samples or with treated and untreated NS samples. ANOVA, analysis of variance; 5HMC, 5-hydroxymethylcytosine; 5MC, 5-methylcytosine; NS, nonstressed; PRS, prenatally stressed.
The effect of clozapine on DNA methylation of Bdnf-ix gene has some specificity for the promoter region because in these same animals, methylation of the Bdnf-ix gene body region, was not influenced by prenatal stress, and the level of methylation was not modified by clozapine. (The % 5MC within the Bdnf-ix body region from +312 bp to +415 bp was: 2.74±0.75 in NS mice receiving vehicle, 2.1±0.080 in NS mice receiving clozapine, 2.86±0.38 in PRS mice receiving vehicle, and 3.0±0.75 in PRS mice receiving clozapine. The % 5HMC at the same Bdnf-ix body region was: 3.59±0.85 in NS mice receiving vehicle, 3.06±0.90 in NS mice receiving clozapine, 2.66±0.53 in PRS mice receiving vehicle, and 2.49±0.22 in PRS mice receiving clozapine; n=6).
We also included Gad2, D2, Htr2a as control promoters to check the specificity of methylation for the above-reported candidate target genes. Gad2 has very low 5MC and 5HMC levels (Figure 3), and D2 and Htr2a receptor genes fail to show enrichment of 5MC and 5HMC in PRS mice (%MeDIP/Input: control D2 1.7±0.28, PRS D2 1.6±0.36. Control Htr2a 5.9±1.1, PRS Htr2a 5.5±1.4; average of six mice per group).
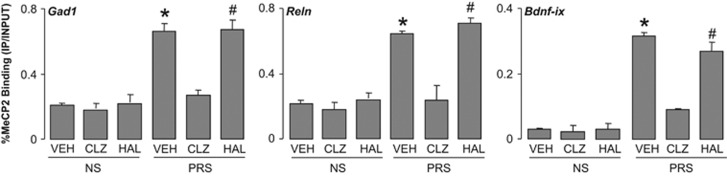
To confirm that specific GABAergic and glutamatergic gene promoters were hypermethylated (5MC+5HMC) and that clozapine reverses promoter hypermethylation in PRS mice, we performed ChIP studies using antibodies specific for MeCP2. As shown in Figure 4, MeCP2 binding to Gad1, Reln and Bdnf-ix promoters was increased in PRS mice. Clozapine, but not haloperidol, strongly reduced the binding of MeCP2 in PRS mice, whereas no statistical effects were detected in NS mice. The increased binding of MeCP2 to the candidate promoters was independent from the level of MeCP2 expression which failed to change in PRS mice.2
Figure 4.
The increased levels of MeCP2 binding to Gad1, Reln and Bdnf-ix promoter regions in the frontal cortex of PRS mice are reduced by treatment with clozapine but not haloperidol. The dose and schedule of drug treatments are described in Figure 2. The data are expressed as mean±s.e.m. One-way ANOVA for MeCP2 binding to Reln (F5,18=4.96, P=0.03), Gad1 (F5,18=6.07, P=0.009), Bdnf-ix (F5,18=4.80, P=0.028). *P<0.05 when vehicle-treated PRS samples are compared with PRS clozapine-treated samples or to vehicle-treated NS samples. #P<0.05 when haloperidol-treated PRS samples are compared with PRS clozapine-treated samples or with treated and untreated NS samples. Student–Newman–Keuls multiple comparisons. ANOVA, analysis of variance; BDNF, brain-derived neurotrophic factor.
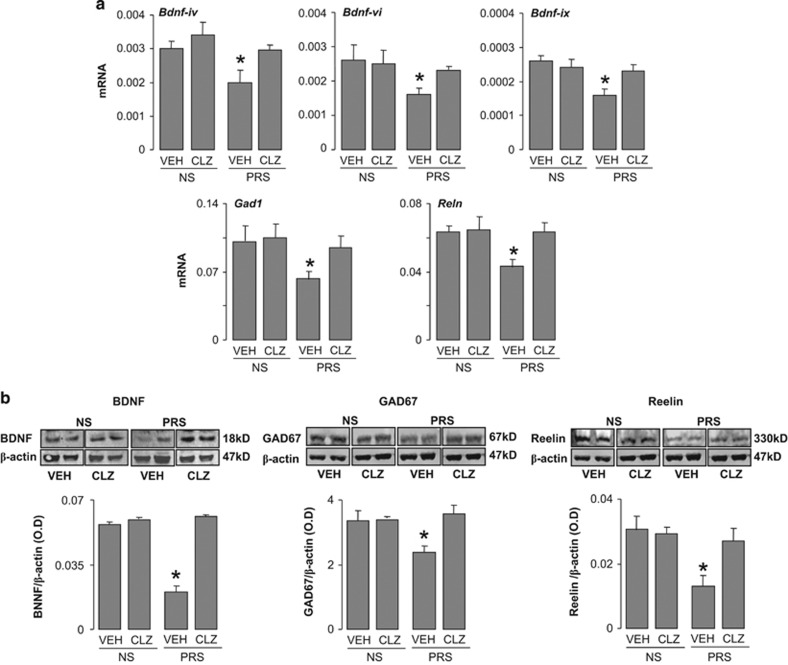
As expected,1, 2 the enrichment of 5MC and 5HMC at Bdnf, Gad1 and Reln promoters in PRS mice was associated with a decrease in corresponding mRNAs (Figure 5a) and proteins (Figure 5b) expression, and this decrease was reduced by 5 days of clozapine treatment.
