Dear editor
Glycated hemoglobin is hemoglobin that has been irreversibly modified by addition of glucose through a non-enzymatic process and provides a weighted average of plasma glucose concentration over the erythrocyte lifespan. HbA1c is a specific glycated hemoglobin that is modified at the N-terminal valine of the Hb beta chains. HbA1c therefore provides a useful estimate of mean glycemia in patients with diabetes that has been shown to be directly related to risks for diabetes complications. Treatment goals for HbA1c have been established, and more recently the test has been recommended for use in diagnosing diabetes (1,2). Therefore, accurate and precise measurement of HbA1c is extremely important. The most common hemoglobin variants worldwide are HbS, HbE, HbC and HbD traits. Previous studies have shown method-specific analytic interference with HbA1c results from these heterozygous hemoglobin variants.
This study was approved by the University of Utah Institutional Review Board. Whole blood samples from individuals homozygous for HbA (n=42) and heterozygous for HbC, HbD, HbE, or HbS trait (n=23, n=41, n=76, n=40, respectively) were collected in EDTA tubes. Samples were frozen at −70°C in small aliquots and shipped on dry ice to 4 sites for HbA1c analysis. Hemoglobin variants were identified by inspection of chromatograms obtained with a Bio-Rad Variant analyzer (Bio-Rad Laboratories, Hercules, CA) using the β Thal Short Program; samples with HbF >5% were excluded. Samples with HbA1c concentrations of 4-12% were included in this study.
The evaluation included four ion-exchange HPLC methods: Variant II Turbo 2.0 (Bio-Rad Laboratories, Hercules, CA), ADAMS A1c HA-8180V (ARKRAY, Japan), and G7 and G8 (Tosoh Biosciences, San Francisco, CA), one boronate affinity method: In2it (Bio-Rad) and one capillary electrophoresis method: Capillarys2 Flex Piercing HbA1c (Sebia, Lisses, France). A boronate affinity HPLC method: ultra2 (Trinity Biotech, Kansas City, MO) was used as the comparative method since it has previously been shown to be unaffected by the presence of hemoglobin variants (3).
An overall test of coincidence of 2 least-squares linear regression lines was performed using SAS Software (SAS Institute, Cary NC) to determine whether the presence of each hemoglobin variant caused a statistically significant difference (P < 0.05) in results relative to the comparative method. Deming regression analysis was performed by using EP evaluator (David G. Rhoads Associates, Kennett Square, PA) to determine whether the presence of these variant traits produced a clinically significant effect on HbA1c results. After correcting for calibration bias by comparing results to those from the homozygous HbAA group, method bias attributable to the presence of hemoglobin variants was evaluated using ±7% relative bias at 6% and 9% HbA1c as evaluation limits for clinically significant bias (i.e., 0.42% at 6% HbA1c and 0.63% at 9% HbA1c).
In previous studies examining potential interferences from hemoglobin variants we used less stringent clinical significance limits of ±10% relative bias at 6 and 9% HbA1c (4,5). However, the overall performance of HbA1c assay methods has improved substantially in recent years. Given that treatment guidelines and many physicians regard a change of 0.5% HbA1c as significant and that HbA1c is now recommended for diagnosing diabetes, clinical requirements have become more stringent. The College of American Pathologists has responded by progressively lowering the acceptable limits for the GH2 whole blood proficiency survey from ±15% of the NGSP target in 2007 (when accuracy-based grading was first adopted) to the current ±7%. We have therefore tightened our acceptable limits for defining clinically significant interference to ±7%.
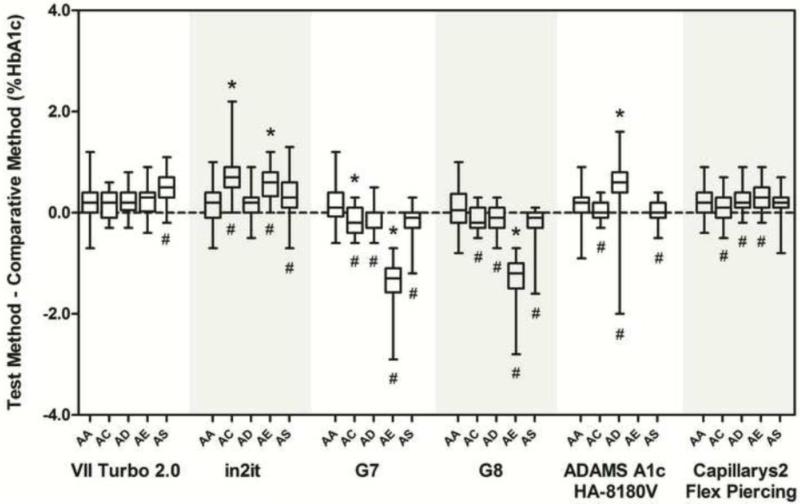
For each method, results for each type of sample were compared with results from the ultra2 method. Box plots of differences for each group of samples by method are shown in Figure 1. We observed statistically significant differences attributable to the presence of HbC and HbE traits for all methods except the Variant II Turbo 2.0. All methods except for the Variant II Turbo 2.0 and in2it showed statistically significant differences in the presence of HbD trait. The presence of HbS trait produced statistically significant differences for all methods except Capillarys2 Flex Piercing. Although these differences were statistically significant, many were very small and not clinically significant.