Figure 5.
(a and b) Reduced expression of Bdnf, Gad1, Reln, mRNAs and proteins in frontal cortex of PRS mice is normalized by treatment with clozapine. (a) Levels of mRNA relative to β-actin for Bdnf-iv, Bdnf-vi, Bdnf-ix, Gad1, Reln and (b) immunoblot of BDNF, GAD67, reelin proteins normalized by β-actin protein levels. Dose and schedule of clozapine treatment are described in Figure 2. The data are expressed as mean±s.e.m. and analyzed by one-way ANOVA followed by Student–Newman–Keuls multiple comparisons. Bdnf-ix mRNA (F3,20=3.45, P=0.026), Bdnf-iv mRNA (F3,20=4.87, P=0.015), Bdnf-vi mRNA (F3,21=3.91, P=0.018), BDNF protein (F3,20=3.33, P=0.04). GAD1 mRNA (F3,23=4.97, P=0.01), Reln mRNA (F3,23=4.68, P<0.05), GAD67protein (F3,19=4.97, P<0.05), reelin protein (F3,23=17.02, P<0.001). *P<0.05 when vehicle-treated PRS samples are compared with PRS clozapine-treated samples or with NS vehicle or clozapine-treated sample. Student–Newman–Keuls multiple comparisons. ANOVA, analysis of variance; BDNF, brain-derived neurotrophic factor; mRNA, messenger RNA; NS, nonstressed; PRS, prenatally stressed.
The strong effect of clozapine on DNA methylation in PRS mice and the virtual lack of this effect in NS mice are not attributable to significant changes in D2, Htr1a or Htr2a receptor expression in the cortex of PRS mice. The mRNA levels of D2 receptor as a percentage relative to β-actin mRNA were 0.120±0.010 in NS mice and 0.135±0.005 in PRS mice (n=6). The mRNA levels for 5Ht2a receptor were 0.135±0.011 in NS mice and 0.105±0.010 in PRS mice (n=6). The mRNA levels for 5Ht1A receptor were 0.115±0.095 in NS mice and 0.120±0.093 in PRS mice (n=6). The mRNA levels of D2, 5Ht2A and 5Ht1A in both NS and PRS mice were not modified by either clozapine or haloperidol treatment.
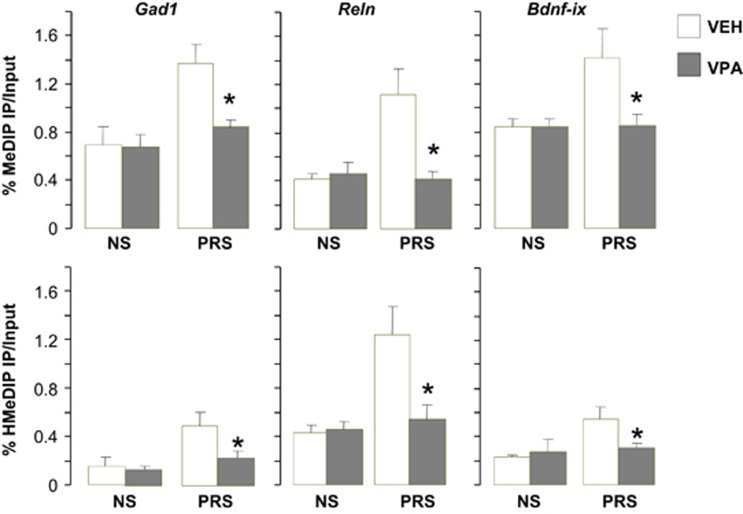
To demonstrate that the DNA demethylation of GABAergic and glutamatergic genes is involved in the restorative effects of clozapine on behavior in PRS mice, we show that the behavioral effects of clozapine, by reducing DNA hypermethylation at candidate gene promoters, were also restored by the histone deacetylase (HDAC) inhibitor VPA. This drug, used in psychiatry to treat BP disorder patients with psychosis, is chemically unrelated to clozapine or haloperidol and fails to bind to monoamines receptors, but instead favors acetylation of histone-3 (H3) lysine 9 and 14, facilitates the opening of chromatin and induces demethylation of promoters presumably activating a DNA demethylating processes.38, 45 As shown in Figure 6, VPA administered for 5 days, in a dose that (70 mg kg−1 subcutaneously) increases H3 acetylation in FC of NS mice38, 39 normalizes the hypermethylation of Gad1, Reln and Bdnf-ix promoters in the FC of PRS mice. We reported previously that at this dose, VPA also normalizes altered behaviors including locomotor hyperactivity, social interaction deficit, pre-pulse inhibition deficit and contextual fear condition deficit.1 These data provide support for the role of promoter hypermethylation at GABAergic and glutamatergic neurons in the pathogenesis of the behavioral deficits in PRS mice.
Figure 6.
Increased levels of 5MC and 5HMC on Gad1, Reln and Bdnf-ix promoter regions in the frontal cortex of PRS mice are reduced by treatment with valproate (VPA) Seventy-five day old mice were treated subcutaneously twice a day for 5 days with vehicle or 70 mg kg−1 valproate. 5MC and 5HMC enrichment was measured on the sixth day, 20 h after the last treatment. No behavioral testing in these animals. Data are expressed as mean±s.e.m. and analyzed with one-way ANOVA followed by Student–Newman–Keuls multiple comparisons. *P<0.05 when vehicle (VEH)-treated PRS samples are compared with PRS VPA-treated samples or with treated or untreated NS samples. ANOVA, analysis of variance; BDNF, brain-derived neurotrophic factor; 5HMC, 5-hydroxymethylcytosine; 5MC, 5-methylcytosine; NS, nonstressed; PRS, prenatally stressed.
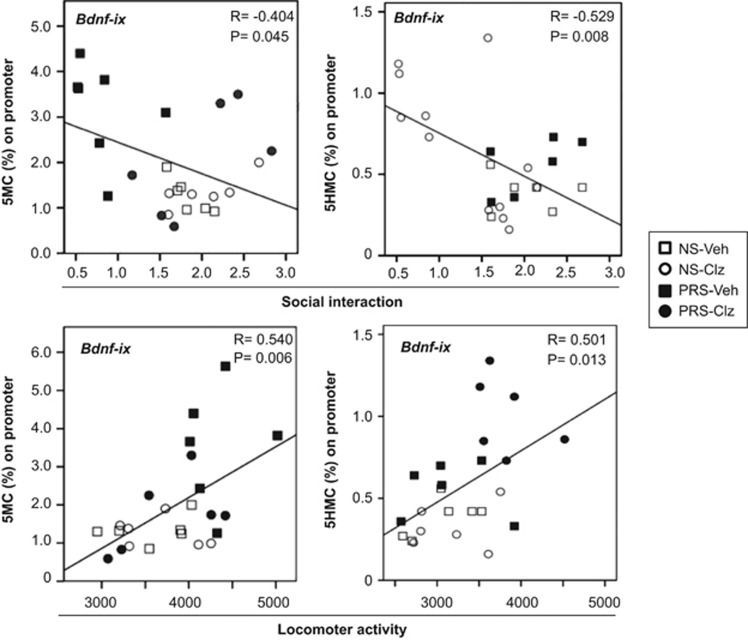
Candidate gene promoter hypermethylation and behavioral deficits are correlated
To establish whether hypermethylation of GABAergic and glutamatergic gene promoters underlie the behavioral abnormalities in PRS mice, we performed correlation analyses between 5MC or 5HMC enrichment at Bdnf-ix, Reln, Gad1 and Gad2 promoters in FC and measurements of social interaction and locomotor activity. As summarized in Table 2 and graphically depicted in Figure 7 for Bdnf-ix, there was a significant negative correlation between candidate gene promoter methylation and hydroxymethylation enrichment and social interaction scores for all of the target genes studied with the exception of Gad2. In addition, there was a significant positive correlation between locomotor activity scores and candidate SZ-related gene promoter methylation or hydroxymethylation enrichment with the exception of 5MC-Gad2, 5HMC-Gad2 and 5HMC-Reln.