Figure 1.
Box plots showing the absolute differences between each assay and the comparative method for each hemoglobin type. The horizontal line inside each box is the median difference between the test and comparative methods. The upper and lower limits of each box correspond to the 25th and 75th percentiles of the differences, respectively. The upper and lower bars represent the maximum and minimum differences between the comparative methods. Differences from HbAA that are statistically significant are indicated (#) below each bar where appropriate; clinically significant differences are indicated (*) above each bar where appropriate.
The number of samples analyzed by each method for each sample type and the average bias at 6% and 9% HbA1c due to the presence of each Hb trait are shown in Table 1. The method bias at 6 and 9% HbA1c attributable to the presence of hemoglobin variants (after correcting for the bias between the test method and the comparative method with HbAA samples) is shown for each Hb trait for each method. Clinically significant differences (>±7% at 6 and/or 9% HbA1c) are indicated. The presence of Hb E trait resulted in a large clinically significant negative bias throughout the range of HbA1c values for the G7 and G8 (as shown previously (5)) and a clinically significant positive bias for the in2it at 9% HbA1c. The ADAMS A1c HA-8180V did not quantify HbA1c in the presence of HbE trait. Samples with HbC trait showed clinically significant positive biases at 6% and HbA1c for the in2it and at 6% HbA1c for the G7. There were no clinically significant differences attributable to the presence of HbS trait for any of the six methods. HbD trait resulted in a clinically significant positive bias at 9% HbA1c for ADAMS A1c HA-8180V method but no effect was seen for the other five methods. In addition, the ADAMS A1c HA-8180V method was not able to reliably detect the presence of HbD trait.
Table 1.
Mean difference between test and comparative methods
| HbC trait | HbD trait | HbE trait | HbS trait | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | n | 6% HbA1c | 9% HbA1c | n | 6% HbA1c | 9% HbA1c | n | 6% HbA1c | 9% HbA1c | n | 6% HbA1c | 9% HbA1c |
| ADAMS A1c HA-8180 | 23 | −0.25 | −0.09 | 31 | 0.17 | 0.64# | 38 | −0.23 | −0.08 | |||
| in2it | 25 | 0.59# | 0.99# | 41 | 0.07 | −0.07 | 76 | 3.34 | 0.70# | 39 | 0.18 | 0.35 |
| VII Turbo 2.0 | 23 | −0.10 | 0.16 | 41 | −0.06 | 0.16 | 76 | 0.00 | 0.04 | 40 | 0.26 | 0.25 |
| Capillarys 2 Flex Piercing | 23 | −0.20 | 0.17 | 34 | −0.03 | 0.29 | 74 | −0.02 | 0.25 | 37 | −0.01 | −0.02 |
| G7 | 23 | −0.46# | −020 | 41 | −0.33 | −0.32 | 76 | −1.45# | −2.01# | 40 | −0.36 | −0.58 |
| GB | 25 | −0.33 | −0.08 | 41 | −0.26 | −0.30 | 76 | −1.37# | −1.94# | 40 | −0.37 | −0.60 |
Deming regression analysis was performed usmg ultra2 as the comparative method. The biases (%HbA1c) for each method at clinical decision cutoffs of 6% and 9% HbA1c were calculated for each Hb trait. To correct for intermethod calibration differences, the mean difference between the method of interest and the comparative method for homozygous HbA samples was subtracted from that calculated for samples containing the variants.
Clinically significant differences (>0.42% or >0.63% HbA1c at 6% or 9% HbA1c respectively).