Table 2. Pearson correlation between 5MC and 5HMC at Gadl, Gad2, Rein and Bdnf-ix promoters and social interaction index or motor activity.
| Promoter methylation | Social interaction index | Motor activity |
|---|---|---|
| 5MC-Gadl | R=−0.552, P=0.003 | R=0.548, P=0.005 |
| 5HMC-Gadl | R=−0.668, P=0.0006 | R=0.370, P=0.055 |
| 5MC-Gad2 | R=−0.017, P=0.937 | R=0.220, P=0.300 |
| 5HMC-Gad2 | R=−0.226, P=0.289 | R=0.305, P=0.148 |
| 5MC-Rein | R=−0.580, P=0.004 | R=0.456, P=0.033 |
| 5HMC-Rein | R=−0.450, P=0.027 | R=0.160, P=0.456 |
| 5MC-Bdnf-ix | R=−0.404, P=0.045 | R=0.540, P=0.006 |
| 5HMC-Bdnf-ix | R=−0.529, P=0.008 | R=0.501, P=0.013 |
Abbreviations: BDNF, brain-derived neurotrophic factor; 5HMC, 5-hydroxymethylcytosine; 5MC, 5-methylcytosine; NS, nonstressed; PRS, prenatally stressed.
The Pearson correlation was established in a cohort of 24 animals including 6 control (NS) mice receiving vehicle, 6 NS mice receiving clozapine, 6 PRS mice receiving vehicle, 6 PRS mice receiving clozapine. Bolded entries show significantly significant P-values.
Figure 7.
Representative example of Pearson correlation between 5MC levels at BDNF-ix promoters in the frontal cortex and social interaction (top) and locomotor activity (bottom) in a cohort of control (NS) mice receiving vehicle (Veh), NS mice receiving clozapine (Clz), PRS mice receiving vehicle and PRS mice receiving clozapine. Pearson correlation analyses show a significant negative correlation between promoter methylation and social interaction and a positive correlation between promoter methylation and locomotor activity. Schedule and doses of treatment are as described in Figure 2. The social interaction activity (ratio of sniffing time to the wire cup with vs without the stranger mouse) and the locomotor activity (total number of interruptions of the horizontal sensors in 15 min) were measured 18 h after the last antipsychotic treatment. 5MC and 5HMC promoter enrichment were measured 2 h after the last behavioral test. BDNF, brain-derived neurotrophic factor; 5HMC, 5-hydroxymethylcytosine; 5MC, 5-methylcytosine; NS, nonstressed; PRS, prenatally stressed.
Interaction of clozapine with promoter methylation mechanisms
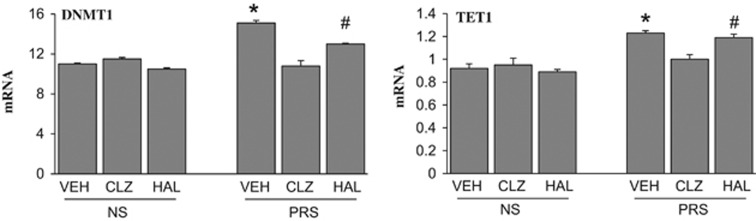
To investigate the mechanism whereby clozapine treatment reverts the increased cytosine methylation levels observed at GABAergic and glutamatergic gene promoters in PRS mice, we focused on the main DNA-modifying enzymes, DNMT1 and TET1. As shown in Figure 8, there was an ~30–40% increase in both DNMT1 and TET1 mRNA in the cortex of young adult (PND 75) PRS mice but these increases were abated by clozapine treatment.
Figure 8.
The increase of DNMT1 and TET1 mRNA in the FC of PRS mice is reduced by clozapine but not by haloperidol. Measurements were performed in the same animals tested in Table 1 and Figure 2. The values of mRNA are expressed as % relative to ActB. The data are expressed as mean±s.e.m. of six mice per group. *P<0.05 when vehicle (VEH)-treated PRS samples are compared with PRS clozapine (CLZ)-treated samples or with treated or untreated NS samples. #P<0.05 when haloperidol (HAL)-treated PRS samples are compared with PRS clozapine-treated samples or to haloperidol-treated NS samples. One-way ANOVA followed by Student–Newman–Keuls multiple-comparison procedures. ANOVA, analysis of variance; DNMT1, DNA-methyltransferase 1; FC, frontal cortex; mRNA, messenger RNA; NS, nonstressed; PRS, prenatally stressed; TET1, ten-eleven methylcytosine dioxygenase 1.
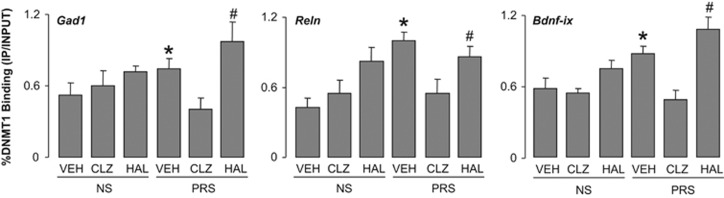
As shown in Figure 9, there was a significant increase in DNMT1 binding to selected Gad1, Reln and Bdnf-ix regulatory regions in PRS mice when compared with 75-day-old NS mice, and this increase was considerably reduced in clozapine-treated mice. Haloperidol failed to reduce the increased DNMT1 binding in PRS mice and actually tended to increase the binding of DNMT1 to target promoters in both the PRS and NS mice.
Figure 9.
The increased DNMT1 binding to Gad1, Reln and Bdnf-ix CpG-rich promoter regions is reduced by treatment with clozapine (CLZ) in the frontal cortex of PRS mice, but not haloperidol (HAL). Dose and schedule of drug treatment are described in Figure 2. The values are the mean±s.e.m. analyzed statistically with one-way ANOVA. DNMT1 binding to Gad1, (F5,30=2.48, P=0.048), Reln (F5,30=6.22, P<0.001), Bdnf-ix (F5,30=6.59 P<0.001). *P<0.05 when vehicle (VEH)-treated PRS samples are compared with PRS clozapine-treated samples or to vehicle-treated NS samples. Student–Newman–Keuls multiple comparisons. #P<0.05 when haloperidol-treated PRS samples are compared with PRS clozapine-treated samples or to vehicle-treated NS samples. Student–Newman–Keuls multiple comparisons. ANOVA, analysis of variance; BDNF, brain-derived neurotrophic factor; DNMT1, DNA-methyltransferase 1; NS, nonstressed; PRS, prenatally stressed.
It has been reported46 that chronic (21 days) treatment with clozapine results in an increased expression of HDAC2 mRNA in mouse FC. However, we failed to detect significant changes in HDAC2 mRNA expression after 5 days clozapine treatment (5 mg kg−1 twice a day) both in NS or PRS mice (values of mRNA expressed as % of β-actin: NS vehicle=0.170±0.028, NS clozapine=0.183±0.029, PRS vehicle=0.209±0.039, PRS clozapine=0.190±0.018; n=6). Taken together, these data suggest that clozapine, unlike haloperidol, may limit DNA methylation, either indirectly by reducing the expression of DNMT and TET, and/or more directly by interfering with the DNMT1 DNA-binding domain.47
Discussion
DNA-methylation is emerging in physiology and pathology as an important epigenetic control mechanism for the regulation of central nervous system function.48 To this end, we have recently directed the focus of our research toward identifying the epigenetic DNA-methylation signature found in the brains of SZ and BP disorder patients, in the brain of offspring of dams stressed during pregnancy (PRS mice).2 We confirm here that young adult PRS mice show behavioral and molecular alterations reminiscent of the epigenetic behavioral and molecular alterations observed in psychotic patients (Table 3). The brain of PRS mice, like that of postmortem SZ and BP disorder patients, is characterized by significant increases in 5MC and 5HMC at promoter regions corresponding to Gad1, Reln and Bdnf (Figures 2 and 3) together with a reduction in the expression of the corresponding mRNAs and proteins (Figures 5a and 5b). Importantly, we show that Gad1, Reln and Bdnf-ix promoter methylation and hydroxymethylation enrichment is correlated with locomotor activity and social interaction index (Table 2 and Figure 7). Taken together, these results and previous reports that there is a significant correlation between altered behavioral phenotypes and Bdnf transcript levels in PRS mice1, 2 suggest that altered DNA methylation dynamics underlie the SZ-like behavioral endophenotypic profiles in these mice.
Table 3. Comparison of molecular and behavioral abnormalities in SZ and BP disorder patients and PRS mice.
| SZ and BP | Reference | PRS mice | Reference | |
|---|---|---|---|---|
| Molecular changes | ||||
| GAD67, RELN, BDNF expression | ↓ | 19, 28, 29 | ↓ | 1, 2 |
| DNMT1, 3A and TET1 expression | ↑ | 19, 29 | ↑ | 1, 2 |
| 5MC and 5HMC enrichment at Gadl, Reln and Bdnf promoters | ↑ | 19, 28, 29 | ↑ | Current paper |
| Behavioral changes | ||||
| Positive symptoms (stereotype behaviors, sensitivity to NMDA receptor antagonists) | ↑ | 34, 36 | ↑ | 1, 2 and current paper |
| Negative symptoms (social interactions) | ↓ | 34, 36 | ↓ | 1, 2 and current paper |
| Cognitive, information processing deficit (PPI, fear conditioning) | ↑ | 34, 36 | ↑ | 1, 2 |
Abbreviations: BDNF, brain-derived neurotrophic factor; BP, bipolar; DNMT1, DNA-methyltransferase 1; 5HMC, 5-hydroxymethylcytosine; 5MC, 5-methylcytosine; NMDA, N-methyl d-aspartate; PPI, pre-pulse inhibition; PRS, prenatally stressed; SZ, schizophrenia; TET1, ten-eleven methylcytosine dioxygenase 1.