The accuracy of HbA1c methods can be adversely affected by the presence of hemoglobin variants. During the present evaluation, we found clinically significant negative biases for the G7 and G8 in the presence of HbE trait. Manufacturer instructions state that HbE trait produces an extra peak on the chromatogram and the results are not reportable; our study confirmed the manufacturer's instructions. However, with both of these methods careful inspection of chromatograms is required as the peak is generally subtle and often not fully resolved from the HbA0 peak. We found a clinically significant negative bias for the G7 (at 6% HbA1c) and clinically significant positive biases for the in2it method at both 6 and 9% HbA1c in the presence of HbC trait. The in2it, which is a boronate affinity method, also showed a clinically significant positive bias with HbE trait at HbA1c. Although boronate affinity methods are generally assumed to be free of interference from the hemoglobin variants tested, we previously observed this phenomenon with several older boronate affinity methods in the case of HbC trait (4). The manufacturer claims that HbE does not interfere with the in2it, but that HbC may cause a small increase in %HbA1c results. It is important to note that unlike the other 5 methods tested, which generally indicate the presence of a hemoglobin variant on the corresponding chromatogram, the in2it gives no such indication since the results do not include a chromatogram. For the ADAMS A1c HA-8180V method, the manufacturer states that abnormal peaks can be detected in the presence of HbD. However, in our study only 3 of the 41 HbD trait samples were detected by this method. There is no information provided by the manufacturer about whether or not Hb variant chromatograms are acceptable or certain chromatographic features are unacceptable. Proper identification of HbD is being further evaluated by the manufacturer. In the present study none of the 6 methods tested showed clinically significant effects from HbS trait. The Variant II Turbo 2.0 and the Capillarys2 Flex Piercing did not show any clinically significant interference with any of the variant traits evaluated.
HbA1c is widely recognized to be a valuable tool for assessing glycemic control in patients with diabetes, and more recently as a diagnostic tool. It is therefore very important that any unacceptable results are detected and not reported. Each new method must be evaluated for Hb variant interference; no generalizations can be made based on method type. Laboratories must be aware of the limitations of their assay method and should consider the prevalence of hemoglobinopathies in their respective patient populations when selecting a HbA1c assay method. Health care providers should also be aware of this potential for interference when interpreting HbA1c results. In cases where an interfering variant is present an alternative HbA1c assay method that does not show interference may need to be used. In cases where red-cell lifespan is altered (e.g. homozygous HbS or HbC) or red-cell glycation is altered (e.g. Hb Raleigh) alternative means of measuring glycemic control using non-hemoglobin-based methods such as glycated albumin or fructosamines may need to be considered.
Acknowledgments
This study was supported in part by the ARUP Institute for Clinical and Experimental Pathology. The author acknowledge Roche for funding, Bio-Rad for sample analysis by the Turbo2.0 and in2it systems, Sebia for supplying reagents for the Capillarys 2 Flex Piercing and ARKRAY for sample analysis by the ADAMS A1c HA-8180. We also thank Dr. Richard Madsen at the University of Missouri (Columbia, MO) for assistance with the statistical analyses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.International Expert Committee International Expert Committee report on the role of the A1C assay in the diagnosis of diabetes. Diabetes Care. 2009;32:1327–34. doi: 10.2337/dc09-9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Diabetes Association Standards of Medical Care in Diabetes. Diabetes Care. 2010;33:S11–S61. doi: 10.2337/dc10-S011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Little RR, Vesper H, Rohlfing CL, Ospina M, Safar-Pour S, Roberts WL. Validation by a mass spectrometric reference method of use of boronate affinity chromatography to measure glycohemoglobin in the presence of hemoglobin S and C traits. Clin Chem. 2005;51(1):264–5. doi: 10.1373/clinchem.2004.043141. [DOI] [PubMed] [Google Scholar]
- 4.Roberts WL, De BK, Brown D, Hanbury CM, Hoyer JD, John WG, et al. Effects of hemoglobin C and S traits on eight glycohemoglobin methods. Clin Chem. 2002;48(2):383–5. [PubMed] [Google Scholar]
- 5.Little RR, Rohlfing CR, Hanson S, Connolly S, Higgins T, Weykamp C, D'Costa M, Luzzi V, Owen WE, Roberts WL. Effects of hemoglobin E and D traits on glycated hemoglobin (HbA1c) Measurements by twenty-three methods. Clin Chem. 2008;54:1277–82. doi: 10.1373/clinchem.2008.103580. [DOI] [PubMed] [Google Scholar]