Patients with SZ and BP disorder often receive behavioral benefits from the use of antipsychotic medications. To address the existence of a possible connection between antipsychotics and improved behavior implicating epigenetic mechanisms, we studied the effect of an atypical antipsychotic (clozapine) vs a typical antipsychotic (haloperidol) on the impaired behavior and altered FC epigenetic profile in PRS mice. In this study, 5 days of treatment with clozapine but not haloperidol, followed by an 18 h washout abolished locomotor hyperactivity and deficits in social interaction displayed by PRS mice (Table 1, Figure 1). In the same animals participating in the behavioral studies, clozapine but not haloperidol, reduced Gad1, Reln and Bdnf promoter hypermethylation and enhanced the transcription of these genes.
To obtain further evidence that the beneficial effect of clozapine on the altered behaviors of PRS mice is associated with the reduction of GABAergic and glutamatergic promoter hypermethylation, we administered VPA, a class 1 HDAC inhibitor that has been reported to normalize the behavioral deficits in PRS mice.1 Like clozapine, VPA induced a decrease of the hypermethylated (5MC and 5HMC) Gad1, Reln and Bdnf gene regulatory regions in FC of PRS mice. Clinically, VPA is extensively used as monotherapy or in conjunction with antipsychotics in the treatment of BP disorder. Reports have further shown that typical or atypical antipsychotics are more potent, more efficacious and less toxic if they are co-administered with VPA,49 although some studies did not report such benefits in SZ patients.50
The precise molecular mechanisms whereby clozapine normalizes altered behaviors and promoter hypermethylation in PRS mice remain to be explored. We suggest that the effects of clozapine are not mediated by blockade of D2 receptor function. In fact, PRS mice express normal levels of D2 receptor mRNA and doses of haloperidol that are capable of blocking D2 receptor function and inducing extrapyramidal side effects, but failed to normalize the altered behavior or to reduce promoter hypermethylation in PRS mice. We can also exclude the hypothesis that the DNA-demethylating action of clozapine is associated with the hyperacetylation of histones 3 and 4, as occurs following the administration of HDAC inhibitors (that is, VPA and MS-275), because clozapine lacks HDAC inhibitory activity38 and actually may increase HDAC2 if administered chronically.46 It was reported that H3K4-methylation, a histone covalent modification associated with active chromatin transcription, is increased at Gad1 and other promoters in FC neurons of mice treated with clozapine but not haloperidol.51 This increase is probably caused by an activation of the mixed-lineage leukemia-1 histone methyltrasferase. Hence, it remains possible that a nuclear action of clozapine on DNA methylation is mediated via chromatin remodeling by histone lysine methylation. We have shown here that clozapine both reduces the overexpression of DNMT1 and TET1 and strongly reverses the binding of DNMT1 to unmethylated target promoters (Figure 9), thus reducing promoter methylation and favoring demethylation. In future studies, it will be important to establish whether clozapine acting at 5HT metabotropic cortical receptors modulates signal transduction mechanisms (for example, the phospholipase C pathway, adenylate cyclase) indirectly interfering with DNMT transcription or with the binding of DNMT1 to promoter or other regulatory regions of target genes.
An important observation of this study is that, despite a potent behavioral and DNA demethylating action in PRS mice, only marginal behavioral and molecular effects of clozapine were detected in NS mice. A possible explanation for clozapine's failure to demethylate target promoters in NS mice is that the dynamic state of DNA methylation is greatly accelerated in PRS mice compared with NS mice. Hence, the steady-state methylation process may be very sensitive to short-term inhibition in PRS mice. This does not necessarily mean that clozapine fails to work on methylation/demethylation processes in normal animals. Similar considerations may apply to the lack of effect of haloperidol. Parametric studies systematically manipulating different aspects of the clozapine and haloperidol administration protocol (varying dose, treatment schedule and so on) need to be conducted in both PRS and NS mice before one can conclude that antipsychotic drugs have no effect on methylation/demethylation in normal control mice.
The remarkable action of clozapine and the failure of haloperidol to decrease DNA methylation beg the question of whether all atypical antipsychotics interfere with DNA methylation dynamics. To answer this question, experiments were carried out in mice in which SZ-related genes were hypermethylated by repeated administration of methionine.25 We found that DNA-demethylation was induced by clozapine and its dibenzodiazepine congeners quetiapine and olanzapine, but not by the chemically unrelated risperidone, also when given in high doses. However, before making definitive conclusions, a more detailed analysis of the effects of several antipsychotic drugs on the epigenetic signature and behavioral phenotype of PRS mice is needed.
Conclusion
Clozapine, the ‘gold standard' antipsychotic, is unique among the typical and atypical antipsychotics in its ability to ameliorate psychosis in patients with treatment-resistant SZ and to reduce the risk of suicide.36, 37 The present study shows that clozapine is capable of correcting behavioral deficits and can induce chromatin remodeling in PRS mice that are resistant to haloperidol treatment. In future studies in PRS mice, we would like to establish whether clozapine has advantages over other antipsychotics in treating cognitive and negative symptoms of SZ because of its unique chromatin remodeling properties.
Collectively, the current and previous studies from our group1, 2 and others11, 12 support the conclusion that the PRS mouse model has construct face validity as an experimental epigenetic model of vulnerability for specific forms of psychopathology (that is, schizoaffective disorders) and can be used to screen potential anti-psychotic drugs for improved clinical efficacy in acting on altered epigenetic mechanisms. Furthermore, the PRS model theoretically has the potential for predicting treatment responses at specific initial stages of the psychopathology with particular attention to early disease detection and possible prophylactic intervention.
Acknowledgments
This work was supported by National Institutes of Health Grant Nos. R01 MH093348 and R01 MH101043 (AG).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Matrisciano F, Tueting P, Dalal I, Kadriu B, Grayson DR, Davis JM et al. Epigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatal stress in mice. Neuropharmacology 2013; 68: 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Dzitoyeva S, Matrisciano F, Tueting P, Grayson DR, Guidotti A. BDNF epigenetic modifications associated with schizophrenia-like phenotype induced by prenatal stress in mice. Biol Psychiatry 2015; 77: 589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry 1987; 44: 660–669. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull 2009; 35: 528–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khashan AS, Abel KM, McNamee R, Pedersen MG, Webb RT, Baker PN et al. Higher risk of offspring for schizophrenia following antenatal maternal exposure to severe adverse life events. Arch Gen Psychiatry 2008; 65: 146–152. [DOI] [PubMed] [Google Scholar]
- Markham JA, Koenig JI. Prenatal stress: role in psychotic and depressive diseases. Psychopharmacology 2011; 214: 89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanoli S, Meyer U. Response to comment on 'Stress in puberty unmasks latent neuropathological consequences of prenatal immune activation in mice". Science 2013; 340: 811. [DOI] [PubMed] [Google Scholar]
- Brown AS, Derkits EJ. Prenatal infection and schizophrenia: a review of epidemiologic and translational studies. Am J Psychiatry 2010; 167: 261–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Os J, Kenis G, Rutten BP. The environment and schizophrenia. Nature 2010; 468: 203–212. [DOI] [PubMed] [Google Scholar]
- Zhang TY, Labonte B, Wen XL, Turecki G, Meaney MJ. Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans. Neuropsychopharmacology 2013; 38: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens HE, Su T, Yanagawa Y, Vaccarino FM. Prenatal stress delays inhibitory neuron progenitor migration in the developing neocortex. Psychoneuroendocrinology 2013; 38: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine R, Zhang J, Stevens HE. Prenatal stress and inhibitory neuron systems: implications for neuropsychiatric disorders. Mol Psychiatry 2014; 19: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Segal M. Prenatal stress affects network properties of rat hippocampal neurons. Biol Psychiatry 2013; 73: 1095–1102. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci 2005; 6: 312–324. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Dong E, Grayson DR, Veldic M et al. GABAergic dysfunction in schizophrenia: new treatment strategies on the horizon. Psychopharmacology 2005; 180: 191–205. [DOI] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet 2008; 82: 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Viana J, Hannon E, Spiers HH, Troakes C, Al-Saraj S et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol 2014; 15: 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka WB, Subburaju S, Benes FM. Circuit-and diagnosis-specific DNA methylation changes at gamma-aminobutyric acid-related genes in postmortem human hippocampus in schizophrenia and bipolar disorder. JAMA Psychiatry 2015; 72: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology 2013; 38: 38–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman JE, Coyle JT, Green RW, Javitt DC, Benes FM, Heckers S et al. Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci 2008; 31: 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Burgos G, Lewis DA. GABA neurons and the mechanisms of network oscillations: implications for understanding cortical dysfunction in schizophrenia. Schizophr Bul 2008; 34: 944–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson DD, Overeem KA, Wolff AR, Williams JM, Abraham WC, Bilkey DK. Association of aberrant neural synchrony and altered GAD67 expression following exposure to maternal immune activation, a risk factor for schizophrenia. Transl Psychiatry 2014; 4: e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F, Tueting P, Maccari S, Nicoletti F, Guidotti A. Pharmacological activation of group-II metabotropic glutamate receptors corrects a schizophrenia-like phenotype induced by prenatal stress in mice. Neuropsychopharmacology 2012; 37: 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston I, Peter CJ, Mitchell A, Straubhaar J, Rogaev E, Akbarian S. Epigenetics in the human brain. Neuropsychopharmacology 2013; 38: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Chen Y, Davis JM, Dong E, Gavin DP et al. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology 2011; 60: 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegame T, Bundo M, Murata Y, Kasai K, Kato T, Iwamoto K. DNA methylation of the BDNF gene and its relevance to psychiatric disorders. J Hum Genet 2013; 58: 434–438. [DOI] [PubMed] [Google Scholar]
- Dong E, Ruzicka WB, Grayson DR, Guidotti A. DNA-methyltransferase1 (DNMT1) binding to CpG rich GABAergic and BDNF promoters is increased in the brain of schizophrenia and bipolar disorder patients. Schizophr Res 2015; 167: 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology 2012; 2: 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Gavin D, Chen Y, Davis J. Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl Psychiatry 2012; 2: e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Tueting P, Grayson DR. Modeling the molecular epigenetic profile of psychosis in prenatally stressed mice. Prog Mol Biol Transl Sci 2014; 128: 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro BM, do Carmo MR, Freire RS, Rocha NF, Borella VC, de Menezes AT et al. Evidence for a progressive microglial activation and increase in iNOS expression in rats submitted to a neurodevelopmental model of schizophrenia: reversal by clozapine. Schizophr Res 2013; 151: 12–19. [DOI] [PubMed] [Google Scholar]
- Meyer U, Knuesel I, Nyffeler M, Feldon J. Chronic clozapine treatment improves prenatal infection-induced working memory deficits without influencing adult hippocampal neurogenesis. Psychopharmacology 2010; 208: 531–543. [DOI] [PubMed] [Google Scholar]
- Basil P, Li Q, Dempster EL, Mill J, Sham PC, Wong CCY et al. Prenatal maternal immune activation causes epigenetic differences in adolescent mouse brain. Transl Psychiatry 2014; 4: e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman JA, Bymaster FP, Meltzer HY, Deutch AY, Duncan GE, Marx CE et al. Antipsychotic drugs: comparison in animal models of efficacy, neurotransmitter regulation, and neuroprotection. Pharmacol Rev 2008; 60: 358–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer HY. Serotonergic mechanisms as targets for existing and novel antipsychotics. Handb Exp Pharmacol 2012; 212: 87–124. [DOI] [PubMed] [Google Scholar]
- Meltzer HY. Update on typical and atypical antipsychotic drugs. Annu Rev Med 2013; 64: 393–406. [DOI] [PubMed] [Google Scholar]
- Meltzer HY, Huang M. In vivo actions of atypical antipsychotic drug on serotonergic and dopaminergic systems. Prog Brain Res 2008; 172: 177–197. [DOI] [PubMed] [Google Scholar]
- Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci USA 2008; 105: 13614–13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Kundakovic M, Satta R, Grayson DR, Costa E. Characterization of the action of antipsychotic subtypes on valproate-induced chromatin remodeling. Trends Pharmacol Sci 2009; 30: 55–60. [DOI] [PubMed] [Google Scholar]
- Montalvo-Ortiz JL, Keegan J, Gallardo C, Gerst N, Tetsuka K, Tucker C et al. HDAC inhibitors restore the capacity of aged mice to respond to haloperidol through modulation of histone acetylation. Neuropsychopharmacology 2014; 39: 1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res 2011; 39: 5015–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res 2004; 32: 4100–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol 2014; 15: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detich N, Bovenzi V, Szyf M. Valproate induces replication independent active DNA demethylation. J Biol Chem 2003; 278: 27586–27592. [DOI] [PubMed] [Google Scholar]
- Kurita M, Holloway T, García-Bea A, Kozlenkov A, Friedman AK, Moreno JL et al. HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat Neuroscience 2012; 15: 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Blackledge NP, Klose RJ. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem Soc Trans 2013; 41: 727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Kennedy AJ, Sweatt JD. DNA methylation and its implications and accessibility for neuropsychiatric therapeutics. Annu Rev Pharmacol Toxicol 2015; 55: 591–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassef AA, Dott SG, Harris A, Brown A, O'Boyle M, Meyer WJ et al. Randomized, placebo-controlled pilot study of divalproex sodium in the treatment of acute exacerbations of chronic schizophrenia. J Clin Psychopharmacol 2000; 20: 357–361. [DOI] [PubMed] [Google Scholar]
- Casey DE, Daniel DG, Tamminga C, Kane JM, Tran-Johnson T, Wozniak P et al. Divalproex ER combined with olanzapine or risperidone for treatment of acute exacerbations of schizophrenia. Neuropsychopharmacology 2008; 34: 1030–1038. [DOI] [PubMed] [Google Scholar]
- Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP et al. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci 2007; 27: 11254–11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.